Abstract
Although ganglioside GD3 levels are highly elevated in malignant melanomas, the role of GD3 in melanomas' malignant properties has not been clearly shown. To investigate this problem, we genetically generated GD3-positive (GD3+) transfectant cells from a GD3-negative (GD3-) mutant line SK-MEL-28-N1 and analyzed the phenotypic changes in the transfected cells. GD3+ cells showed markedly increased cell growth and invasive characteristics. Two bands that underwent stronger tyrosine phosphorylation in GD3+ cell lines than in controls after treatment with FCS were found with molecular masses of 130 and 68 kDa. They were identified as p130Cas and paxillin by sequential immunoprecipitation. Their roles in cell growth and invasion were analyzed with a small interfering RNA (siRNA) approach. Cell growth, as analyzed by BrdUrd uptake, was strongly suppressed in GD3+ cells to near the levels of GD3- cells when treated with siRNA for p130Cas but not when treated with siRNA for paxillin. However, treatment with siRNAs of either p130Cas or paxillin resulted in the marked suppression of the invasive activity of GD3+ cells almost to the levels of control cells. These results suggested that these two molecules function as effectors of GD3-mediated signaling, leading to such malignant properties as rapid cell growth and invasion.
Keywords: small interfering RNA, sialyltransferase, phosphorylation
Malignant melanoma is very difficult to cure because of its rapid growth, high tendency of metastasis, rigorous invasion into surrounding tissues, and resistance to chemotherapy and/or radiation (1). Therefore, a number of therapeutic approaches to overcome the malignant properties of the disease have been attempted. By identifying melanoma-specific antigens (2-4), it has been possible to apply antibody therapies (5) and induce melanoma-specific cytotoxic T cells (6). Vaccination therapy with melanoma-associated gangliosides combined with a unique adjuvant has also been studied (7).
In particular, ganglioside GD3, which is widely expressed on melanomas (8, 9), has been a common target of these studies. Anti-GD3 mAb has been used in clinical trials in melanoma patients (5). Based on the expression pattern of GD3 in normal and malignant cells, its role in cell proliferation and/or activation has been postulated for a long time. For example, GD3 is expressed almost exclusively among gangliosides in the early stage of the brain development of mice and rats (10). GD3 is also detected in the T cell malignant leukocytes, such as acute lymphoblastic leukemia cells (11-13), adult T cell leukemia cells (14), and activated normal human T lymphocytes (5, 15). However, no clear function of GD3 in the malignant tumor cells has been demonstrated, although anti-GD3 antibody was reported to suppress the cell growth of cultured melanoma cells (16), and a GD3-negative cell isolate, SK-MEL-28-N1, was found to have slower growth and tumorigenic properties than the parent cell line (17).
In this study, we have generated GD3+ transfectant cells from the GD3- mutant of SK-MEL-28 designated N1 (17) by using cloned GD3 synthase cDNA (18), and we have analyzed the phenotypic changes in GD3+ transfectant cells and molecular mechanisms for GD3-mediated biosignals. Subsequently, we identified p130Cas and paxillin as effector molecules involved in the increased cell growth and/or invasion activity observed in the GD3+ transfectant cells. To our knowledge, this report is the first to clearly elucidate the mechanisms whereby GD3 expression influences the malignant properties of melanoma cells.
Materials and Methods
Antibodies. Anti-rabbit IgG conjugated with horseradish peroxidase (HRP) was purchased from Cell Signaling Technology (Beverly, MA). Anti-phosphotyrosine mAb (PY20) and anti-paxillin (mouse mAb IgG1) were from Transduction Laboratories (Lexington, KY). Anti-p130Cas (rabbit IgG C-20) was from Santa Cruz Biotechnology. Anti-mouse IgG conjugated with HRP was from Amersham Pharmacia Biotech.
Reagents. Protein G-Sepharose or A-Sepharose beads were from Amersham Pharmacia Biosciences.
Cell Cultures. SK-MEL-28-N1 was established as described in ref. 17 and maintained in DMEM supplemented with 7.5% FCS at 37°C in a humidified atmosphere containing 5% CO2.
Construction of a cDNA Expression Vector. Human α2,8-sialyltransferase cDNA clone pD3T-31 (18) was digested by XhoI and inserted into the XhoI site of pMIKneo (provided by K. Maruyama, Tokyo Medical and Dental University, Tokyo) to obtain the pMIKneo/D3T-31.
Gene Transfection and Selection. SK-MEL-28-N1 cells used for cDNA transfection were plated in a 60-mm plastic plate (Falcon). The cDNA expression vectors were transfected with Effectene (Qiagen, Valencia, CA). Stable transfectants were selected in the presence of 400 μg/ml G418 (Sigma).
Flow Cytometry. The cell surface expression of GD3 and other gangliosides was analyzed by flow cytometry (Becton Dickinson) as described in ref. 19. Control samples were prepared by using nonrelevant mAbs with the same subclasses for individual mAbs.
Preparation of Cell Lysates. Cells (2 × 105) were plated in a 6-cm dish and serum-starved for 12 h before the treatment with FCS or recombinant proteins such as human platelet-derived growth factor, basic FGF, or EGF. Cells were treated with FCS or recombinant proteins for 0-60 min at 37°C. Then, cells were lysed with a lysis buffer (20 mM Tris·HCl, pH 7.5/150 mM NaCl/1 mM Na2EDTA/1 mM EGTA/1% Triton X-100/2.5 mM sodium pyrophosphate/1 mM β-glycerophosphate/1 mM Na3VO4/1 μg/ml leupeptin/1 mM PMSF), and insoluble materials were removed by centrifugation at 4°C at 10,000 × g for 10 min.
Western Immunoblotting (IB). Lysates were separated by SDS/PAGE using 7.5-12% gels. The separated proteins were transferred onto an Immobilon-P membrane (Millipore). Blots were blocked with BSA in PBS containing 0.05% Tween 20. The membrane was first probed with primary antibodies. After being washed, the blots were incubated with goat anti-rabbit IgGs or goat anti-mouse IgGs conjugated with horseradish peroxidase (1:2,000). After the membranes were washed, bound conjugates were visualized with an enhanced chemiluminescence detection system (PerkinElmer). To analyze the band intensities in IB, bands were scanned with photoshop 6.0 (Adobe Systems, San Jose, CA) and quantified by using nih image 1.61.
Immunoprecipitation (IP). The lysates were immunoprecipitated with mAb or polyclonal antibody bound to protein G-Sepharose or A-Sepharose beads at 4°C for 75 min. The beads were washed five times with a washing buffer (50 mM Tris·HCl, pH 7.5/150 mM NaCl/1 mM MgCl2/0.5% Nonidet P-40/1 mM Na3VO4) and finally resuspended in 20 μl of 2× SDS sample buffer. The precipitated proteins were separated by SDS/PAGE and then immunoblotted.
3-(4,5-Dimethylthiazol-2-yl)-2,5-diphenyl Tetrazolium Bromide (MTT) Assay. Cells (4 × 103) were seeded in 98-well plates. MTT assay was performed by assessing the reduction of MTT to formazan based on the absorbance at 590 nm using an ELISA reader Immuno Mini NJ-2300 (System Instruments, Tokyo).
In Vitro Invasion Assay. Invasion assays with a Boyden chamber were performed as described in ref. 19. In brief, Matrigel (Becton Dickinson) was diluted with ice-cold PBS (100 μg/ml), and 0.6 ml was added to each Falcon 3093 filter (polyethylene terephthalate membrane, 8-mm pore size, 23.1-mm diameter) and left to be polymerized overnight. The membrane was reconstituted with serum-free medium. The lower chamber (6-well plate, Falcon 3502) was filled with the culture medium with or without serum. Cells (1-8 × 105 per well) were added to serum-free medium in the upper chamber and incubated for 24 h, and then the cells on the surface of the filter were stained with Giemsa and counted under microscopy.
BrdUrd Assay. Cells grown on a 60-well plate were incubated in the presence of BrdUrd for 14 h with a cell proliferation kit (Amersham Pharmacia Biosciences) according to the manufacturer's instructions and then fixed with acid-ethanol for 30 min. The cells were immunostained with anti-BrdUrd antibody and Alexa Fluor 546-conjugated secondary antibody (Molecular Probes). The BrdUrd-positive cells were observed by laser microscopy, and the percentage of BrdUrd-positive cells was calculated.
Inhibition of p130Cas and Paxillin Expression by Small Interfering RNA (siRNA). Human melanoma cells were plated at 70-80% confluency in 3.5-cm or 6-cm cell culture dishes and were cultured overnight. They were transiently transfected in Optimem I medium (Invitrogen) with Lipofectamine 2000 reagent (Invitrogen) with control (fluorescein-labeled luciferase GL2 duplex, Dharmacon Research, Lafayette, CO), anti-p130Cas siRNAs, or paxillin siRNAs (100 nM) following the manufacturer's instructions. Four hours later, the Optimem I medium was replaced by regular culture medium. Effects of down-regulation on p130Cas or paxillin were assessed at 48 h after the transfection with IB. For invasion assay, cells were collected after 2 days of the transfection and replated.
Statistical Analysis. Statistical significance of data was determined by using Student's t test.
Results
Generation of GD3+ Clones of SK-MEL-28-N1 with Transfection of GD3 Synthase cDNA. After the transfection of SK-MEL-28-N1 cells with the expression vector pMIKneo/D3T-31, two GD3+ clones (G5 and G11) were derived. Two clones transfected with pMIKneo alone (V5 and V9) were also isolated. These GD3+ clones showed reduced GM3 expression and low levels of GD2, whereas control cells expressed only GM3 (see the supporting information, which is published on the PNAS web site). Using these clones, the effects of GD3 expression on cell proliferation and invasion were determined.
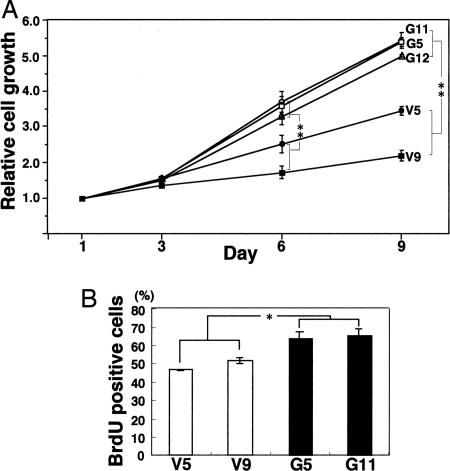
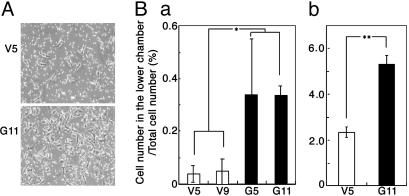
Effects of GD3 Expression on Cell Proliferation and Invasion. The proliferation of the GD3+ transfectant cells and the control cells were compared by using two methods. In MTT assay, the GD3+ cells showed markedly increased cell growth. The cell numbers of these transfectants were much higher than those of the control cells at day 9 of culture (Fig. 1A). In BrdUrd uptake, GD3+ cells showed significantly higher growth rate than the control cells (Fig. 1B). Invasion activity, as analyzed by the Boyden chamber, was also markedly increased in the GD3+ cells (Fig. 2A) under both conditions examined: i.e., no serum (Fig. 2Ba) and serum added (Fig. 2Bb) in the lower chamber.
Fig. 1.
Effects of GD3 expression on cell proliferation. (A) Cells (4 × 103) were seeded in 98-well plates. At day 1, 3, 6, and 9 of culture, MTT assay was performed. The experiments were repeated three times with similar results. (B) Cells were cultured with BrdUrd for 14 h and stained with BrdUrd antibody as described in Materials and Methods. *, P < 0.05; **, P < 0.01.
Fig. 2.
Invasion activity of GD3 synthase gene transfectant cells. Invasion activity of the transfectants was analyzed by using a Boyden chamber as described in Materials and Methods. (A) Micrographs at 200× magnification of a vector control transfectant (V5) and GD3+ transfectant (G11) showing migrated cells to the lower surface of the filter. (B) Summaries of the invasion assay. The ratios of cell number in the lower chamber/total cell number applied (in percent) are shown. V5 and V9 are the vector controls, and G5 and G11 are the GD3 synthase gene transfectant cells. Results when the lower chamber was filled with serum-free medium (Ba) or with FCS-containing medium (Bb) are shown. *, P < 0.05; **, P < 0.01.
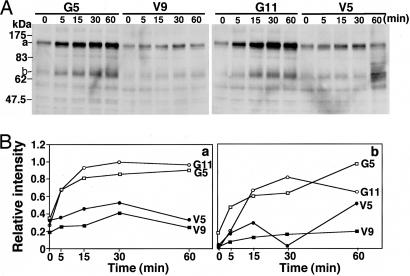
Tyrosine Phosphorylation of p130 and p68. To analyze critical proteins involved in the increased proliferation and invasion of GD3+ cells, IB with PY20 was performed by using lysates from cells treated with FCS or various recombinant growth factors. When cells were treated with FCS, two bands appeared with increasing intensities from 5 min and reached a plateau at 30 min after the stimulation. The intensity of two tyrosine-phosphorylated proteins was much stronger in GD3+ cells than in the controls, and their molecular masses were ≈130 kDa (Fig. 3 A and Ba) and 68 kDa (Fig. 3 A and Bb). No bands with higher intensity in GD3+ cells were detected with any known stimulatory factors examined (see the supporting information).
Fig. 3.
Tyrosine-phosphorylated proteins appearing after FCS treatment. (A) IB with PY20 was performed by using lysates from cells treated with FCS after serum starvation for 12 h. (B) Bands in A were quantitated by a scanner, and the relative intensities of the bands were plotted. Two tyrosine-phosphorylated proteins showed molecular masses of ≈130 (a) and 68 (b) kDa.
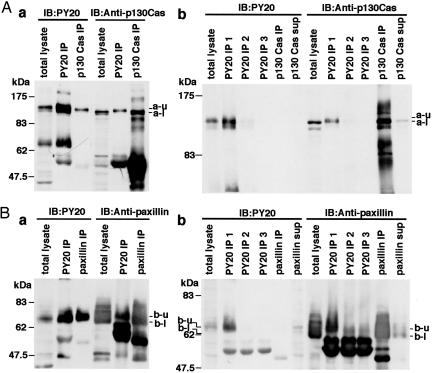
Identification of p130 as p130Cas and Identification of p68 as Paxillin. The two tyrosine-phosphorylated bands (130 and 68 kDa) were suspected to be p130Cas and paxillin, respectively, based on their migration rates in SDS/PAGE. To clarify this point, IP and sequential IB were performed. In the IB with PY20, both the immunoprecipitate with PY20 and that with anti-p130Cas showed bands with similar mobility (Fig. 4Aa). In contrast, in the IB with anti-p130Cas, p130 bands precipitated with anti-p130Cas as well as those in the total lysate were detected at the faster-migrating site compared with a band precipitated with PY20 (a-u and a-l in Fig. 4Aa). For the immunodepletion of phosphotyrosine-containing proteins, the lysates were repeatedly precipitated with PY20, and then the resultant supernatant was precipitated with anti-p130Cas.
Fig. 4.
Identification of p130 as p130Cas and identification of p68 as paxillin. IP/IB was performed to identify the two tyrosine-phosphorylated components using PY20 and anti-p130Cas (A) and PY20 and anti-paxillin (B) antibodies. (A) Immunoprecipitates with PY20 (PY20 IP) and those with anti-p130Cas (p130Cas IP) together with total lysate were immunoblotted by using PY20 (a Left) or anti-p130Cas (a Right). The bands at ≈130 kDa were designated a-u (upper) and a-l (lower). Then, immunodepletion of phosphotyrosine-containing proteins was performed before IP/IB as follows. The lysates were repeatedly immunoprecipitated with PY20, and the individual immunoprecipitates were designated PY20 IP1, PY20 IP2, and PY20 IP3. The resultant supernatant was immunoprecipitated with anti-p130Cas (p130Cas IP). (B) Immunoprecipitates with PY20 and those with anti-paxillin antibody were immunoblotted as performed for p130Cas in A to identify the 68-kDa band to be paxillin (Ba). As was performed for p130Cas, the identification of the 68-kDa band as paxillin was performed with sequential IP (Bb).
In the IB of these immunoprecipitates, PY20 IP1 showed a band that could be detected with similar mobility both with PY20 and with anti-p130Cas antibody. In PY20 IP3, no band could be detected, indicating the complete immunodepletion of phosphotyrosine-containing proteins with PY20 antibody (Fig. 4Ab Left). In contrast, a definite band of p130Cas could be detected in p130Cas IP, as in the total lysate, at the faster-migrating site, compared with that in PY20 IP1 with anti-p130Cas antibody (Fig. 4Ab Right). These results indicated that the band at 130 kDa detected with PY20 was p130Cas, and only a small portion of p130Cas was tyrosine-phosphorylated. As for paxillin, the identification of the 68-kDa band as paxillin was achieved just as p130Cas (Fig. 4B), showing that only a small portion of paxillin was tyrosine-phosphorylated.
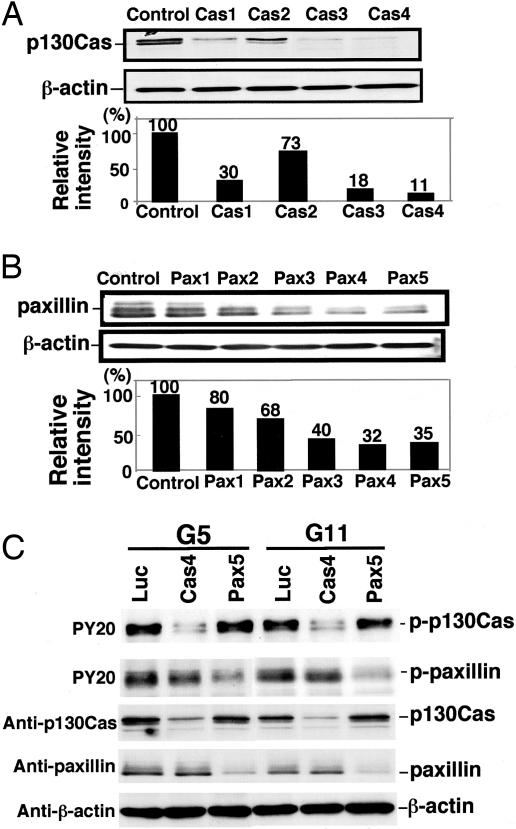
Selection of siRNA for Specific Knock-Down of p130Cas and Paxillin. To analyze the involvement of p130Cas and paxillin in cell proliferation and invasion, we used a siRNA-based approach. We first tested the ability of siRNAs to down-regulate p130Cas and paxillin proteins in SK-MEL-28-N1 with IB. Transfection efficiency of siRNAs as analyzed with fluorescein-labeled luciferase was >90-95% constantly. Among effective siRNAs, highly effective siRNAs (Cas4, Cas3, Pax5, and Pax4) were selected for the following experiments (Fig. 5 A and B). The transfection with control siRNA had no effect on p130Cas or paxillin levels. Specific knock-down of p130Cas and paxillin with individual siRNAs was confirmed with IB. As shown in Fig. 5C, Cas4 suppressed the protein level of p130Cas but not that of paxillin. Pax5 suppressed the protein level of paxillin but not that of p130Cas. Tyrosine-phosphorylated proteins also underwent specific effects with the siRNAs.
Fig. 5.
siRNAs for the knock-down of p130Cas and paxillin. (A) p130Cas suppression after transfection of SK-MEL-28-N1 with siRNAs Cas1-Cas4 was examined by IB as described in Materials and Methods. Actin served as a control for equal loading. The percentage of remaining p130Cas in cells transfected with anti p130Cas-siRNAs was calculated as a percentage of p130Cas in cells transfected with the control siRNA. (B) Paxillin suppression was assessed as performed for p130Cas in A with siRNAs Pax1-Pax5 by IB. The percentage of remaining paxillin in cells transfected with anti-paxillin siRNAs was calculated. (C) Suppression of tyrosine phosphorylation in G5 and G11 after the transfection of siRNAs. Cell lysates after the transfection with Cas4 or Pax5 were served for IB using PY20, anti-p130Cas, anti-paxillin, and anti-β-actin as indicated.
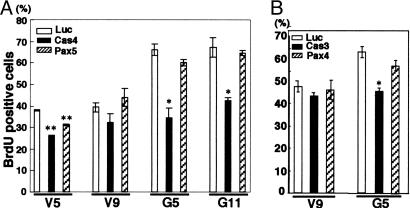
Effects of Knock-Down of p130Cas and Paxillin on BrdUrd Uptake. To analyze the effects of transient knock-down of p130Cas and paxillin with siRNA on BrdUrd uptake, siRNA Cas4 and Pax5 were transfected into the GD3+ cells and the vector controls, and then the ratio of BrdUrd-positive cells was counted. The GD3+ cells showed a significantly higher ratio of BrdUrd uptake than the controls (Fig. 1B). Both the GD3+ cells and the controls showed no definite differences in the ratio of BrdUrd-positive cells between siRNAs for luciferase and Pax5 (Fig. 6A). In contrast, both the GD3+ cells and the control cells transfected with siRNA Cas4 showed a lower ratio of BrdUrd uptake than those transfected with luciferases. The GD3+ cells with siRNA Cas4 showed a much decreased ratio of BrdUrd uptake compared with the control cells, suggesting that p130Cas plays a key role in cell proliferation but paxillin does not. These results were further confirmed by using siRNAs for other sites (Cas3 and Pax4) with similar effects (Fig. 6B). Because the knock-down of paxillin was not enough, repeated treatments of cells with Pax5 or combined use of Pax5 and Pax4 were examined, resulting in better suppression of paxillin protein (see the supporting information). However, BrdUrd uptake was not affected even in these conditions.
Fig. 6.
BrdUrd assay to examine the role of p130Cas and paxillin in cell proliferation. (A) The cells were analyzed for BrdUrd uptake after treatment with either Cas4 or Pax5 siRNAs as described in Materials and Methods, and the results were compared with those of control siRNA (Luc). (B) Similar experiments were performed using other siRNAs (i.e., Cas3 or Pax4), showing similar results. *, P < 0.05; **, P < 0.01.
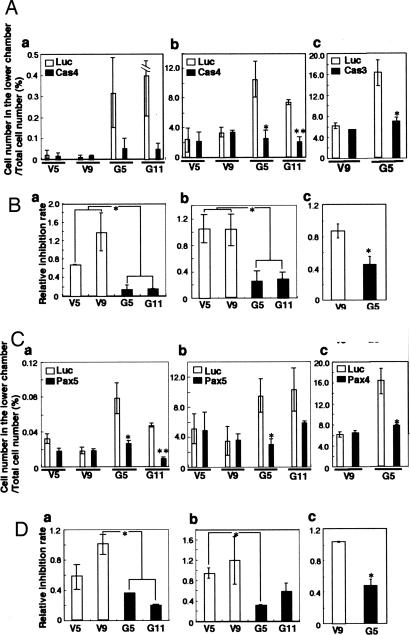
Involvement of p130Cas and Paxillin in Invasion. After the transfection of SK-MEL-28-N1 transfectant cells with siRNAs, invasion assays were performed with the Boyden chamber method. The invasion activity was markedly reduced by the knock-down of p130Cas and paxillin in GD3+ cells, although the invasion activity showed no change in GD3- controls (Fig. 7). Using the lower chamber filled with serum-free medium, the relative invasion activities were 0.68 and 1.38 for p130Cas-suppressed GD3- cells and 0.14 and 0.12 for p130Cas-suppressed GD3+ cells (Fig. 7Ba). When the lower chamber was filled with serum-containing medium, the relative invasion activities were 1.04, 1.04, 0.26, and 0.29 in V5, V9, G5, and G11, respectively (Fig. 7Bb). Similar effects were also observed with Cas3 in V9 and G5 (Fig. 7 Ac and Bc). When paxillin was suppressed with Pax5, the relative invasion activities were 0.57, 0.98, 0.34, and 0.19 in V5, V9, G5, and G11, respectively, using the lower chamber without serum (Fig. 7Da). Using the lower chamber with serum, the relative invasion activities were 0.95, 1.19, 0.32, and 0.59 in V5, V9, G5, and G11, respectively (Fig. 7Db). Similar effects were also observed with Pax4 in V9 and G5 (Fig. 7 Cc and Dc). These results indicated that p130Cas and paxillin were involved in the invasion activity only in GD3+ cells and suggested the possibility that GD3 expression caused the enhancement of invasion activity by means of the activation of p130Cas and paxillin.
Fig. 7.
Suppression of the invasion activity with siRNA in GD3 synthase gene transfectant cells. (A) Invasion activity of each transfectant was investigated after 2 days of transfection with the control and Cas4 or Cas3 siRNA. (a) The lower chamber was filled with serum-free medium. (b and c) The lower chamber was filled with culture medium containing FCS. (B) Relative inhibition rate of invasion activity. Relative inhibition rate was calculated as the ratio of the number of invaded siRNA Cas4- or siRNA Cas3-treated cells to the control (Luc) siRNA-treated cells. (C) Invasion activity of each transfectant was investigated after transfection with the control (Luc) and Pax5 or Pax4 siRNA. (a) The lower chamber was filled with serum-free medium. (b and c) The lower chamber was filled with culture medium containing FCS. (D) Relative inhibition rate of invasion activity was shown as the ratio of the number of invaded siRNA Pax5- or siRNA Pax4-treated cells to the control siRNA-treated cells. *, P < 0.05; **, P < 0.01.
Discussion
GD3 is a melanoma-associated antigen (3, 4, 8, 9) and has been used as a target for the immune therapy of melanomas (5). The facts that GD3 is highly expressed in melanomas in comparison with melanocytes, is exclusively expressed in the early developmental stage of the fetal brain (10), and is highly expressed in activated T lymphocytes and in T cell malignant tumor cells (12-15) suggest that GD3 is involved in tumor phenotypes, such as enhanced proliferation and frequent metastasis. There have been a number of reports indicating that GD3 is associated with cell adhesion to the extracellular matrix (20), increased cell growth (17), and invasion activity (21). In fact, anti-GD3 antibodies appeared to suppress cultured melanoma growth (16, 22), and GD3-lacking mutants of SK-MEL-28 showed markedly reduced cell proliferation (17), supporting its role in cell growth. Suppression of GD3 expression with antisense GD3 synthase cDNA also resulted in the reduction of tumor cell phenotypes such as growth, migration, metastasis, and angiogenesis in F11 cells (23, 24). However, molecular mechanisms for the enhancement of malignant properties by GD3 have never been demonstrated.
In this study, we established a set of melanoma cell lines with or without GD3 expression by using a GD3-deficient mutant of SK-MEL-28 that was generated by the treatment of the parent cell with anti-GD3 mAb and complement (17). Reexpression of GD3 with GD3 synthase cDNA provided cell lines that enabled us to identify the phenotypic changes due to GD3 expression in the transfectant cells and also to investigate the molecular mechanisms for the enhanced malignant properties correlated with GD3 expression. Because only low GD2 expression appeared and low GM3 expression disappeared in the transfectants, these cells seemed to be useful to analyze the roles of GD3. As expected, cell growth and invasion activity were markedly enhanced after the induction of GD3 expression. Moreover, two proteins (p130Cas and paxillin) were identified as the candidates for the effectors through which GD3 brings about the biological features of melanoma cells. Suppression of these two molecules with siRNA clearly demonstrated their roles in the increased cell growth and/or invasion in GD3+ cells: i.e., p130Cas is involved in both the cell growth and invasion, and paxillin is involved in invasion in these cells. The fact that siRNAs of either p130Cas or paxillin sufficiently suppressed the increased levels of growth and/or invasion in GD3+ cells suggested that the increased cell growth and/or invasion in GD3+ cells was dependent mainly on these two molecules. In contrast, GD3- cells were scarcely affected by these treatments. These clear differential effects of siRNA encourage us to expect that these agents could have therapeutic application for melanoma patients. Analysis of the phosphorylation levels of p130Cas and paxillin in other melanoma cell lines and melanocytes also revealed much higher intensities of these molecules in tumor lines (see the supporting information).
p130Cas was originally defined as a highly tyrosine-phosphorylated molecule during transformation by p60v-src, as well as by v-Crk, forming stable complexes with these oncoproteins, suggesting that it was a direct signal mediator of v-Crk (25). Then, it was reported that the binding of melanoma growth stimulatory activity (MGSA), a CXC chemokine to the class II IL-8 receptor (IL-8RB) (26, 27), enhanced the tyrosine phosphorylation of p130Cas and a 70-kDa protein (28). These results suggest that some growth factor or factors in FCS bind to IL-8RB and induce the tyrosine phosphorylation of p130Cas and paxillin. In a preliminary experiment, MGSA (Groα) stimulation of GD3+ transfectant cells induced tyrosine phosphorylation of the two molecules with a different time course from that with FCS. Furthermore, GD3+ cells expressed IL-8RB at a minimal level. Therefore, we need to consider factors other than MGSA, although it may partially contribute as an autocrine factor to the signals triggered with FCS as reported previously (29, 30).
p130Cas is now considered to be an important adaptor molecule in a variety of biological processes, including cell adhesion (31), migration (32), growth factor stimulation (33), and cytokine receptor engagement (28). It is well known that p130Cas is implicated in many different receptor signaling pathways and interacts with a wide variety of molecules (34). Above all, association with focal adhesion kinase (FAK) and Src family kinases should be important in the development of malignant phenotypes (35). In the null mutant mice (36), p130Cas was shown to be essential in actin filament assembly as well as in the development of heart and blood vessels. Therefore, it seems reasonable to think that p130Cas plays an essential role in the increased growth and invasion in GD3+ transfectant cells, whatever the extrinsic stimulant is. In our IP of p130Cas, it was shown that only a small portion of p130Cas was tyrosine-phosphorylated. This active form might be translocated from cytoplasm to the plasma membrane, as reported (25). Furthermore, the membrane fraction toward which p130Cas moved might be glycolipid-enriched microdomains (37), where GD3 is also enriched in the transfectants. If so, interaction between p130Cas and GD3 in glycolipid-enriched microdomains could be crucial in the phenotypic changes of GD3+ cells.
Paxillin is a multidomain protein that primarily localizes to cell adhesion sites at the extracellular matrix (ECM) called focal adhesions (38). Focal adhesions form a structure link between the ECM and the actin cytoskeleton and are also important sites of signal transduction. Focal adhesion proteins, including paxillin, serve as a point of convergence for signals resulting from stimulation of various growth factor receptors (39). Thus, it also seems reasonable that paxillin is involved in the increased invasion of GD3+ cells. The paxillin and FAK complex plays roles in association with extracellular signal-regulated kinase (ERK) in DNA synthesis (40), survival, and motility (41) and in morphogenesis (42). Correspondingly, GD3+ cells showed higher phosphorylation levels of ERK1/2 and Akt (see the supporting information).
The results with inhibitors of mitogen-activated protein kinase kinase or phosphatidylinositol 3-kinase (PI3-K) suggested that only Akt activation is involved in the process of tyrosine phosphorylation of p130Cas and paxillin, although both the ERK1/2 and PI3-K/Akt pathways seem to be involved in the changes of GD3+ cells. An important issue to be investigated is how these pathways interact with FAK/p130Cas/paxillin in the signal crosstalk.
In the immunocytostaining, colocalization of GD3 and p130Cas or GD3 and paxillin at the leading edges of melanoma cells was observed (see the supporting information). Here, cells seemed to be affected by signals both through unidentified growth factor/receptor(s) and through integrins. Although focal adhesions consisting of integrins, FAK, p130Cas, Src family kinases, and paxillin are considered to be formed based on the interaction between integrin and the extracellular matrix (43), resulting in the migration and fillopodia/lamellipodia formation (44), growth factor/receptors are also involved under proper coordination (45). Coordination of adhesion and growth factor signals might be facilitated by the close physical proximity of key molecules in the signaling cascades (39) and might be regulated by the docking molecules called “platform” (39), such as FAK (46) in the focal adhesion complex.
The remaining issue is how GD3 regulates the input of extrinsic signals on the cell surface. One possibility is that it functions through interactions with integrins as suggested by Cheresh et al. (20). Also, Hakomori (37) proposed interactions between ganglioside and integrin as one of patterns for signal regulation in the membrane microdomain on the cell surface. We have also suggested the possibility that ganglioside GM2 modulates integrin function (47). Simultaneously, GD3 might also interact with unidentified growth factor receptor(s) and enhance the receptor function, as we demonstrated in the study of a rat pheochromocytoma cell line, PC12 (48). The elucidation of molecules associating with GD3 in GD3+ cells is critical.
As for known growth factors effective in the stimulation of the growth in melanocytes/melanomas, basic FGF, platelet-derived growth factor, and EGF have been demonstrated to be responsible. However, these known factors did not show increased tyrosine phosphorylation in any proteins of GD3+ transfectant cells in our study (see Fig. 9 and Table 1). In turn, the treatment of GD3+ transfectant cells with FCS showed markedly increased tyrosine phosphorylation and enabled us to identify p130Cas and paxillin as actual effectors involved in inducing some malignant features of GD3+ melanoma cells. Although these effectors were discovered with FCS treatment, results of the following function analyses suggested that they are critical molecules in the growth and/or invasion of GD3+ melanomas, regardless of the real stimulants. These results strongly suggested that p130Cas and paxillin are widely involved in the malignant properties of melanoma cell lines and melanoma tissues (28). Thus, expression levels and tyrosine phosphorylation levels of these molecules in melanoma cell lines and melanoma tissues deserve to be investigated.
Supplementary Material
Acknowledgments
We thank Ms. T. Mizuno and Ms. Y. Nakayasu for technical assistance. This work was supported by a Grant-in-Aid for Scientific Research on Priority Areas (14082102) and a grant for Core Research for Evolutional Science and Technology from the Ministry of Education, Culture, Sports, Science, and Technology of Japan.
Author contributions: K.H., Keiko Furukawa, M.U., K.O.L., and Koichi Furukawa designed research; K.H., Keiko Furukawa, T. Hayashi, T. Hattori, J.N., H.N., T.O., H.M., H.H., T.U., and Koichi Furukawa performed research; K.H., Keiko Furukawa, T. Hayashi, T. Hattori, J.N., H.N., H.M., and Koichi Furukawa contributed new reagents/analytic tools; K.H., Keiko Furukawa, T. Hayashi, T. Hattori, T.O., H.H., M.U., T.U., K.O.L., and Koichi Furukawa analyzed data; and K.H., Keiko Furukawa, and Koichi Furukawa wrote the paper.
Abbreviations: MTT, 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyl tetrazolium bromide; IP, immunoprecipitation; IB, immunoblotting; FAK, focal adhesion kinase; siRNA, small interfering RNA.
References
- 1.Furukawa, K. & Lloyd, K. O. (1990) in Human Melanoma: From Basic Research to Clinical Application, ed. Ferrone, S. (Springer, Heidelberg), pp. 15-30.
- 2.Lloyd, K. O., Travassos, L. R., Takahashi, T. & Old, L. J. (1979) J. Natl. Cancer Inst. 63, 623-634. [DOI] [PubMed] [Google Scholar]
- 3.Dippold, W. G., Lloyd, K. O., Li, L. T., Ikeda, H., Oettgen, H. F. & Old, L. J. (1980) Proc. Natl. Acad. Sci. USA 77, 6114-6118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Pukel, C. S., Lloyd, K. O., Travassos, L. R., Dippold, W. G., Oettgen, H. F. & Old, L. J. (1982) J. Exp. Med. 155, 1133-1147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Houghton, A. N., Mintzer, D., Cordon-Cardo, C., Welt, S., Fliegel, B., Vadhan, S., Carswell, E., Melamed, M. R., Oettgen, H. F. & Old, L. J. (1985) Proc. Natl. Acad. Sci. USA 82, 1242-1246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Brichard, V., Van Pel, A., Wolfel, T., Wolfel, C., De Plaen, E., Lethe, B., Coulie, P. & Boon, T. (1993) J. Exp. Med. 178, 489-495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Livingston, P. O., Adluri, S., Helling, F., Yao, T. J., Kensil, C. R., Newman, M. J. & Marciani, D. (1994) Vaccine 12, 1275-1280. [DOI] [PubMed] [Google Scholar]
- 8.Portoukalian, J., Zwingelstein, G. & Dore, J. F. (1979) Eur. J. Biochem. 94, 19-23. [DOI] [PubMed] [Google Scholar]
- 9.Carubia. J. M., Yu, R. K., Macala, L. J., Kirkwood, J. M. & Varga, J. M. (1984) Biochem. Biophys. Res. Commun. 120, 500-504. [DOI] [PubMed] [Google Scholar]
- 10.Yu, R. K., Macala, L. J., Taki, T., Weinfield, H. M. & Yu, F. S. (1988) J. Neurochem. 50, 1825-1829. [DOI] [PubMed] [Google Scholar]
- 11.Siddiqui, B., Buehler, J., DeGregorio, M. W. & Macher, B. A. (1984) Cancer Res. 44, 5262-5265. [PubMed] [Google Scholar]
- 12.Merritt, W. D., Casper, J. T., Lauer, S. J. & Reaman, G. H. (1987) Cancer Res. 47, 1724-1730. [PubMed] [Google Scholar]
- 13.Okada, M., Furukawa, K., Yamashiro, S., Yamada, Y., Haraguchi, M., Horibe, K., Kato, K., Tsuji, Y. & Furukawa, K. (1996) Cancer Res. 56, 2844-2848. [PubMed] [Google Scholar]
- 14.Furukawa, K., Akagi, T., Nagata, Y., Yamada, Y., Shimotohno, K., Cheung, N. K., Shiku, H. & Furukawa, K. (1993) Proc. Natl. Acad. Sci. USA 90, 1972-1976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Yamashiro, S., Okada, M., Haraguchi, M., Furukawa, K., Lloyd, K. O., Shiku, H. & Furukawa, K. (1995) Glycoconj. J. 12, 894-900. [DOI] [PubMed] [Google Scholar]
- 16.Dippold, W. G., Knuth, A. & Meyer zum Buschenfelde, K. H. (1984) Cancer Res. 44, 806-810. [PubMed] [Google Scholar]
- 17.Nakano, J., Raj, B. K., Asagami, C. & Lloyd, K. O. (1996) J. Invest. Dermatol. 107, 543-548. [DOI] [PubMed] [Google Scholar]
- 18.Haraguchi, M., Yamashiro, S., Yamamoto, A., Furukawa, K., Takamiya, K., Lloyd, K. O., Shiku, H. & Furukawa, K. (1994) Proc. Natl. Acad. Sci. USA 91, 10455-10459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Yoshida, S., Fukumoto, S., Kawaguchi, H., Sato, S., Ueda, R. & Furukawa, K. (2001) Cancer Res. 61, 4244-4252. [PubMed] [Google Scholar]
- 20.Cheresh, D. A., Pierschbacher, M. D., Herzig, M. A. & Mujoo, K. (1986) J. Cell Biol. 102, 688-696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Iliopoulos, D., Ernst, C., Steplewski, Z., Jambrosic, J. A., Rodeck, U., Herlyn, M., Clark, W. H., Jr., Koprowski, H. & Herlyn, D. (1989) J. Natl. Cancer Inst. 81, 440-444. [DOI] [PubMed] [Google Scholar]
- 22.Miyazaki, H., Shiku, H. & Furukawa, K. (1996) Int. J. Oncol. 9, 241-245. [DOI] [PubMed] [Google Scholar]
- 23.Zeng, G., Gao, L., Birkle, S. & Yu, R. K. (2000) Cancer Res. 60, 6670-6676. [PubMed] [Google Scholar]
- 24.Zeng, G., Gao, L. & Yu, R. K. (2000) Int. J. Cancer 88, 53-57. [DOI] [PubMed] [Google Scholar]
- 25.Sakai, R., Iwamatsu, A., Hirano, N., Ogawa, S., Tanaka, T., Mano, H., Yazaki, Y. & Hirai, H. (1994) EMBO J. 13, 3748-3756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lee, J., Horuk, R., Rice, G. C., Bennett, G. L., Camerato, T. & Wood, W. I. (1992) J. Biol. Chem. 267, 16283-16287. [PubMed] [Google Scholar]
- 27.Cheng, Q. C., Han, J. H., Thomas, H. G., Balentien, E. & Richmond, A. (1992) J. Immunol. 148, 451-456. [PubMed] [Google Scholar]
- 28.Schraw, W. & Richmond, A. (1995) Biochemistry 34, 13760-13767. [DOI] [PubMed] [Google Scholar]
- 29.Sauvaigo, S., Fretts, R. E., Riopelle, R. J. & Lagarde, A. E. (1986) Int. J. Cancer 37, 123-132. [DOI] [PubMed] [Google Scholar]
- 30.Schadendorf, D., Moller, A., Algermissen, B., Worm, M., Sticherling, M. & Czarnetzki, B. M. (1993) J. Immunol. 151, 2667-2675. [PubMed] [Google Scholar]
- 31.Nojima, Y., Morino, N., Mimura, T., Hamasaki, K., Furuya, H., Sakai, R., Sato, T., Tachibana, K., Morimoto, C., Yazaki, Y. & Hirai, H. (1995) J. Biol. Chem. 270, 15398-15402. [DOI] [PubMed] [Google Scholar]
- 32.Cary, L. A., Han, D. C., Polte, T. R., Hanks, S. K. & Guan, J. L. (1998) J. Cell Biol. 140, 211-221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ribon, V. & Saltiel, A. R. (1996) J. Biol. Chem. 271, 7375-7380. [DOI] [PubMed] [Google Scholar]
- 34.O'Neill, G. M., Fashena, S. J. & Golemis, E. A. (2000) Trends Cell Biol. 10, 111-119. [DOI] [PubMed] [Google Scholar]
- 35.Polte, T. R. & Hanks, S. K. (1997) J. Biol. Chem. 272, 5501-5509. [DOI] [PubMed] [Google Scholar]
- 36.Honda, H., Oda, H., Nakamoto, T., Honda, Z., Sakai, R., Suzuki, T., Saito, T., Nakamura, K., Nakao, K., Ishikawa, T., et al. (1998) Nat. Genet. 19, 361-365. [DOI] [PubMed] [Google Scholar]
- 37.Hakomori, S. (2002) Proc. Natl. Acad. Sci. USA 99, 10231-10233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Turner, C. E. (2000) J. Cell Sci. 23, 4139-4140. [DOI] [PubMed] [Google Scholar]
- 39.Turner, C. E. (2000) Nat. Cell Biol. 2, E231-E236. [DOI] [PubMed] [Google Scholar]
- 40.Ruhl, M., Johannsen, M., Atkinson, J., Manski, D., Sahin, E., Somasundaram, R., Riecken, E. O. & Schuppan, D. (1999) Exp. Cell Res. 250, 548-557. [DOI] [PubMed] [Google Scholar]
- 41.Subauste, M. C., Pertz, O., Adamson, E. D., Turner, C. E., Junger, S. & Hahn, K. M. (2004) J. Cell Biol. 165, 371-381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ishibe, S., Joly, D., Liu, Z. X. & Cantley, L. G. (2004) Mol. Cell 16, 257-267. [DOI] [PubMed] [Google Scholar]
- 43.Zaidel-Bar, R., Ballestrem, C., Kam, Z. & Geiger, B. (2003) J. Cell Sci. 116, 4605-4613. [DOI] [PubMed] [Google Scholar]
- 44.Aznavoorian, S., Stracke, M. L., Parsons, J., McClanahan, J. & Liotta, L. A. (1996) J. Biol. Chem. 271, 3247-3254. [DOI] [PubMed] [Google Scholar]
- 45.Moro, L., Dolce, L., Cabodi, S., Bergatto, E., Erba, E. B., Smeriglio, M., Turco, E., Retta, S. F., Giuffrida, M. G., Venturino, M., et al. (2002) J. Biol. Chem. 277, 9405-9414. [DOI] [PubMed] [Google Scholar]
- 46.Hsia, D. A., Mitra, S. K., Hauck, C. R., Streblow, D. N., Nelson, J. A., Ilic, D., Huang, S., Li, E., Nemerow, G. R., Leng, J., et al. (2003) J. Cell Biol. 160, 753-767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Chen, H. H., Fukumoto, S., Furukawa, K., Nakao, A., Akiyama, S., Urano, T. & Furukawa, K. (2003) Int. J. Cancer 103, 169-176. [DOI] [PubMed] [Google Scholar]
- 48.Fukumoto, S., Mutoh, T., Hasegawa, T., Miyazaki, H., Okada, M., Goto, G., Furukawa, K., Urano, T. & Furukawa, K. (2000) J. Biol. Chem. 275, 5832-5838. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.