Abstract
Gaucher disease results from an autosomal recessive deficiency of the lysosomal enzyme glucocerebrosidase. The glucocerebrosidase gene is located in a gene-rich region of 1q21 that contains six genes and two pseudogenes within 75 kb. The presence of contiguous, highly homologous pseudogenes for both glucocerebrosidase and metaxin at the locus increases the likelihood of DNA rearrangements in this region. These recombinations can complicate genotyping in patients with Gaucher disease and contribute to the difficulty in interpreting genotype-phenotype correlations in this disorder. In the present study, DNA samples from 240 patients with Gaucher disease were examined using several complementary approaches to identify and characterize recombinant alleles, including direct sequencing, long-template polymerase chain reaction, polymorphic microsatellite repeats, and Southern blots. Among the 480 alleles studied, 59 recombinant alleles were identified, including 34 gene conversions, 18 fusions, and 7 downstream duplications. Twenty-two percent of the patients evaluated had at least one recombinant allele. Twenty-six recombinant alleles were found among 310 alleles from patients with type 1 disease, 18 among 74 alleles from patients with type 2 disease, and 15 among 96 alleles from patients with type 3 disease. Several patients carried two recombinations or mutations on the same allele. Generally, alleles resulting from nonreciprocal recombination (gene conversion) could be distinguished from those arising by reciprocal recombination (crossover and exchange), and the length of the converted sequence was determined. Homozygosity for a recombinant allele was associated with early lethality. Ten different sites of crossover and a shared pentamer motif sequence (CACCA) that could be a hotspot for recombination were identified. These findings contribute to a better understanding of genotype-phenotype relationships in Gaucher disease and may provide insights into the mechanisms of DNA rearrangement in other disorders.
Introduction
In mammalian cells, DNA rearrangement occurs by chromosomal pairing, breakage, and crossover between two nonsister chromatids during meiosis and is an essential cellular process. However, recombination between homologous sequences of DNA may also introduce harmful mutations resulting from the unequal pairing of chromosomes. The frequency of nonequal chromosomal pairing and DNA rearrangement is influenced by the degree of DNA sequence homology that exists between corresponding segments on the two chromosomes. The occurrence of homologous recombination can be enhanced by the presence of highly repetitive DNA sequence families, low-copy repeats, a processed or nonprocessed pseudogene sequence, or the duplication of a region within the locus (Chen et al. 1997; Purandare and Patel 1997; Strachan and Read 1999; Stankiewicz and Lupski 2002a). Misalignment and recombination of homologous sequences cause DNA rearrangements that lead to genetic disorders and include deletions, duplications, inversions, fusions, and gene conversions (Mazzarella and Schlessinger 1998; Emanuel and Shaikh 2001; Hurles 2001; Inoue et al. 2001; Stankiewicz and Lupski 2002b). Human disorders resulting from DNA rearrangements due to structural features of the genome have been referred to as “genomic disorders” (Lupski 1998). Analyzing the results of homologous recombination in human genes can provide insights into the mechanisms of DNA rearrangement and may also enhance our understanding of factors contributing to the complexity seen in specific disorders. In the present study, we have evaluated the frequency and possible phenotypic consequences of crossover events and gene conversion occurring between the glucocerebrosidase (GBA) gene and its nonprocessed pseudogene located on the same chromosome.
The human GBA gene (GenBank accession number J03059), located on chromosome 1q21, has a pseudogene (GenBank accession number J03060) that shares 96% exonic sequence homology, located 16 kb downstream of the functional gene (Horowitz et al. 1989; Winfield et al. 1997). The 1q21 region is gene rich, with seven genes and two pseudogenes located in close proximity (GenBank accession number AF023263) (Winfield et al. 1997). The gene for metaxin (MTX) is located immediately downstream of the GBA pseudogene and is convergently transcribed, encoding a protein of 317 amino acids that appears to be part of a preprotein import complex in the outer membrane of mitochondria (Armstrong et al. 1997). MTX also has a pseudogene located between the GBA gene and pseudogene, immediately adjacent to the 3′ end of the functional GBA gene (Long et al. 1996). These genes can serve as a model to demonstrate the effects of unequal pairing and recombination on gene function.
Gaucher disease (MIM 230800), the autosomal recessive inherited deficiency of lysosomal glucocerebrosidase (EC 3.2.1.45), presents with many diverse clinical phenotypes (Stone et al. 2000; Beutler and Grabowski 2001). On the basis of the presence and rate of progression of neurological symptoms, Gaucher disease has been divided into type 1 (nonneuronopathic), type 2 (acute neuronopathic), and type 3 (subacute neuronopathic) forms. The gene for glucocerebrosidase is organized into 11 exons coding for 498 amino acids, and, to date, >200 different mutations have been identified (Beutler and Gelbart 1998; Hodanova et al. 1999; Koprivica et al. 2000; Orvisky et al. 2002; GeneDis Databank). Studies of genotype-phenotype correlation indicate that clinically similar patients have many different genotypes and that patients sharing the same PCR-defined point mutations can have a spectrum of clinical manifestations (Koprivica et al. 2000; Zhao and Grabowski 2002). Most mutations resulting in Gaucher disease are point mutations that are distributed throughout the gene, with a majority clustered between exons 8 and 11. Several missense mutations, particularly N370S and L444P, are encountered with an increased frequency. Other mutations include frameshifts, deletions (with or without insertions), and splice-site mutations.
Another important group of mutations result from homologous recombination between the GBA gene and pseudogene. Patients with a recombinant allele containing three single point mutations—two that introduce the amino acid substitutions L444P and A456P, and a silent mutation, V460V, in exon 10 (Rec NciI)—were first reported in 1990 (Eyal et al. 1990; Latham et al. 1990). This allele is referred to as “recombinant 1” (Rec 1) in the present study. Subsequently, another recombinant allele was described, encompassing the same three changes with an additional amino acid substitution, D409H, in exon 9 (Rec TL or Rec 2) (Eyal et al. 1990; Zimran and Horowitz 1994). Homologous recombination resulting in a fusion that includes the 5′ segment of the GBA functional gene extending from exon 1 to intron 9, with the remainder of the sequence corresponding to the pseudogene, was shown by Zimran et al. (1990). Strasberg et al. (1994) first reported homozygosity for a recombinant allele in a patient with lethal type 2 Gaucher disease. Another type of mutant allele, a 55-bp deletion in exon 9 resulting from gene conversion from the pseudogene sequence, has also been reported (Walley and Harris 1993; Beutler et al. 1995; Tayebi et al. 1996b). Furthermore, recombinant alleles that include the 55-bp deletion, D409H, L444P, A456P, and V460V (Rec 3) have been identified, in which the site of crossover is located within the region extending from the end of intron 8 to the beginning of exon 9 (Hatton et al. 1997; Tayebi et al. 1998). Several reports have described fusion alleles in which the recombination occurred in the upstream portion of the gene, including introns 2, 3, and 6, introducing a larger segment of pseudogene into the recombinant allele (Reissner et al. 1998; Cormand et al. 2000; Filocamo et al. 2000). Patients with a crossover site in the 3′ UTR of the GBA gene or the adjacent MTX pseudogene have also been identified (Stone et al. 2000; Tayebi et al. 2000, 2001).
To improve upon previous genotype-phenotype studies, we performed detailed molecular analyses of the GBA gene mutations in 480 alleles from clinically diverse patients with Gaucher disease. Because the high sequence homology between genes and pseudogenes at this locus increases the likelihood of recombination, we set out to determine the frequency, type, location, and mechanisms of recombination and whether different recombinant alleles had different phenotypic consequences.
Subjects and Methods
Patients
DNA samples from 240 patients with Gaucher disease were analyzed, including 155 patients with type 1, 37 patients with type 2, and 48 patients with type 3 Gaucher disease. Medical records were reviewed. DNA was also collected from selected family members. The patients and relatives provided informed consent through a protocol approved by the National Institute of Mental Health institutional review board. The series also included several cell lines purchased from the National Institute of General Medical Sciences Human Genetic Mutant Cell Repository.
DNA Preparation
High-molecular-weight DNA was isolated from lymphocytes, cultured fibroblasts, and/or EBV transformed lymphoblast cell lines from normal, affected, and carrier individuals, by standard protocols (Sambrook et al. 1989).
Mutation Analysis
Genomic DNA samples were screened for five common point mutations—N370S, L444P, R463C, c.84-85insG, and IVS2+1G→A—and the 55-bp deletion, through use of PCR, the amplification refraction mutation system (ARMS), restriction-enzyme digestion, and direct sequencing methods, as described by Mistry et al. (1992) and Tayebi et al. (1996a, 1997). After this screening, DNA from any patient with an unidentified mutation, a homozygous genotype, or a mutant allele that derived from pseudogene sequence was subjected to further analysis.
To confirm the presence of the mutations identified by screening and to search for unknown mutations, each of the 11 exonic regions of the GBA gene, including flanking intronic sequences and a 1-kb fragment of the 5′ UTR, were selectively amplified using PCR primers that were designed to amplify the functional GBA gene but not the pseudogene (Reissner et al. 1998; Stone et al. 2000). Direct sequencing of the PCR products in both directions was performed by cycle sequencing using the fluorescent dideoxy termination method according to standard protocols. Fluorometric sequences were analyzed using the 373A DNA Sequencer (Applied Biosystems).
Long-template PCR was performed in selected cases, as described by Tayebi et al. (1996a, 1998). The bands corresponding to the recombinant alleles were isolated and sequenced to confirm the recombination and the site of crossover.
Southern Blot Analyses
Genomic DNA (10–15 μg) was isolated from patients and normal control individuals and was digested overnight at 37°C with the restriction enzymes SstII, SspI, EcoRI, or HincII. The digested DNAs were electrophoresed on a 0.6% I.D.NA (FMC Bioproducts) agarose gel for 16–18 h (for SspI and HincII) or for 40–48 h (for the restriction enzymes SstII and EcoRI). The gels were transferred to a supported nitrocellulose filter (Schleicher and Schuell) and were hybridized with 32P-labeled GBA cDNA or MTX cDNA probes.
In addition, Southern blots prepared with HincII-digested DNA were hybridized to three shorter GBA cDNA probes. The first probe was a 250-bp fragment encompassing exons 2–4, which was amplified using the forward primer 5′-CAGGCAGTGTCGTGGGCATC-3′ and the reverse primer 5′-CAGAACAGAAGTTCCAGAAA-3′ (annealing temperature 60°C). A second probe, a 476-bp fragment covering exons 4–7, was amplified with the forward primer 5′-CTGCTGCTCTCAACATCCTT-3′ and the reverse primer 5′-TGTTGAGTGGATACCCCTTC-3′ (annealing temperature 60°C), and a third 762-bp fragment, containing exons 7–11, was amplified using the forward primer 5′-TTCCTGGATGCCTATGCTGA-3′ and the reverse primer 5′-GCTCCTCTAAGGATGTCCC-3′ (annealing temperature 60°C). Southern blots of HincII-digested DNA were also hybridized to an MTX cDNA probe.
Analysis of Polymorphic Sites
Genotypes at two polymorphic microsatellite sites, a dinucleotide (CT) repeat located upstream of the GBA gene and a tetranucleotide (AAAT) repeat located downstream of the GBA gene, were determined as described by Lau et al. (1999).
Analysis of Motif Sequences
To identify DNA regions representing shared motif sequences at specific crossover sites in the GBA gene, all sequences surrounding the approximate sites of recombination were aligned using the GCG and Sequencher software programs (Gene Codes).
Nomenclature for Recombinant Alleles
The approximate initiation point for a gene conversion or reciprocal recombination event was determined by examination of known mismatches between the functional gene and pseudogene sequences. The crossover region was defined as the sequence between the last mismatch corresponding to the functional GBA gene and the first mismatch corresponding to pseudogene. In the case of gene conversions, the length of converted sequence was determined by identifying where the functional gene sequence resumed.
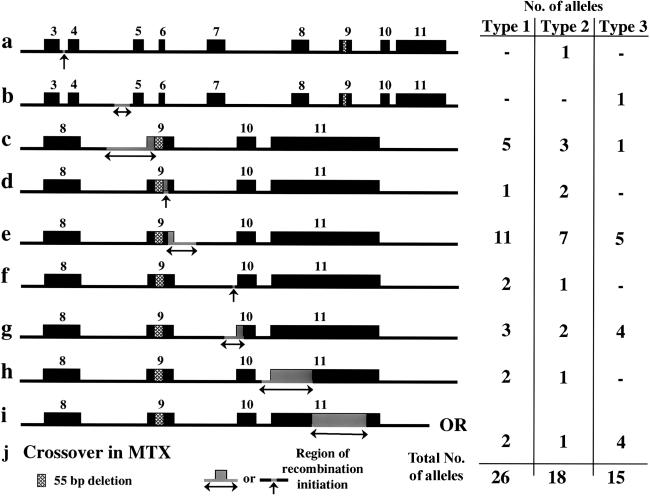
The different recombinant alleles are referred to as Rec 1–7, depending upon the exonic site at which pseudogene sequence was first detected (table 1). Recombinant alleles resulting from a nonreciprocal event such as gene conversion are further defined with an “a.” These alleles carried only a small converted segment originating from the pseudogene (table 2). Recombinant alleles resulting from reciprocal recombination are designated with a “b” (table 3). These alleles may carry either a fusion between the gene and pseudogene or a duplicated allele.
Table 1.
Recombinant Alleles Resulting from Crossover or Gene Conversion between the GBA Gene and Pseudogene
| Recombination | Description |
| Rec 1a or b | The allele carries mutations L444P, A456P, and V460V. Crossover sites include Ex9-Int-9, Int-9 or Int9-Ex10. (See fig. 1e, 1f, and 1g). |
| Rec 2a or b | The allele carries mutations D409H, L444P, A456P, and V460V. Crossover site is in Ex9. (See fig. 1d). |
| Rec 3a or b | The allele carries the 55-bp deletion in exon 9, as well as point mutations D409H, L444P, A456P, and V460V. Crossover site is in the Int8-Ex9 region (see fig. 1c). |
| Rec 4a or b | The allele carries pseudogene sequence beginning from intron 3. Crossover site is in Int3 (see fig. 1a). |
| Rec 5a or b | The allele carries the pseudogene sequence beginning from intron 4. Crossover site is in Int4 (see fig. 1b). |
| Rec 6a or b | The allele carries the pseudogene sequence beginning from intron 10 or has a duplicated pseudogene with a fusion between metaxin and pseudometaxin. Crossover site is in the Int10-Ex11 region (see fig. 1h). |
| Rec 7a or b | The allele carries the pseudogene sequence beginning from the 3′ UTR or has a duplicated pseudogene with a fusion between MTX and pseudometaxin. Crossover site is in the 3′ UTR (see fig. 1i) or in MTX. |
Table 2.
Genotype/Phenotype Data on Patients with an Allele Resulting from Nonreciprocal Recombination[Note]
| DiseaseType andPatientNumber | Ethnic Background | Age atDiagnosis/Evaluation(years) | Genotypea | Site WhereGeneConversionStartsb | MaximumPossibleSize ofConvertedSequence(bp) | Southern Blot Results | Clinical Features |
| Type 1: | |||||||
| 1 | European American | 2/3 | N370S/Rec 1a |
Ex9-Int9 | 824 | Normal | Hepatosplenomegaly, anemia, thrombocytopenia |
| 2 | Ashkenazi | NA | N370S/c.1263-1317del-a |
Int8-Ex9 | 288 | Missing exon 9 site on HincII | Hepatosplenomegaly, anemia, thrombocytopenia |
| 3 | Hispanic | 13/48 | N370S/Rec 1a |
Ex9-Int9 | 824 | Normal | Splenectomy, hepatomegaly, bone disease |
| 4 | African American | 14/15 | c.222-224delTAC/Rec 1a+Rec 6b |
Ex9-Int9 | 824 | Pseudogene duplication | Hepatosplenomegaly, anemia, thrombocytopenia |
| 5 | Hispanic | 10/34 | N370S/Rec 1a |
Ex9-Int9 | 824 | Normal | Splenectomy, hepatomegaly, bone disease |
| 6 | European American | 4/8 | N370S/c.1263-1317del-a |
Int8-Ex9 | 288 | Missing exon 9 site on HincII | Hepatosplenomegaly |
| 7 | African American | 2/5 | V352L/Rec 1a+Rec7b |
Ex9-Int9 | 824 | Pseudogene duplication | Splenectomy, hepatomegaly |
| 8 | European American | 6/21 | N370S/Rec 1a |
Int9-Ex10 | 475 | Normal | Hepatosplenomegaly, thrombocytopenia, bone disease |
| 9 | European American | 8/28 | N370S/L444Pa |
Int9 | 194 | Normal | Splenectomy, hepatomegaly, bone disease |
| 10 | French | NA | N370S/Rec 1a |
Int9 | 824 | Normal | Parkinsonism |
| 11 | African American | 1/1 | N117D/Rec 1a |
Int9-Ex10 | 424 | Normal | Splenectomy, bone disease, hepatomegaly, anemia, thrombocytopenia |
| 12 | European American | 13/16 | N370S/Rec 1a |
Ex9-Int9 | 824 | Normal | Hepatosplenomegaly, thrombocytopenia |
| 13 | NA | NA | N370S/c.1263-1317del-a |
Int8-Ex9 | 288 | NA | NA |
| 14 | French | 24 | N370S/c.1263-1317del-a |
Int8-Ex9 | 288 | Missing exon 9 site on HincII | Thrombocytopenia, splenectomy, parkinsonism |
| 15 | NA | NA | N370S/Rec 1a |
Ex9–Int9 | 824 | Normal | NA |
| Type 2: | |||||||
| 16 | European American | 5 mo | L444P/Rec 1a |
Int9-Ex10 | 424 | Normal | Hepatosplenomegaly, seizures |
| 17 | Greek | Birth | H225Q/Rec 3a |
Int8-Ex9 | 849 | Normal | Hepatosplenomegaly, seizures, opisthotonus |
| 18 | NA | Birth | Rec 1a/Rec la |
Ex9-Int9 | 824 | Normal | Hydrops |
| 19 | Lebanese | Birth | Rec 1a/Rec 1a |
Ex9-Int9 | 824 | Normal | Hydrops |
| 20 | Australian | Birth | R257Q/c.1263-1317del-a |
Int8-Ex9 | 288 | Normal | Ichthyosis, hepatosplenomegaly |
| 21 | European American | Birth | Rec 2a/Rec 2a |
Ex9 | 1193 | Missing exon 9 site on HincII | Hydrops |
| 22 | European American | NA | L444P/Rec 1a |
Int9-Ex10 | 424 | Normal | NA |
| 23 | European American | NA | R285H/c. 1263-1317del-a |
Int8-Ex9 | 288 | Missing exon 9 site on HincII | NA |
| 24 | European American | 11 mo | L444P/Rec 1a |
Ex9-Int9 | 824 | Normal | Hepatosplenomegaly, seizures, opisthotonus |
| Type 3: | |||||||
| 25 | European American | 1/6 | D409H/Rec 1a |
Ex9-Int9 | 824 | Normal | Hepatosplenomegaly, cardiac fibrosis, abnormal horizontal saccades |
| 26 | European American | 4/28 | F216Y/c.1263-1317del-a |
Int8-Ex9 | 288 | Missing exon 9 site on HincII | Renal disease, bone disease, myoclonic epilepsy |
| 27 | European American | 4/13 | R463C/Rec 1a+Rec7b |
Ex9-Int9 | 824 | Pseudogene duplication | Abnormal horizontal saccades, splenectomy, hepatomegaly, bone disease |
| 28 | Ashkenazi/Sephardic | 5/9 | V394L/Rec 1a |
Int9-Ex 10 | 824 | Normal | Abnormal horizontal saccades, hepatosplenomegaly, myoclonic epilepsy, developmental delay, lung disease |
| 29 | African American | 38/40 | N188S/Rec 1a |
Int9-Ex10 | 824 | Normal | Hepatosplenomegaly, abnormal horizontal saccades, myoclonic epilepsy |
| 30 | French | 1/4 | V394L/Rec 1a |
Int9-Ex10 | 824 | Normal | Hepatosplenomegaly, abnormal horizontal saccades, myoclonic epilepsy, developmental delay |
| 31 | Mexican | 4/6 | F213I/L444Pa |
Ex9-Int9 | 510 | Normal | Hepatosplenomegaly, abnormal horizontal saccades, myoclonic epilepsy, developmental delay |
Note.— NA = not available.
Alleles discussed in this table are underlined.
Refer to figure 1 for precise description of sites.
Table 3.
Genotype/Phenotype Data on Patients with an Allele Resulting from Reciprocal Recombination[Note]
| Disease Type andPatient Number | Ethnic Background | Age atDiagnosis/Evaluation(years) | Genotypea | Site of Crossoverb | Southern Blot Results | Clinical Features |
| Type 1: | ||||||
| 1 | European American | 1/12 | N370S/Rec 2b |
Ex9 | Fusion | Hepatosplenomegaly, anemia, thrombocytopenia |
| 2 | African American | 2/5 | R170C/L444P+Rec 7b |
3′ UTR or MTX | Pseudogene duplication | Hepatosplenomegaly, anemia, thrombocytopenia, bone disease |
| 3 | European American | 3/18 | R463C/Rec 1b |
Ex9-Int9 | Fusion | Splenectomy, hepatomegaly, thrombocytopenia, renal disease |
| 4 | European American | 1/7 | N370S/Rec 1b |
Int9-Ex10 | Fusion | Hepatosplenomegaly |
| 5 | African American | 14/15 | c.222-224delTAC/Rec 1a+Rec 6b |
Int10-Ex11 | Pseudogene duplication | Hepatosplenomegaly, anemia, thrombocytopenia |
| 6 | African American | 2/5 | V352L/Rec 1a+Rec 7b |
3′ UTR or MTX | Pseudogene duplication | Splenectomy, hepatomegaly, bone disease |
| 7 | African American | 13/22 | c.153-154insTACAGC/Rec 1b |
Ex9-Int9 | Fusion | Hepatosplenomegaly, anemia, thrombocytopenia, bone disease |
| 8 | NA | NA | N370S/Rec 1b |
Ex9-Int9 | Fusion | NA |
| 9 | European American | 30/30 | N370S/Rec 3b |
Int8-Ex9 | Fusion | Hepatosplenomegaly, bone disease, thrombocytopenia |
| 10 | European American | 5/28 | N370S/Rec 1b |
Int9-Ex10 | Fusion | Hepatosplenomegaly, thrombocytopenia |
| 11 | European American | 10/21 | R463C/Rec 1b |
Ex9-Int9 | Fusion | Splenectomy, anemia, thrombocytopenia, liver disease |
| Type 2: | ||||||
| 12 | European American | 6 mo | R257Q/L444P+Rec 6b |
Int10-Ex11 | Fusion | Hepatosplenomegaly, seizures |
| 13 | Ashkenazi | Fetus | IVSl0+2T→G/Rec 4b |
Int3 | Fusion | Hydrops |
| 14 | African American | 8 mo | E41K/Rec 1b |
Int9 | Fusion | Hepatosplenomegaly, seizures, developmental delay |
| 15 | European American | NA | Rec 1b/Rec 1b |
Ex9-Int9 | Fusion | NA |
| 16 | Mexican | NA | L444P+E326K/L444P+Rec 7b |
MTX | Fusion in MTX | Hepatosplenomegaly, opisthotonus |
| Type 3: | ||||||
| 17 | Korean | 22/22 | N188S/Rec 5b |
Int4 | Fusion | Myoclonic epilepsy |
| 18 | European American | 3/13 | R463C/Rec 1b |
Ex9-Int9 | Fusion | Hepatosplenomegaly, abnormal horizontal saccades |
| 19 | European American | 4/13 | R463C/Rec 1a+Rec 7b |
3′ UTR or MTX | Pseudogene duplication | Hepatomegaly, abnormal horizontal saccades, splenectomy, bone disease |
| 20 | European American | 18/45 | L444P/D409H+Rec 7b |
MTX | Pseudogene duplication | Parkinsonism, splenectomy |
| 21 | European American | 7/28 | N188S/Rec 1b |
Int9-Ex10 | Fusion | Abnormal horizontal saccades, myoclonic epilepsy, dementia |
| 22 | European American | 5/9 | R463C/Rec 1b |
Ex9-Int9 | Fusion | Hepatomegaly, splenectomy, lung disease, bone disease, abnormal horizontal saccades |
| 23 | European American | 1/1 | L444P/L444P+Rec 7b |
3′ UTR or MTX | Pseudogene duplication | Hepatosplenomegaly, abnormal horizontal saccades, developmental delay |
| 24 | European American | 1/4 | G202R/L444P+Rec 7b |
3′ UTR or MTX | Pseudogene duplication | Abnormal horizontal saccades, developmental delay, myoclonic epilepsy |
Note.— NA = not available.
Alleles discussed in this table are underlined.
Refer to figure 1 for precise description of sites.
The site at which conversion begins in the “a” group can be similar to the site of crossover observed in the “b” group (tables 1, 2, and 3). For example, although Rec 1a and Rec 1b both have sequence in exon 10 derived from the pseudogene, Rec 1a results from a gene conversion and Rec 1b is a fusion allele. Because the beginning and extent of a crossover or gene conversion can be detected only by the identification of pseudogene sequence that is not homologous to that of the functional gene, the location of the initiation of the recombination is not exact. Although seven different groups of recombinations are defined, 10 different sites of recombination are described, as shown in figure 1.
Figure 1.
Recombination initiation sites and allele frequencies. The initiation regions for recombination events were determined by comparing mismatches between the GBA functional gene and pseudogene sequences. The region where a recombination starts is defined as the sequence between the last mismatch containing functional gene sequence and the first mismatch carrying pseudogene sequence. The schematics show the exonic structure of GBA, with the different initiation regions indicated in gray, with arrows below. The number of alleles found in patients with type 1, 2, or 3 Gaucher disease with either reciprocal or nonreciprocal recombinations starting within the given region are shown next to each schematic map. The abbreviations for each initiation region are used in tables 1, 2, and 3. a, Int3; the recombination starts within 26 bp in intron 3. b, Int4; the recombination starts within 98 bp in intron 4. c, Int8-Ex9; the recombination starts within 288 bp encompassing the intron 8/exon 9 junction. d, Ex9; the recombination starts within 23 bp of exon 9 following the 55 bp that are deleted in pseudogene. e, Ex9-Int9; the recombination starts within 186 bp encompassing the exon 9/intron 9 junction. f, Int9; the recombination starts within 30 bp at the end of intron 9. g, Int9-Ex10; the recombination starts within 126 bp encompassing the intron 9/exon 10 junction. h, Int10-Ex11; the recombination starts within 300 bp encompassing the intron 10/exon 11 junction. i, 3′ UTR; the recombination starts within 304 bp in the 3′ UTR. j, MTX (not shown); the recombination starts within the MTX pseudogene.
Results
We analyzed 480 alleles from 240 patients with Gaucher disease. This analysis included mutation detection and total and/or partial sequencing of the 11 exons and flanking intronic regions in 422 alleles. In ∼20% of these alleles, the sequencing focused on the region from exon 8 to exon 11, whereas, for the rest, the entire gene was sequenced. All patients with mutation L444P or any other pseudogene-derived mutation were analyzed by Southern blots. Moreover, patients with a homozygous genotype were examined by long PCR template and/or Southern blots, and, when possible, parental DNA was studied to exclude the possibility of a deletion on one allele. The final result was the identification of 59 recombinant alleles, representing 12% of the alleles studied, and the type, frequency, and distribution of recombinant alleles among patients with different types of Gaucher disease are summarized in table 4. Twenty-two percent of the patients evaluated had at least one recombinant allele present. However, since Southern blots and full sequencing were not performed on every proband, this may be an underrepresentation of the total number.
Table 4.
Summary of Recombinant Allele Frequencies
|
No. of Alleles for EachType of Recombination |
||||||
| GaucherDisease Type | No. of Patients/No. of Alleles Examined | No. (%) of Patients withRecombinant Alleles | No. (%) of Recombinant Allelesa | Gene Conversion | Fusion | Duplication |
| 1 | 155/310 | 24 (16) | 26 (8) | 15 | 8 | 3 |
| 2 | 37/74 | 14 (38) | 18 (24) | 12 | 6 | 0 |
| 3 | 48/96 |
14 (29) |
15 (16) |
7 |
4 |
4 |
| Total | 240/480 | 52 (22) | 59 (12) | 34 | 18 | 7 |
Several patients had more than one recombination on the same allele.
Sequencing data showed that a significant number of the patients with point mutations previously identified by PCR screening for individual mutations actually carried recombinant alleles with more than one point mutation derived from the pseudogene sequence. The sequence was examined carefully, focusing on every site at which the sequence for the gene and pseudogene were known to differ, and the length of the pseudogene/functional gene substitutions and the site of crossover or gene conversion were estimated (tables 1, 2, and 3).
To pinpoint the crossover site and the type of recombination for the remaining recombinant alleles, long PCR results and Southern blot analyses were examined. Long PCR indicated that several patients had smaller fusion alleles. The fusion alleles detected by this method resulted from recombination between the GBA gene and its pseudogene, with crossover sites located in the 5′ regions of the gene. Since the GBA pseudogene has large deletions in introns 2, 4, 6, and 7 (Horowitz et al. 1989), long PCR demonstrates two separate bands, which represent the normal allele (6.6 kb) and a shorter recombinant allele (∼5 kb). By examining the long PCR results, two fusion alleles, designated as “Rec 4b” and “Rec 5b” (table 3; patients 13 and 17), were identified with a crossover in the 5′ region of the gene. Sequencing of the mutant band demonstrated the specific crossover sites.
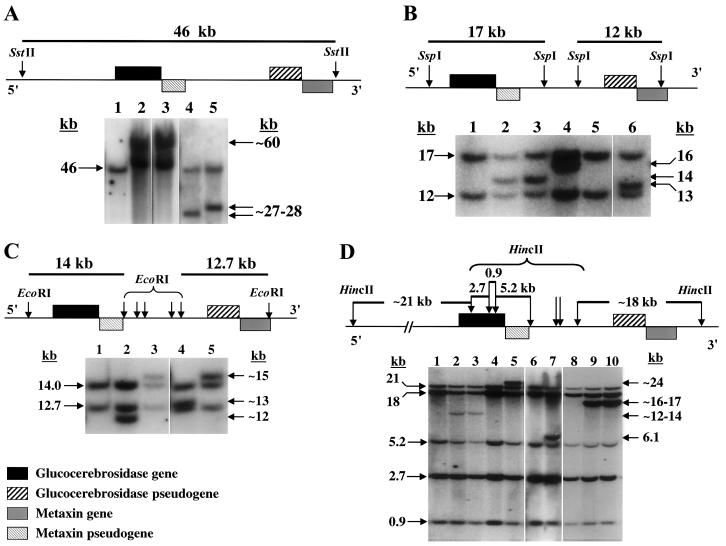
In general, alleles with reciprocal recombinations including fusions and duplications could be detected by Southern blots using the restriction enzymes SstII, SspI, and EcoRI (fig. 2A, 2B, and 2C). On the other hand, recombinations resulting from gene conversion had a normal pattern on Southern blots. The restriction enzyme HincII, which cleaves at several sites within the GBA gene but not the pseudogene (fig. 2D), was used to localize the crossover region. Three short probes (from exons 2–4, 4–7, and 7–11) were hybridized to Southern blots of HincII-digested DNA, which enabled the identification of the site of crossover in better detail in some cases.
Figure 2.
Southern blots of the GBA gene and pseudogene. Genomic DNAs were digested using four different restriction enzymes and were hybridized to a human GBA cDNA probe. A representative selection of patients with different recombinant alleles is shown in each panel. Included are schematic maps showing the normal restriction sites. Samples in each lane are identified by the table, patient number, and the recombinant allele they demonstrate (i.e., T2-2, Rec 7b is table 2, patient 2 with Rec 7b). The approximate sizes of the abnormal bands are shown to the right of each blot. A, SstII-digested DNA samples: lane 1, normal control demonstrating a single 46-kb band; lane 2, T3-2, Rec 7b; lane 3, T3-20, Rec 7b; lane 4, T3-13, Rec 4b; and lane 5, T3-14, Rec 1b. B, SspI-digested DNA samples: lane 1, normal control with 17- and 12-kb bands; lane 2, T3-11, Rec 1b; lane 3, T3-1, Rec 2b; lane 4, T3-20, Rec 7b; lane 5, T2-20, c.1263-1317del-a; and lane 6, T3-17, Rec 5b. C, EcoRI-digested DNA samples: lane 1, normal control with 14- and 12.7-kb bands; lane 2, T3-24, Rec 7b; lane 3, T3-12, Rec 6b; lane 4, T3-13, Rec 4b; and lane 5, T3-14, Rec1b. D, HincII-digested DNA samples: lanes 1, 6, and 8, normal controls showing 21-, 18-, 5.2-, 2.7-, and 0.9-kb bands; lane 2, T3-16, Rec7b; lane 3, T3-11, Rec 1b; lane 4, T2-5, Rec 1a; lane 5, T3-13, Rec 4a; lane 7, T2-20, c.1263-1317del-a; lane 9, T3-2, Rec 7b; and lane 10, T3-5, Rec 6b.
Alleles Resulting from Gene Conversion Events
Table 2 summarizes the molecular and clinical data in patients with type 1, 2, and 3 Gaucher disease who had gene conversion events or nonreciprocal recombinations. Thirty-four (58%) of the 59 recombinant alleles fell in this category, including 58% of all recombinant alleles in patients with type 1, 67% in patients with type 2, and 47% in patients with type 3 (table 4). A wide range of mutations was encountered on the second allele, as shown in table 2. In all 31 patients with these alleles, the site of gene conversion appeared to begin between exon 8 and intron 9 (fig. 1). It is possible that the gene conversion data is an underestimate, since not all intronic regions and, in some cases, not all of the 5′ exonic regions were sequenced. For the recombinant alleles in table 2, the maximum possible length of the converted sequence ranged between 194 bp and 1,193 bp. The Southern blot results for this group were normal except for the six patients who carried the 55-bp deletion in exon 9, where a HincII site is lost (see representative example, fig. 2D). There were two alleles carrying L444P (table 2; patients 9 and 31), in which the mutation appears to have resulted from a gene conversion event and not as an individual point mutation, since pseudogene sequence was also present in intron 9. Patient 9, with type 1 disease, carried only one pseudogene mismatch, whereas patient 31, with type 3 disease, had two sites corresponding to pseudogene sequence in intron 9.
Alleles Resulting from Reciprocal Recombination
Table 3 describes the alleles resulting from reciprocal recombination that carried either a fusion between the GBA gene and pseudogene or a duplication. For each of the 25 alleles identified (42% of all recombinants), Southern blots using the four restriction enzymes confirmed the reciprocal recombination (see fig. 2 for representative examples). Reciprocal recombinations were found in 42% of recombinant alleles from patients with type 1, 33% from patients with type 2, and 53% from patients with type 3 disease (table 4).
Seven alleles carried a duplication of the GBA pseudogene and a fusion between the MTX gene and pseudometaxin, resulting from a crossover at the 3′ region of the gene (Tayebi et al. 2000). The site of crossover was identified by sequencing and by Southern blots using the restriction enzymes HincII and EcoRI (fig. 2C and 2D). In three of these patients, the presence of a duplication was confirmed through studies of the tetranucleotide repeat polymorphism ITG6.2, located in the region between the GBA gene and pseudogene. In these individuals, three allelic forms (318 bp, 322 bp, and 326 bp) were found. The site of crossover for these recombinant alleles was pinpointed by sequencing and is referred to as “Rec 6b” (crossover initiates between Int10 and Ex11) or “Rec 7b” (crossover within a 304-bp sequence at the 3′ UTR or between MTX and pseudometaxin). Southern blots prepared using HincII-digested DNA were also hybridized to an MTX cDNA probe (data not shown). The results confirmed the duplication and fusion alleles found in the 3′ region.
Although 18 fusion alleles were identified, the size of the fusion product seen on the Southern blots varied because of different sites of crossover (fig. 2). The sites were mostly clustered between intron 8 and exon 11 of the GBA gene, with the exception of two alleles in which the site of crossover was in intron 3 (table 3, patient 13) or intron 4 (table 3, patient 17) and two unique fusion alleles resulting from crossovers within intron 10 to exon 11 or within pseudometaxin (table 3, patients 12 and 16). The latter two alleles both had fusions between the MTX gene and pseudogene. One patient (table 3, patient 15) was homozygous for a fusion allele in which the site of crossover occurred within the last 186 bp of exon 9 or in the beginning of intron 9. Homozygosity for a fusion allele has not previously been reported. Among the patients with reciprocal crossover, there was also a wide range of mutations present on the second allele.
Nine alleles with a reciprocal recombination in the 3′ region of the GBA gene, two with fusions and seven with duplications, also carried a point mutation or another recombinant allele. An analysis of parental DNA showed that the 3′ region recombinations (Rec 6b and Rec 7b) were located on the allele with L444P or Rec 1a, except for one case (table 3; patient 20) in which Rec 7b and D409H were on the same allele. To determine which was the original mutation on these alleles and whether the first mutation increases the likelihood of a second mutation, studies of several successive generations will be needed.
Sites of Recombination and Motif Sequences
The examination of the 59 recombinant alleles by Southern blots and detailed sequencing led to the identification of 10 different sites of crossover or gene conversion. An accurate identification of the site where the crossover event occurred was dependent upon the degree of gene/pseudogene homology in a specific region. Seven of the 10 sites were located between intron 8 and the 3′ UTR (fig. 1).
The sequences within the regions surrounding the 10 sites were compared to identify one or more possible motif sequences that might predispose to crossover. A pentamer sequence (CACCA) was found in five of the crossover regions and was identified in 35 of 59 recombinant alleles. A similar tetramer sequence (CACC) was found at two additional sites in 11 patient alleles. The remaining two sites without this sequence shared a different tetramer sequence (TGGG), which was found in 13 recombinant alleles.
Correlation with Clinical Data
The ethnic or racial backgrounds of patients carrying recombinant alleles were quite varied. Of note, although nine recombinant alleles were identified in 10 African American patients with Gaucher disease, the recombinations included both fusions and gene conversions, suggesting that this was not due to a founder effect.
The majority of the 24 patients with type 1 Gaucher disease carrying recombinant alleles had significant disease manifestations. Twenty-one of the subjects whose clinical data were available were diagnosed by adolescence, and a majority had hepatosplenomegaly and cytopenia. Eight had undergone splenectomy and seven were described as having bone disease. Seventeen of these patients had mutation N370S on their second allele. Although the patients with genotype N370S/Rec had a wide range of disease severity, the severity in those with recombinant alleles resulting from reciprocal recombination did not differ significantly from those with gene conversions.
Among the 14 patients with type 2 Gaucher disease, nine had recombinant alleles resulting from gene conversion. In the three cases in which there was homozygosity for a gene conversion allele (table 2, patients 18, 19, and 21), death resulted from hydrops fetalis, although the three patients did not all share the same site of recombination. Three other patients with L444P/Rec1a (table 2, patients 16, 22, and 24) lived for approximately one year. Of the five patients with fusion alleles, one had a hydrops fetalis phenotype. This fetus (table 3, patient 13) carried a fusion allele that resulted from a crossover in intron 3, along with a splice site mutation in exon 10 on the second allele. Another patient was found to be homozygous for a fusion allele (table 3, patient 15). One infant with a classic type 2 phenotype had genotype L444P/L444P (table 3, patient 16), a genotype not normally associated with type 2 Gaucher disease. However, each L444P allele also had a second alteration that might have a modifying effect on the phenotype. One carried the presumed polymorphism E326K (Park et al. 2002a) and the other a 3′-region duplication within the sequence for pseudometaxin (Rec 7b) (Tayebi et al. 2000).
Among the 14 patients with type 3 Gaucher disease carrying recombinant alleles, six had a recombination resulting from gene conversion, seven as a result of reciprocal recombination, and one patient had both types of recombinant alleles. Clinically, patients with alleles resulting from both reciprocal and nonreciprocal recombinations were similar, with a diagnosis in early childhood in all but three patients. Of note, half of the subjects with either type of recombinant allele had myoclonic epilepsy associated with their other disease manifestations. Mutation N188S was found together with recombinant alleles in three patients with type 3 Gaucher disease (table 2, patient 29; and table 3, patients 17 and 21). While each of the three had a different recombinant allele, all three patients were adults with uncontrolled myoclonic epilepsy. The two patients with genotype V394L/Rec 1a (table 2, patients 28 and 30) shared a similar phenotype, with progressive myoclonic epilepsy in childhood. Three patients with type 3 disease, one with R463C/Rec 1a (table 2, patient 27) and two with R463C/Rec 1b (table 3, patients 18 and 22), had aggressive visceral and skeletal disease and slowing of the horizontal saccades as their principal neurologic manifestation. However, two other patients with R463C/Rec 1b (table 3, patients 3 and 11) had a type 1 phenotype with no neurologic symptoms.
Discussion
Homologous recombination, both reciprocal and nonreciprocal, has been implicated as the cause of mutations in many human disorders. Repeated DNA sequences can be one of the forces driving recombination and have been classified on the basis of the location, distribution and length of the repeat sequence, and the number of repeats (Purandare and Patel 1997). Alu sequences, a specific class of repeat sequences, are one of the major sources of DNA rearrangement and are implicated in several genetic disorders (Brooks et al. 2001; Martinez et al. 2001; Batzer and Deininger 2002; Lutskiy et al. 2002). Another cause of recombination is the presence of duplicated gene sequences or “duplicons,” which can mediate local deletions, duplications, fusions, inversions, and gene conversion (Ji et al. 2000; Eichler 2001). Duplicons can consist of tandemly repeated sequences or low-copy repeat sequences (LCRs), including gene clusters, gene fragments, pseudogenes, or repeat elements flanking unique sequence. LCRs are usually 10–400 kb in size and exhibit 95%–98% homology (Stankiewicz and Lupski 2002a). The likelihood of DNA rearrangement is increased when repeated fragments are located near one another on the same chromosome but are not directly adjacent. Recombinant alleles resulting in human disease can be categorized by the type of recombination, either nonreciprocal or reciprocal. This distinction proved very useful in our characterization of recombinations at the GBA gene locus.
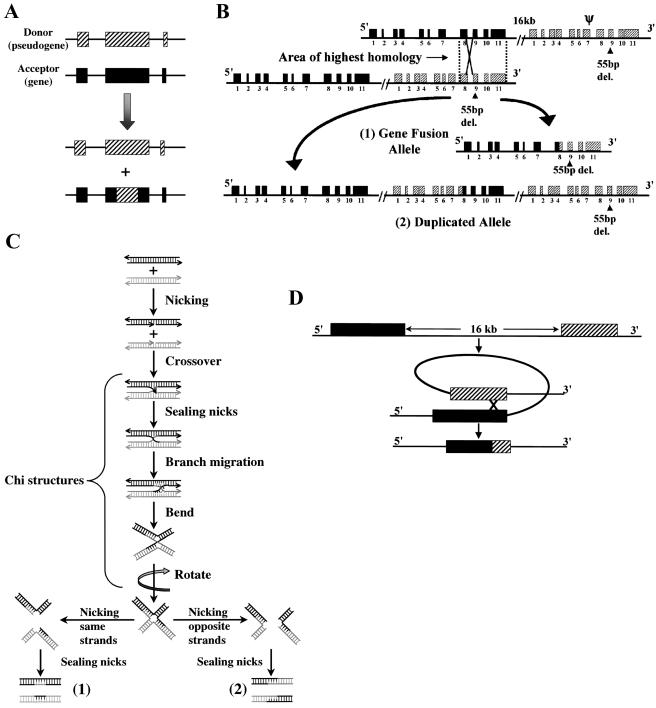
The nonreciprocal exchange of homologous sequence, also known as gene conversion, has been extensively examined in lower eukaryotes, in which, unlike mammalian systems, the results of meiosis can be easily recovered and studied. In nonreciprocal genetic exchange, the transient pairing of two similar, although not identical, genes or alleles is followed by conversion of one sequence to that of the other. The donor sequence is not changed, but the acceptor sequence is altered by the incorporation of sequence copied from the donor sequence (fig. 3A) (Strachan and Read 1999).
Figure 3.
Illustrations of proposed mechanisms for recombination events occurring between the GBA gene and its pseudogene. A, Allelic gene conversion. This is a nonreciprocal sequence exchange, facilitated by sequence homology between allelic genes. The donor sequence is unaltered, but the acceptor sequence is changed by the incorporation of regions copied from the donor sequence. B, Reciprocal crossover between homologous regions. This event results in two possible gene rearrangements. One is a fusion between the gene and its pseudogene and a deletion of the intergenic region shown in (1). The second is a partial duplication of the pseudogene and gene sequences, which are fused together (2). C, Holliday model. This is a mechanism of recombination that includes nicking and reunion between two homologous sequences. Nicking occurs in the same location in both homologous sequences (i.e., in the GBA gene and pseudogene). Crossover, exchanges, and sealing nicks create chi structures. Branch migration and breaking in some base pairs in the four strands increase the chance of bending, rotation, and exchange between strands, resulting in two possible gene conversion–like alleles (1) and two possible fusion-like alleles (2). Figure adapted from Weaver and Hedrick (1989). D, Intramolecular crossover. This is another proposed model for recombination between the GBA gene and pseudogene on the same chromosome, resulting in a fusion allele. In this case, the intergenic region and portions of the gene and pseudogene are removed as an extrachromosomal fragment and are lost.
In general, gene conversion results from nonreciprocal sequence exchange between either nonallelic or allelic DNA sequences. Gene conversion facilitated by nonallelic tandem repeat sequences has been implicated in some clustered human gene families, including the globins (Papadakis and Patrinos 1999) and immunoglobulins (Sitnikova and Su 1998). Gene conversion involving allelic exchanges is more likely to occur when the gene in question has a nearby pseudogene or is part of a gene cluster located on the same chromosome, as in the HLA genes (Blanco-Gelaz et al. 2001), 21-β hydroxylase deficiency (Tusie-Luna and White 1995), and polycystic kidney disease (Watnick et al. 1998; Bogdanova et al. 2001). Allelic exchanges are a likely mechanism for the DNA rearrangements observed in the GBA locus.
Although gene conversion cannot easily be distinguished from double crossover events, double crossovers occurring in very close proximity would not normally be expected. The sizes of the converted sequences identified in this series, ranging from 190 bp to 1,193 bp, are presumably far too short to be the result of a double crossover. Liskay et al. (1987) demonstrated, in cultured mammalian cells, that when duplicated chromosomal sequences had regions of shared homology ranging between 295 bp and 1.8 kb in length, gene conversion events were efficient, with the rate being directly proportional to the extent of homology.
Our results demonstrate that the regions where the GBA gene and pseudogene share the greatest sequence homology are those with the highest rate of gene conversion (table 2). The sequence homology shared between the GBA gene and its pseudogene in the region between intron 8 and the 3′ UTR is ∼98% and includes five segments of exact identity, each >200 bp in length.
The second major type of homologous exchange, reciprocal recombination, can arise by several possible mechanisms. Gene rearrangement during meiosis or, rarely, mitosis, can occur by direct crossover between identical or very similar DNA sequences. This usually involves the breakage of a pair of homologous chromosomes and a rejoining of the fragments to generate new recombinant strands.
Tandemly repeated genes or gene clusters are subject to reciprocal rearrangements leading to disease. For example, reciprocal recombination events between the clustered, highly homologous red and green visual pigment genes at the color vision locus at Xq28 introduce deletions or fusions, resulting in red/green color blindness (Vollrath et al. 1988; Jagla et al. 2002). In the thalassemias, crossover between duplicated sequences at the β-globin gene cluster on 11p15.5 cause the deletion of the β gene, resulting in β-thalassemia, while the same type of crossover in the α-globin gene cluster on 16p13.3 can cause α-thalassemia (Hattori 1999; Papadakis and Patrinos 1999; Waye et al. 2001). Fusion genes resulting from unequal crossing over are also seen in the β-globin gene cluster (Metzenberg et al. 1991).
Reciprocal DNA rearrangements involving LCRs can cause duplications and deletions associated with diseases. Nonallelic homologous recombination between two LCRs on 17p12 sharing 98.7% sequence homology can result in a 1.4-Mb duplication found in Charcot Marie Tooth disease type IA, and a deletion of the region causes hereditary neuropathy with liability to pressure palsies (Lopes et al. 1999; Ji et al. 2000; Hurles 2001; Stankiewicz and Lupski 2002b). Likewise, recombination between complex LCRs at the proximal and distal regions of 17p11.2 can result in the deletion of a 4-Mb genomic segment in patients with Smith-Magenis syndrome or a duplication of the region in patients with dup(17)(p11.2 p11.2) syndrome (Lucas et al. 2001; Park et al. 2002b; Shaw et al. 2002).
In a similar fashion, the presence of a pseudogene can increase the likelihood of reciprocal recombination and, hence, disease. Unequal crossover or unequal sister chromatid exchange between a functional gene and a related pseudogene can result in deletion of the functional gene or the formation of fusion genes carrying a segment derived from the pseudogene (Purandare and Patel 1997; Timms et al. 1997; Lobato et al. 1998; Stone et al. 2000). In steroid 21-hydroxylase deficiency and Hunter syndrome, most of the mutations arise as a result of such sequence exchanges between the functional gene and a very closely related pseudogene (Birot et al. 1996; Karsten et al. 1997; Bunge et al. 1998; Wedell 1998; Dracopoulou-Vabouli et al. 2001).
As demonstrated in the present study, recombinations between the GBA gene and its pseudogene, resulting in gene fusions or duplications, provide a prime example of the consequences of reciprocal crossover (fig. 3B). Fusion alleles with the 5′ sequence deriving from the gene and 3′ sequence from the pseudogene were the most prevalent. Moreover, in several instances, recombination occurred as a result of crossover between the nearby gene and pseudogene for MTX.
Another mechanism of recombination demonstrated in simple organisms that may be relevant to Gaucher disease is the Holliday junction model, which occurs during meiosis. In figure 3C, the different steps of the Holliday model observed in plasmid DNA are shown (Weaver and Hedrick 1989; Fu et al. 1994). An essential step in this model occurs when the two homologous strands cross over and the new branches are sealed by DNA ligase in a form called the “chi structure” or “half chiasma.” The branch in the half chiasma can migrate in either direction simply by breaking old base pairs and forming new ones. The chi structure is first shown with two of its strands crossed, but a 180° rotation of either the upper or the lower uncrossed strands can give rise to different alleles, such as fusion alleles and gene conversion–like alleles (a short sequence of the donor strand can be inserted into an acceptor sequence). The Holliday junction mechanism has also been used to explain recombinant alleles in yeast (Petes 2001).
A third possible mechanism to explain reciprocal fusions within the GBA locus is intramolecular crossover (fig. 3D) (Weaver and Hedrick 1989). When two regions sharing similar sequence are located nearby on one chromosome but are separated by ⩾10 kb, the chance of an intramolecular crossover with a loop formation occurring during the cell cycle is increased. As a result, one daughter cell carries an allele with a fusion between the gene and pseudogene, and the other daughter cell carries a small extrachromosomal loop consisting of a short fragment of gene and pseudogene and the intergenic region (fig. 3D).
In higher organisms, clearly one cannot demonstrate directly which mechanism occurred during the cell cycle. However, extrapolating from prokaryotes and yeast, several possible mechanisms can be considered. The gene conversion alleles identified in this study could be explained either by the classic gene conversion model or the Holliday junction model. Likewise, the fusion alleles encountered could arise either by direct crossover, intramolecular recombination or by the Holliday junction model.
Crossover hotspot regions in prokaryotes and eukaryotes have been analyzed, and some of the factors that regulate or initiate recombination have been identified (Bell et al. 1998; Wahls 1998; Petes 2001). In a number of different human disorders caused by DNA rearrangements, several short DNA sequences or repeat sequences have been identified at the crossover break points. It is hypothesized that these sequences might represent a motif that leads to a recombination hotspot (Karsten et al. 1997; Bunge et al. 1998; Aarskog and Vedeler 2001; Kutsche et al. 2002). The motif sequence (CACCA) identified in the present study may be helpful in demonstrating other recombination sites in the GBA gene.
Earlier genotype/phenotype studies in Gaucher disease led to an appreciation that while there are certain generalizations regarding the ability to predict phenotype from genotype, many exceptions remain. On the basis of the present study, PCR-based genotyping is clearly not adequate to accurately describe the mutant alleles in at least 22% of the patients with Gaucher disease analyzed. This observation is of utmost importance to laboratories engaged in molecular diagnostics for Gaucher disease.
Based upon the current genotype/phenotype analyses, it appears that recombinant alleles are essentially null alleles. Homozygosity for a recombinant allele results in very early lethality, usually in utero or in the first days of life. Genotype L444P/recombinant allele is associated with classic type 2 Gaucher disease, although homozygosity for L444P results in a milder phenotype. Likewise, genotypes V394L/Rec 1a, N188S/Rec 5b, and N188S/Rec1a were encountered in patients with type 3 Gaucher disease and myoclonic epilepsy, although homozygosity for V394L and N188S have been seen in patients with type 1 disease. These examples suggest a dose response, in which one copy of a point mutation together with a null allele is more detrimental than two copies of the missense allele.
The current study demonstrates the frequency and the type of recombinations in a broad population of patients with Gaucher disease. Both gene conversion and reciprocal recombination occur at this locus far more frequently than previously appreciated. Although it could not be established whether different mechanisms or sites of recombination made specific contributions to phenotype, it can be concluded that a recombinant allele has a deleterious effect, regardless of how it arose. The one exception were patients who carried the 3′ duplication or Rec 7b. While this recombination alone did not appear to result in Gaucher disease, it still could potentially have a modifying effect on phenotype.
The region surrounding the human GBA gene on chromosome 1 is particularly gene rich, with seven genes and two pseudogenes located within ∼85 kb of sequence (Winfield et al. 1997). It is particularly interesting to speculate that some of the recombinations occurring within this locus could potentially disrupt other nearby genes with phenotypic consequences. Certainly, this gene-rich region provides fertile ground for a deeper study of the mechanisms of recombination in the human genome.
Acknowledgments
The authors thank the many patients, family members, and referring physicians who have contributed greatly to this study. We acknowledge Dr. Brian Martin for the synthesis of oligonucleotide primers and Dr. Erich Roessler for helpful discussions. The secretarial assistance of Marie Hall is also gratefully acknowledged.
Electronic-Database Information
Accession numbers and URLs for data presented herein are as follows:
- GenBank, http://www.ncbi.nlm.nih.gov/Genbank/ (for human GBA [accession number J03059], the human GBA pseudogene [accession number J03060], and the sequences of genes surrounding human GBA [accession number AF023263])
- Human Genetic Disease Databank (GeneDis), http://life2.tau.ac.il/GeneDis/ (for compilation of mutations in human glucocerebrosidase)
- Online Mendelian Inheritance in Man (OMIM), http://www.ncbi.nlm.nih.gov/Omim/ (for Gaucher disease [MIM 230800], Gaucher disease type 2 [MIM 230900], and Gaucher disease type 3 [MIM 231000])
References
- Aarskog NK, Vedeler CA (2001) Recombination breakpoints in the Charcot-Marie-Tooth 1A repeat sequence in Norwegian families. Acta Neurol Scand 104:97–100 [DOI] [PubMed] [Google Scholar]
- Armstrong LC, Komiya T, Bergman BE, Mihara K, Bornstein P (1997) Metaxin is a component of a preprotein import complex in the outer membrane of the mammalian mitochondrion. J Biol Chem 272:6510–6518 [DOI] [PubMed] [Google Scholar]
- Batzer MA, Deininger PL (2002) Alu repeats and human genomic diversity. Nat Rev Genet 3:370–379 [DOI] [PubMed] [Google Scholar]
- Bell SJ, Chow YC, Ho JY, Forsdyke DR (1998) Correlation of chi orientation with transcription indicates a fundamental relationship between recombination and transcription. Gene 216:285–292 [DOI] [PubMed] [Google Scholar]
- Beutler E, Grabowski GA (2001) Gaucher disease. In: Scriver CR, Beaudet AL, Valle D, Sly WS (eds) The metabolic and molecular bases of inherited disease. McGraw-Hill, New York, pp 3635–3668 [Google Scholar]
- Beutler E, Gelbart T (1998) Hematologically important mutations: Gaucher disease. Blood Cells Mol Dis 24:2–8 [DOI] [PubMed] [Google Scholar]
- Beutler E, Gelbart T, Demina A, Zimran A, LeCoutre P (1995) Five new Gaucher disease mutations. Blood Cells Mol Dis 21:20–24 [DOI] [PubMed] [Google Scholar]
- Birot AM, Bouton O, Froissart R, Maire I, Bozon D (1996) IDS gene-pseudogene exchange responsible for an intragenic deletion in a Hunter patient. Hum Mutat 8:44–50 [DOI] [PubMed] [Google Scholar]
- Blanco-Gelaz MA, Lopez-Vazquez A, Garcia-Fernandez S, Martinez-Borra J, Gonzalez S, Lopez-Larrea C (2001) Genetic variability, molecular evolution, and geographic diversity of HLA-B27. Hum Immunol 62:1042–1050 [DOI] [PubMed] [Google Scholar]
- Bogdanova N, Markoff A, Gerke V, McCluskey M, Horst J, Dworniczak B (2001) Homologues to the first gene for autosomal dominant polycystic kidney disease are pseudogenes. Genomics 74:333–341 [DOI] [PubMed] [Google Scholar]
- Brooks EM, Branda RF, Nicklas JA, O'Neill JP (2001) Molecular description of three macro-deletions and an Alu-Alu recombination-mediated duplication in the HPRT gene in four patients with Lesch-Nyhan disease. Mutat Res 476:43–54 [DOI] [PubMed] [Google Scholar]
- Bunge S, Rathmann M, Steglich C, Bondeson ML, Tylki-Szymanska A, Popowska E, Gal A (1998) Homologous nonallelic recombinations between the iduronate-sulfatase gene and pseudogene cause various intragenic deletions and inversions in patients with mucopolysaccharidosis type II. Eur J Hum Genet 6:492–500 [DOI] [PubMed] [Google Scholar]
- Chen KS, Manian P, Koeuth T, Potocki L, Zhao Q, Chinault AC, Lee CC, Lupski JR (1997) Homologous recombination of a flanking repeat gene cluster is a mechanism for a common contiguous gene deletion syndrome. Nat Genet 17:154–163 [DOI] [PubMed] [Google Scholar]
- Cormand B, Diaz A, Grinberg D, Chabas A, Vilageliu L (2000) A new gene-pseudogene fusion allele due to a recombination in intron 2 of the glucocerebrosidase gene causes Gaucher disease. Blood Cells Mol Dis 26:409–416 [DOI] [PubMed] [Google Scholar]
- Dracopoulou-Vabouli M, Maniati-Christidi M, Dacou-Voutetakis C (2001) The spectrum of molecular defects of the CYP21 gene in the Hellenic population: variable concordance between genotype and phenotype in the different forms of congenital adrenal hyperplasia. J Clin Endocrinol Metab 86:2845–2848 [DOI] [PubMed] [Google Scholar]
- Eichler EE (2001) Recent duplication, domain accretion and the dynamic mutation of the human genome. Trends Genet 17:661–669 [DOI] [PubMed] [Google Scholar]
- Emanuel BS, Shaikh TH (2001) Segmental duplications: an “expanding” role in genomic instability and disease. Nat Rev Genet 2:791–800 [DOI] [PubMed] [Google Scholar]
- Eyal N, Wilder S, Horowitz M (1990) Prevalent and rare mutations among Gaucher patients. Gene 96:277–283 [DOI] [PubMed] [Google Scholar]
- Filocamo M, Bonuccelli G, Mazzotti R, Giona F, Gatti R (2000) Identification of a novel recombinant allele in three unrelated Italian Gaucher patients: implications for prognosis and genetic counseling. Blood Cells Mol Dis 26:307–311 [DOI] [PubMed] [Google Scholar]
- Fu TJ, Tse-Dinh YC, Seeman NC (1994) Holliday junction crossover topology. J Mol Biol 236:91–105 [DOI] [PubMed] [Google Scholar]
- Hatton CE, Cooper A, Whitehouse C, Wraith JE (1997) Mutation analysis in 46 British and Irish patients with Gaucher's disease. Arch Dis Child 77:17–22 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hattori Y (1999) [Globin gene mutation is a model of genetic abnormalities]. Rinsho Byori 47:244–251 [PubMed] [Google Scholar]
- Hodanova K, Hrebicek M, Cervenkova M, Mrazova L, Veprekova L, Zeman J (1999) Analysis of the beta-glucocerebrosidase gene in Czech and Slovak Gaucher patients: mutation profile and description of six novel mutant alleles. Blood Cells Mol Dis 25:287–298 [PubMed] [Google Scholar]
- Horowitz M, Wilder S, Horowitz Z, Reiner O, Gelbart T, Beutler E (1989) The human glucocerebrosidase gene and pseudogene: structure and evolution. Genomics 4:87–96 [DOI] [PubMed] [Google Scholar]
- Hurles ME (2001) Gene conversion homogenizes the CMT1A paralogous repeats. BMC Genomics 2:11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inoue K, Dewar K, Katsanis N, Reiter LT, Lander ES, Devon KL, Wyman DW, Lupski JR, Birren B (2001) The 1.4-Mb CMT1A duplication/HNPP deletion genomic region reveals unique genome architectural features and provides insights into the recent evolution of new genes. Genome Res 11:1018–1033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jagla WM, Jagle H, Hayashi T, Sharpe LT, Deeb SS (2002) The molecular basis of dichromatic color vision in males with multiple red and green visual pigment genes. Hum Mol Genet 11:23–32 [DOI] [PubMed] [Google Scholar]
- Ji Y, Eichler EE, Schwartz S, Nicholls RD (2000) Structure of chromosomal duplicons and their role in mediating human genomic disorders. Genome Res 10:597–610 [DOI] [PubMed] [Google Scholar]
- Karsten SL, Lagerstedt K, Carlberg BM, Kleijer WJ, Zaremba J, Van Diggelen OP, Czartoryska B, Pettersson U, Bondeson ML (1997) Two distinct deletions in the IDS gene and the gene W: a novel type of mutation associated with the Hunter syndrome. Genomics 43:123–129 [DOI] [PubMed] [Google Scholar]
- Koprivica V, Stone DL, Park JK, Callahan M, Frisch A, Cohen IJ, Tayebi N, Sidransky E (2000) Analysis and classification of 304 mutant alleles in patients with type 1 and type 3 Gaucher disease. Am J Hum Genet 66:1777–1786 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kutsche K, Ressler B, Katzera HG, Orth U, Gillessen-Kaesbach G, Morlot S, Schwinger E, Gal A (2002) Characterization of breakpoint sequences of five rearrangements in L1CAM and ABCD1 (ALD) genes. Hum Mutat 19:526–535 [DOI] [PubMed] [Google Scholar]
- Latham T, Grabowski GA, Theophilus BD, Smith FI (1990) Complex alleles of the acid β-glucosidase gene in Gaucher disease. Am J Hum Genet 47:79–86 [PMC free article] [PubMed] [Google Scholar]
- Lau EK, Tayebi N, Ingraham LJ, Winfield SL, Koprivica V, Stone DL, Zimran A, Ginns EI, Sidransky E (1999) Two novel polymorphic sequences in the glucocerebrosidase gene region enhance mutational screening and founder effect studies of patients with Gaucher disease. Hum Genet 104:293–300 [DOI] [PubMed] [Google Scholar]
- Liskay RM, Letsou A, Stachelek JL (1987) Homology requirement for efficient gene conversion between duplicated chromosomal sequences in mammalian cells. Genetics 115:161–167 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lobato MN, Aledo R, Meseguer A (1998) High variability of CYP21 gene rearrangements in Spanish patients with classic form of congenital adrenal hyperplasia. Hum Hered 48:216–225 [DOI] [PubMed] [Google Scholar]
- Long GL, Winfield S, Adolph KW, Ginns EI, Bornstein P (1996) Structure and organization of the human metaxin gene (MTX) and pseudogene. Genomics 33:177–184 [DOI] [PubMed] [Google Scholar]
- Lopes J, Tardieu S, Silander K, Blair I, Vandenberghe A, Palau F, Ruberg M, Brice A, LeGuern E (1999) Homologous DNA exchanges in humans can be explained by the yeast double-strand break repair model: a study of 17p11.2 rearrangements associated with CMT1A and HNPP. Hum Mol Genet 8:2285–2292 [DOI] [PubMed] [Google Scholar]
- Lucas RE, Vlangos CN, Das P, Patel PI, Elsea SH (2001) Genomic organization of the approximately 1.5 Mb Smith-Magenis syndrome critical interval: transcription map, genomic contig, and candidate gene analysis. Eur J Hum Genet 9:892–902 [DOI] [PubMed] [Google Scholar]
- Lupski JR (1998) Genomic disorders: structural features of the genome can lead to DNA rearrangements and human disease traits. Trends Genet 14:417–422 [DOI] [PubMed] [Google Scholar]
- Lutskiy MI, Jones LN, Rosen FS, Remold-O'Donnell E (2002) An Alu-mediated deletion at Xp11.23 leading to Wiskott-Aldrich syndrome. Hum Genet 110:515–519 [DOI] [PubMed] [Google Scholar]
- Martinez J, Dugaiczyk LJ, Zielinski R, Dugaiczyk A (2001) Human genetic disorders, a phylogenetic perspective. J Mol Biol 308:587–596 [DOI] [PubMed] [Google Scholar]
- Mazzarella R, Schlessinger D (1998) Pathological consequences of sequence duplications in the human genome. Genome Res 8:1007–1021 [DOI] [PubMed] [Google Scholar]
- Metzenberg AB, Wurzer G, Huisman TH, Smithies O (1991) Homology requirements for unequal crossing over in humans. Genetics 128:143–161 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mistry PK, Smith SJ, Ali M, Hatton CS, McIntyre N, Cox TM (1992) Genetic diagnosis of Gaucher's disease. Lancet 339:889–892 [DOI] [PubMed] [Google Scholar]
- Orvisky E, Park JK, Parker A, Walker JM, Martin BM, Stubblefield BK, Uyama E, Tayebi N, Sidransky E (2002) The identification of eight novel glucocerebrosidase (GBA) mutations in patients with Gaucher disease. Hum Mutat 19:458–459 [DOI] [PubMed] [Google Scholar]
- Papadakis MN, Patrinos GP (1999) Contribution of gene conversion in the evolution of the human beta-like globin gene family. Hum Genet 104:117–125 [DOI] [PubMed] [Google Scholar]
- Park JK, Tayebi N, Stubblefield BK, LaMarca ME, MacKenzie JJ, Stone DL, Sidransky E (2002a) The E326K mutation and Gaucher disease: mutation or polymorphism? Clin Genet 61:32–34 [DOI] [PubMed] [Google Scholar]
- Park SS, Stankiewicz P, Bi W, Shaw C, Lehoczky J, Dewar K, Birren B, Lupski JR (2002b) Structure and evolution of the Smith-Magenis syndrome repeat gene clusters, SMS-REPs. Genome Res 12:729–738 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petes TD (2001) Meiotic recombination hot spots and cold spots. Nat Rev Genet 2:360–369 [DOI] [PubMed] [Google Scholar]
- Purandare SM, Patel PI (1997) Recombination hot spots and human disease. Genome Res 7:773–786 [DOI] [PubMed] [Google Scholar]
- Reissner K, Tayebi N, Stubblefield BK, Koprivica V, Blitzer M, Holleran W, Cowan T, Almashanu S, Maddalena A, Karson EM, Sidransky E (1998) Type 2 Gaucher disease with hydrops fetalis in an Ashkenazi Jewish family resulting from a novel recombinant allele and a rare splice junction mutation in the glucocerebrosidase locus. Mol Genet Metab 63:281–288 [DOI] [PubMed] [Google Scholar]
- Sambrook J, Fritsch EF, Maniatis T (1989) Molecular cloning: a laboratory manual. Cold Spring Harbor, New York pp 9.14–9.22 [Google Scholar]
- Shaw CJ, Bi W, Lupski JR (2002) Genetic proof of unequal meiotic crossovers in reciprocal deletion and duplication of 17p11.2. Am J Hum Genet 71:1072–1081 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sitnikova T, Su C (1998) Coevolution of immunoglobulin heavy- and light-chain variable-region gene families. Mol Biol Evol 15:617–625 [DOI] [PubMed] [Google Scholar]
- Stankiewicz P, Lupski JR (2002a) Genome architecture, rearrangements and genomic disorders. Trends Genet 18:74–82 [DOI] [PubMed] [Google Scholar]
- ——— (2002b) Molecular evolutionary mechanisms for genomic disorders. Curr Opin Genet Dev 12:312–319 [DOI] [PubMed] [Google Scholar]
- Stone DL, Tayebi N, Orvisky E, Stubblefield B, Madike V, Sidransky E (2000) Glucocerebrosidase gene mutations in patients with type 2 Gaucher disease. Hum Mutat 15:181–188 [DOI] [PubMed] [Google Scholar]
- Strachan T, Read AP (1999) Human molecular genetics 2. Wiley-Liss, New York [Google Scholar]
- Strasberg PM, Skomorowski MA, Warren IB, Hilson WL, Callahan JW, Clarke JT (1994) Homozygous presence of the crossover (fusion gene) mutation identified in a type II Gaucher disease fetus: is this analogous to the Gaucher knock-out mouse model? Biochem Med Metab Biol 53:16–21 [DOI] [PubMed] [Google Scholar]
- Tayebi N, Callahan M, Madike V, Stubblefield BK, Orvisky E, Krasnewich D, Fillano JJ, Sidransky E (2001) Gaucher disease and parkinsonism: a phenotypic and genotypic characterization. Mol Genet Metab 73:313–321 [DOI] [PubMed] [Google Scholar]
- Tayebi N, Cushner SR, Kleijer W, Lau EK, Damschroder-Williams PJ, Stubblefield BK, Den Hollander J, Sidransky E (1997) Prenatal lethality of a homozygous null mutation in the human glucocerebrosidase gene. Am J Med Genet 73:41–47 [DOI] [PubMed] [Google Scholar]
- Tayebi N, Cushner S, Sidransky E (1996a) Differentiation of the glucocerebrosidase gene from pseudogene by long-template PCR: implications for Gaucher disease. Am J Hum Genet 59:740–741 [PMC free article] [PubMed] [Google Scholar]
- Tayebi N, Park J, Madike V, Sidransky E (2000) Gene rearrangement on 1q21 introducing a duplication of the glucocerebrosidase pseudogene and a metaxin fusion gene. Hum Genet 107:400–403 [DOI] [PubMed] [Google Scholar]
- Tayebi N, Reissner KJ, Lau EK, Stubblefield BK, Klineburgess AC, Martin BM, Sidransky E (1998) Genotypic heterogeneity and phenotypic variation among patients with type 2 Gaucher's disease. Pediatr Res 43:571–578 [DOI] [PubMed] [Google Scholar]
- Tayebi N, Stern H, Dymarskaia I, Herman J, Sidransky E (1996b) 55-base pair deletion in certain patients with Gaucher disease complicates screening for common Gaucher alleles. Am J Med Genet 66:316–319 [DOI] [PubMed] [Google Scholar]
- Timms KM, Bondeson ML, Ansari-Lari MA, Lagerstedt K, Muzny DM, Dugan-Rocha SP, Nelson DL, Pettersson U, Gibbs RA (1997) Molecular and phenotypic variation in patients with severe Hunter syndrome. Hum Mol Genet 6:479–486 [DOI] [PubMed] [Google Scholar]
- Tusie-Luna MT, White PC (1995) Gene conversions and unequal crossovers between CYP21 (steroid 21-hydroxylase gene) and CYP21P involve different mechanisms. Proc Natl Acad Sci USA 92:10796–10800 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vollrath D, Nathans J, Davis RW (1988) Tandem array of human visual pigment genes at Xq28. Science 240:1669–1672 [DOI] [PubMed] [Google Scholar]
- Wahls WP (1998) Meiotic recombination hotspots: shaping the genome and insights into hypervariable minisatellite DNA change. Curr Top Dev Biol 37:37–75 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walley AJ, Harris A (1993) A novel point mutation (D380A) and a rare deletion (1255del55) in the glucocerebrosidase gene causing Gaucher's disease. Hum Mol Genet 2:1737–1738 [DOI] [PubMed] [Google Scholar]
- Watnick TJ, Gandolph MA, Weber H, Neumann HP, Germino GG (1998) Gene conversion is a likely cause of mutation in PKD1. Hum Mol Genet 7:1239–1243 [DOI] [PubMed] [Google Scholar]
- Waye JS, Eng B, Patterson M, Carcao MD, Chang L, Olivieri NF, Chui DH (2001) Identification of two new alpha-thalassemia mutations in exon 2 of the alpha1-globin gene. Hemoglobin 25:391–396 [DOI] [PubMed] [Google Scholar]
- Weaver RF, Hedrick PW (1989) The mechanism of recombination. In: Kane K (ed) Genetics. Wm C. Brown, Dubuque, IA, pp 184–189 [Google Scholar]
- Wedell A (1998) Molecular genetics of congenital adrenal hyperplasia (21-hydroxylase deficiency): implications for diagnosis, prognosis and treatment. Acta Paediatr 87:159–164 [DOI] [PubMed] [Google Scholar]
- Winfield SL, Tayebi N, Martin BM, Ginns EI, Sidransky E (1997) Identification of three additional genes contiguous to the glucocerebrosidase locus on chromosome 1q21: implications for Gaucher disease. Genome Res 7:1020–1026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao H, Grabowski GA (2002) Gaucher disease: perspectives on a prototype lysosomal disease. Cell Mol Life Sci 59:694–707 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zimran A, Horowitz M (1994) RecTL: a complex allele of the glucocerebrosidase gene associated with a mild clinical course of Gaucher disease. Am J Med Genet 50:74–78 [DOI] [PubMed] [Google Scholar]
- Zimran A, Sorge J, Gross E, Kubitz M, West C, Beutler E (1990) A glucocerebrosidase fusion gene in Gaucher disease: implications for the molecular anatomy, pathogenesis, and diagnosis of this disorder. J Clin Invest 85:219–222 [DOI] [PMC free article] [PubMed] [Google Scholar]