Abstract
In Saccharomyces cerevisiae, Rub1p, like ubiquitin, is conjugated to proteins. Before protein conjugation, the carboxyl-terminal asparagine residue of Rub1p is removed. Rub1p conjugation is dependent on the carboxyl-terminal processing enzyme Yuh1p, whereas Rub1p lacking the asparagine residue is conjugated without Yuh1p. Thus, Yuh1p is the major processing enzyme for Rub1p.
Rub1p is a ubiquitin-like protein conjugated to the cullin Cdc53p in the yeast Saccharomyces cerevisiae (7, 9, 11). Cdc53p is a subunit of the ubiquitin ligases (E3s) known as SCFs (SKP1-Cullin-F-box complexes) (12, 23). SCFs are RING finger-containing complexes necessary for the ubiquitinylation of cell cycle regulators and transcription factors (4, 18-20). The SCFs target these substrates for degradation via the 26S proteasome (4, 18-20). Although the function of Rub1p conjugation remains elusive, altered expression of SCF proteins can be lethal in the absence of Rub1p conjugation to Cdc53p (9).
Rub1p and ubiquitin conjugation are enzymatically related. Rub1p is activated by an E1 enzyme composed of a heterodimer of Enr2/Ula1p and Uba3p. Enr2/Ula1p and Uba3p resemble the amino-terminal and carboxyl-terminal portions of the ubiquitin-activating enzyme Uba1p, respectively (9, 11). Rub1p forms a thioester linkage with Uba3p (11). Next, Rub1p is transferred to the Rub1p-conjugating enzyme Ubc12p, a homolog of the ubiquitin-conjugating enzymes (11). Enr2/Ula1p, Uba3p, and Ubc12p are necessary for Rub1p modification of Cdc53p. Yeast cells containing a conditional allele of SKP1 are also deficient for Rub1p conjugation to Cdc53p, even under permissive conditions (9). The Rub1p attachment site on Cdc53p is likely to be lysine 760 based on the attachment site in cullin-2 of NEDD8, the mammalian homolog of Rub1p (21). Removal of a portion of Cdc53p, including lysine 760, does not cause inviability in yeast cells; however, these cells resemble mutants lacking enzymes of the Rub1p pathway (9).
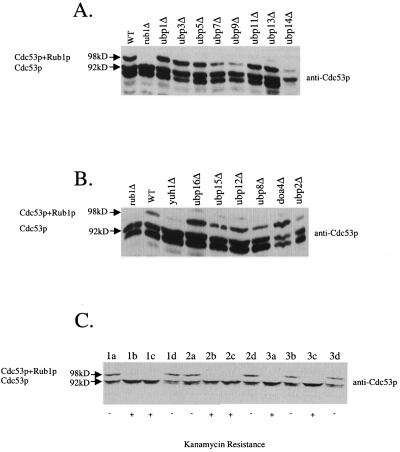
Rub1p is 53% similar to ubiquitin (9). Therefore, we reasoned that the enzymes known as ubiquitin hydrolases might also act on Rub1p. In S. cerevisiae, there are 16 genes that encode such proteins, termed UBP1 through UBP16 (1). Deletion mutants of these genes, except for UBP6 and UBP10, were purchased from Research Genetics (Table 1) and screened for the ability to conjugate Rub1p to Cdc53p. Yeast cells were grown at 30°C on yeast extract-peptone-dextrose medium by standard procedures (2), and cell extracts were prepared as previously described (5, 12). Extracts from cells with mutations in the ubiquitin hydrolases contained Rub1p conjugated to Cdc53p (Fig. 1A and B). Rub1p-Cdc53p conjugates were also detected in cells with mutations in UBP6 (B. Linghu and M. G. Goebl, unpublished observations). Yeast cells also contain Yuh1p, a homolog of the ubiquitin carboxyl-terminal hydrolases which remove small adducts from ubiquitin (10). In cells with mutations in YUH1, Rub1p-Cdc53p conjugates were virtually absent (Fig. 1B). To verify that the loss of Rub1p conjugation to Cdc53p is due to the loss of YUH1, a yuh1Δ strain (R16911) in which YUH1 is replaced by a kanamycin resistance gene was mated to FY23, the YUH1 wild-type strain. This diploid was sporulated, and tetrads were dissected and examined for kanamycin resistance (G418 concentration, 200 μg/ml) and Rub1p conjugation to Cdc53p. Kanamycin-resistant cells derived from isolated spores failed to conjugate Rub1p to Cdc53p (Fig. 1C). These results indicate that the YUH1 gene is critical for Rub1p conjugation to Cdc53p.
TABLE 1.
Yeast strains used in this study
| Strain | Genotype | Source |
|---|---|---|
| FY23 | MATaura3-52 trp1-Δ63 leu2-Δ1 | Fred Winston |
| FY24 | MATα ura3-52 trp1-Δ63 leu2-Δ1 | Fred Winston |
| FY78 | MATahis3-Δ200 | Fred Winston |
| BL2 | MATα | Segregant of FY24 × FY78 |
| BY4741 | MATahis3Δ1 leu2Δ0 met15Δ0 ura3Δ0 | Research Genetics |
| BY4742 | MATα his3Δ1 leu2Δ0 lys2Δ0 ura3Δ0 | Research Genetics |
| BY4743 | BY4741 × BY4742 | Research Genetics |
| R4073 | rub1::Km in BY4741 | Research Genetics |
| R2825 | enr2::Km in BY4741 | Research Genetics |
| R5214 | ubc12::Km in BY4741 | Research Genetics |
| R5485 | uba3::Km in BY4741 | Research Genetics |
| YPH1172 | MATaura3-52 lys2-801 ade2-101 trp1-Δ63 his3-Δ200 leu2-Δ1 skp1Δ1::TRP1 skp1-3::LEU2 CFIII(CEN3.L. YPH983) HIS3 SUP11 | Phillip Hieter |
| R16911 | yuh1::Km in BY4742 | Research Genetics |
| R33819 | ubp1::Km/ubp1::Km in BY4743 | Research Genetics |
| R32380 | ubp2::Km/ubp2::Km in BY4743 | Research Genetics |
| R36148 | ubp3::Km/ubp3::Km in BY4743 | Research Genetics |
| R34044 | doa4::Km/doa4::Km in BY4743 | Research Genetics |
| R35842 | ubp5::Km/ubp5::Km in BY4743 | Research Genetics |
| R32315 | ubp7::Km/ubp7::Km in BY4743 | Research Genetics |
| R30809 | ubp8::Km/ubp8::Km in BY4743 | Research Genetics |
| R36404 | ubp9::Km/ubp9::Km in BY4743 | Research Genetics |
| R36014 | ubp11::Km/ubp11::Km in BY4743 | Research Genetics |
| R31228 | ubp12::Km/ubp12::Km in BY4743 | Research Genetics |
| R33093 | ubp13::Km/ubp13::Km in BY4743 | Research Genetics |
| R33195 | ubp14::Km/ubp14::Km in BY4743 | Research Genetics |
| R30892 | ubp15::Km/ubp15::Km in BY4743 | Research Genetics |
| R32755 | ubp16::Km/ubp16::Km in BY4743 | Research Genetics |
| JL268 | MATα ade2 leu2 his3 ura3 trp1 rub1::TRP1 | Judy Callis |
| BLA1 | MATα his3 trp1 ura3 leu2 lys2 | Segregant of R16911 × FY23 |
| BLA2 | MATayuh1::Km ura3 leu2 lys2 | Segregant of R16911 × FY23 |
| BLA3 | MATα trp1 ura3 leu2 | Segregant of R16911 × FY23 |
| BLA4 | MATayuh1::Km his3 ura3 leu2 | Segregant of R16911 × FY23 |
| BLB1 | MATα trp1 ura3 leu2 lys2 | Segregant of R16911 × FY23 |
| BLB2 | MATayuh1::Km ura3 leu2 | Segregant of R16911 × FY23 |
| BLB3 | MATα yuh1::Km his3 ura3 leu2 lys2 | Segregant of R16911 × FY23 |
| BLB4 | MATahis3 trp1 ura3 leu2 | Segregant of R16911 × FY23 |
| BLC1 | MATahis3 trp1 ura3 leu2 lys2 | Segregant of R16911 × FY23 |
| BLC2 | MATayuh1::Km trp1 ura3 leu2 | Segregant of R16911 × FY23 |
| BLC3 | MATα yuh1::Km ura3 leu2 lys2 | Segregant of R16911 × FY23 |
| BLC4 | MATα his3 ura3 leu2 | Segregant of R16911 × FY23 |
| BLF1 | MATα yuh1::Km rub1::TRP1 trp1 ura3 leu2 | Segregant of BLC2 × JL268 |
| BLF7 | MATα yuh1::Km rub1::TRP1 ade2 trp1 ura3 leu2 | Segregant of BLC2 × JL268 |
FIG. 1.
Western blot analysis of cells with mutated genes encoding the putative ubiquitin hydrolases, using antibodies to Cdc53p. Cdc53p and Cdc53p-Rub1p conjugates are marked by arrows at 92 and 98 kDa, respectively. (A and B) Cell extracts were prepared from the indicated mutant strains and subjected to SDS-PAGE and Western blot analysis using antibodies to Cdc53p. Wild-type (WT; strain BL2) and rub1Δ mutant cell extracts were also loaded to serve as positive and negative controls for Rub1p conjugation.Mutant strain designations: rub1Δ, strain R4073; ubp1Δ, strain R33819; ubp2Δ, strain R32380; ubp3Δ, strain R36148; doa4(ubp4)Δ, strain R34044; ubp5Δ, strain R35842; ubp7Δ, strain R32315; ubp8Δ, strain R30809; ubp9Δ, strain R36404; ubp11Δ, strain R36014; ubp12Δ, strain R31228; ubp13Δ, strain R33093; ubp14Δ, strain R33195; ubp15Δ, strain R30892; ubp16Δ, strain R32755; yuh1Δ, strain R16911. All strains were congenic with BY4743 and were diploids, with both wild-type alleles of the indicated gene replaced by the KanMX gene, except for BL2 and R16911, which were haploids. (C) Cell extracts were prepared from spore colonies of three tetrads (denoted by numerals 1 through 3) from a cross between R16911 and FY23 and subjected to SDS-PAGE and Western blot analysis using antibodies to Cdc53p. The four spore colonies from each tetrad are denoted by letters a through d. Whether the spore colony was sensitive (−) or resistant (+) to G418 (kanamycin) is indicated below each lane.
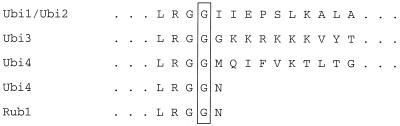
In all known ubiquitin-like conjugations, the carboxyl-terminal glycine is attached via an isopeptide linkage to a lysine on a substrate protein (6, 16). However, the initial gene product made from each ubiquitin gene and RUB1 has a carboxyl-terminal extension (14, 15) (Fig. 2). Therefore, removal of these extensions is necessary before ubiquitin or Rub1p is suitable for substrate conjugation.
FIG. 2.
Junctions between the carboxyl termini of mature ubiquitin and Rub1p and the carboxyl extensions of the immature proteins from S. cerevisiae. The carboxyl-terminal glycine of each processed molecule is boxed. Dots at the amino-terminal ends represent the remaining amino acid residues of ubiquitin and Rub1p. Dots at the carboxyl-terminal ends represent the remaining amino acid residues of the carboxyl tails.
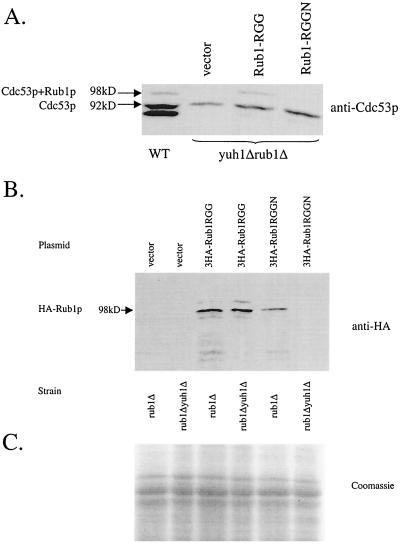
If the function of Yuh1p were to remove the carboxyl-terminal asparagine of Rub1p, then expression of a RUB1 gene encoding a Rub1p lacking the carboxyl-terminal asparagine would suppress the loss of Rub1p conjugation to Cdc53p in yuh1Δ strains. Therefore, we transformed a yuh1Δ rub1Δ strain with two plasmids, pRub1RGG, which expresses a RUB1 gene encoding a mutant Rub1p lacking the carboxyl-terminal asparagine, and pRub1RGGN, which expresses wild-type RUB1 (details of plasmid construction are available upon request) (17). These plasmids are derived from p426ADH and PCR DNA fragments of RUB1 (13). The RUB1 gene is expressed from the yeast ADH1 promoter. Cell extracts were obtained from these transformants and subjected to sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) and Western blot analysis with anti-Cdc53 (5, 12). Expression of the Rub1p lacking asparagine allowed the production of a protein that comigrates with Rub1p-Cdc53p conjugates at 98 kDa and was detectable with anti-Cdc53 (Fig. 3). This protein was not detected in yuh1Δ cells containing p426ADH or pRub1RGGN expressing wild-type RUB1 (Fig. 3).
FIG. 3.
Rub1p lacking asparagine is conjugated to Cdc53p in the absence of Yuh1p. Plasmids were created by standard techniques. (A) Cell extracts were prepared from the yeast strains described below and subjected to SDS-PAGE and Western blot analysis using antibodies to Cdc53p. Cdc53p and Cdc53p-Rub1p conjugates are marked by arrows at 92 and 98 kDa, respectively. Yeast strains included the wild-type strain FY23 (WT) and the yuh1Δrub1Δ mutant strain BLF7. (B) Cell extracts were prepared from yeast strain JL268 or BLF7 transformed with the plasmids indicated below and subjected to SDS-PAGE and Western blot analysis using antibodies to the HA epitope. Cdc53p-Rub1p conjugates are marked by an arrow at 98 kDa. Lanes: vector, extracts from cells transformed with p426ADH; 3HA-Rub1RGG and 3HA-Rub1RGGN, extracts from cells transformed with plasmids p3HA-Rub1RGG and p3HA-Rub1RGGN, respectively. (C) The whole-cell extracts used for Western blot analysis in panel B were stained with Coomassie blue. In all panels, the vector is p426ADH, Rub1RGGN is the wild-type Rub1, and Rub1RGG is Rub1 lacking COOH-asparagine.
To verify that Rub1p lacking asparagine is attached to Cdc53p in the absence of Yuh1p, we transformed the rub1Δ and rub1Δ yuh1Δ strains with plasmids p3HA-Rub1-RGG and p3HA-Rub1-RGGN. These plasmids, derived from p426ADH, again provided the ADH1 promoter to express RUB1 encoding a fusion protein of three hemagglutinin (HA) epitopes at the amino terminus of mutant Rub1p lacking asparagine or wild-type Rub1p, respectively. Cell extracts were prepared from the transformants and subjected to SDS-PAGE and Western blot analysis using anti-HA. Whereas HA-Rub1p-Cdc53p conjugates lacking asparagine were detected regardless of the presence of Yuh1p, wild-type HA-Rub1p was only found conjugated to Cdc53p in the presence of Yuh1p (Fig. 3). While a search was conducted for synthetic interactions between YUH1 and the SCF genes, none were detected. However, since our initial report of this phenomenon, we have subsequently found the synthetic interactions to vary greatly among strains (M. Goebl and J. Callis, unpublished observations).
A mammalian homolog of Yuh1p, UCH-L3, processes NEDD8 in vitro (22). Our results suggest that a function of these UCH-like enzymes is to process the Rub1p and NEDD8 proteins. Another mammalian homolog, UCH-L1, is highly expressed in neural tissue (3), and mice lacking both UCH-L1 and UCH-L3 have severe neurological disorders that lead to early death (8). We therefore propose that NEDD8 processing and cullin modification are critical for the development and function of neural tissue.
Acknowledgments
We thank Jana Narasimhan for critical reading of the manuscript and the Goebl laboratory staff for comments.
This work was supported by grants to J.C. from the U.S. Department of Energy (DE-FG03-00ER15056) and to M.G.G. from the National Science Foundation (MCB-0091317).
REFERENCES
- 1.Amerik, A. Y., S. J. Li, and M. Hochstrasser. 2000. Analysis of the deubiquitinating enzymes of the yeast Saccharomyces cerevisiae. Biol. Chem. 381:981-992. [DOI] [PubMed] [Google Scholar]
- 2.Burke, D., D. Dawson, and T. Stearns. 2000. Methods in yeast genetics: a Cold Spring Harbor Laboratory course manual. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 3.Day, I. N., L. J. Hinks, and R. J. Thompson. 1990. The structure of the human gene encoding protein gene product 9.5 (PGP9.5), a neuron-specific ubiquitin C-terminal hydrolase. Biochem. J. 268:521-524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Feldman, R. M. R., C. C. Correll, K. B. Kaplan, and R. J. Deshaies. 1997. A complex of Cdc4p, Skp1p, and Cdc53p/cullin catalyzes ubiquitination of the phosphorylated CDK inhibitor Sic1p. Cell 91:221-230. [DOI] [PubMed] [Google Scholar]
- 5.Goebl, M. G., L. Goetsch, and B. Byers. 1994. The Ubc3 (Cdc34) ubiquitin-conjugating enzyme is ubiquitinated and phosphorylated in vivo. Mol. Cell. Biol. 14:3022-3029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hershko, A., and A. Ciechanover. 1998. The ubiquitin system. Annu. Rev. Biochem. 67:425-479. [DOI] [PubMed] [Google Scholar]
- 7.Hochstrasser, M. 1998. There's the Rub: a novel ubiquitin-like modification linked to cell cycle regulation. Genes Dev. 12:901-907. [DOI] [PubMed] [Google Scholar]
- 8.Kurihara, L. J., T. Kikuchi, K. Wada, and S. M. Tilghman. 2001. Loss of UCH-L1 and UCH-L3 leads to neurodegeneration, posterior paralysis, and dysphagia. Hum. Mol. Genet. 10:1963-1970. [DOI] [PubMed] [Google Scholar]
- 9.Lammer, D., N. Mathias, J. M. Laplaza, W. Jiang, Y. Liu, J. Callis, M. G. Goebl, and M. Estelle. 1998. Modification of yeast Cdc53p by the ubiquitin-related protein Rub1p affects function of the SCFCdc4 complex. Genes Dev. 12:914-926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Larsen, C. N., B. A. Krantz, and K. D. Wilkinson. 1998. Substrate specificity of deubiquitinating enzymes: ubiquitin C-terminal hydrolases. Biochemistry 37:3358-3368. [DOI] [PubMed] [Google Scholar]
- 11.Liakopoulos, D., G. Doenges, K. Matuschewski, and S. Jentsch. 1998. A novel protein modification pathway related to the ubiquitin system. EMBO J. 17:2208-2214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Mathias, N., S. L. Johnson, M. Winey, A. E. M. Adams, L. Goetsch, J. R. Pringle, B. Byers, and M. G. Goebl. 1996. Cdc53p acts in concert with Cdc4p and Cdc34p to control the G1-to-S-phase transition and identifies a conserved family of proteins. Mol. Cell. Biol. 16:6634-6643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Mumberg, D., R. Müller, and M. Funk. 1995. Yeast vectors for the controlled expression of heterologous proteins in different genetic backgrounds. Gene 156:119-122. [DOI] [PubMed] [Google Scholar]
- 14.Ökaynak, E., D. Finley, and A. Varshavsky. 1984. The ubiquitin gene: head-to-tail repeats encoding polyubiquitin precursor protein. Nature 312:663-666. [DOI] [PubMed] [Google Scholar]
- 15.Ökaynak, E., D. Finley, M. J. Solomon, and A. Varshavsky. 1987. The yeast ubiquitin genes: a family of natural gene fusions. EMBO J. 6:1429-1439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Pickart, C. M. 2001. Mechanisms underlying ubiquitination. Annu. Rev. Biochem. 70:503-533. [DOI] [PubMed] [Google Scholar]
- 17.Sambrook, J., and D. W. Russell. 2001. Molecular cloning: a laboratory manual, 3rd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 18.Seol, J. H., R. M. R. Feldman, W. Zachariae, A. Shevchenko, C. C. Correll, S. Lyapina, Y. Chi, M. Galova, J. Claypool, S. Sandmeyer, K. Nasmyth, A. Shevchenko, and R. J. Deshaies. 1999. Cdc53/cullin and the essential Hrt1 RING-H2 subunit of SCF define a ubiquitin ligase module that activates the E2 enzyme Cdc34. Genes Dev. 13:1614-1626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Skowrya, D., K. L. Craig, M. Tyers, S. J. Elledge, and J. W. Harper. 1997. F-box proteins are receptors that recruit phosphorylated substrates to the SCF ubiquitin-ligase complex. Cell 91:209-219. [DOI] [PubMed] [Google Scholar]
- 20.Skowrya, D., D. M. Koepp, T. Kamura, M. N. Conrad, R. C. Conaway, J. W. Conaway, S. J. Elledge, and J. W. Harper. 1999. Reconstitution of G1 cyclin ubiquitination with complexes containing SCFGrr1 and Rbx1. Science 284:662-665. [DOI] [PubMed] [Google Scholar]
- 21.Wada, H., E. T. H. Yeh, and T. Kamitani. 1999. Identification of NEDD8-conjugation site in human cullin-2. Biochem. Biophys. Res. Commun. 257:100-105. [DOI] [PubMed] [Google Scholar]
- 22.Wada, H., K. Kito, L. S. Caskey, E. T. H. Yeh, and T. Kamitani. 1998. Cleavage of the C-terminus of NEDD8 by UCH-L3. Biochem. Biophys Res. Commun. 251:688-692. [DOI] [PubMed] [Google Scholar]
- 23.Willems, A. R., S. Lanker, E. E. Patton, K. L. Craig, T. F. Nason, N. Mathias, R. Kobayashi, C. Wittenberg, and M. Tyers. 1996. Cdc53 targets phosphorylated G1 cyclins for degradation by the ubiquitin proteolytic pathway. Cell 86:453-463. [DOI] [PubMed] [Google Scholar]