Abstract
In an attempt to define the distinctive Wolf-Hirschhorn syndrome (WHS) phenotype, and to map its specific clinical manifestations, a total of eight patients carrying a 4p16.3 microdeletion were analyzed for their clinical phenotype and their respective genotypes. The extent of each individual deletion was established by fluorescence in situ hybridization, with a cosmid contig spanning the genomic region from MSX1 (distal half of 4p16.1) to the subtelomeric locus D4S3359. The deletions were 1.9–3.5 Mb, and all were terminal. All the patients presented with a mild phenotype, in which major malformations were usually absent. It is worth noting that head circumference was normal for height in two patients (those with the smallest deletions [1.9 and 2.2 Mb]). The currently accepted WHS critical region (WHSCR) was fully preserved in the patient with the 1.9-Mb deletion, in spite of a typical WHS phenotype. The deletion in this patient spanned the chromosome region from D4S3327 (190 b4 cosmid clone included) to the telomere. From a clinical point of view, the distinctive WHS phenotype is defined by the presence of typical facial appearance, mental retardation, growth delay, congenital hypotonia, and seizures. These signs represent the minimal diagnostic criteria for WHS. This basic phenotype maps distal to the currently accepted WHSCR. Here, we propose a new critical region for WHS, and we refer to this region as “WHSCR-2.” It falls within a 300–600-kb interval in 4p16.3, between the loci D4S3327 and D4S98-D4S168. Among the candidate genes already described for WHS, LETM1 (leucine zipper/EF-hand-containing transmembrane) is likely to be pathogenetically involved in seizures. On the basis of genotype-phenotype correlation analysis, dividing the WHS phenotype into two distinct clinical entities, a “classical” and a “mild” form, is recommended for the purpose of proper genetic counseling.
Introduction
Wolf-Hirschhorn syndrome (WHS [MIM 194190]) is a segmental aneusomy syndrome caused by a partial 4p deficiency. Many patients carry a deletion of several megabases that results in severe mental and growth retardation, major malformations and seizures, and a characteristic facial appearance with wide forehead, large and protruding eyes, hypertelorism, prominent glabella, down-turned mouth, and micrognathia. In the last few years, several patients have been reported who have WHS and in whom a 4p16.3 microdeletion can be detected only by molecular probes on apparently normal chromosomes. Patients in this group usually present with a milder phenotype that lacks congenital malformations (Gandelman et al. 1992; Johnson et al. 1994; Clemens et al. 1996; Lindeman-Kusse et al. 1996; Fang et al. 1997; Partington et al. 1997; Wieczorek et al. 2000; Zollino et al. 2000).
As a result of overlapping deletion analysis, the area currently regarded as the WHS critical region (WHSCR) is restricted to the 165-kb interval on 4p16.3, defined by the loci D4S166 (cosmid 174g8 included) and D4S3327 (cosmid 19h1 included) (Wright et al. 1997). It was suspected that a few pleiotropic genes—or just one gene—residing in this region may be critically responsible for the WHS phenotype, acting as transcriptional regulators of other genes. Three different genes have been independently described as candidates for WHS. WHSC1 (Stec et al. 1998 [MIM 602952])—identified by cosmids 19h1, 190b4, and, in part, 184d6—overlaps the distal boundary of the WHSCR. WHSC2 (Wright et al. 1999 [MIM 606026]), which falls entirely within the WHSCR, is identified by cosmid 96a2. The WHSCR flanking gene LETM1 (leucine zipper/EF-hand–containing transmembrane [MIM 604407]), which is involved in Ca2+ signaling, was recently described as an excellent candidate gene for WHS, in particular for seizures (Endele et al. 1999). It is identified by cosmid 75b9 and PAC clone 184O23, and it maps distally to the WHSCR. Actually, most WHS-associated deletions, either terminal or interstitial, are much larger than the WHSCR, usually including all the candidate genes. Rauch et al. (2001) recently reported a unique patient who had a small interstitial deletion restricted to the WHSCR and in whom the LETM1 gene was preserved. As pointed out by Rauch et al. (2001), the patient presented with an atypical WHS phenotype that included normal height, questionable WHS facial appearance, and, more importantly, no seizures. Another large interstitial deletion—resulting in an atypical WHS phenotype, with no seizures—established the distal boundary of the WHSCR (Somer et al. 1995; Wright et al. 1997). The LETM1 gene was also preserved in this patient.
Using the approach of genotype-phenotype correlation analysis in eight informative patients, we characterize the distinctive WHS clinical signs that represent the minimal diagnostic criteria for this condition, and we map this basic phenotype outside the currently defined WHSCR.
Patients and Methods
Patients
Eight patients with WHS carrying a 4p microdeletion (four boys and four girls), aged 1–15 years, were analyzed for their clinical phenotype and their respective genotypes. Clinical evaluation in all patients included heart and kidney ultrasonography and electroencephalographic examination, and brain magnetic resonance imaging (MRI) was performed in most. Individual clinical signs are summarized in table 1. Patients 13, 14, and 15 were reported elsewhere among a large series of patients (Zollino et al. 2000). A more detailed clinical description is in order for patient MG, and a lymphoblastoid cell line of this patient is available.
Table 1.
Clinical and Molecular Data Related to Patients with WHS and Microdeletions[Note]
| Patientand Sex | Age atDelivery(wk) | BirthWeight(g) | Age atExamination(years) | GrowthDelay | CharacteristicFace | MR | Seizures | Microcephaly | CleftPalate | Age at LanguageAcquisition, Walking(mo)a | DeletionSize(Mb) |
| 43, F | 38 | 2,075 | 1 | + | + | + | +b | + | + | N/A | 3.5c |
| 44, M | 40 | 2,430 | 11 | + | + | + | + | + | − | N/A | 3.5 |
| 13, M | 40 | 2,230 | 12 | + | + | + | + | + | − | 20 | 2.6 |
| 46, F | 40 | 2,650 | 1 | + | + | + | + | + | + | N/A | 2.6 |
| 14, M | 40 | 2,600 | 10 | + | + | + | + | + | − | 28 | 2.5d |
| 26, F | 39 | 2,600 | 7 | + | + | + | + | + | − | 48 | 2.5 |
| 15, F | 40 | 2,600 | 10 | + | + | + | + | − | − | 60 | 2.2 |
| MG, M | 40 | 2,500 | 1 | + | + | + | + | − | − | N/A | 1.9 |
Note.— Patients are listed according to the extent of the deletion.
N/A = not available because of very early age at examination.
Electroencephalographic anomalies.
der(4)(pter→p16.3:10p15.3→pter).
+ dup(4)(p16.1p16.3).
Patient MG
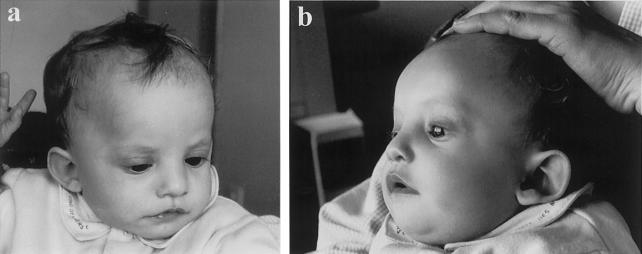
Patient MG is 12 mo old at the time of writing and is the fifth child of healthy, nonconsanguineous parents. The father was 42 and the mother 39 years old at the time of his birth. Family history was unremarkable. The mother underwent amniocentesis, and results showed a normal male karyotype. Intrauterine growth retardation was apparent during and after the 6th mo of pregnancy. MG was born at term by normal delivery, with a birth weight of 2,500 g, a length of 48 cm, and an occipitofrontal circumference (OFC) of 32 cm. A diagnosis of congenital hypotonia was established at an early age: sucking was poor; the infant could not raise his head until age 4 mo; and at 12 mo he was still unable to sit unsupported. The postnatal course was characterized by persistent failure to thrive, with gastroesaphageal reflux in the first 9 mo. At the age of 9 mo, MG experienced his first episode of generalized seizures, with four relapses in 1 wk. Subsequently, seizures were controlled by anticonvulsant therapy. The boy was also noted to have an unusual facial appearance. This, together with failure to thrive and congenital hypotonia and seizures, prompted a referral to one of us (M.Z.), with the tentative diagnosis of WHS. When we first saw him, at the age of 9 mo, length was 62 cm (≪3rd centile), weight was 5.5 kg (≪3rd centile), and OFC was 41 cm (<2nd centile) and proportionate to length. Facial appearance was undoubtedly that of WHS. The infant presented with high forehead, frontal bossing, prominent glabella, hypertelorism, large and protruding eyes, down-turned corners of the mouth, and abnormally shaped and low-set ears (fig. 1). His interaction with the environment was very good: he was interested in toys and had good eye contact. No major malformations were detected. Ultrasound examination of heart and kidneys was reported to show normal results, and results of a brain MRI were normal. Electroencephalographic examination at age 9 mo showed sequences of parieto-occipital sharp waves and high-voltage waves with superimposed spikes. The diagnosis of WHS was confirmed by many clinicians experienced in the field.
Figure 1.
Frontal (a) and lateral (b) view of patient MG. Note the typical WHS facial appearance, with high forehead, frontal bossing, large eyes, high nasal bridge, down-turned mouth, and dysmorphic ears.
Conventional and Molecular Cytogenetics
Prometaphase chromosome analysis (600–800 bands) was performed on peripheral blood lymphocytes by R(RBG) banding. At least 20 metaphase spreads were scored for each patient.
FISH analyses were performed on standard chromosomes, as described elsewhere (Lichter et al. 1990), with a set of overlapping cosmids spanning the chromosome region from the distal half of 4p16.1 to the telomere. In each patient, FISH analyses were performed with the subtelomeric probe for D4S3359 (Vysis) and with probes 19h1 and 174g8, which delimit, distally and proximally, the currently defined WHSCR (Wright et al. 1997). Individual patients were analyzed further with a series of cosmid probes that were specifically selected from the following: MSX1, 228a7, 21f12, 247f6, 241c2, 185f6, 139h8, 212a9, 79f5, 95e6, 161c2, 33c6, 124h12 (10d12), 108f12, 141a8, 190b4, pC385.12, pC678, IS28, and CD2. In patient MG (who has a 1.9-Mb deletion), FISH was performed with the following probes: 27h9, 96a2, and 184d6, in addition to the above-mentioned 19h1, 174g8, 190b4, pC385.12, and pC678 and the subtelomeric probe for D4S3359. Cosmids tested in individual patients are listed in table 2. Parents’ chromosomes were analyzed with probe pC678 (for D4S96, which was found to be deleted in all patients).
Table 2.
Cosmids Tested by FISH[Note]
|
Patientb |
||||||||||
| Locus | Cosmid | Distancefrom 4pter(Mb)a | 44 | 43 | 46 | 13 | 14 | 26 | 15 | MG |
| S3359 | Tel | − | − | − | − | − | − | − | − | |
| S90 | CD2 | − | ||||||||
| IS28 | − | |||||||||
| S96 | pC678 | − | − | − | − | − | − | − | − | |
| S98FGFR3 | pC385.12 | 1.8 | − | − | ||||||
| 184d6 | − | |||||||||
| S3327 | 190b4 | 1.9 | − | − | − | − | ||||
| 19h1 | 1.9 | − | − | − | − | − | − | − | + | |
| 96a2 | + | |||||||||
| 27h9 | + | |||||||||
| 174g8 | − | − | − | − | − | − | − | + | ||
| 141a8 | 2.2 | − | ||||||||
| 108f12 | 2.2 | − | − | +/− | + | |||||
| 124h12 | 2.2 | + | ||||||||
| S43 | 33c6 | 2.25 | − | − | − | − | − | − | + | |
| 161c2 | 2.35 | − | ||||||||
| 95e6 | 2.4 | − | ||||||||
| 79f5 | 2.5 | − | − | +/− | ||||||
| 212a9 | 2.5 | + | ||||||||
| 139h8 | 2.6 | − | + | |||||||
| 185f6 | 2.6 | +/− | + | |||||||
| 241c2 | 2.7 | + | + | |||||||
| S182 | 247f6 | 2.8 | + | + | + | + | ||||
| S180 | 21f12 | 3.3 | − | − | + | + | + | + | ||
| S81 | 228a7 | 3.7 | + | + | + | + | ||||
| MSX1 | MSX1 | 4.7 | + | + | + | + | + | |||
Note.— Coincident evaluations refer to overlapping cosmids (Baxendale et al. 1993).
Distances were approximated to within 50 kb.
+/− = a weak fluorescent signal on one chromosome 4 and a strong signal on the homologous one.
Results
Chromosomes
On conventional cytogenetic examination, all the patients had apparently normal chromosomes. By FISH, a terminal 4p16.3 microdeletion was detected in each patient. The microdeletion varied in size from 1.9 Mb (190b4 cosmid included) to 3.5 Mb (21f12 cosmid included). Deletion sizes were established according to Ensembl mapping and the mapping reported by Baxendale et al. (1993). In patients 13, 14, and 15, the deletion size was slightly different from the size reported elsewhere (Zollino et al. 2000), on the basis of the updated physical map.
It is worth noting that, on FISH results, the currently defined WHSCR was included in the deletion interval in seven patients, but it was fully preserved in one patient (table 2). The WHS-producing 4p deletion was a de novo event in seven patients. A partial 4p duplication was also detected in one of them (patient 14), as previously reported (Zollino et al. 1999). In one patient, the 4p deletion was the result of an unbalanced segregation of a t(4;10)(p16.3;p15.3) maternal translocation (patient 43).
Genotype-Phenotype Correlations
All the patients reported here had a relatively mild WHS phenotype. The average age at independent walking was ∼30 mo; major malformations were absent in most patients; and in two patients head circumference was within normal limits, relative to height (patients 15 and MG, who carried a 2.2-Mb and a 1.9-Mb deletion, respectively). In particular, patients 13 and 44 (aged 12 and 11 years, respectively) presented with very mild mental delay: they were able to form complex sentences with a rich vocabulary and could read and write. Productive language was limited to about 20 words in patient 14, but his comprehension and his interaction with the environment were fairly good. The more severe delay is most likely due to the double chromosome imbalance (dup/del 4p) detected in this patient. Patients 15 and 26 had no language and presented with a more severe motor delay: they walked unsupported at the ages of 5 and 4 years, respectively. However, the microdeletion detected in these patients correlated with normal head circumference (for height) in patient 15 and with the absence of major malformations in both. Although patients 43 and 46 were observed at a very young age (12 mo), lively and sociable behavior was evident.
Cleft palate occurred in association with deletions of 3.5 and 2.6 Mb, in patients 43 and 46, respectively, who both presented with mild psychomotor delay. Phenotypic variability was still evident among the present patients. Clinical manifestations shared by all the patients included characteristic facial appearance, psychomotor retardation, growth delay, and seizures (table 1). However, the severity of this phenotype was much less than that observed in patients with larger deletions (Wilson et al. 1981; Wieczorek et al. 2000; Zollino et al. 2000).
Discussion
The WHS phenotype is characterized by a particular facial appearance, with “Greek helmet” profile, severe growth delay, microcephaly, profound mental retardation, congenital hypotonia, midline defects, such as cleft palate and hypospadias, iris coloboma, congenital heart defects, renal abnormalities, and seizures (Wilson et al. 1981). A 4p deletion of several megabases is easily detectable by conventional cytogenetics in association with this phenotype. However, in the last few years, molecular cytogenetics allowed the detection of several 4p16.3 microdeletions in apparently normal chromosomes (Gandelman et al. 1992; Johnson et al. 1994; Clemens et al. 1996; Lindeman-Kusse et al. 1996; Fang et al. 1997; Partington et al. 1997; Wieczorek et al. 2000; Zollino et al. 2000). These small deletions usually result in a mild phenotype lacking major malformations.
Elsewhere we have observed that genotype-phenotype correlations in WHS mostly depend on the extent of the deletion, with 3.5 Mb representing some sort of discriminating size (Zollino et al. 2000). Physical distances from the telomere in the patients reported elsewhere are now slightly modified, in accordance with updated 4p maps. Here, we report a clinical-molecular study in a total of eight patients with microdeletions, three of whom were described elsewhere (Zollino et al. 2000). The deletion size varied in these patients, from 1.9 to 3.5 Mb. All of them presented with characteristic facial appearance, growth delay, mental retardation, congenital hypotonia, and seizures. With the exception of cleft palate—which occurred in the patients with 3.5-Mb (patient 43) and 2.6-Mb (patient 46) deletions, who both presented with mild psychomotor delay—other major malformations were not detected. Head circumference was normal for height, in the two patients who carried the smallest deletions (1.9 and 2.2 Mb, respectively).
WHS can be considered a contiguous gene syndrome, with an increasing number, as well as increasing severity, of clinical manifestations, depending on the extent of the deletion. Most clinical signs have been tentatively mapped (Estabrooks et al. 1995). However, we can now modify Estabrooks and colleagues' phenotypic map relative to “cleft palate” and “microcephaly,” moving the distal limit of their genomic region to a distance of 2.6 and 2.2 Mb from the telomere, respectively.
When patients carrying a large deletion are compared with those carrying a microdeletion, the shared clinical manifestations include characteristic facial appearance, congenital hypotonia, mental retardation, growth delay, and seizures. These signs define the basic WHS phenotype and should be regarded as minimal diagnostic criteria for this condition. According to this view, the critical genomic region for WHS would be the smallest region whose haploinsufficiency gives rise to this basic phenotype. The currently recognized WHSCR is restricted to a 165-kb interval in 4p16.3, defined by the loci D4S166 (cosmid 174g8 included) proximally and D4S3327 (cosmid 19h1 included) distally (Wright et al. 1997). It was inferred that a few genes, or just one, residing in this region can be responsible for the WHS phenotype, possibly by acting as transcriptional regulators of other genes. Three candidate genes have been independently reported. WHSC1 (Stec et al. 1998), which for two-thirds of its length overlaps with the distal half of the WHSCR, is included in cosmids 19h1, 190b4, and, partially, 184D6. WHSC2 (Wright et al. 1999) falls entirely within the WHSCR and is included in cosmid 96a2. The flanking gene LETM1 was recently described as a candidate gene for seizures (Endele et al. 1999). It lies just distal to the WHSCR and is identified by cosmid 75b9 and PAC clone 184O23. The ubiquitous protein encoded by LETM1 is a member of the superfamily of EF-hand Ca2-banding proteins that respond to changes in the intracellular calcium level. Since impaired Ca2+ homeostasis in nerve cells has been suggested to correlate with neurodegenerative disorders and seizures (Burgess et al. 1997), LETM1 is an excellent candidate gene for neuromuscular impairment in WHS.
The WHS-producing deletions are usually larger than the WHSCR, including all the candidate genes. The patient with a small interstitial deletion restricted to the WHSCR reported by Rauch et al. (2001) presented with an atypical WHS phenotype, and his facial appearance, although suggestive of WHS, was thought to reflect resemblance to the parents. More importantly, he had no seizures. Therefore, a diagnosis of WHS is questionable in this case. The patient reported by Rauch et al. (2001) was hemizygous for WHSC2 and WHSC1, and LETM1 was fully preserved.
The distal boundary of the WHSCR was established to occur at the level of cosmid 19h1, as a result of the molecular analysis of a large interstitial deletion (Somer et al. 1995; Wright et al. 1997). However, this rearrangement also resulted in atypical WHS phenotype, with no seizures, and LETM1 was fully preserved. Patient MG in our series presented with a rather typical WHS phenotype, including marked growth delay, characteristic facial gestalt, psychomotor delay, and seizures with typical EEG pattern (Sgrò et al. 1995). Yet, on FISH analysis, the currently recognized WHSCR was apparently fully preserved. The commercially available FISH probe for WHS, provided by Vysis, also failed to identify the microdeletion. By combining the manufacturer’s information and 4p16.3 physical maps, we conclude that the Vysis probe appears to span a 90-kb interval of the WHSCR, including cosmids 19h1 and 96a2. A partial deletion of 19h1 in patient MG cannot be completely ruled out on the basis of FISH analyses. However, it can be safely concluded that this patient is hemizygous for WHSC1, since the 5′ half of the gene (cosmids 190b4 and 184d6) is included in the deletion interval. The patient of Rauch et al. was also hemizygous for WHSC1, yet he did not have even the basic WHS phenotype. Although the clinical description of the patient initially reported by Somer et al. (1995) and again by Wright et al. (1997) is not very detailed, he is currently accepted as having WHS. However, we considered him atypical, like the patient of Rauch et al. (2001), because of the absence of seizures. When we compare patient MG with the two above-mentioned patients with interstitial deletions (Somer et al. 1995; Rauch et al. 2001), we must conclude that the full WHS phenotype maps outside the WHSCR and is distal to it.
Fang et al. (1997) described a patient with a typical WHS phenotype, including seizures, who had an interstitial deletion of ∼1.9 Mb, with distal breakpoint between loci D4S96 and D4S168 (Fang et al. 1997). Wright et al. (1996) reported a 10-mo-old patient with WHS who carried an interstitial deletion (also reviewed by Fang et al. 1997) whose distal breakpoint occurred between D4S168 and D4S98, proximal to that found in Fang and colleagues' patient. Relevant WHS-associated deletions are shown in figure 2.
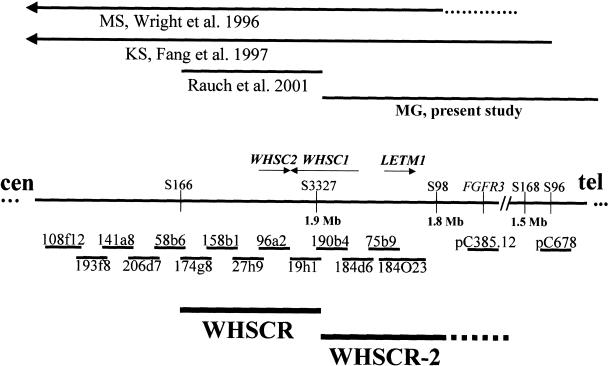
Figure 2.
Genomic organization of the WHS region in chromosome 4p16.3. With the exception of the PAC clone 184O23, all the reported probes are cosmids, most of which were used in our analysis for FISH. A contig including all the WHS candidate genes (WHSC2, WHSC1, and LETM1) is indicated. Note that WHSC2 is identified by cosmid 96a2; WHSC1 by cosmids 19h1, 190b4, and, in part, 184d6; and LETM1 by cosmid 75b9 and by PAC clone 184O23. Relevant 4p deletions are indicated (top).
We conclude that the distinctive WHS phenotype maps within a 300–600-kb interval in 4p16.3, between loci D4S3327 (patient MG) and D4S98–D4S168 (Wright et al. 1996; also reviewed by Fang et al. [1997]) (fig. 2). The newly proposed WHSCR, which we designate “WHSCR-2,” is contiguous distally with the currently defined WHSCR. At a molecular level, the WHSC1 gene overlaps the two regions, merging its 5′ half into WHSCR-2 and merging its 3′ half into WHSCR. However, as noted above, haploinsufficiency of this gene does not account for the entire basic WHS phenotype. Most likely, haploinsufficiency of WHSC1 accounts for some facial dysmorphisms, thus providing evidence that WHS is a contiguous gene syndrome.
With respect to WHS candidate genes, WHSCR-2 includes, in addition to the 5′ moiety of WHSC1, the entire LETM1. Given this evidence, WHSC2 should no longer to be considered a candidate gene for this condition. Rauch and colleagues' patient was hemizygous for WHSC1 but not for LETM1. LETM1 turns out to be the only candidate gene for seizures, as suggested elsewhere (Endele et al. 1999; Rauch et al. 2001). Thus, the hypothesis that typical WHS could be a single-gene disorder is unlikely. We think that the full WHS phenotype results from the haploinsufficiency of several different genes, some of which, like LETM1, may directly correlate with specific clinical signs. Additional candidate genes should be sought within the newly defined critical region. The same consideration applies to the diagnostic test. On the basis of overlapping analyses of either terminal or interstitial deletions associated with a typical WHS phenotype, D4S98 (FGFR3) appears to be the most specific haploinsufficiency locus for the molecular diagnosis of WHS.
Finally, the evidence from the patients reported here suggests that small deletions tend to be associated with a milder phenotype. To confirm this conclusion, we reviewed a total of 28 patients with microdeletions (including the present patients) (Gandelman et al. 1992; Johnson et al. 1994; Clemens et al. 1996; Lindeman-Kusse et al. 1996; Fang et al. 1997; Partington et al. 1997; Wieczorek et al. 2000) (table 3). Although some clinical descriptions are incomplete, there is substantial evidence that small deletions usually result in a milder phenotype with respect to both the occurrence of major malformations and the degree of mental retardation. Only 3 of 28 patients with microdeletions were severely retarded. With the exception of two patients, who presented with cleft palate and mild psychomotor delay, major malformations were not detected. On the other hand, phenotypic variability is largely assumed to characterize the WHS, possibly in relation to other still unknown genetic factors.
Table 3.
WHS-Associated Microdeletions: Clinical Data[Note]
| ClinicalManifestation | No. Affected/Total Patients | % |
| Characteristic face | 28/28 | 100 |
| Growth retardation | 28/28 | 100 |
| Microcephaly | 26/28 | 93 |
| Seizures | 26/27a | 96 |
| Mild congenital hypotonia | 22/24 | 91 |
| Mild-to-moderate mental retardation | 24/27 | 89 |
| Severe mental retardation | 3/27 | 11 |
| Congenital heart disease | 0/28 | 0 |
| Renal abnormalities | 0/28 | 0 |
| Cleft palate | 2/28 | 7 |
Note.— Microdeletions associated with an unbalanced translocation were considered only when an estimate of the deletion size was provided.
A 10-mo-old patient with no seizures was excluded from this evaluation.
For the purpose of proper genetic diagnosis and counseling, we recommend dividing WHS into a “classical” and a “mild” form, depending on the severity of the clinical presentation, which, in turn, is strongly correlated with the size of the deletion.
Acknowledgments
We are grateful to Tracy J. Wright and to Anita Rauch, for providing most cosmids used for FISH. We thank all the families with WHS and the Italian Association for WHS. This work was supported by Cofinanziamento 2001–MIUR.
Electronic-Database Information
Accession numbers and URLs for data presented herein are as follows:
- Ensembl Human Map, http://www.ensembl.org (for map of chromosome 4p)
- Online Mendelian Inheritance in Man (OMIM), http://www.ncbi.nlm.nih.gov/Omim/ (for WHS [MIM 194190], WHSC1 [MIM 602952], WHSC2 [MIM 606026], and LETM1 [MIM 604407])
References
- Baxendale S, MacDonald ME, Mott R, Francis F, Lin C, Kirby SF, James M, Zehetner G, Hummerich H, Valdes J, Collins FS, Deaven LJ, Gusella JF, Lehrach H, Bates GP (1993) A cosmid contig and high resolution restriction map of the 2 megabase region containing the Huntington’s disease gene. Nat Genet 4:181–186 [DOI] [PubMed] [Google Scholar]
- Burgess DL, Jones JM, Meisler MH, Noebels JL (1997) Mutation in the Ca2 channel β-subunit gene Cchb4 is associated with ataxia and seizures in the lethargic (lh) mouse. Cell 88:385–392 [DOI] [PubMed] [Google Scholar]
- Clemens M, Martsolf JT, Rogers JG, Mowery-Rushton P, Surti U, McPherson E (1996) Pitt-Rogers-Danks syndrome: the result of a 4p microdeletion. Am J Med Genet 66:95–100 [DOI] [PubMed] [Google Scholar]
- Endele S, Fuhry M, Pak SJ, Zabel BU, Winterpacht A (1999) LETM1, a novel gene encoding a putative EF-hand Ca2+-banding protein, flanks the Wolf-Hirschhorn syndrome (WHS) critical region and is deleted in most WHS patients. Genomics 60:218–225 [DOI] [PubMed] [Google Scholar]
- Estabrooks LL, Rao KW, Driscoll DA, Crandall BF, Dean JCS, Ikonen E, Korf B, Aylsworth S (1995) Preliminary phenotypic map of chromosome 4p16 based on 4p deletions. Am J Med Genet 57:581–586 [DOI] [PubMed] [Google Scholar]
- Fang YY, Bain S, Haan EA, Eyre HJ, MacDonald M, Wright TJ, Altherr MR, Riess O, Sutherland G, Callen DF (1997) High resolution characterization of an interstitial deletion of less than 1.9 Mb at 4p16.3 associated with Wolf-Hirschhorn syndrome. Am J Med Genet 71:453–457 [PubMed] [Google Scholar]
- Gandelman KY, Gibson L, Meyer MS, Yang-Feng TL (1992) Molecular definition of the smallest region of deletion overlap in the Wolf-Hirschhorn syndrome. Am J Hum Genet 51:571–578 [PMC free article] [PubMed] [Google Scholar]
- Johnson WP, Altherr MR, Blake JM, Keppen LD (1994) FISH detection of Wolf-Hirschhorn syndrome: exclusion of D4F26 as critical site. Am J Med Genet 52:70–74 [DOI] [PubMed] [Google Scholar]
- Lichter P, Chang CJ, Call K, Hermanson G, Evans GA, Housman D, Ward DC (1990) High resolution mapping of human chromosome 11 by in situ hybridization with cosmid clones. Science 247:64–69 [DOI] [PubMed] [Google Scholar]
- Lindeman-Kusse MC, Van Haeringen A, Hoorweg-Nijman JJG, Brunner HG (1996) Cytogenetic abnormalities in two new patients with Pitt-Rogers-Danks phenotype. Am J Med Genet 66:104–112 [DOI] [PubMed] [Google Scholar]
- Partington MW, Fagan K, Soubjaki V, Turner G (1997) Translocations involving 4p16.3 in three families: deletion causing the Pitt-Rogers-Danks syndrome and duplication resulting in a new overgrowth syndrome. J Med Genet 34:719–728 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rauch A, Schellmoser S, Kraus C, Dorr HG, Trautmann U, Altherr MR, Pfeiffer RA, Reis A (2001) First known microdeletion within the Wolf-Hirschhorn-syndrome critical region refines genotype-phenotype correlation. Am J Med Genet 99:338–342 [DOI] [PubMed] [Google Scholar]
- Sgrò V, Riva E, Canevini MP, Colamaria V, Rottoli A, Minotti L, Canger R, Dalla Bernardina B (1995) 4p− Syndrome: a chromosomal disorder associated with a particular EEG pattern. Epilepsia 36:1206–1214 [DOI] [PubMed] [Google Scholar]
- Somer M, Peippo M, Keinanen M (1995) Controversial findings in two patients with commercially available probe D4S96 for the Wolf-Hirschhorn syndrome. Am J Hum Genet 57:A127 [Google Scholar]
- Stec I, Wright TJ, van Ommen G-JB, de Boer PAJ, van Haeringen A, Moorman FM, Altherr MR, den Dunnen JT (1998) WHSC1, a 90 kb SET domain–containing gene, expressed in early development and homologous to a Drosophila dysmorphy gene maps in the Wolf-Hirschhorn syndrome critical region and is fused to IgH in t(4;14) multiple myeloma. Hum Mol Genet 7:1071–1082 [DOI] [PubMed] [Google Scholar]
- Wieczorek D, Krause M, Majewski F, Albrecht B, Horn D, Riess O, Gillessen-Kaesbach G (2000) Effect of the size of the deletion and clinical manifestation in Wolf-Hirschhorn syndrome: analysis of 13 patients with a de novo deletion. Eur J Hum Genet 8:519–526 [DOI] [PubMed] [Google Scholar]
- Wilson MG, Towner JW, Coffin GS, Ebbin AJ, Siris E, Brager P (1981) Genetic and clinical studies in 13 patients with the Wolf-Hirschhorn syndrom [del(4p)]. Hum Genet 59:297–307 [DOI] [PubMed] [Google Scholar]
- Wright TJ, Costa JL, Naranjo C, Francis-West P, Altherr MR (1999) Comparative analysis of a novel gene from the Wolf-Hirschhorn/Pitt-Rogers-Danks syndrome critical region. Genomics 59:203–212 [DOI] [PubMed] [Google Scholar]
- Wright TJ, Denison K, Johnson V, Zackai E, Altherr M (1996) High resolution analysis of the Wolf-Hirschhorn syndrome region on chromosome 4p16.3. Proceedings of the Fourth International Workshop on Human Chromosome 4 Mapping. San Francisco, November 1996 [Google Scholar]
- Wright TJ, Ricke DO, Denison K, Abmayr S, Cotter PD, Hirschhorn K, Keinanen M, McDonald-McGinn D, Somer M, Spinner N, Yang-Feng T, Zackai E, Altherr MR (1997) A transcript map of the newly defined 165 kb Wolf-Hirschhorn syndrome critical region. Hum Mol Genet 6:317–324 [DOI] [PubMed] [Google Scholar]
- Zollino M, Di Stefano C, Zampino G, Mastroiacovo P, Wright TJ, Sorge G, Selicorni A, Tenconi R, Zappalà A, Battaglia A, Di Rocco M, Palka G, Pallotta R, Altherr M, Neri G (2000) Genotype-phenotype correlations and clinical diagnostic criteria in Wolf-Hirschhorn syndrome. Am J Med Genet 94:254–261 [DOI] [PubMed] [Google Scholar]
- Zollino M, Wright TJ, Di Stefano C, Tosolini A, Battaglia A, Altherr MR, Neri G (1999) “Tandem” duplication of 4p16.1p16.3 chromosome region associated with 4p16.3pter molecular deletion resulting in Wolf-Hirschhorn syndrome phenotype. Am J Med Genet 82:371–375 [PubMed] [Google Scholar]