Abstract
Bardet-Biedl syndrome (BBS) is a genetically heterogeneous disorder, the primary features of which include obesity, retinal dystrophy, polydactyly, hypogenitalism, learning difficulties, and renal malformations. Conventional linkage and positional cloning have led to the mapping of six BBS loci in the human genome, four of which (BBS1, BBS2, BBS4, and BBS6) have been cloned. Despite these advances, the protein sequences of the known BBS genes have provided little or no insight into their function. To delineate functionally important regions in BBS2, we performed phylogenetic and genomic studies in which we used the human and zebrafish BBS2 peptide sequences to search dbEST and the translation of the draft human genome. We identified two novel genes that we initially named “BBS2L1” and “BBS2L2” and that exhibit modest similarity with two discrete, overlapping regions of BBS2. In the present study, we demonstrate that BBS2L1 mutations cause BBS, thereby defining a novel locus for this syndrome, BBS7, whereas BBS2L2 has been shown independently to be BBS1. The motif-based identification of a novel BBS locus has enabled us to define a potential functional domain that is present in three of the five known BBS proteins and, therefore, is likely to be important in the pathogenesis of this complex syndrome.
Introduction
Bardet-Biedl syndrome (BBS [MIM 209900]) is a pleiotropic disorder of likely developmental origin, in which the primary features of retinal dystrophy, obesity, polydactyly, gonadal, and renal malformations, as well as developmental delay, manifest with substantial inter- and intrafamilial variability (Beales et al. 1999). Despite an initial expectation that BBS might represent a single-locus disorder, linkage studies established substantial genetic heterogeneity in BBS. To date, six loci have been mapped in the human genome with evidence for at least one more unmapped locus (Kwitek-Black et al. 1993; Leppert et al. 1994; Sheffield et al. 1994; Carmi et al. 1995; Young et al. 1999; Katsanis et al. 2000; Beales et al. 2001). Furthermore, the identification of the first three BBS genes revealed increased complexity in the mode of trait transmission, given that, in some families, three mutations at two loci were shown to be required for pathogenesis (Katsanis et al. 2001a, 2002; Badano and Katsanis 2002).
The identification of the first three BBS genes (BBS2, BBS4, and BBS6) provided limited information about their cellular role (Katsanis et al. 2000; Slavotinek et al. 2000; Mykytyn et al. 2001; Nishimura et al. 2001). BBS6 shows significant similarity to group II chaperonins (Stone et al. 2000), whereas the BBS4 sequence contains fewer functional clues, as it exhibits modest similarity to TPR-containing proteins such as O-linked N-acetylglucosamine (O-GlcNAc) transferases from different species (Nishimura et al. 2001; Wells et al. 2001; Katsanis et al. 2001b). By contrast, BBS2 encodes a 721–amino acid protein of unknown function and, with the exception of a putative coiled-coil domain between residues 332–365, computational analyses have not revealed any other known motifs. Furthermore, alignment of the human BBS2 peptide sequence to its orthologs from various species has provided limited information about functionally important residues because of the substantial conservation across the entire BBS2 protein family.
We hypothesized that identifying more distantly related members of the BBS2 family might facilitate the recognition of critical domains, as these likely would be subjected to elevated evolutionary pressure. We report here the identification of two novel genes with moderate similarity to BBS2, which we initially termed “BBS2L1” and “BBS2L2” (BBS2-like 1 and 2). We also demonstrate that mutations in BBS2L1 cause BBS, thereby defining a novel locus for this syndrome, BBS7. During the course of this work, the second BBS2 paralog, BBS2L2, was shown independently to be BBS1, the major BBS locus, and to share modest similarity over 192 amino acids to BBS2 (Mykytyn et al. 2002). These data point to the first direct link between three of the five known BBS genes, by virtue of the presence of a novel protein motif whose disruption may commonly result in the same clinical phenotype.
Patients and Methods
Families with Bardet-Biedl Syndrome
Individuals with Bardet-Biedl syndrome were ascertained on the basis of criteria established elsewhere (Beales et al. 1999). For all kindreds, interviews were conducted, and a genealogy was constructed. With informed consent, one of us (R.A.L.) obtained and reviewed all available medical records to verify phenotypic features and secure the diagnosis.
Genetic Analyses
DNA was extracted from venous lymphocytes; families were genotyped with published markers from the respective critical intervals, and haplotypes were constructed. A genome screen with the 10-cM Linkage Map Set (Perkin Elmer) was applied to suitable pedigrees; alleles were analyzed and LOD scores were tabulated as described elsewhere (Katsanis et al. 2000). Marker order, heterozygosity values, and relative distances were obtained from the Genome Database and by BLAT analysis of the June 2001 version of the draft human genome sequence (build 30).
Genomic Structure of BBS7 and Mutation Screening
We aligned the cDNA sequence of both BBS7 isoforms to genomic sequence by BLAT and established the presence of 19 exons. We extracted 100–300 bp of sequence flanking each coding exon and designed amplicons that span both exons and intronic splice junctions as described by Katsanis et al. (2000). Amplified PCR products from patients, relatives, and unrelated but ethnically matched control individuals were purified with the Exo-SAP cleanup kit (USB) and sequenced with dye-primer chemistry, using an ABI 377 automated sequencer (Applied Biosystems). We aligned the resulting sequences and evaluated mutations with the Sequencher sequence alignment program (Gene codes).
Expression Studies
To determine the size and tissue distribution of BBS7, we amplified a 217-bp fragment from the 3′ UTR and probed a northern blot that contained polyA+ mRNA from 12 adult human tissues. To validate the presence of two alternative 3′ end isoforms and to investigate the tissue distribution of each mRNA species, we amplified a 298-bp 3′ UTR fragment of the short BBS7 isoform and a 314-bp 3′ UTR fragment of the long BBS7 isoform from 17 adult and 8 fetal tissues.
Results
Identification of Two Distant Paralogs of BBS2
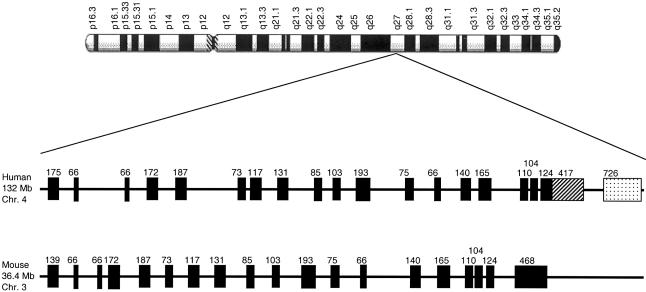
To identify proteins with limited similarity to BBS2, we used the human and zebrafish BBS2 peptide sequence to search the conceptual translation of the human subset of the EST database (dbEST). We identified numerous ESTs with 25%–40% similarity to BBS2, which we assembled into five discrete contigs, B2lc1–5. Alignment of the consensus nucleotide sequence of each contig indicated that B2lc1 and B2lc3 were 99.8% identical across an overlapping region of 124 bp and were, therefore, integrated into a single 1,048-bp sequence. However, the absence of a bona fide start methionine or a stop codon implied that our sequence represented a partial cDNA. To obtain the full-length transcript, we performed additional BLAST searches of dbEST and the human draft genomic sequence coupled with exon prediction analyses. Combining additional EST sequence with genomic information, we assembled a single contig of 2,580 nucleotides (GenBank accession number AF521643). Given the strong Kozak consensus around the first methionine, with a stop codon 21 bp upstream and an AATAAA polyadenylation signal 12 bp before the 3′ end of our sequence, we concluded that we had likely determined the full-length sequence of this novel gene. We named this transcript “BBS2L1,” as it exhibits 42.5% similarity between residues 147 and 398 of BBS2; it contains a single 672–amino acid ORF from bp 149–2164, encoded by 19 exons, and maps to chromosome 4q27 (fig. 1).
Figure 1.
Mapping and genomic structure of BBS2L1 and its murine ortholog. The position and size of each exon of the human and mouse transcript are shown (black boxes). In the human transcript, two different splice variants of exon 18 are shown. In the short isoform, the 124-bp–long exon 18 is contiguous with an additional 417-bp 3′ UTR (box with diagonal dashes), whereas, in the long isoform, this exon is spliced with a new 726-bp exon that encodes an additional 44 residues and a different 3′ UTR (dotted box). We subsequently termed this locus “BBS7” because of the presence of pathogenic mutations that segregated with BBS.
Contigs Bl2c2, 4, and 5 did not align directly to each other. However, BLAT analysis of human draft genomic sequence (Kent 2002) indicated that all three contigs map within 20 kb of each other and likely represent different regions of the same transcript, which we termed “BBS2L2.” During the progress of these studies, a sequence identical to BBS2L2 was reported to harbor pathogenic mutations in patients with BBS and shown to correspond to the major BBS locus, BBS1 (Mykytyn et al. 2002).
Identification of the Murine Ortholog of BBS2L1
To extend our evolutionary studies of the BBS2 protein family, we deduced the sequence of the mouse Bbs2l1 ortholog by using the human BBS2L1 nucleotide sequence to query the mouse subdivision of dbEST, as well as the mouse draft genomic sequence. We identified 14 mouse ESTs that exhibited 82%–93% identity with BBS2L1 at the nucleotide level. Assembly of all murine cDNA sequences and alignment to BBS2L1 revealed three independent contigs that corresponded to different, nonoverlapping regions of the human gene: a 706-bp 5′ contig, a middle 996-bp contig, and a 597-bp 3′ contig. Further EST assembly bridged the middle and 3′ contigs, leaving an ∼900-bp gap. To obtain the full-length mouse cDNA, we amplified this gap by RT-PCR and sequenced the resulting products. The final assembled 2,594-bp murine sequence encodes a 716–amino acid peptide that exhibits 94.1% similarity and 91.5% identity to the human BBS2L1 protein. This suggested that we had identified the ortholog of BBS2L1, which we termed “Bbs2l1” (GenBank accession number AF521645).
BBS2L1 Utilizes Differentially Expressed Alternative 3′ Termini
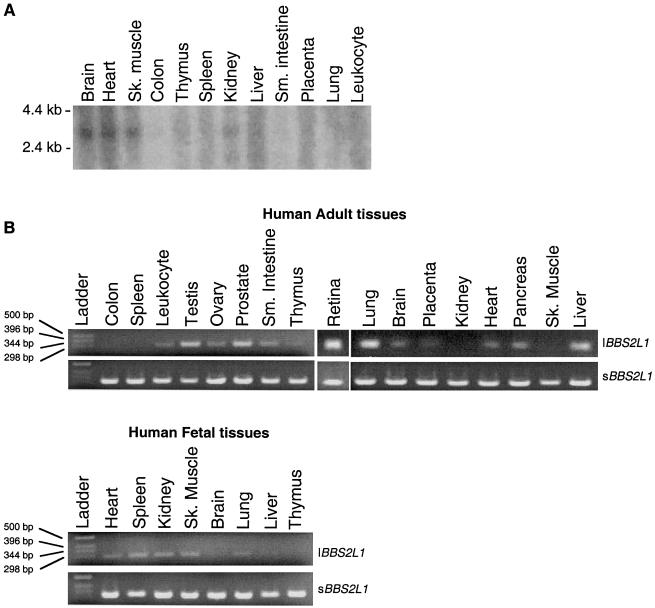
To confirm that we had identified the full-length sequence of BBS2L1 and to investigate its pattern of expression, we performed northern blotting and RT-PCR on adult and fetal human tissues. We detected a BBS2L1 message of the expected size in a wide range of tissues (fig. 2a). However, when we compared the sequence of BBS2L1 with its murine ortholog, we observed that the mouse transcript had a different 3′ end, and we considered the possibility of alternative splicing. Using the unique mouse 3′ end to search dbEST and genomic sequence, we determined a putative alternative 3′ end for BBS2L1, which yields an mRNA that encodes an additional 44 residues and terminates in a discrete 3′ UTR. To ensure that this sequence was not a computational artifact, we performed RT-PCR and determined that this additional BBS2L1 sequence was present in multiple tissues but with a pattern distinct from the previously recognized expression pattern of BBS2L1 (fig. 2b).
Figure 2 .
Expression profile of BBS7. A, Northern blotting of a human 3′ UTR probe showing an ∼2.7-kb species expressed in low-to-moderate levels in multiple human tissues. B, RT-PCR analysis of the two alternatively spliced isoforms in 17 adult and 8 fetal tissues. The short isoform (sBBS2L1) is ubiquitously expressed, whereas the long isoform (lBBS2L1) is expressed in numerous, but not all, tissues.
BBS2L1 Defines a Novel BBS Locus, BBS7
BBS2L1 maps to 4q27, a region of the genome not associated previously with any known BBS locus. However, given the similarity between BBS2L1 and BBS2, we hypothesized that BBS2L1 mutations may also cause BBS, a concept reinforced by the identification of pathogenic alterations in BBS1 (Mykytyn et al. 2002), the only other known BBS2 paralog. To test this possibility, we screened all exons and splice junctions of both splice variants of BBS2L1 in patients from 84 independent families of primarily European ancestry who had BBS. Since complex inheritance has been demonstrated in BBS, whereby mutations at more than one locus can be necessary for pathogenesis (Katsanis et al. 2001a), we did not preselect the family cohort on the basis of linkage or mutational data from the other established loci.
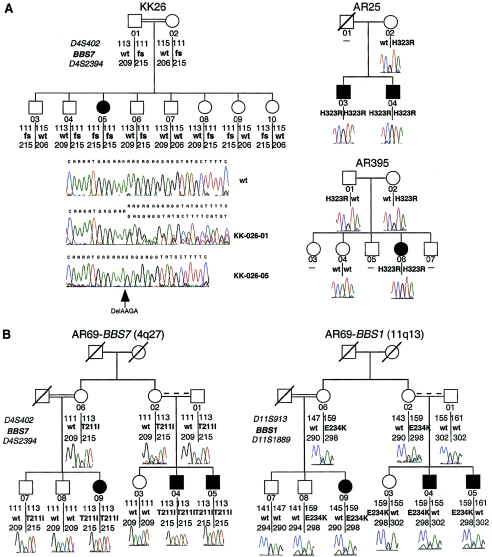
We identified potentially pathogenic mutations in three pedigrees. In pedigrees AR25 and AR395, we found a homozygous H323R alteration in exon 10 that segregated with the disorder (fig. 3A). In a third pedigree, AR69, we detected a T211I homozygous alteration that also segregated with the disorder in both arms of the family (fig. 3B), including individual –09, who is the offspring of a consanguineous mating. Given these data, we hypothesized that dysfunction of BBS2L1 may cause BBS, especially since, in addition to consistent segregation of each mutant allele with the disorder under a Mendelian recessive model, neither change was found in 192 ethnically matched control chromosomes.
Figure 3.
Mutations in BBS7 associated with BBS. A, Recessive inheritance in BBS7. The sole affected individual, –05, in consanguineous family KK26 harbors a homozygous 4-bp deletion in exon 7 that is predicted to lead to premature termination in exon 9 (K237fsX296), thereby abolishing ∼64% of the wild-type protein. Likewise, in two unrelated pedigrees (AR25 and AR395), all patients are homozygous for an H323R allele. Individuals unavailable for collection are shown with dashes. B, Complex inheritance in BBS7. Consanguineous pedigree AR69 harbors a homozygous T211I missense mutation in BBS7 and a heterozygous E234K mutation in BBS1. All patients carry all three mutant alleles, and genetic analyses across BBS1 indicate that this family is unlikely to carry other mutations at that locus, since affected individuals –04 and –05 have inherited different paternal chromosomes. Note that asymptomatic sibling –03 has identical BBS1 haplotypes to patient –04, including the heterozygous E234K mutation.
The H323R change is predicted to affect local charge, and the T211I alteration is likely to affect either hydrophobicity or the ability to form hydrogen bonds at this residue. However, the paucity of functional data for the protein left the formal possibility that these alterations represent rare benign polymorphisms. Therefore, we examined genomewide genotypes generated at an ∼10-cM resolution from nine consanguineous pedigrees of Saudi Arabian origin, each of whom had been excluded from harboring recessive mutations in all known loci by haplotype and sequence analysis. In pedigree KK26, we identified a >5-cM region of homozygosity, on 4q26–q27, that encompassed the BBS2L1 genomic locus. Performing additional linkage studies, we established that only the affected individual –05 was homozygous across a region extending at least 2.6 cM proximally and >3 cM distal to BBS2L1, and we derived a multipoint LOD score of 1.8 at θ=0.001 for D4S408, which lies 2.6 cM proximal to BBS2L1. Although not statistically significant by itself, this LOD score peaked at the simulated theoretical maximum for this family, and, given our previous mutational data, we screened KK26 for BBS2L1. We identified a homozygous 4-bp deletion that abolishes the lysine at position 237 in exon 7 and which, by conceptual translation, results in premature termination in exon 9, at residue 296 (K237fsX296). This alteration segregated with BBS in this family, as only the affected individual, –05, was homozygous for the deletion (fig. 3A). The fact that this alteration eliminates nearly 65% of the predicted protein, its absence from 288 control chromosomes, including 96 chromosomes of normal, unrelated Saudi individuals, and our previous mutational data, we concluded that BBS2L1 represents a novel BBS locus, which we term “BBS7.”
Potential Complex Inheritance in BBS7
Recent data support the existence of non-Mendelian inheritance in some families with BBS, whereby three or more mutant alleles at two loci either may be required for pathogenesis (triallelic inheritance) or may modify the phenotype (Katsanis et al. 2001a, 2002; Badano and Katsanis 2002). The mutations we found in all four BBS7 pedigrees satisfy the segregation criteria for a recessive disorder. Nonetheless, we conducted genetic and mutational analyses across the other four known BBS genes to investigate the possibility that some families with BBS7 mutations might also harbor pathogenic variants at other loci. We found no additional pathogenic alterations in families AR25, AR395, and KK26. In family AR69, however, we detected an E234K alteration in exon 8 of BBS1 that was present in all affected individuals (fig. 3B). Although the functional significance of this substitution cannot be assayed at present, the severe structural consequences of this alteration and its absence from 192 ethnically matched control chromosomes imply that AR69 may harbor three pathogenic alleles. However, since no asymptomatic individuals have two mutant BBS7 alleles, it is not possible to establish whether three mutations are necessary in this family to cause the disease, whether a third mutation in BBS1 modifies the phenotype, or whether the third mutation is only coincidental. Nonetheless, our data indicate that, like BBS2, BBS4, and BBS6, BBS7 may also interact genetically with other loci to produce the BBS phenotype. A recent evaluation of a small BBS1 patient cohort suggested that mutations at this locus are inherited in an exclusively recessive manner (Mykytyn et al. 2002). However, our data indicate that BBS1 mutations might have a synergistic effect with BBS7, although evaluation of larger patient cohorts will be required to test this hypothesis.
Domain Sharing between BBS1, BBS2, and BBS7
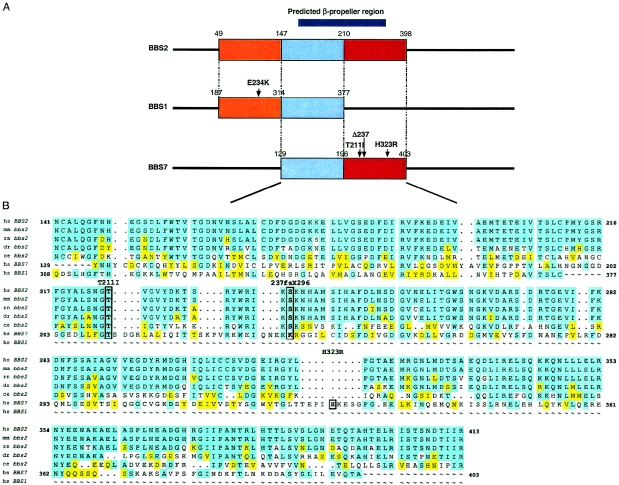
BBS7 exhibits similarity with a 252–amino acid region of BBS2, between residues 147 and 398 (fig. 4). SMART analyses (Schultz et al. 2000) of this region in BBS2 detected a hypothetical structural classification of proteins (SCOP) (Murzin et al. 1995; Lo Conte et al. 2002) domain that lies in the conserved area between residues 171–315 and is predicted to encode a six-bladed β-propeller structure. Local alignment of BBS1, BBS2, and BBS7 indicate that both BBS1 and BBS7 contain partially overlapping portions of this SCOP domain (fig. 4a), raising the possibility that dysfunction of this module could yield a common mechanism to the phenotype caused by mutations at each locus.
Figure 4.
A potential domain shared between multiple BBS proteins. A, Schematic representation of the region of overlap between the three proteins. The region of homology between BBS1 and BBS2 is shown in orange; the homology between BBS2 and BBS7 only is shown in red, whereas the area of overlap among all three proteins is depicted in light blue. Numbers correspond to amino acid residues; the predicted β-propeller, as well as the mutations in BBS1 and BBS7, are also shown (scale is approximate). Note that the potential BBS1 triallelic mutation E234K maps in a region shared between BBS1 and BBS2. B, Local alignment of BBS2 orthologs with homologous regions in BBS7 and BBS1. The positions of the BBS7 mutations are also shown; the threonine 211 residue is completely conserved and forms part of this potential domain, whereas the 4-bp deletion at the lysine 237 residue of BBS7 abolishes most of the region shared uniquely with BBS2. hs = Homo sapiens; mm = Mus musculus; rn = Rattus norvegicus; dr = Danio rerio; and ce = Caenorhabditis elegans.
Discussion
We report here the identification of the gene for BBS7, which defines the seventh locus (the fifth one cloned) for this disorder. Analysis of the BBS7 peptide sequence does not offer any clues as to its cellular function, since it neither resembles known molecules nor contains domains of known function. However, comparison of BBS7 with the other BBS proteins suggests a potential structural link between BBS1, BBS2, and BBS7: a distinct substructure shared between the three molecules. Further analyses of the human genomic sequence did not indicate the presence of other proteins with this motif. Thus, BBS1, BBS2, and BBS7 may belong to a distinct subfamily of proteins, mutations in any of which lead to the same clinical entity. We detected by BLAST analysis some similarity between this region and two members of the integrin family (integrin 9 and 11) and the Escherichia coli transcriptional regulator zraR (data not shown). The biological significance of this observation remains to be determined. Nevertheless, the identification of this module is likely to be valuable in determining the roles of these BBS proteins and their involvement in the pathogenesis of this phenotype.
With the exception of BBS1 and BBS2, which account for 40% and 16% of BBS mutations, respectively (Bruford et al. 1997; Katsanis et al. 1999; Beales et al. 2001), the remaining BBS loci have a small contribution to BBS (Beales et al. 2001; Katsanis et al. 2002). BBS7 is no exception, since only 4 of 84 independent families carry pathogenic alterations at this locus (3.5%), a frequency similar to that of BBS4 (Katsanis et al. 2002). Our data also indicate that ∼40% of our patients do not carry mutations in any of the five known BBS genes. Although the formal possibility exists that noncoding mutations or genomic rearrangements may account for the remainder of mutations, high-density haplotype analysis across all seven loci has excluded ∼19% of our pedigrees from carrying recessive mutations at any of these loci. We therefore suggest that at least one more BBS locus remains to be identified in the human genome.
Given the substantial genetic heterogeneity in BBS, it is likely that identifying minor loci by traditional linkage approaches will become increasingly difficult. In those instances, domain-based computational approaches, such as the one presented here or the evaluation of functional candidate genes (such as BBS-interacting proteins, for example), may prove to be a successful means of identifying additional transcripts involved with this disorder.
Genetic heterogeneity and clinical pleiotropy are also potentially significant factors in modeling the transmission of the disease trait in BBS. From our data and previous studies, it appears that non-Mendelian inheritance may sometimes exert an effect on all BBS genes cloned to date, albeit at varying frequency. Of the four families with BBS7 mutations, one family (AR69) segregates a third, potentially pathogenic, BBS1 allele. However, since none of the unaffected sibs are homozygous mutant for BBS7, it is equally likely that the third mutant allele is either required for pathogenesis or that it modifies the phenotype. This genetic interaction between all BBS loci potentially underlies a functional interaction, as dictated by the principles of nonallelic noncomplementation (Badano and Katsanis 2002). The premise that three of the BBS genes (BBS1, BBS2, and BBS7) encode the same motif may even suggest that these proteins can substitute for each other with varying degrees of efficiency. Functional studies are required to investigate this possibility, ascertain the effects of the alterations found in patients, and establish why, in some instances, three mutant alleles are required for pathogenesis.
Acknowledgments
We thank all participating families, for their continued and willing participation, and Dr. John Cavender, for providing some clinical observations of family KK26. We also thank Dr. Philip Beales for his critique of this manuscript. This work was supported by the March of Dimes (to J.R.L. and N.K.) and the Foundation Fighting Blindness (to J.R.L. and R.A.L.). R.A.L. is a Senior Scientific Investigator of Research to Prevent Blindness, New York.
Electronic-Database Information
Accession numbers and URLs for data presented herein are as follows:
- BLAT search engine, http://genome.ucsc.edu/cgi-bin/hgBlat?command=start (for aligning cDNA and genomic sequences)
- GenBank, http://www.ncbi.nlm.nih.gov/ (for accession numbers AF521643 [human BBS7 short splice isoform], AF521644 [human BBS7 long splice isoform], and AF521645 [mouse Bbs7]) [Google Scholar]
- Genome Database, http://www.gdb.org/ (for microsatellite amplification primers)
- Online Mendelian Inheritance in Man (OMIM), http://www.ncbi.nlm.nih.gov/Omim/ (for BBS [MIM 209900]) [PubMed]
- SMART Protein Motif database, http://smart.embl-heidelberg.de/ (for protein motif analysis)
References
- Badano JL, Katsanis N (2002) Beyond Mendel: an evolving view of human genetic disease transmission. Nat Rev Genet 3:779–789 [DOI] [PubMed] [Google Scholar]
- Beales PL, Elcioglu N, Woolf AS, Parker D, Flinter FA (1999) New criteria for improved diagnosis of Bardet-Biedl syndrome: results of a population survey. J Med Genet 36:437–446 [PMC free article] [PubMed] [Google Scholar]
- Beales PL, Katsanis N, Lewis RA, Ansley SJ, Elcioglu N, Raza J, Woods MO, Green JS, Parfrey PS, Davidson WS, Lupski JR (2001) Genetic and mutational analyses of a large multiethnic Bardet-Biedl cohort reveal a minor involvement of BBS6 and delineate the critical intervals of other loci. Am J Hum Genet 68:606–616 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bruford EA, Riise R, Teague PW, Porter K, Thomson KL, Moore AT, Jay M, Warburg M, Schinzel A, Tommerup N, Tornqvist K, Rosenberg T, Patton M, Mansfield DC, Wright AF (1997) Linkage mapping in 29 Bardet-Biedl syndrome families confirms loci in chromosomal regions 11q13, 15q22.3-q23, and 16q21. Genomics 41:93–99 [DOI] [PubMed] [Google Scholar]
- Carmi R, Rokhlina T, Kwitek-Black AE, Elbedour K, Nishimura D, Stone EM, Sheffield VC (1995) Use of a DNA pooling strategy to identify a human obesity syndrome locus on chromosome 15. Hum Mol Genet 4:9–13 [DOI] [PubMed] [Google Scholar]
- Katsanis N, Ansley SJ, Badano JL, Eichers EE, Lewis RA, Hoskins BE, Scambler PJ, Davidson WS, Beales PL, Lupski JR (2001a) Triallelic inheritance in Bardet-Biedl syndrome, a Mendelian recessive disorder. Science 293:2256–2259 [DOI] [PubMed] [Google Scholar]
- Katsanis N, Beales PL, Woods MO, Lewis RA, Green JS, Parfrey PS, Ansley SJ, Davidson WS, Lupski JR (2000) Mutations in MKKS cause obesity, retinal dystrophy and renal malformations associated with Bardet-Biedl syndrome. Nat Genet 26:67–70 [DOI] [PubMed] [Google Scholar]
- Katsanis N, Eichers ER, Ansley SJ, Lewis RA, Kayserili H, Hoskins BE, Scambler PJ, Beales PL, Lupski JR (2002) BBS4 is a minor contributor to Bardet-Biedl syndrome and may also participate in triallelic inheritance. Am J Hum Genet 71:22–29 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katsanis N, Lewis RA, Stockton DW, Mai PMT, Baird L, Beales PL, Leppert M, Lupski JR (1999) Delineation of the critical interval of Bardet-Biedl syndrome 1 (BBS1) to a small region of 11q13 through linkage and haplotype analysis of 91 pedigrees. Am J Hum Genet 65:1672–1679 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katsanis N, Lupski JR, Beales PL (2001b) Exploring the molecular basis of Bardet-Biedl syndrome. Hum Mol Genet 10:2293–2299 [DOI] [PubMed] [Google Scholar]
- Kent WJ (2002) BLAT—the BLAST-like alignment tool. Genome Res 12:656–664 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwitek-Black AE, Carmi R, Duyk GM, Buetow KH, Eldebour K, Parvati R, Yandava CN, Stone EM, Sheffield VC (1993) Linkage of Bardet-Biedl syndrome to chromosome 16q and evidence for non-allelic genetic heterogeneity. Nat Genet 5:392–396 [DOI] [PubMed] [Google Scholar]
- Leppert M, Baird L, Anderson KL, Otterud B, Lupski JR, Lewis RA (1994) Bardet-Biedl syndrome is linked to DNA markers on chromosome 11q and is genetically heterogeneous. Nat Genet 7:108–112 [DOI] [PubMed] [Google Scholar]
- Lo Conte L, Brenner SE, Hubbard TJ, Chothia C, Murzin AG (2002) SCOP database in 2002: refinements accommodate structural genomics. Nucleic Acids Res 30:264–267 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murzin AG, Brenner SE, Hubbard T, Chothia C (1995) SCOP: a structural classification of proteins database for the investigation of sequences and structures. J Mol Biol 247:536–540 [DOI] [PubMed] [Google Scholar]
- Mykytyn K, Braun T, Carmi R, Haider NB, Searby CC, Shastri M, Beck G, Wright AF, Iannaccone A, Elbedour K, Riise R, Baldi A, Raas-Rothchild A, Gorman SW, Duhl DM, Jacobson SG, Casavant T, Stone EM, Sheffield VC (2001) Identification of the gene that, when mutated, causes the human obesity syndrome BBS4. Nat Genet 28:188–191 [DOI] [PubMed] [Google Scholar]
- Mykytyn K, Nishimura DY, Searby CC, Shastri M, Yen HJ, Beck JS, Braun T, Streb LM, Cornier AS, Cox GF, Fulton AB, Carmi R, Luleci G, Chandrasekharappa SC, Collins FS, Jacobson SG, Heckenlively JR, Weleber RG, Stone EM, Sheffield VC (2002) Identification of the gene (BBS1) most commonly involved in Bardet-Biedl syndrome, a complex human obesity syndrome. Nat Genet 31:435–438 [DOI] [PubMed] [Google Scholar]
- Nishimura DY, Searby CC, Carmi R, Elbedour K, Maldergem LV, Fulton AB, Lam BL, Powell BR, Swiderski RE, Bugge KE, Haider NB, Kwitek-Black AE, Ying L, Duhl DM, Gorman SW, Heon E, Iannaccone A, Bonneau D, Biesecker LG, Jacobson SG, Stone EM, Sheffield VC (2001) Positional cloning of a novel gene on chromosome 16q causing Bardet-Biedl syndrome (BBS2). Hum Mol Genet 10:865–874 [DOI] [PubMed] [Google Scholar]
- Schultz J, Copley RR, Doerks T, Ponting CP, Bork P (2000) SMART: a web-based tool for the study of genetically mobile domains. Nucleic Acids Res 28:231–234 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheffield VC, Carmi R, Kwitek-Black A, Rokhlina T, Nishimura D, Duyk GM, Elbedour K, Sunden SL, Stone EM (1994) Identification of a Bardet-Biedl syndrome locus on chromosome 3 and evaluation of an efficient approach to homozygosity mapping. Hum Mol Genet 3:1331–1335 [DOI] [PubMed] [Google Scholar]
- Slavotinek AM, Stone EM, Mykytyn K, Heckenlively JR, Green JS, Heon E, Musarella MA, Parfrey PS, Sheffield VC, Biesecker LG (2000) Mutations in MKKS cause Bardet-Biedl syndrome. Nat Genet 26:15–16 [DOI] [PubMed] [Google Scholar]
- Stone DL, Slavotinek A, Bouffard GG, Banerjee-Basu S, Baxevanis AD, Barr M, Biesecker LG (2000) Mutation in a gene encoding a putative chaperonin causes McKusick-Kaufman syndrome. Nat Genet 25:79–82 [DOI] [PubMed] [Google Scholar]
- Wells L, Vosseller K, Hart GW (2001) Glycosylation of nucleocytoplasmic proteins: signal transduction and O-GlcNAc. Science 291:2376–2378 [DOI] [PubMed] [Google Scholar]
- Young T-L, Penney L, Woods MO, Parfrey PS, Green JS, Hefferton D, Davidson WS (1999) A fifth locus for Bardet-Biedl syndrome maps to 2q31. Am J Hum Genet 64:900–904 [DOI] [PMC free article] [PubMed] [Google Scholar]