Abstract
The distal arthrogryposes (DAs) are a group of disorders characterized by multiple congenital contractures of the limbs. We previously mapped a locus for DA type 2B (DA2B), the most common of the DAs, to chromosome 11. We now report that DA2B is caused by mutations in TNNI2 that are predicted to disrupt the carboxy-terminal domain of an isoform of troponin I (TnI) specific to the troponin-tropomyosin (Tc-Tm) complex of fast-twitch myofibers. Because the DAs are genetically heterogeneous, we sought additional candidate genes by examining modifiers of mutant Drosophila isoforms of TnI. One of these modifiers, Tm2, encodes tropomyosin, another component of the Tc-Tm complex. A human homologue of Tm2, TPM2, enocodes β-tropomyosin and maps to the critical interval of DA type 1 (DA1). We discovered that DA1 is caused by substitution of a highly conserved amino acid residue in β-tropomyosin. These findings suggest that DAs, in general, may be caused by mutations in genes encoding proteins of the contractile apparatus specific to fast-twitch myofibers. This provides a new opportunity to directly study the etiology and pathogenesis of multiple-congenital-contracture syndromes.
Introduction
A child with an isolated congenital contracture (e.g., clubfoot) is born once in every 200–500 live births, and ∼1 of every 3,000 children is born with two or more body areas affected by congenital contractures (i.e., arthrogryposis). Nevertheless, the etiology and pathogenesis of congenital contractures in neurologically normal children remains unclear in the majority of cases, most of which are sporadic. However, it is clear that the etiology of clubfoot has a genetic component (Andrade et al. 1998), and autosomal dominant segregation of contractures has been well documented (Hall et al. 1982; Bamshad et al. 1996a; Krakowiak et al. 1998). In the mid-1990s, to facilitate the identification of genes causing contractures, we revised the definition and classification of a group of heritable disorders called the “distal arthrogryposes” (DAs) (Bamshad et al. 1996b). In general, the DAs are characterized by nonprogressive, congenital contractures of two or more different body areas without primary neurological and/or muscle disease that affects limb function. Features common to all DAs include a consistent pattern of distal joint involvement, limited proximal joint involvement, an autosomal dominant inheritance pattern, and widely variable expressivity. Ten different DAs were recognized and classified hierarchically according to the proportion of features they share with one another (Bamshad et al. 1996b).
The prototypic DA is DA type 1 (DA1 [MIM 108120]), which is primarily characterized by camptodactyly and clubfoot, although the shoulders and hips may also be affected. The phenotype of DA1 is similar to that of another DA called “Freeman-Sheldon syndrome” (FSS [MIM 193700]). In contrast to DA1, the phenotype of FSS includes scoliosis, a very small oral orifice (often only a few millimeters in diameter at birth), H-shaped dimpling of the chin, deep nasolabial folds, and blepharophimosis (narrow palpebral fissures) (Freeman and Sheldon 1938). In some individuals, however, distinguishing between the diagnosis of DA1 and FSS can be difficult. Moreover, in some extended families, different individuals have been assigned a diagnosis of either DA1 or FSS (Klemp and Hall 1995).
In 1997, we described individuals with characteristics of both DA1 and FSS (Krakowiak et al. 1997). Their features included a triangular face, downward-slanting palpebral fissures, prominent nasolabial folds, a small mouth, small mandible, cervical webbing, severe camptodactyly, ulnar deviation, and clubfoot or calcaneovalgus deformities (fig. 1). We classified this disorder as variant FSS, or DA2B (MIM 601680), whereas individuals meeting the full diagnostic criteria for FSS (Bamshad et al. 1996b) were referred to as having classical FSS, or DA2A. Subsequently, we mapped a locus for DA2B to chromosome 11p15.5 within a few hundred kilobases of the telomere (Krakowiak et al. 1997).
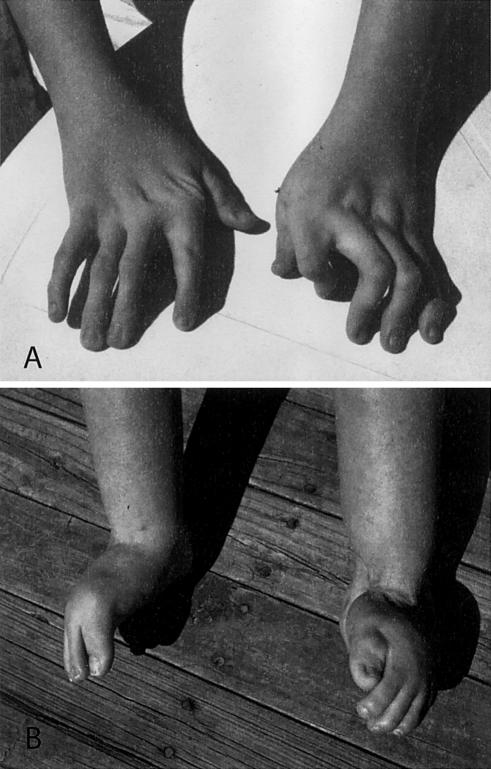
Figure 1.
Typical malformations observed in individuals with DA2B. A, Hands characterized by camptodactyly and ulnar deviation. B, Feet showing camptodactyly accompanied by calcaneovalgus deformities and a vertical talus or clubfoot (not shown).
We now report that DA2B is caused by mutations in TNNI2, encoding an isoform of troponin I (TnI) specific to fast-twitch myofibers. Homologues of TNNI2 have been studied in a wide variety of model organisms, including Drosophila, chick, rabbit, and mouse models (Mullen and Barton 2000). In Drosophila, TnI mutants with abnormalities of skeletal muscle architecture causing wing and limb defects have been available for many decades (Vigoreaux 2001). These mutants have hypercontracted intercostal flight muscles (IFM), a condition that causes protracted elevation of the wings, reduced frequency of wing beats, and an awkward, irregular gait (Beall and Fryberg 1991; Barbas et al. 1993). As a consequence, the TnI locus was named “wings up A” (wup A) (Hotta and Benzer 1972), although the name was subsequently changed to “heldup” (Deak 1977; Deak et al. 1982).
Because of the indispensability of the IFM for flight, the flightlessness phenotype has been a useful screening device for identification of genes involved in myogenesis. Many of the genes identified encode components of the troponin-tropomyosin-actin complex. This led, in part, to the search for genetic interactions among alleles of these loci, and a handful of dominant suppressors of heldup mutants have been identified (Vigoreaux 2001). Recently, a serine-to-phenylalanine substitution at amino acid residue 185 of tropomyosin, encoded by a mutant allele (D53) of tropomyosin-2 (Tm2), was found to be a suppressor of heldup2 (Naimi et al. 2001). Genetic suppressors are often associated with their own mutant phenotype, and D53 homozygotes are no exception, exhibiting a significant reduction in flight ability in comparison with the wild type. The human homologue of Tm2 (TPM2) maps to chromosome 9p11.3 in the critical interval for DA1 (Bamshad et al. 1994). Moreover, the protein encoded by TPM2 (β-tropomyosin) interacts directly with TnI. Consequently, we considered TPM2 a candidate gene for other DAs. Screening of TPM2 led to the identification of a mutation causing DA1. Thus, mutant alleles encoding TnI and tropomyosin in Drosophila are both associated with reduced movement and morphological defects, and they encode proteins that interact with one another. Mutations in both of the human homologues cause multiple-congenital-contracture syndromes.
Material and Methods
Clinical Status
All studies were performed with the approval of the institutional review board of the University of Utah and the General Counsel of the Shriners Hospitals for Children. After informed consent was obtained, living members of each kindred were evaluated by review of their medical history, completion of a questionnaire, and physical examination. An individual was considered to have a diagnosis of DA1 if two or more of the major clinical manifestations were present (Bamshad et al. 1996b). For DA2B, at least two major features plus two minor features were required for diagnosis (Krakowiak et al. 1997). Major diagnostic criteria of the upper limbs included ulnar deviation, camptodactyly, hypoplastic and/or absent flexion creases, and overriding fingers at birth. Major diagnostic criteria of the lower limbs included talipes equinovarus, calcaneovalgus deformities, a vertical talus, and metatarsus varus. Minor diagnostic criteria include a triangular face, downward-slanting palpebral fissures, attached ear lobules, a small mouth, small mandible, arched palate, cervical webbing, and short stature.
For DA1, a total of 10 patients with familial disease and 4 patients with sporadic disease were tested; for DA2B, 16 patients with familial disease and 18 patients with sporadic disease were examined. Fourteen patients with classical FSS (5 familial and 9 sporadic) were also screened for mutations in TNNI2 and TPM2.
Mutation Analysis of TNNI2 and TPM2
Genomic DNA was prepared from peripheral lymphocytes and/or Epstein-Barr virus–transformed lymphoblastoid cell lines, using standard techniques. Genomic DNA sequences were amplified according to the Qiagen HotStarTaq protocol in 1× buffer (10 mM Tris [pH 8.3], 40 mM KCl, 1.5 mM MgCl2, and 1× Q solution). The reaction was performed with 25 ng of template genomic DNA, 200 μM of dNTPs, 10 pmol of each primer, and 0.625 U of HotStarTaq DNA polymerase in a total reaction volume of 25 μl. Samples were cycled 30 times in an MJ Research DNA Engine Tetrad, using a standard three-step PCR profile with an initial denaturing step at 94°C for 15 min and a final extension step at 72°C for 10 min. Annealing temperatures and primer sequences can be found in A and B (online only). PCR products of TNNI2 were purified on a 2% agarose gel, and PCR products of TPM2 were purified using Qiaquick columns (Qiagen). Purified PCR products were sequenced using ABI BigDye Terminator, version 2.0, reagent. Sequenced products were loaded on an ABI 377 automated sequencer and analyzed by Sequencing Analysis 3.4.1 and Sequencher 4.1 software (Genecodes). The forward and reverse strand of exons 1–8 of TNNI2 and exons 1–9 of TPM2, including the flanking splice-recognition sequences, were analyzed.
Table A.
Primers Used to Amplify TNNI2 Exons
| Exon andTroponin Primer | Primer Sequences | PCR ProductSize(bp) | AnnealingTemperature(°C) |
| 1: | |||
| 1A | 5′-AGCCCAACTCGCCGCTGCCG-3′ | 248 | 60 |
| 1B | 5′-TAATACCTGCCTCTCGCTA-3′ | 248 | 60 |
| 2/3: | |||
| 2/3A | 5′-ACAGGCATCACGGTGGCAA-3′ | 310 | 65 |
| 2/3B | 5′-TGTGACCTATGACCTATGCT3-3′ | 310 | 65 |
| 4/5: | |||
| 4/5A | 5′-AGGCTCTGCTGGTCCCG-3′ | 408 | 65 |
| 4/5B | 5′-TGCTGCTGGGGAGGCGGGGT-3′ | 408 | 65 |
| 6: | |||
| 6A | 5′-ACCCCGCCTCCCCAGCAGGA-3′ | 271 | 65 |
| 6B | 5′-GCCACCGCCTACCCACCT-3′ | 271 | 65 |
| 7: | |||
| 7A | 5′-TGGGTAGGCGGTGGCCCA-3′ | 310 | 68 |
| 7B | 5′-AGGGTAGGTGTGGTGGTGCT-3′ | 310 | 68 |
| 8: | |||
| 8A | 5′-ATAAGTGGGTGAGCCTGA-3′ | 409 | 58 |
| 8B | 5′-TTCCCTCCCAACCTGAAAT-3′ | 409 | 58 |
Table B.
Primers Used to Amplify TPM2 Exons
| Exon and Primer | Primer Sequences | PCR ProductSize(bp) | AnnealingTemperature(°C) |
| 1: | |||
| TPM2E1F | 5′-AAGGGAGGCGGTCCCCAGTGGGT-3′ | 379 | 68 |
| TPM2E1R | 5′-ATGGCAGCGGCCCACCCTTGCCCTA-3′ | 379 | 68 |
| 2: | |||
| TPM2E2F | 5′-TACACACACCGCGTCTCCATT-3′ | 276 | 53 |
| TPM2E2R | 5′-AATCCTCTTATTCCCATA-3′ | 276 | 53 |
| 3: | |||
| TPM2E3F | 5′-ATCCTCTGGAGCCTCTCTGAT-3′ | 212 | 63 |
| TPM2E3R | 5′-TTCGATGACCTTCATTCCTCT-3′ | 212 | 63 |
| 4: | |||
| TPM2E4F | 5′-TGATGAGAGCGAGAGGTGGTCA-3′ | 218 | 55 |
| TPM2E4R | 5′-ACTGGGCATGTTGCAGGCT-3′ | 218 | 55 |
| 5: | |||
| TPM2E5F | 5′-AGCCTGCAACATGCCCAGT-3′ | 171 | 63 |
| TPM2E5R | 5′-AGGGCCAATGGGCTCACTTGTCA-3′ | 171 | 63 |
| 6: | |||
| TPM2E6F | 5′-TGACAAGTGAGCCCATTGGCCCT-3′ | 277 | 63 |
| TPM2E6R | 5′-TGAGAAAGAGCAGCCTGCGGT-3′ | 277 | 63 |
| TPM2E6bF | 5′-ATCTCCACTGTCTGGCTTCG-3′ | 194 | 60 |
| TPM2E6bR | 5′-CTCACCTCCCTCCCCTGT-3′ | 194 | 60 |
| 7: | |||
| TPM2E7F | 5′-ATCTGTGGTAGCGCTGTACTGAT-3′ | 250 | 63 |
| TPM2E7R | 5′-ACAGACTACTGCCTAAGTCAA-3′ | 250 | 63 |
| 8: | |||
| TPM2E8F | 5′-TTGACTTAGGCAGTAGTCTGT-3′ | 192 | 63 |
| TPM2E8R | 5′-ACAGACTGATAATCACGGCAGGT-3′ | 192 | 63 |
| 9: | |||
| TPM2E9F | 5′-AATGCATGTGTCTTCCTTGA-3′ | 279 | 63 |
| TPM2E9R | 5′-AGAGGCTAGTAACATCAGT-3′ | 279 | 63 |
| TPM2E9a1F | 5′-GGGGTAGAGATGGGGAGAAG-3′ | 353 | 60 |
| TPM2E9a1R | 5′-TCATTGAGTGCGTTGTCCAG-3′ | 353 | 60 |
| TPM2E9a2F | 5′-AGGCCATTAGCGAGGAACTG-3′ | 248 | 60 |
| TPM2E9a2R | 5′-GGGTGGAAGGGGATAGGTAAAG-3′ | 248 | 60 |
| TPM2E9a3F | 5′-AAATTGCCAACATTGCACAG-3′ | 138 | 60 |
| TPM2E9a3R | 5′-GGGTGTCAGTAAGTGCCAGGG-3′ | 138 | 60 |
The presence of each mutation was confirmed in at least one affected individual in each kindred by cloning a PCR-amplified product into pCR2.1 plasmid by use of the TA cloning kit (Invitrogen), according to manufacturer’s recommendations. For each individual, 10 transformed clones and 3 control clones were screened. Plasmid DNA was isolated with a Qiaprep miniprep kit (Qiagen) and was subjected to direct sequencing, as described above. In all other family members, mutations were detected by restriction digestion. In TNNI2, a G→A mutation at position 521 in exon 8 creates a novel MspI restriction site, and the C→T nonsense mutation at bp 466 of TNNI2 eliminates a BfuAI restriction site. A C→G missense mutation at position 271 in exon 3 of TPM2 destroys a novel SacII restriction site. Accordingly, each of these exons—and the regions flanking it—was amplified from affected and unaffected family members and was then digested, with the appropriate restriction enzyme, and gel fractionated. For each mutation, 140 control chromosomes were also screened. Haplotypes of TNNI2 were constructed by genotyping each affected individual and his or her parents for three microsatellite markers: D11S1984, D11S4893, and D11S1923.
Results
Two different mutations in TNNI2 were found in 4 (∼10%) of 34 kindreds with DA2B (fig. 2). The first mutation found was a G→A missense mutation at bp 521 that causes an arginine-to-glutamine substitution at amino acid residue 174 (R174Q). Several factors suggest that this mutation is probably disease causing. First, the R174Q mutation results in the substitution of an amino acid residue that is highly conserved in all isoforms of TnI and between mice and humans (fig. 3). Second, this change was found in two unrelated families with DA2B but was not found in 140 control chromosomes. Third, in pedigree K2 (fig. 2), the R174Q missense mutation arises de novo and causes DA2B in all of the children who inherited it (data not shown).
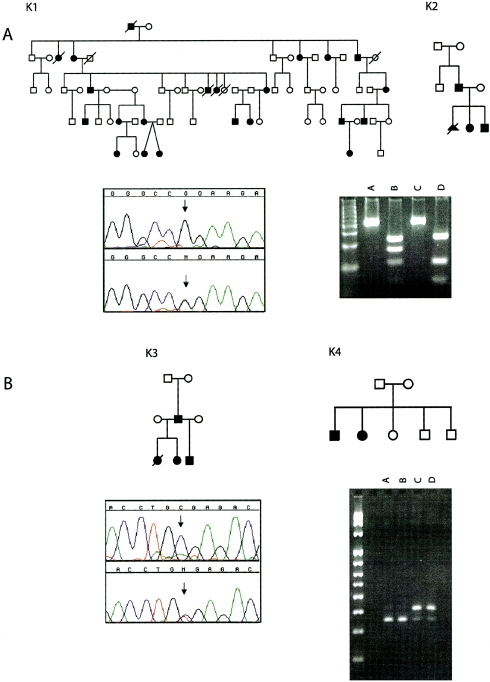
Figure 2.
Electropherograms of mutations in exon 8 of TNNI2. A, G→A missense mutation at position 521 in exon 8 of TNNI2 in kindreds K1 and K2. The mutation creates a novel MspI restriction site. Digests of TNNI2 amplicons from an affected individual (lane B) fractionate into four fragments (243 bp, 166 bp, 110 bp, and 56 bp), whereas only three fragments (243 bp, 110 bp, and 56 bp) are observed in an unaffected individual (lane D). Lanes A and C, Undigested control samples. B, C→T nonsense mutation at bp 466 of exon 8 of TNNI2 in kindreds K3 and K4. The mutation eliminates a BfuAI restriction site. Digests of TNNI2 amplicons from two affected sibs in kindred K4 (lanes C and D) fractionate into two fragments (409 bp and 344 bp), whereas only one fragment (344 bp) is found in the unaffected parents (lanes A and B). The 65-bp product cannot be observed on this gel.
Figure 3.
Alignment of genes encoding isoforms of TnI specific to fast-twitch muscle fibers (TNNI2) in humans and mice, slow-twitch muscle fibers (TNNI1), and cardiac muscle (TNNI3). The positions of the R156Ter and R174Q mutations are indicated.
The second mutation found in TNNI2 was a C→T nonsense mutation at bp 466 that is predicted to encode a mutant TnI lacking 26 amino acid residues of the carboxy-terminus. In kindred K3, this mutation arose de novo in the affected father and was subsequently transmitted to all of his offspring, each of whom is affected and has a different mother (fig. 2). The segregation of this mutation in kindred K4 suggests either that it arose de novo twice or that mosaicism for this mutation exists in one of the phenotypically normal parents. However, the level of mosaicism may be too low to be detected in lymphocytes and/or may exist in cell populations that were not tested (e.g., gonadal tissue). In each of the kindreds in which we found a disease-causing mutation in TNNI2, the mutation arose on a different haplotype background (data not shown). In addition, these mutations were found on both maternally and paternally derived chromosomes from different kindreds.
Patients with sporadic DA2B and those belonging to families with only an affected parent-child pair, which are uninformative for linkage studies, were also screened for mutations in TNNI2 by sequencing all of the exons and one of the known regulatory regions of TNNI2 (Mullen and Barton 2000). No disease-causing mutations were found. To look for large deletions that may not be detected via sequencing of genomic regions, we screened for allelic dropout, using a battery of microsatellites that bracket TNNI2. In addition, we used long-range PCR to amplify genomic regions of ∼2–3 kb spanning entire exons and introns, and we looked for amplicons of aberrant size; none were found. In these patients, DA2B may be caused by mutations in regulatory regions of TNNI2 that were not screened, or DA2B may be genetically heterogeneous.
TPM2 was screened for mutations in 14 probands with DA1. Only a single mutation in TPM2 was found. This was a C→G missense mutation at bp 271 that results in an arginine-to-glycine substitution at amino acid residue 91 (R91G) (fig. 4). It was found in the proband and affected family members of the kindred originally used to map DA1 to chromosome 9. This mutation is predicted to cause a nonconservative amino acid substitution of a residue that is invariant among mouse, chick, and jellyfish homologues of TPM2 (fig. 5). Of the remaining 13 probands with DA1 who either had sporadic disease or belonged to families with only an affected parent and child, screening TNNI2 by sequencing of all of its exons did not reveal any additional disease-causing mutations.
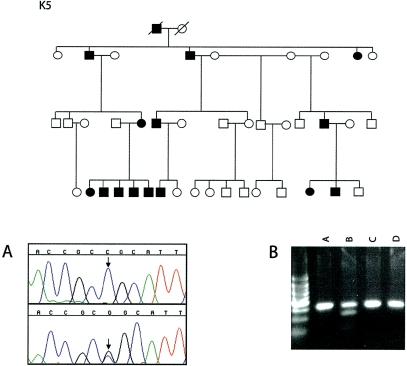
Figure 4.
Top, Pedigree of family K5. Bottom, electropherogram of a mutation on exon 3 and digests of amplicons. A, Electropherogram of a C→G missense mutation at position 271 in exon 3 of TPM2. The mutation creates a novel SacII restriction site in kindred K5. B, Digests of TPM2 amplicons from an affected individual. The digests from this individual separate into three fragments (320 bp, 218 bp, and 102 bp) (lane B), whereas amplicons from an unaffected individual remain undigested (lane D). Lanes A and C, Undigested control amplicons.
Figure 5.
Alignment of isoforms of β-tropomyosin from six animal species. The R91G substitution replaces an arginine residue that is invariant among species.
Fourteen individuals with classical FSS were screened for mutations in TNNI2 and TPM2. No disease-causing mutations were found.
Discussion
DA2B and DA1 are caused by mutations in TNNI2 and TPM2, respectively. Mutations were found in a total of 5 patients with DA from a collection of 48 probands that were screened, and all of these patients had familial disease. This includes a kindred in which two affected children have normal parents. This finding may explain why DA appears to sometimes segregate in an autosomal recessive pattern, an inference based on anecdotal reports of multiple affected sibs born to unaffected parents (Hall 1985). No mutations were found in any patients with sporadic DA, suggesting that the etiology of congenital contractures in patients with sporadic disease may be different from that in patients with familial disease.
We suggested elsewhere that DA1 is a genetically heterogeneous condition (Bamshad et al. 1994). However, this was based on the observation that linkage to chromosome 9 was excluded in a second family with DA1 that was later reclassified as having DA2B (family T [Bamshad et al. 1994]), and linkage data from other available families with DA1 do not exclude the locus on chromosome 9 (M.B., J.C.C., and L.B.J., unpublished data). Thus, it is not clear whether DA1 is genetically heterogeneous or there are mutations in regions of TPM2 that have not been screened. It has been hypothesized elsewhere that DA2B and classical FSS may be caused by mutations in the same gene (Klemp and Hall 1995; Krakowiak et al. 1997). The hypothesis was based, in part, on the observation that some individuals with DA2B, including some in the kindred used to map DA2B to chromosome 11p15.5 (K1 in fig. 2), had phenotypes similar to those of individuals with classical FSS. No mutations were found in TNNI2 in any of the probands with classical FSS; thus, its etiology remains unknown.
Missense mutations in TPM2 have been reported in two probands with nemaline rod myopathy (Donner et al. 2002). One individual had a Q147P substitution and presented with difficulty walking and weakness at 12 years of age. Another had an E117K substitution and presented with severe hypotonia, mild facial weakness, and ankle contractures at 4 years of age. None of the individuals with DA1 caused by a mutation in TPM2 had abnormal results on neurological examination, nor were nemaline rods found on histological analysis of the skeletal muscle of several individuals. Thus, although some clinical findings are common to both disorders, the phenotypes are easy to distinguish from one another. Interestingly, nemaline rod myopathy can also be caused by mutations in TPM3, which encodes γ-tropomyosin and TNNI1, a homologue of TNNI2, both of which are expressed predominately in slow-twitch myofibers (Johnston et al. 2000). The different phenotypes caused by mutations in genes encoding tropomyosins and troponins may provide further insights about the functional properties of these proteins.
The mechanism by which mutations in TPM2 and TNNI2 cause multiple congenital contractures is unclear. In all vertebrates, the contraction of skeletal muscle is dependent on an increase in the intracellular concentration of Ca+2 ions that produces a conformational change in the contractile machinery of the muscle and thus muscle contraction (Clark et al. 2002). The sensor/regulator of Ca+2 ion levels in skeletal muscle is the troponin complex, which is composed of three subunits, troponin T (TnT), troponin C (TnC), and TnI. TnI binds actin-tropomyosin and prevents muscle contraction by inhibiting actomyosin ATPase activity. TnC binds Ca+2 ions, leading to a conformational change that relieves TnI inhibition on actomyosin ATPase activity. Yet, its sensitivity to Ca+2 ions appears to be determined, in part, by the carboxy-terminal residues of TnI (Digel et al. 2001; Burton et al. 2002).
Mutations in the TNNI2 paralogue, TNNI3, encoding a cardiac-specific isoform of TnI, can cause a cardiomyopathy by either a dominant-negative effect or haploinsufficiency (Seidman and Seidman 2001). In TNNI3, substitution of amino acid residues homologous to those juxtaposed on either side of R174Q in TNNI2 result in diminished sensitivity to Ca+2 ions and a reduction in contractile force leading to the development of a cardiomyopathy (Watkins et al. 1995; Murphy et al. 2001). For example, a glycine-to-serine substitution of amino acid residue 203 results in decreased maximal contractile force and diminished sensitivity to Ca+2 ions (Burton et al. 2002). Similarly, deletion of the 17 carboxy-terminal amino acid residues of TnI, thought to occur in the heart during myocardial stunning, results in diminished sensitivity to Ca+2 ions and reduced contractility (McDonough et al. 1999). Transgenic mice expressing a gene encoding this truncated protein also develop diminished responsiveness to Ca+2 ions, diminished contractility, and ventricular dilatation—recapitulating the stunned myocardium (Murphy et al. 2000). The R156Ter mutation predicted to delete the 26 carboxy-terminal amino acid residues of TnI specific to fast-twitch myofibers might have a comparable effect. Accordingly, we hypothesize that both substitution and deletion mutants of the carboxy-terminus of fast-twitch–specific TnI result in diminished responsiveness to Ca+2 ions, leading to reduced muscle contraction and congenital contractures.
It is also possible that the contractures observed in individuals with DA2B are caused by perturbation of a nontraditional function of TnI. The TnI isoform encoded by TNNI2 has recently been discovered to bind to bFGF, to inhibit capillary endothelial cell proliferation (Feldman and Rouleau 2002), and to be a potent inhibitor of angiogenesis in human cartilage (Moses et al. 1999). Other proteins of the contractile apparatus (e.g., actin) are also found in cellular compartments that are different from their conventional location, although their function in these compartments is unknown.
β-Tropomyosin is a coiled-coil protein that binds head to tail along the length of the actin filament. It is negatively charged overall, and the R91G substitution is predicted to cause a local reduction in surface charge that may alter both the local conformation of the coiled coil and its flexural rigidity (fig. 6). This could influence its interactions with actin, which are thought to be critical for thin-filament regulation (Squire and Morris 1998). Glycine residues in porcine cardiac and rabbit skeletal α-tropomyosins appear to be compatible with the overall helical character of the protein suggested by low-resolution crystal structures (Whitby and Phillips 2000). A glycine side chain might, however, create an irregularity in the coiled coil, resulting in a region of increased flexibility that affects thin-filament regulation. Interestingly, a missense mutation in TPM3 that causes nemaline rod myopathy encodes a γ-tropomyosin molecule that folds abnormally and exhibits reduced sensitivity of actomyosin ATPase activity in the presence of Ca+2 ions (Moraczewska et al. 2000). Thus, the reduced sensitivity of actomyosin ATPase to Ca+2 ions may be the mechanism that causes congenital contractures in individuals with DA1.
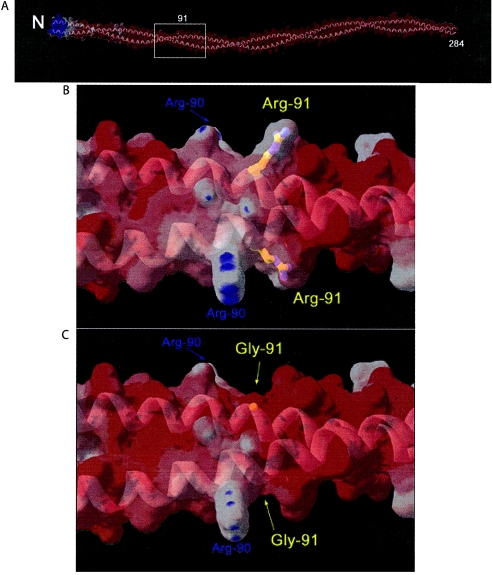
Figure 6.
A, Homology model of human β-tropomyosin dimer with a molecular surface colored by relative surface electrostatic charge. The model is based on the 7-Å crystal structure of porcine cardiac α-tropomyosin dimer (Protein Database entry 1C1G). A partially transparent molecular surface is shown colored by electrostatic charge: red indicates regions dominated by negative charge; white indicates areas that are neutral; blue indicates positive charge. Relatively few positively charged arginine and lysine surface amino acid side chains are found outside the N-terminal region. The sparse positively charged amino acids are largely neutralized by the large number of surface negatively charged aspartate and glutamate amino acids, which creates the dominant negatively charged surface character of the molecule. A ribbon model (gray) shows the characteristic coiled-coil fold of tropomyosin and is visible through the partially transparent molecular surface. “N” indicates the relatively positively charged N-terminal region, which is important for end-to-end polymerization, and “284” indicates the C-terminus. Box indicates the region of the molecule surrounding residue 91 shown in detail (panels B and C). B, Detailed view of the area of surrounding residue 91. A ball-and-stick model of the Arg91 side chain is colored by atom type and is visible through the partially transparent molecular surface. The positive charge provided by Arg90 and Arg91 is flanked by negative charge, characteristic of the molecule as a whole. C, Detailed view of the area surrounding residue 91 with R91G mutation. Mutation to glycine neutralizes the positive charge at that position, leading to an increase in local negatively charged character, seen as an increase in red color in panel C relative to panel B. Figures were generated with the program Swiss PDB Viewer.
The hypothesis that some DA disorders have a myopathic origin has been tendered for two decades (Hall et al. 1982), yet many affected individuals have never undergone a muscle biopsy, and the findings, if any, in those who do undergo biopsy are typically nonspecific. Moreover, the site of the muscle biopsy is commonly chosen for its convenience (e.g., quadriceps) and is usually not a muscle rich in fast-twitch myofibers. Of the existing anecdotal reports of the muscle histology of children affected with various DAs, a single case report is noteworthy. It describes a paucity of fast-twitch myofibers in an individual with a phenotype resembling DA2B (Vanek et al. 1986). Our results suggest that further study of muscles rich in fast-twitch myofibers in children with DA1 and DA2B is warranted.
Although the expression of TnI isoforms is restricted among adult muscle fiber types, the spatial expression patterns of these genes during development is not (Zhu et al. 1995). In the developing skeletal muscle of mice, isoforms of TnI are first expressed in the myotome of somites at 9.5 days post conception (dpc). The slow-twitch isoform is expressed first in all newly formed myotubes, regardless of the future fiber type. As primary myotubes develop into secondary myotubes (14–16 dpc), expression of the fast-twitch isoform of TnI increases. The timing of this switch between isoforms is intriguing, because, in our experience with half a dozen patients for whom a diagnosis of DA2B was established prenatally, the limbs are often normal during the first 18–20 wk of gestation. It is typically over the next 6–8 wk that the contractures of the hands and feet become recognizable via ultrasonography. Although there are no data on the temporal and spatial expression patterns of TnI isoforms in humans, the onset of expression of a mutant TNNI2 during myogenesis appears to coincide with our ability to detect fetal contractures in DA2B.
In newborn mice, the expression of slow-twitch TnI is stronger in “deep” muscles containing predominately slow-twitch fibers (e.g., soleus), whereas expression of fast-twitch TnI is stronger in more-peripheral muscles (e.g., tibialis anterior and extensor digitorum longus) (Zhu et al. 1995). The homologous peripheral muscles in humans also contain predominately fast-twitch myofibers and control the movements of body areas affected in individuals with DA2B. In contrast, expression of TPM2 is lower in fast-twitch myofibers than in slow-twitch myofibers. However, the expression of β-tropomyosin protein is higher in fast-twitch myofibers (Pieple and Wieczorek 2000). Thus, the spatial expression pattern of TPM2 also reflects the peripheral distribution of contractures in individuals with DA1.
In 1991, Beall and Fyrberg discovered that the original heldup allele (i.e., heldup2), a troponin mutant, was caused by an alanine-to-valine substitution. A variety of heldup alleles with phenotypes varying in severity from embryonic lethal to mild abnormalities of flight performance and jump ability have subsequently been described. No substantial differences were noted between the phenotypes of the individuals with different alleles of TNNI2, although each of these mutant alleles disrupts the same domain of TnI. The IFM of many heldup mutants show nearly normal morphogenesis but subsequently begin to hypercontract, leaving only stumps near their attachment points in newly emerged flies. This recapitulates the reports of misplaced, hypoplastic, or absent tendons in some individuals with DA (Hall et al. 1982), as well as in those in whom we identified mutations in TNNI2.
The spatial and temporal expression patterns of genes encoding proteins of the Tc-tropomyosin-actin complex in fast-twitch fibers may not be regulated independently in patients with DA. For example, a lack of TnT in Drosophila results in a secondary reduction of tropomyosin and actin (Fyrberg et al. 1990). This indicates that a feedback mechanism tightly coordinates the expression of interrelated genes and proteins. Accordingly, abnormalities of other proteins of the Tc-tropomyosin-actin complex in fast-twitch myofibers may cause other multiple-congenital-contracture disorders. We suspect that the development of DA may be a final common outcome of mutations that alter the function and stoichiometry of any one of the proteins of the Tc-tropomyosin-actin complex in fast-twitch fibers. This would be analogous to the observation that mutations in genes encoding many of the isoforms of the Tc-tropomyosin-actin complex in cardiac muscle can cause a cardiomyopathy.
Acknowledgments
We would like to thank the families for their participation, generosity, and patience. We are indebted to the Freeman-Sheldon Parents Support Group for their enthusiasm and perseverance. We would like to thank J. Hall, K. Flanigan, and two anonymous reviewers, for discussion, review, and comments. This project was completed with the support of the Shriners Hospitals for Children and National Institutes of Health grant PHS M01-00064 to the General Clinical Research Center at the University of Utah.
Electronic-Database Information
Accession numbers and the URL for data presented herein are as follows:
- Online Mendelian Inheritance in Man (OMIM) http://www.ncbi.nlm.nih.gov/Omim/ (for DA1 [MIM 108120], DA2B [MIM 601680], and FSS [MIM 193700])
References
- Andrade MD, Barnholtz JS, Amos CI, Lockmiller C, Scott A, Risman M, Hecht J (1998) Segregation analysis of idiopathic talipes equinovarus in a Texan population. Am J Med Genet 79:97–102 [DOI] [PubMed] [Google Scholar]
- Bamshad M, Bohnsack JF, Jorde LB, Carey JC (1996a) Distal arthrogryposis type 1: clinical analysis of a large kindred. Am J Med Genet 65:282–285 [DOI] [PubMed] [Google Scholar]
- Bamshad M, Jorde LB, Carey JC (1996b) A revised and extended classification of the distal arthrogryposes. Am J Med Genet 65:277–281 [DOI] [PubMed] [Google Scholar]
- Bamshad M, Watkins WS, Zenger RK, Bohnsack JF, Carey JC, Otterud B, Krakowiak PA, Robertson M, Jorde LB (1994) A gene for distal arthrogryposis type 1 maps to the pericentromeric region of chromosome 9. Am J Hum Genet 55:1153–1158 [PMC free article] [PubMed] [Google Scholar]
- Beall CJ, Fyrberg E (1991) Muscle abnormalities in Drosophila melanogaster heldup mutants are caused by missing or aberrant troponin-I isoforms. J Cell Biol 114:941–951 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barbas JA, Galceran J, Torroja L, Prado A, Ferrus A (1993) Abnormal muscle development in the heldup mutant of Drosophila melanogaster is caused by a splicing defect affecting selected troponin I isoforms. Mol Cell Biol 13:1433–1439 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burton D, Abdulrazzak H, Knott A, Elliott K, Redwood C, Watkins H, Marston S, Ashley C (2002) Two mutations in troponin I that cause hypertrophic cardiomyopathy have contrasting effects on cardiac muscle contractility. Biochem J 362:443–451 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clark KA, McElhinny AS, Beckerle MC, Gregorio CC (2002) Striated muscle cytoarchitecture: an intricate web of form and function. Annu Rev Cell Dev Biol 18:637–706 [DOI] [PubMed] [Google Scholar]
- Deak II (1977) Mutations in Drosophila melanogaster that affect muscles. J Embryol Exp Morphol 40:885–895 [PubMed] [Google Scholar]
- Deak II, Bellamy PR, Bienz M, Dubuis Y, Fenner E, Gollin M, Rahmi A, Ramp T, Reinhardt CA, Cotton B (1982) Mutations affecting the indirect flight muscles of Drosophila melanogaster. J Embryol Exp Morphol 69:61–81 [PubMed] [Google Scholar]
- Digel J, Abugo O, Kobayashi T, Gryczynski Z, Lakowicz JR, Collins JH (2001) Calcium- and magnesium-dependent interactions between the C-terminus of troponin I and the N- terminal, regulatory domain of troponin C. Arch Biochem Biophys 387:243–249 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donner K, Ollikainen M, Ridanpää M, Christen H-J, Goebel HH, de Visser M, Pelin K, Wallgren-Pettersson C (2002) Mutations in the β-tropomysin (tPM2) gene: a rare cause of nemaline myopathy. Neuromuscul Disord 12:151–158 [DOI] [PubMed] [Google Scholar]
- Feldman L, Rouleau C (2002) Troponin I inhibits capillary endothelial cell proliferation by interaction with the cell’s bFGF receptor. Microvasc Res 63:41–49 [DOI] [PubMed] [Google Scholar]
- Freeman E, Sheldon J (1938) Cranio-carpotarsal dystrophy: undescribed congenital malformation. Arch Dis Child 13:277–283 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fyrberg, E, Fyrberg CC, Beall C, Saville DL (1990) Drosophila melanogaster troponin-T mutations engender three distinct syndromes of myofibrillar abnormalities. J Mol Biol 216:657–675 [DOI] [PubMed] [Google Scholar]
- Hall JG (1985) Genetic aspects of arthrogryposis. Clin Orthop 194:44–53 [PubMed] [Google Scholar]
- Hall JG, Reed SD, Green G (1982) The distal arthrogryposes: delineation of new entities—review and nosologic discussion. Am J Med Genet 11:185–239 [DOI] [PubMed] [Google Scholar]
- Hotta Y, Benzer S (1972) Mapping of behaviour in Drosophila mosaics. Nature 240:527–535 [DOI] [PubMed] [Google Scholar]
- Johnston JJ, Kelley RI, Crawford TO, Morton DH, Agarwala R, Koch T, Schäffer AA, Francomano CA, Biesecker LG (2000) A novel nemaline myopathy in the Amish caused by a mutation in troponin TI. Am J Hum Genet 67:814–821 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klemp EL, Hall JG (1995) Dominant distal arthrogryposis in a Maori family with marked variability of expression. Am J Med Genet 55:414–419 [DOI] [PubMed] [Google Scholar]
- Krakowiak PA, Bohnsack JF, Carey JC, Bamshad M (1998) Clinical analysis of a variant of Freeman-Sheldon syndrome (DA2B). Am J Med Genet 76:93–98 [DOI] [PubMed] [Google Scholar]
- Krakowiak PA, O’Quinn JR, Bohnsack JF, Watkins WS, Carey JC, Jorde LB, Bamshad M (1997) A variant of Freeman-Sheldon syndrome maps to 11p15.5-pter. Am J Hum Genet 60:426–432 [PMC free article] [PubMed] [Google Scholar]
- McDonough JL, Arrell DK, Van Eyk JE (1999) Troponin I degradation and covalent complex formation accompanies myocardial ischemia/reperfusion injury. Circ Res 84:9–20 [DOI] [PubMed] [Google Scholar]
- Moraczewska J, Greenfield NJ, Liu Y, Hitchock-DeGregori SE (2000) Alteration of tropomyosin function and folding by a nemaline myopathy-causing mutation. Biophys J 79:3217–3225 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moses MA, Wiederschain D, Wu I, Fernandez CA, Ghazizadeh V, Lane WS, Flynn E, Sytkowski A, Tao T, Langer R (1999) Troponin I is present in human cartilage and inhibits angiogenesis. Proc Natl Acad Sci USA 96:2645–2650 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mullen AJ, Barton PJR (2000) Structural characterization of the human fast skeletal muscle troponin I gene (TNNI2). Gene 242:313–320 [DOI] [PubMed] [Google Scholar]
- Murphy AM, Kögler H, Georgakopoulos G, McDonough JL, Kass DA, Van Eyk JE, Marbán E (2000) Transgenic mouse model of stunned myocardium. Science 287:488–491 [DOI] [PubMed] [Google Scholar]
- Naimi B, Harrison A, Cummins M, Nongthomba U, Clark S, Canal I, Ferrus A, Sparrow JC (2001) A tropomyosin-2 mutation suppresses a troponin I myopathy in Drosophila. Mol Biol Cell 12:1529–1539 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pieples K, Wieczorek DF (2000) Tropomyosin 3 increases striated muscle isoform diversity. Biochemistry 39:8291–8297 [DOI] [PubMed] [Google Scholar]
- Seidman JG, Seidman C (2001) The genetic basis for cardiomyopathy: from mutation identification to mechanistic paradigms. Cell 104:557–567 [DOI] [PubMed] [Google Scholar]
- Squire JM, Morris EP (1998) A new look at thin filament regulation in vertebrate skeletal muscle. FASEB J 12:761–771 [DOI] [PubMed] [Google Scholar]
- Vanek J, Janda J, Amblerová V, Flosan F (1986) Freeman-Sheldon syndrome: a disorder of congenital myopathic origin. J Med Genet 23:231–236 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vigoreaux JO (2001) Genetics of the Drosophila flight muscle myofibril: a window into the biology of complex systems. Bioessays 23:1047–1063 [DOI] [PubMed] [Google Scholar]
- Watkins H, McKenna WJ, Thierfelder L, Suk HJ, Anan R, O’Donoghue A, Spirito P, Matsumori A, Moravec CS, Seidman JG, Seidman CE (1995) Mutations in the genes for cardiac troponin T and α-tropomyosin in hypertrophic cardiomyopathy. N Engl J Med 332:1058–1064 [DOI] [PubMed] [Google Scholar]
- Whithy FG, Phillips GN Jr (2000) Crystal structure of tropomyosin at 7 angstroms resolution. Proteins 38:49–59 [PubMed] [Google Scholar]
- Zhu L, Lyons GE, Juhasz O, Joya JE, Hardeman EC, Wade R (1995) Developmental regulation of troponin I isoform genes in striated muscles of transgenic mice. Dev Biol 169:487–503 [DOI] [PubMed] [Google Scholar]