Abstract
Ellis-van Creveld syndrome (EvC) is an autosomal recessive skeletal dysplasia. Elsewhere, we described mutations in EVC in patients with this condition (Ruiz-Perez et al. 2000). We now report that mutations in EVC2 also cause EvC. These two genes lie in a head-to-head configuration that is conserved from fish to man. Affected individuals with mutations in EVC and EVC2 have the typical spectrum of features and are phenotypically indistinguishable.
Ellis-van Creveld syndrome (MIM 225500) is a recessive chondrodysplasia characterized by short limbs and short ribs, postaxial polydactyly, and dysplastic nails and teeth. In addition to the chondroectodermal phenotype, congenital heart defects, most commonly an atrio-ventricular septal defect, are often observed.
Two years ago, we identified mutations in a novel gene in patients with EvC (Ruiz-Perez et al. 2000). Subsequently, we have screened the 21 EVC coding exons in 58 patients, who gave informed consent, and identified mutations in 13. In these 13 patients, we identified both parental mutations; in the remaining 45 cases, we have not detected any mutations. That is, we have not identified any patients with EvC who have a mutation on only one allele. cDNA analysis of fibroblast RNA from three of these patients was normal, excluding aberrant splicing or gene rearrangements that would not have been detected by our strategy of sequencing genomic DNA.
These observations raised the possibility of genetic heterogeneity. Six consanguineous pedigrees lacking EVC mutations were consistent with localization to the EVC region by microsatellite analysis, suggesting that the putative second gene might be in the same region. The six probands were homozygous for a large area telomeric to 164F16P2, and we reviewed the evidence for involvement of other genes in the region. MSX1—a good candidate gene, on the basis of the phenotype of the Msx1 mouse knockout (dental abnormalities, cleft palate, and abnormalities of nail development) (Jumlongras et al. 2001)—had been excluded as the causative gene in the Amish (Ide et al. 1996), but the possibility remained that it was a second EvC gene.
Heterozygous MSX1 mutations have been identified in families with Witkop syndrome (MIM 189500) (Jumlongras et al. 2001) and in families with dental agenesis with or without clefting (Vastardis et al. 1996; van den Boogaard et al. 2000). We considered whether this could be analogous to the milder phenotype we have observed in individuals with Weyers acrodental dysostosis who have heterozygous EVC mutations (Ruiz-Perez et al. 2000). However, we did not identify any mutations after sequencing MSX1 in these six consanguineous families.
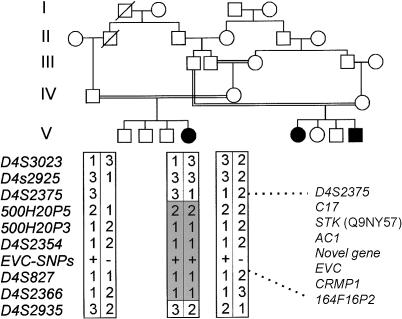
Subsequently, we have identified an affected individual from a consanguineous Gypsy pedigree who has a shorter region of homozygosity, excluding MSX1, and who does not have an EVC mutation. A simplified pedigree is shown in figure 1. Given the small size of the homozygous region and the multiple consanguinity in this ethnic group, it is likely that the common ancestor is from an earlier generation. From this pedigree, D4S2375 is the telomeric marker; the centromeric marker for the gene locus, 164F16P2, is based on haplotyping we reported in a Brazilian pedigree (Ruiz-Perez et al. 2000).
Figure 1.
Haplotype analysis of the consanguineous pedigree with the shortest region of homozygosity. Informative markers and haplotypes are shown from telomere to centromere beneath the affected individual and her parents, showing the region of homozygosity between D4S2375 and D4S2935. Analysis of the other six pedigrees places the centromeric limit for the gene locus at 164F16P2 (data not shown). The gene order between D4S2375 and 164F16P2 is also shown.
Interrogation of the human and mouse databases showed the following genes in the region of interest:C17 (which encodes a cytokine-like protein), a serine threonine kinase gene (Q9NY57), AC1, a predicted novel gene, EVC, and CRMP1. We began systematic screening of these genes. However, our interest was directed to the novel gene by the recent finding that it is mutated in bovine chondrodysplastic dwarfism (Takeda et al. 2002). As the deposited cDNA sequence (GenBank accession number NM_147127) did not contain the transcription start site, we performed 5′ RACE (Gene Racer Kit, Invitrogen) in adult human kidney RNA, using gene-specific primers. The RACE product, confirmed by RT-PCR, revealed an additional GC-rich exon containing an inframe AUG with loss of the open reading frame 21 nucleotides upstream. This additional sequence has homology to the N-terminus of the murine orthologue.
We aligned the extended cDNA (AY185210) against human genomic sequence (Venter et al. 2001; Celera Public Database; Ensembl Genome Browser) to design primers to amplify the 22 coding exons in the seven probands. We identified a frameshift mutation (3660delC) in exon 22 in the Gypsy family. The clinical features in affected individuals in this family included atrioventricular septal defects, mesomelic limb shortening with genu valgum, polydactyly with nail dysplasia, multiple oral frenulae, and dysplastic teeth. Two affected individuals in earlier generations are reported to have had postaxial polydactyly and congenital heart defects but absence of other features of EvC.
Mutation analysis of the Brazilian pedigree (Oliveira da Silva et al. 1980) identified a five-nucleotide insertion in exon 1, 198insGGCGG. The clinical features in this family include atrial septal defect, short limb dysplasia with genu valgum, polydactyly, multiple oral frenulae, oligodontia, and dysplastic teeth.
We found another frameshift (2056insC) in exon 14 in a child of second-cousin parents who presented with short limb dysplasia, short ribs, and postaxial polydactyly of the hands. Her nails and teeth were dysplastic, with hyperplasia of the alveolar ridges and multiple oral frenulae. She had a patent arterial duct but no other cardiac abnormality. We detected a C1195T transition in exon 10 that introduced a nonsense codon (R399X) in a stillborn child who had a ventricular septal defect and short limbs with postaxial polydactyly of the hands. Permission for post mortem was refused, but radiographs showed typical changes of EvC, with short limbs and a classical pelvic configuration. We found another transition (C1855T) in exon 12 that causes a nonsense codon (Q619X) in an Ecuadorian family with two affected daughters. Both had disproportionate short stature with acromesomelic limb shortening and short ribs. In the hands, there was postaxial polydactyly with nail dysplasia and fusion of the hamate and capitate. Both had oligodontia with dysplastic teeth and multiple oral frenulae. One of the girls had genu valgum. The five homozygous truncating mutations in patients with EvC, taken together with the null phenotype observed in cattle, provide compelling evidence that this is a second EvC gene (EVC2).
In addition to these five truncating mutations (fig. 2), we identified a missense change, I283R, in exon 7 that was not present in 100 normal control chromosomes. This nonpolar (isoleucine) to basic (arginine) amino acid change is at a residue that is conserved in the mouse, human, and bovine protein. There were two affected offspring in the family with this mutation; both had ostium primum atrial septal defects with short limb dysplasia, postaxial polydactyly of the hands, and nail and tooth dysplasia.
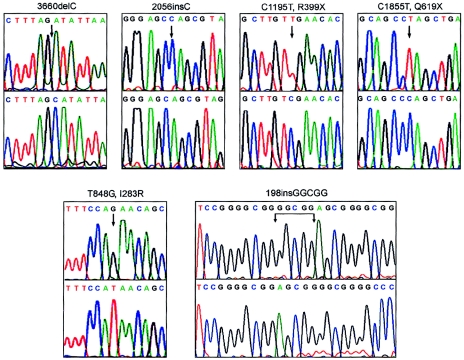
Figure 2.
Identification of EVC2 mutations. For each mutation, mutant genomic sequence is shown above corresponding wild-type sequence. Numbering starts at the translation start site in AY185210. The first two mutations are a single-base deletion (exon 22) and a single-base insertion (exon 14) that rapidly introduce stop codons. 3660delC is the mutation in the pedigree in figure 1. The C1195T (exon 10) and C1855T (exon 12) transitions introduce stop codons. T848G (exon 7) results in arginine replacing an isoleucine residue. The five-nucleotide insertion, 198 insGGCGG in exon 1, was found in the Brazilian pedigree described by Oliveira da Silva et al. (1980).
We have not identified a mutation in the remaining consanguineous pedigree. The proband in this pedigree had an atrioventricular septal defect, short limbs, short ribs, postaxial polydactyly, and an absent alveolar sulcus. Radiographs showed short long bones with a typical pelvic configuration. We have amplified and sequenced both EVC and EVC2 cDNA from the proband; the transcripts are normally spliced, excluding splicing mutations and gene rearrangements. We have not excluded the possibility of a regulatory mutation in this family. As there is a 1/16 probability of homozygosity by descent at any locus in the offspring of first cousins, it is also possible that the homozygosity is coincidental and that this family does not have an EVC or EVC2 mutation.
Chondrodysplastic dwarfism in Japanese brown cattle is an autosomal recessive disorder with shortening of long bones. Moritomo and colleagues (1992) reported abnormalities at the epiphyseal plate of the humerus, ulna, radius, metacarpus, femur, tibia, and metatarsus in affected calves, but the spheno-occipital synchondrosis, vertebrae, costochondral junction, iliac crest, and the articular epiphyseal cartilage of the long bones were comparable to control calves. Given the rib shortening in EvC, it is interesting that the costochondral junction in affected cattle was not different from that of controls. There have been no reports of cardiovascular malformations in affected cattle. Takeda and colleagues (2002) identified two mutations in limbin (LBN), the bovine orthologue of EVC2. Affected animals are either compound heterozygotes or homozygous for a frameshifting mutation. The murine orthologue is strongly expressed in proliferating chondrocytes, osteoblasts, and osteoclasts (Takeda et al. 2002).
EVC and EVC2 are arranged in a divergent configuration with transcription start sites separated by 2,624 bp in human (on the basis of alignment of cDNAs AF216184 and AY185210 against genomic sequence) (Venter et al. 2001; Celera Public Database; Ensembl Genome Browser) and 1,647 bp in mouse (from alignment of cDNAs AJ250841 and BC037473 with genomic sequence). Recent analysis indicates that such head-to-head configurations may be a common feature of the human genome (Adachi and Lieber 2002), and there are examples where coregulation by a single promoter with bidirectional activity has been shown (Shimada et al. 1989; Platzer et al. 1997). Likewise, the expression of EVC and EVC2 could be coordinated by the same promoter or shared elements of overlapping promoters. We are aware of one previous example where mutations in genes in such a configuration have led to the same phenotype; mutations in both ABCG5 and ABCG8 cause sitosterolemia (Berge et al. 2000; Lu et al. 2001). ABCG5 and ABCG8 are highly homologous members of the ATP-binding cassette family. In the case we are presenting, there are no obvious similarities between EVC and EVC2, and, other than the predicted transmembrane domains, there are no motifs giving clues to their function.
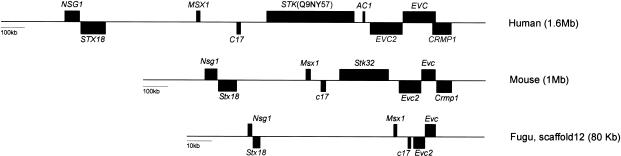
Analysis of the human, mouse, and pufferfish genome sequence indicates that EVC and EVC2 lie within a syntenic region with conserved gene order and transcription orientation encompassing genes NSG1, STX18, MSX1, C17, EVC, and EVC2 (fig. 3). Synteny in Fugu is lost outside these boundaries but extends farther when human and mouse sequences are compared. Given the phenotypic similarities associated with mutations in Msx1 and EVC-EVC2, it is intriguing that their order and orientation have been conserved over 450 million years of evolution (fig. 3).
Figure 3.
Scale representation of the syntenic chromosomal region shared by human, mouse, and pufferfish at the Ellis-van Creveld locus. Orientation of the human short arm of chromosome 4 and mouse chromosome 5 is from telomere to centromere; Fugu scaffolds lack chromosomal assignation. The size of the syntenic region for each species appears in brackets, and genes are represented by full boxes above or below the line, depending on whether their direction of transcription is toward the centromere or telomere, respectively. CRMP1, the 3′ end of which overlaps with the 3′ end of EVC in human (Ruiz-Perez et al. 2000), is not in an equivalent position in pufferfish. The serine threonine kinase is not found in Fugu, and interrogation of the mouse and Fugu databases (DOE Joint Genome Institute; The Fugu Genomics Project) does not detect AC1 orthologues. NSG1 is a neuron-specific protein (Carlock et al. 1996), and STX18 is a member of the syntaxin family of protein receptors that are involved in vesicle docking (Hatsuzawa et al. 2000). This diagram is based on August 2002 data freeze of the human, mouse, and Fugu genome assemblies.
In conclusion, mutations in EVC and EVC2 lead to an indistinguishable phenotype. We are assessing the frequency of EVC and EVC2 mutations in a patient panel to assess their relative contributions in the pathogenesis of EvC and whether, between them, these two genes account for all cases of EvC.
Acknowledgments
This work was funded by the Medical Research Council, British Heart Foundation, and European Commission (QLG1-2001-02188).
Electronic-Database Information
Accession numbers and URLs for data presented herein are as follows:
- Celera Public Database, http://public.celera.com/cds/login.cfm (for EVC2 exon 1 and sequence between EVC and EVC2)
- DOE Joint Genome Institute, http://www.jgi.doe.gov/
- Ensembl Genome Browser, http://www.ebi.ac.uk/ensembl/index.html
- The Fugu Genomics Project, http://fugu.hgmp.mrc.ac.uk/
- GenBank, http://www.ncbi.nlm.nih.gov/GenBank/index.html (for sequence of limbin [accession number NM_147127] and cDNA alignment [AF216184 and AY185210])
- Online Mendelian Inheritance in Man (OMIM), http://www.ncbi.nlm.nih.gov/Omim/ (for EvC [MIM 225500] and Witkop syndrome [MIM 189500])
References
- Adachi N, Lieber MR (2002) Bidirectional gene organization: a common architectural feature of the human genome. Cell 109:807–809 [DOI] [PubMed] [Google Scholar]
- Berge KE, Tian H, Graf GA, Yu L, Grishin NV, Schultz J, Kwiterovich P, Shan B, Barnes R, Hobbs HH (2000) Accumulation of dietary cholesterol in sitosterolemia caused by mutations in adjacent ABC transporters. Science 290:1771–1775 [DOI] [PubMed] [Google Scholar]
- Carlock L, Vo T, Lorincz M, Walker PD, Bessert D, Wisniewski D, Dunbar JC (1996) Variable subcellular localization of a neuron-specific protein during NTera 2 differentiation into post-mitotic human neurons. Brain Res Mol Brain Res 42:202–212 [DOI] [PubMed] [Google Scholar]
- Hatsuzawa K, Hirose H, Tani K, Yamamoto A, Scheller RH, Tagaya M (2000) Syntaxin 18, a SNAP receptor that functions in the endoplasmic reticulum, intermediate compartment, and cis-Golgi vesicle trafficking. J Biol Chem 275:13713–13720 [DOI] [PubMed] [Google Scholar]
- Ide SE, Ortiz de Luna RI, Francomano CA, Polymeropoulos MH (1996) Exclusion of the MSX1 homeobox gene as the gene for the Ellis van Creveld syndrome in the Amish. Hum Genet 98:572–575 [DOI] [PubMed] [Google Scholar]
- Jumlongras D, Bei M, Stimson JM, Wang WF, DePalma SR, Seidman CE, Felbor U, Maas R, Seidman JG, Olsen BR (2001) A nonsense mutation in MSX1 causes Witkop syndrome. Am J Hum Genet 69:67–74 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu K, Lee MH, Hazard S, Brooks-Wilson A, Hidaka H, Kojima H, Ose L, Stalenhoef AF, Mietinnen T, Bjorkhem I, Bruckert E, Pandya A, Brewer HB Jr, Salen G, Dean M, Srivastava A, Patel SB (2001) Two genes that map to the STSL locus cause sitosterolemia: genomic structure and spectrum of mutations involving sterolin-1 and sterolin-2, encoded by ABCG5 and ABCG8, respectively. Am J Hum Genet 69:278–290 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moritomo Y, Ishibashi T, Miyamoto H (1992) Morphological changes of epiphyseal plate in the long bone of chondrodysplastic dwarfism in Japanese brown cattle. J Vet Med Sci 54:453–459 [DOI] [PubMed] [Google Scholar]
- Oliveira da Silva E, Janovitz D, de Alberquerque SC (1980) Ellis-van Creveld syndrome: report of 15 cases in an inbred kindred. J Med Genet 17:349–356 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Platzer M, Rotman G, Bauer D, Uziel T, Savitsky K, Bar-Shira A, Gilad S, Shiloh Y, Rosenthal A (1997) Ataxia-telangiectasia locus: sequence analysis of 184 kb of human genomic DNA containing the entire ATM gene. Genome Res 7:592–605 [DOI] [PubMed] [Google Scholar]
- Ruiz-Perez VL, Ide SE, Strom TM, Lorenz B, Wilson D, Woods K, King L, Francomano C, Freisinger P, Spranger S, Marino B, Dallapiccola B, Wright M, Meitinger T, Polymeropoulos MH, Goodship J (2000) Mutations in a new gene in Ellis-van Creveld syndrome and Weyers acrodental dysostosis. Nat Genet 24:283–286 [DOI] [PubMed] [Google Scholar]
- Shimada T, Fujii H, Lin H (1989) A 165-base pair sequence between the dihydrofolate reductase gene and the divergently transcribed upstream gene is sufficient for bidirectional transcriptional activity. J Biol Chem 264:20171–20174 [PubMed] [Google Scholar]
- Takeda H, Takeda H, Takami M, Oguni T, Tsuji T, Yoneda K, Sato H, Ihara N, Itoh T, Kata SR, Mishina Y, Womack JE, Moritomo Y, Sugimoto Y, Kunieda T (2002) Positional cloning of the gene LIMBIN responsible for bovine chondrodysplastic dwarfism. Proc Natl Acad Sci USA 99:10549–10554 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van den Boogaard MJ, Dorland M, Beemer FA, van Amstel HK (2000) MSX1 mutation is associated with orofacial clefting and tooth agenesis in humans. Nat Genet 24:342–343 [DOI] [PubMed] [Google Scholar]
- Vastardis H, Karimbux N, Guthua SW, Seidman JG, Seidman CE (1996) A human MSX1 homeodomain missense mutation causes selective tooth genesis. Nat Genet 13:417–421 [DOI] [PubMed] [Google Scholar]
- Venter JC, Adams MD, Myers EW, Li PW, Mural RJ, Sutton GG, Smith HO, et al (2001) The sequence of the human genome. Science 291:1304–1351 [DOI] [PubMed] [Google Scholar]