Abstract
We have demonstrated that the breakpoints of the constitutional t(11;22) are located at palindromic AT-rich repeats (PATRRs) on 11q23 and 22q11. As a mechanism for this recurrent translocation, we proposed that the PATRR forms a cruciform structure that induces the genomic instability leading to the rearrangement. A patient with neurofibromatosis type 1 (NF1) had previously been found to have a constitutional t(17;22) disrupting the NF1 gene on 17q11. We have localized the breakpoint on 22q11 within the 22q11-specific low-copy repeat where the breakpoints of the constitutional t(11;22)s reside, implying a similar palindrome-mediated mechanism for generation of the t(17;22). The NF1 gene contains a 195-bp PATRR within intron 31. We have isolated the junction fragments from both the der(17) and the der(22). The breakpoint on 17q11 is close to the center of the PATRR. A published breakpoint of an additional NF1-afflicted patient with a constitutional t(17;22) is also located close to the center of the same PATRR. Our data lend additional support to the hypothesis that PATRR-mediated genomic instability can lead to a variety of translocations.
The constitutional t(11;22) is the only known recurrent non-Robertsonian translocation. The recurrent nature of the rearrangement implicates a specific genomic structure at the t(11;22) breakpoints. Translocation breakpoints of t(11;22) cases have been cloned, and all the breakpoints are located within palindromic AT-rich repeats (PATRRs) on 11q23 and 22q11 (Kurahashi et al. 2000; Edelmann et al. 2001; Tapia-Paez et al. 2001). The majority of individuals with the t(11;22) have breakpoints at the center of the PATRRs, suggesting that the center of the PATRR is susceptible to double-strand-breaks (DSBs) leading to the translocation (Kurahashi and Emanuel 2001a). Indeed, translocation-specific PCR can detect a high frequency of de novo t(11;22) occurrences in normal sperm samples (Kurahashi and Emanuel 2001b).
Neurofibromatosis type 1 (NF1 [MIM 162200]) is an autosomal-dominant inherited disorder characterized particularly by cafe-au-lait spots and fibromatous tumors of the skin. The NF1 gene was positionally cloned by two groups, one of which utilized an NF1 case with a constitutional karyotype of t(17;22)(q11;q11) (Viskochil et al. 1990; Wallace et al. 1990). We have further analyzed the breakpoints of this original patient with t(17;22) (Ledbetter et al. 1989).
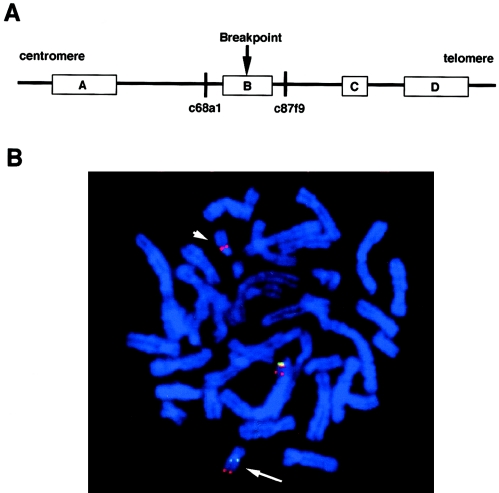
The chromosome 17 breakpoint of the patient with NF1 had been mapped elsewhere using a somatic cell hybrid that carries the der(22) of the patient (O'Connell et al. 1989). Subsequently, the NF1 gene was identified, and the translocation was shown to disrupt the gene (Viskochil et al. 1990). The chromosome 22 breakpoint of the patient was also mapped into the region typically deleted in patients with DiGeorge syndrome, using this somatic cell hybrid (Budarf et al. 1996). To more precisely localize the breakpoint within 22q11, FISH was performed on metaphase preparations from a cell line derived from the patient (P89-75L). We used multiple cosmids located on 22q11 and isolated from the LL22NCO3 cosmid library. Signals of c68a1 were detected on the der(22), whereas those of c87f9 were detected on the der(17). Since c68a1 and c87f9 contain the proximal and distal markers flanking low-copy repeat 22B (LCR22B) (Shaikh et al. 1999, 2000), the breakpoint of the patient was localized within LCR22B (fig. 1). Since there is still a 90-kb unclonable contig gap within LCR22B in the draft sequence of the human genome, sequence of the breakpoint region cannot be obtained (Dunham et al. 1999).
Figure 1.
Localization of the 22q11 breakpoint of the patient with t(17;22). A, Map of the 22q11 breakpoint region. LCR-22s are indicated by boxes. Location of probes used for FISH analysis are indicated by the vertical bars. B, FISH results on metaphase chromosomes of the t(17;22) patient. Both c68a1 (N41, red) and c87f9 (ZNF74, green) are hybridized simultaneously to metaphase chromosomes. As a control, cos82 (22q13, red) was used to mark distal 22q. Signals for c87f9 and cos82 are observed on the der(17) (arrow), whereas the c68a1 signal is observed remaining on the der(22) (arrow head). This indicates that the breakpoint is located between these markers in LCR22B. The other chromosome with signal for all three probes is the normal chromosome 22.
The breakpoint of the constitutional t(11;22) has been mapped within LCR22B (Funke et al. 1999; Shaikh et al. 1999), and sequence analysis of t(11;22) junction fragments demonstrated that the 22q11 breakpoint of the t(11;22) is located within a PATRR. Indeed, LCR22B appears to be a hotspot for various translocations involving other autosomes (Kurahashi et al. 2000), suggesting that the PATRR within LCR22B is large and quite unstable. The t(17;22) has also been hypothesized to be mediated by PATRRs on 17q11 and 22q11.
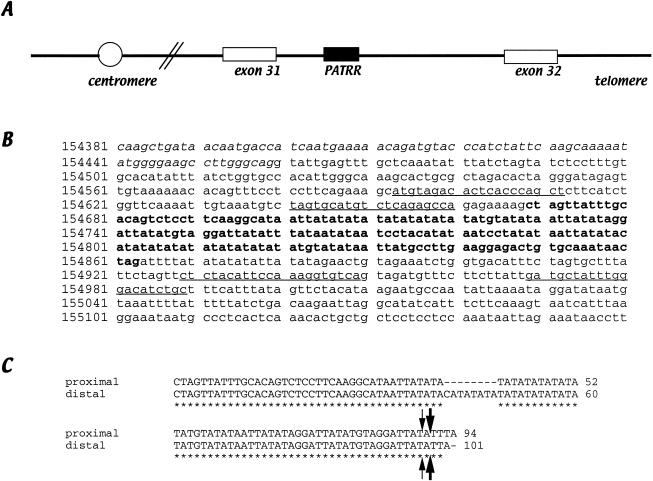
To isolate the chromosome 17–breakpoint region of the t(17;22), the entire genomic sequence of the NF1 gene (GenBank accession number AC004526) was surveyed for PATRRs using the PALINDROME software. Thus, we identified a PATRR within intron 31 of the gene, 209 bp downstream of the end of exon 31 (fig. 2A). The PATRR comprises a nearly perfect palindrome of 195 bp (fig. 2B). Homology between the proximal and distal arms is 97% (fig. 2C). The distal arm has an extra 8 bp in addition to the sequence of the proximal arm, and there is no obvious spacer region at the center of the PATRR. The AT content of the PATRR is as high as 80%. A relatively GC-rich region is located on both ends of the arms of the palindrome, the local GC content of which is 55% in 29 bp. The structure of the PATRR appears quite similar to that of the 11q23 and 22q11 PATRRs.
Figure 2.
The chromosome 17 PATRR. A, The PATRR is located 209 bp downstream of exon 31 within the NF1 gene. B, Genomic sequence of intron 31. Exonic sequence is displayed in italics. The 195-bp PATRR is shown in bold. PCR primers used for this study are underlined. C, Sequence comparison of the proximal and distal arms of the PATRR by ClustalW. Asterisks indicate identical nucleotides between the proximal and distal arms. Thick arrows indicate the breakpoints of the patient with t(17;22), whereas thin arrows indicate that of the patient described by Kehrer-Sawatski et al. (1997).
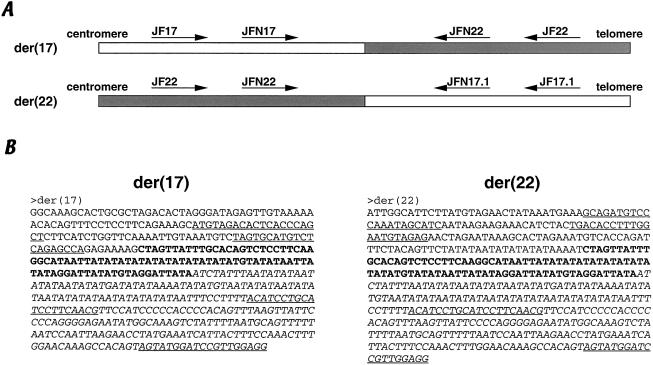
To isolate the junction fragments of the der(17) and the der(22) of the patient with t(17;22), PCR was performed using one primer flanking the chromosome 17 PATRR and another chromosome 22 primer flanking the chromosome 22 PATRR (fig. 3). The latter primer had been used for isolation of t(11;22) junction fragments (Kurahashi et al. 2000). Both the der(17) and the der(22) junction fragments were successfully PCR amplified, indicating that the translocation breakpoints were located within the PATRRs of chromosomes 17 and 22. The breakpoint sequences of the der(17) and the der(22) are nearly identical to each other (fig. 3B), as was the case in almost all instances of the t(11;22). This implies that the translocation occurs at the center of the PATRRs on 17q11 and 22q11, with or without a symmetrical central deletion. Indeed, the chromosome 17 breakpoint is located near the center of the PATRR (fig. 2C). Provided that the original chromosome 17 involved in the t(17;22) had a similar sequence to that in the database, there should be a symmetric 7-bp deletion at the center of the PATRR. The chromosome 22 sequences of the t(17;22) junction fragments were identical to those of the t(11;22) reported elsewhere, suggesting that the same PATRR on 22q11 is involved (Kurahashi and Emanuel 2001a). Since there is no homology between the sequences of the 17q11 and 22q11 breakpoints, including the flanking regions, the PATRR is the only common feature of the breakpoint regions.
Figure 3.
Junction fragments of the t(17;22). A, Structure of the junction fragments of the der(17) and the der(22). White boxes indicate chromosome 17, and gray boxes indicate chromosome 22. PCR primers are indicated by arrows. B, Sequence of the junction fragments. Each sequence is shown from chromosome 17 to chromosome 22. Chromosome-17 PATRR is indicated by boldface, whereas chromosome 22 PATRR is indicated by italics. PCR primers are underlined.
The junction fragment sequence of another NF1-affected patient with a t(17;22) had been reported elsewhere (Kehrer-Sawatski et al. 1997). We have also mapped the breakpoint of Kehrer-Sawatski's patient in the chromosome 17 PATRR. The breakpoint of the patient is one nucleotide distal to the breakpoints of our patient from the center of the PATRR, and a symmetric 9-bp deletion is also observed (fig. 2C). Thus, it is likely that these translocations arose independently by a common mechanism involving the PATRR.
To estimate the frequency of de novo t(17;22)s, translocation-specific PCR (fig. 3A) was performed using sperm samples obtained from four healthy donors, after obtaining informed consent. To increase the sensitivity, nested PCR reactions were performed. PCRs for the der(17) and the der(22) were performed on 44 aliquots of each of four sperm DNA samples. These PCR reactions failed to detect any de novo translocation-specific products (data not shown). Thus, the calculated frequency for de novo t(17;22) rearrangements in male meiosis is <7.0×10-7.
In the present study, we demonstrate that this constitutional t(17;22) is also mediated by similar PATRRs on chromosomes 17 and 22, as proposed by Edelmann et al. (2001). Palindromic DNA is unstable in prokaryotes and eukaryotes, inducing both homologous and illegitimate recombination. It is generally assumed that palindromic DNA forms a cruciform structure that is unstable and could lead to a translocation. The data presented here add support to this proposed mechanism. The mechanism of PATRR-mediated DSB induction is still unknown. Perhaps the cruciform structure blocks progression of the replication fork and generates free ends that must be repaired (Leach 1994). Alternatively, a conformation-specific endonuclease may cleave at the center of the PATRR, although the key enzyme which catalyzes cleavage of the PATRR has not yet been identified. Cruciform structures resemble Holliday junctions, which may be a target for Holliday junction resolvase. Symmetrical deletion at the center of the PATRR and lack of homology between the sequences of the 17q11 and 22q11 breakpoint regions are reminiscent of repair by nonhomologous end joining (NHEJ). Recent study indicates that the Mre11 protein, which is essential for NHEJ, is necessary for repair of palindrome-mediated DSBs (Lobachev et al. 2002).
We demonstrated elsewhere that a long AT-rich region with relatively GC-rich ends is characteristic of the PATRRs on chromosomes 11 and 22 (Kurahashi and Emanuel 2001a). We have proposed that the AT richness of the PATRR contributes to strand separation at physiological temperatures and that the relatively GC-rich end of the PATRR contributes to the stable intrastrand complementary interaction of the PATRR, which may induce formation of a cruciform structure. The chromosome 17 PATRR also has these two characteristics, which may contribute to the genomic instability of the PATRR.
The size of the PATRR can presumably affect its stability, influencing the frequency of the translocation. The 11q23 PATRR is 445 bp long and has a nearly complete palindromic structure (Kurahashi and Emanuel 2001a). In contrast, the chromosome 17 PATRR in the database has a size of 195 bp, about half that of the 11q23 PATRR. Although the PATRR on 22q11 is located within one of the unclonable gaps in the human genome sequence, it is presumed that there is a larger PATRR than that on 11q23 or that there are multiple copies of the PATRR on 22q11. Although the t(11;22) is a somewhat rare condition, the t(17;22) appears to be much less common. The low frequency of the t(17;22) may reflect that PATRR instability is size dependent.
Although the mouse Nf1 gene has a similar exon-intron structure to that of humans, the mouse gene has no PATRR in the corresponding intron (data not shown). This implies that the chromosome-17 PATRR arose recently in the evolution of mammals. On the other hand, the short polymorphic version of the chromosome 11 PATRR appears to have an asymmetric central deletion, which may be derived from a longer version of the PATRR (Kurahashi and Emanuel 2001a). Long palindromic sequences are removed from the genome of transgenic mice with asymmetric insertion or deletion, which stabilizes the palindrome (Collick et al. 1996; Akgun et al. 1997; Lewis 1999). Alu-inverted repeats, which are highly homologous, are substantially underrepresented in the genome relative to direct repeats, as though they also may have been eliminated from the genome (Lobachev et al. 2000). These data suggest that inverted repeats, like these chromosome-11, -17, and -22 PATRRs, are generated but are eliminated within a short time during evolution because of their instability in the genome. Thus, studies of the PATRR might lead to elucidation of the evolutionary dynamics of the genome.
Acknowledgments
The authors thank Dr. D. H. Ledbetter, for patient samples for this study; Sara Dutton Sackett, for laboratory assistance; and Dr. M. Taniguchi, for valuable discussions and careful review of the manuscript. These studies were supported in part by CA39926, DC02027, and HD26979 from the National Institutes of Health and by funds provided by the Charles E. H. Upham Chair in Pediatrics (to B.S.E.).. The studies were also supported, in part, by a grant-in-aid for Scientific Research from the Ministry of Education, Science, Sports and Culture of Japan (to H.K.).
Electronic-Database Information
Accession numbers and URLs for data presented herein are as follows:
- ClustalW, http://www.ebi.ac.uk/clustalw/ (for software for pairwise alignment of sequences)
- GenBank, http://www.ncbi.nlm.nih.gov/GenBank/ (for NF1 genomic sequence [accession number AC004526])
- Online Mendelian Inheritance in Man (OMIM), http://www.ncbi.nlm.nih.gov/Omim/ (for NF1 [MIM 162200]) [PubMed]
- PALINDROME, http://bioweb.pasteur.fr/seqanal/interfaces/palindrome.html (for software for identification of palindromic sequence)
References
- Akgun E, Zahn J, Baumes S, Brown G, Liang F, Romanienko PJ, Lewis S, Jasin M (1997) Palindrome resolution and recombination in the mammalian germ line. Mol Cell Biol 17:5559–5570 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Budarf ML, Eckman B, Michaud D, McDonald T, Gavigan S, Buetow KH, Tatsumura Y, Liu Z, Hilliard C, Driscoll D, Goldmuntz E, Meese E, Zwarthoff EC, Williams S, McDermid H, Dumanski JP, Biegel J, Bell CJ, Emanuel BS (1996) Regional localization of over 300 loci on human chromosome 22 using a somatic cell hybrid mapping panel. Genomics 35:275–288 [DOI] [PubMed] [Google Scholar]
- Collick A, Drew J, Penberth J, Bois P, Luckett J, Scaerou F, Jeffreys A, Reik W (1996) Instability of long inverted repeats within mouse transgenes. EMBO J 15:1163–1171 [PMC free article] [PubMed] [Google Scholar]
- Dunham I, Shimizu N, Roe BA, Chissoe S, Hunt AR, Collins JE, Bruskiewich R, et al (1999) The DNA sequence of human chromosome 22. Nature 402:489–495 [DOI] [PubMed] [Google Scholar]
- Edelmann L, Spiteri E, Koren K, Pulijaal V, Bialer MG, Shanske A, Goldberg R, Morrow BE (2001) AT-rich palindromes mediate the constitutional t(11;22) translocation. Am J Hum Genet 68:1–13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Funke B, Edelmann L, McCain N, Pandita RK, Ferreira J, Merscher S, Zohouri M, Cannizzaro L, Shanske A, Morrow BE (1999) Der(22) syndrome and velo-cardio-facial syndrome/DiGeorge syndrome share a 1.5-Mb region of overlap on chromosome 22q11. Am J Hum Genet 64:747–758 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kehrer-Sawatzki H, Haussler J, Krone W, Bode H, Jenne DE, Mehnert KU, Tummers U, Assum G (1997) The second case of a t(17;22) in a family with neurofibromatosis type 1: sequence analysis of the breakpoint regions. Hum Genet 99:237–247 [DOI] [PubMed] [Google Scholar]
- Kurahashi H, Emanuel BS (2001a) Long AT-rich palindromes and the constitutional t(11;22) breakpoint. Hum Mol Genet 10:2605–2617 [DOI] [PubMed] [Google Scholar]
- ——— (2001b) Unexpectedly high rate of de novo constitutional t(11;22) translocations in sperm from normal males. Nat Genet 29:139–140 [DOI] [PubMed] [Google Scholar]
- Kurahashi H, Shaikh TH, Hu P, Roe BA, Emanuel BS, Budarf ML (2000) Regions of genomic instability on 22q11 and 11q23 as the etiology for the recurrent constitutional t(11;22). Hum Mol Genet 9:1665–1670 [DOI] [PubMed] [Google Scholar]
- Leach DR (1994) Long DNA palindromes, cruciform structures, genetic instability and secondary structure repair. Bioessays 16:893–900 [DOI] [PubMed] [Google Scholar]
- Ledbetter DH, Rich DC, O'Connell P, Leppert M, Carey JC (1989) Precise localization of NF1 to 17q11.2 by balanced translocation. Am J Hum Genet 44:20–24 [PMC free article] [PubMed] [Google Scholar]
- Lewis SM (1999) Palindromy is eliminated through a structure-specific recombination process in rodent cells. Nucleic Acids Res 27:2521–2528 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lobachev KS, Gordenin DA, Resnick MA (2002) The Mre11 complex is required for repair of hairpin-capped double-strand breaks and prevention of chromosome rearrangements. Cell 108:183–193 [DOI] [PubMed] [Google Scholar]
- Lobachev KS, Stenger JE, Kozyreva OG, Jurka J, Gordenin DA, Resnick MA (2000) Inverted Alu repeats unstable in yeast are excluded from the human genome. EMBO J 19:3822–3830 [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Connell P, Leach RJ, Ledbetter DH, Cawthon RM, Culver M, Eldridge JR, Frej AK, Holm TR, Wolff E, Thayer MJ, Schafer AJ, Fountain JW, Wallace MR, Collins FS, Skolnick MH, Rich DC, Fournier REK, Baty BJ, Carey JC, Leppert MF, Lathrop GM, Lalouel JM, White R (1989) Fine structure DNA mapping studies of the chromosomal region harboring the genetic defect in neurofibromatosis type I. Am J Hum Genet 44:51–57 [PMC free article] [PubMed] [Google Scholar]
- Shaikh TH, Budarf ML, Celle L, Zackai EH, Emanuel BS (1999) Clustered 11q23 and 22q11 breakpoints and 3:1 meiotic malsegregation in multiple unrelated t(11;22) families. Am J Hum Genet 65:1595–1607 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shaikh TH, Kurahashi H, Saitta SC, O'Hare AM, Hu P, Roe BA, Driscoll DA, McDonald-McGinn DM, Zackai EH, Budarf ML, Emanuel BS (2000) Chromosome 22-specific low copy repeats and the 22q11.2 deletion syndrome: genomic organization and deletion endpoint analysis. Hum Mol Genet 9:489–501 [DOI] [PubMed] [Google Scholar]
- Tapia-Paez I, Kost-Alimova M, Hu P, Roe BA, Blennow E, Fedorova L, Imreh S, Dumanski JP (2001) The position of t(11;22)(q23;q11) constitutional translocation breakpoint is conserved among its carriers. Hum Genet 109:167–177 [DOI] [PubMed] [Google Scholar]
- Viskochil D, Buchberg AM, Xu G, Cawthon RM, Stevens J, Wolff RK, Culver M, Carey JC, Copeland NG, Jenkins NA, White R, O'Connell P (1990) Deletions and a translocation interrupt a cloned gene at the neurofibromatosis type 1 locus. Cell 62:187–192 [DOI] [PubMed] [Google Scholar]
- Wallace MR, Marchuk DA, Andersen LB, Letcher R, Odeh HM, Saulino AM, Fountain JW, Brereton A, Nicholson J, Mitchell AL, Brownstein BH, Collins FS (1990) Type 1 neurofibromatosis gene: identification of a large transcript disrupted in three NF1 patients. Science 249:181–186 [DOI] [PubMed] [Google Scholar]