Abstract
To investigate the potential involvement of genome architecture in nonrecurrent chromosome rearrangements, we analyzed the breakpoints of eight translocations and 18 unusual-sized deletions involving human proximal 17p. Surprisingly, we found that many deletion breakpoints occurred in low-copy repeats (LCRs); 13 were associated with novel large LCR17p structures, and 2 mapped within an LCR sequence (middle SMS-REP) within the Smith-Magenis syndrome (SMS) common deletion. Three translocation breakpoints involving 17p11 were found to be located within the centromeric α-satellite sequence D17Z1, three within a pericentromeric segment, and one at the distal SMS-REP. Remarkably, our analysis reveals that LCRs constitute >23% of the analyzed genome sequence in proximal 17p—an experimental observation two- to fourfold higher than predictions based on virtual analysis of the genome. Our data demonstrate that higher-order genomic architecture involving LCRs plays a significant role not only in recurrent chromosome rearrangements but also in translocations and unusual-sized deletions involving 17p.
Introduction
The molecular bases of recurrent interstitial chromosomal deletions and duplications have been uncovered only recently. Most of these rearrangements result from meiotic homologous recombination between nonallelic copies of low-copy repeats (LCRs). The involvement of genome architectural features in susceptibility to rearrangements resulting in disease traits appears to be a general phenomenon. These conditions have been referred to as “genomic disorders” (Lupski 1998, 2003). The number of recognized genomic disorders continues to rise (Emanuel and Shaikh 2001; Inoue and Lupski 2002; Stankiewicz and Lupski 2002a;) simultaneously with the discovery, by computational analysis, that LCRs (>10 kb; >95%–97% identity) comprise 5%–10% of the human genomic sequence (Bailey et al. 2001). Thus, the human genome is laden with clusters of LCRs, most of which appear to have evolved during primate speciation (Stankiewicz and Lupski 2002b), making it likely that genome rearrangements will continue to be recognized as playing a major role in human disease and potentially in the evolution of the human species.
We were interested in investigating the molecular basis of nonrecurrent chromosome rearrangements, such as translocations and unusual-sized deletions, that occurred in genomic regions in which recurrent rearrangements have been identified previously. To elucidate the molecular mechanisms, we analyzed extensively the rearrangement breakpoints. Because of a wealth of information with regard to the complete genome sequence (International Human Genome Sequencing Consortium [IHGSC] 2001; Venter et al. 2001), structure of LCRs, and knowledge of higher-order genome architecture (Bi et al. 2002; Inoue and Lupski 2002; Park et al. 2002; Stankiewicz and Lupski 2002a), we focused on breakpoints within proximal 17p in patients with nonrecurrent rearrangements.
The gene-rich and highly unstable human genomic region 17p11.2-p12 has been found to be rearranged in a variety of different structural chromosome aberrations. The same ∼1.4-Mb genomic fragment within chromosome 17p12 is duplicated and deleted, respectively, in patients with Charcot-Marie-Tooth type 1A disease (CMT1A) and in patients with hereditary neuropathy with liability to pressure palsies (HNPP) (Chance et al. 1994; Reiter et al. 1996). This genomic segment is flanked by two ∼24-kb LCRs, termed the “proximal CMT1A-REP” and the “distal CMT1A-REP” (fig. 1), which serve as substrates for nonallelic homologous recombination (NAHR) (Pentao et al. 1992; Reiter et al. 1997). The same LCRs/NAHR-based mechanism results in del(17)(p11.2p11.2), causing Smith-Magenis syndrome (SMS [MIM 182290]) and the newly described duplication dup(17)(p11.2p11.2) syndrome (Chen et al. 1997; Potocki et al. 2000). On the basis of the identification of recurrent junction fragments by pulsed-field gel electrophoresis (PFGE), these genomic disorders were shown to be caused by the reciprocal deletion and duplication, respectively, of the same ∼4-Mb genomic region in chromosome 17p11.2 in 80%–90% of patients. The rearranged segment is flanked by the proximal SMS-REP (∼256 kb) and the distal SMS-REP (∼176 kb) LCRs (Chen et al. 1997; Potocki et al. 2000; Park et al. 2002); a third LCR copy, the middle SMS-REP (∼241 kb), maps between them and is inverted in orientation (fig. 1) (Park et al. 2002).
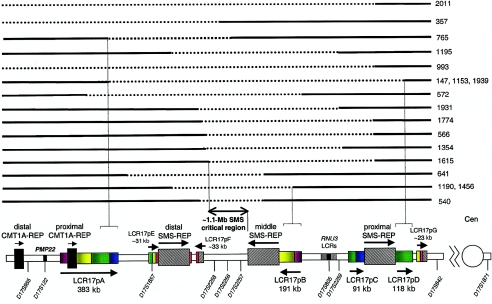
Figure 1.
Breakpoint analysis of unusual-sized deletions. Proximal chromosome 17p is depicted at the bottom, showing the size, position, and orientation of LCRs. Dashed horizontal lines represent the genomic segment deleted for 18 different patients, and solid horizontal lines depict the retained genomic material, with the patient number shown to the right. The LCR17p structures are depicted in colors, to better represent their homology and orientation with respect to each other; the closed arrowheads represent the orientation of the LCR17p subunits. Selected breakpoints involving the LCR17ps are shown as vertical dashed lines. The horizontal line flanked by open arrowheads (below the genomic segments) depicts the SMS critical region; the common deletion (80%–90% of patients with SMS) occurs between proximal and distal SMS-REP copies. Note that the distal deletion breakpoints in patients 357, 993, and 2011 map outside the analyzed genomic region and thus were not included in the calculation of the percentage of chromosome breakpoints associated with LCRs in proximal 17p. Only >20-kb LCRs are depicted. The map is not to scale.
In addition to the CMT1A-REP and SMS-REP LCRs, we recently described a novel large LCR family termed “LCR17p” (Park et al. 2002). These LCRs are localized in 17p11.2-p12 around the proximal CMT1A-REP (∼383-kb LCR17pA), adjacent to the middle SMS-REP on the centromeric side (∼191-kb LCR17pB), and flanking the proximal SMS-REP (∼91-kb LCR17pC and ∼118-kb LCR17pD) (fig. 1). An ancestral genomic interval syntenic to the LCR17pA has been shown to be involved in the origin of an evolutionary chromosome translocation t(4;19) in Gorilla gorilla (Stankiewicz et al. 2001).
The majority of the chromosome aberrations reported (Brewer et al. 1998, 1999) appear to have random breakpoints, whereas, for recurrent interstitial deletions and reciprocal duplications, the breakpoints are associated with particular genomic architectural features (e.g., LCRs, AT-rich palindromes, and fragile sites) that mediate the recurrence of the aberrations (Lupski 1998; Shaffer and Lupski 2000; Emanuel and Shaikh 2001; Richards 2001; Inoue and Lupski 2002; Stankiewicz and Lupski 2002a). To investigate whether nonrecurrent chromosomal rearrangements in proximal 17p represent random events or reflect susceptibilities to DNA rearrangements due to genome architecture, we analyzed the breakpoints of 18 unusual-sized deletions and eight balanced and unbalanced chromosome translocations involving proximal 17p.
Material and Methods
Chromosome Rearrangements
On the basis of the absence of the ∼1.1-Mb junction fragment in PFGE analysis (Chen et al. 1997), we selected 16 patients with SMS who have unusual-sized deletions (patients 147, 540, 566, 572, 641, 993, 1153, 1190, 1195, 1354, 1456, 1615, 1774, 1931, 1939, and 2011) and two patients without the major features of SMS with deletions involving the SMS-common deletion chromosome region (patients 357 and 765) for this study. In addition, eight cell lines from patients with balanced or unbalanced chromosome translocations involving 17p11 were studied (table 1). Peripheral blood samples from patients and family members were obtained after informed consent.
Table 1.
Summary of Findings in Patients with Chromosome Translocations Involving Proximal 17p
|
Breakpoints |
||||
| Patient | Karyotype | Chromosome 17 | Partner Chromosome | Clinical Information |
| GM03119a | 46,XX,t(9;17)(p22;q11.1) | Within D17Z1 in 17q11.1 (fig.4E) | Not mapped | A clinically normal woman with history of several spontaneous abortions |
| GM02836a | 46,XY,t(9;17)(q12;p11)[61%]/46,XY,t(9;17)(q12;p11),ins(3;1)(q21;q25q44)[39%] | Between RP11-728E14 and D17Z1 (fig. 4D) | Not mapped | A clinically normal individual |
| TIC90 | 46,XX,t(9;17)(q34.1;p11.2)mat, del(9)(q22.32q33.2) | Within two overlapping BAC clones: CTD-2354J3 and RP11-311F12 | Between RP11-489N22 and RP11-88G17 | An 11-year-old girl with Gorlin syndrome and features of nail-patella syndrome |
| UK | 46,XY,t(1;17)(p36.3;p11.2) | Distal-most 1/3 portion of clone RP11-344E13 | Within two overlapping PAC clones RP1-453P22 and RP1-505B13 | A family in which nonsyndromic mental retardation and an apparently balanced reciprocal translocation segregated in eight individuals over three generations (Hussain et al. 2000) |
| 1071 | 46,XX,t(X;17)(p22.3;p11.2) | Between RP11-728E14 and D17Z1 | Distal to the most subtelomeric BAC clone RP11-215A12 | A clinically normal 2-year-old girl, in whom balanced chromosome abnormality was found prenatally during amniocentesis (advanced maternal age) |
| 1183 | 46,XY,t(2;17)(p25.3;p11.1) | Within distal-most ∼1/4 portion of D17Z1 in 17p11.1 | Within two overlapping BAC clones RP11-455M16 and RP11-163G21 | A 9-year-old boy with mental retardation, in whom the balanced chromosome abnormality was found prenatally (advanced maternal age); a diagnosis of SMS has been excluded |
| 1307 | 46,XY/46,XY,der(X)t(X;17)(p22.1;p11.1), ∼50% mosaic | Within the middle of D17Z1 | Not mapped | A patient with clinical and electrophysiological features of the CMT1A, in whom an extra PMP22 gene resulted from a rare unbalanced translocation of 17p to the X chromosome (King et al. 1998) |
| 1576 | 46,XY,der(17)t(10;17)(q26.3;p11.2) | Within PAC clone RP1-48J12, at the centromeric end of the distal SMS-REP (fig 4F, 7) | Within overlapping BAC clones RP13-137A17 and RP13-439H18 | A 5.5-year-old boy with the features of partial trisomy 17p and monosomy 10q26.3qter, including CMT1A |
The skin fibroblast cell lines in patients GM03119 and GM02836 were purchased from Coriell Cell Repositories.
Chromosome Breakpoint Mapping
The BAC and PAC clones used for the chromosome breakpoint mapping were identified on the physical maps of the regions of interest (Inoue et al. 2001; Bi et al. 2002; National Center for Biotechnology Information Home Page; UCSC Genome Bioinformatics Web site). Interspersed repeat sequences within the downloaded DNA sequence of the clones were eliminated by RepeatMasker (RepeatMasker Web Server) and were analyzed using Sequencher (Gene Codes) and NCBI BLAST (NCBI BLAST Home Page). The BAC/PAC clones were purchased from the BACPAC Resources Center and Research Genetics, and DNA was prepared from each through use of the PSI Clone BAC DNA kit (Princeton Separations) according to the manufacturer’s instructions.
FISH
FISH was performed on metaphase and interphase cells of peripheral blood lymphocytes, Epstein-Barr virus–transformed peripheral blood lymphoblasts, and skin fibroblasts, as described by Shaffer et al. (1997). For chromosome 17 centromere identification, a directly labeled SpectrumGreen centromeric probe, D17Z1 (Vysis), was used.
FISH Screening for the SMS Common Deletions
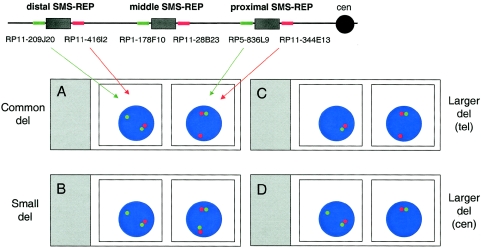
A FISH assay with probes flanking the SMS-REPs was developed. For probes, we used proximal and distal SMS-REP–flanking BAC/PAC clones: RP11-344E13 or RP11-98L14 (adjacent to the proximal SMS-REP on the centromeric side), RP5-836L9 (adjacent to the proximal SMS-REP on the telomeric side), RP11-416I2 (centromeric to the distal SMS-REP), and RP11-209J20 (telomeric and adjacent to the distal SMS-REP) (fig. 2). Clones flanking proximal SMS-REP were used in FISH analysis, and those flanking distal SMS-REP were used concurrently in a separate chamber on the same slide (fig. 2). When the smaller-sized deletion was identified, the middle SMS-REP–flanking BAC/PAC clones RP11-28B23 (centromeric to the middle SMS-REP) and RP1-178F10 (telomeric to the middle SMS-REP) were cohybridized, using FISH in interphase cells, to determine whether the middle SMS-REP was involved.
Figure 2.
Schematic representation of a dual-color interphase FISH assay developed to screen for common, A, versus unusual-sized SMS deletions. The map of chromosome 17p11.2 with the placement of the FISH probes for one chromosome homologue is shown at the top of the figure. The proximal SMS-REP–flanking clones BAC RP11-344E13 and PAC RP5-836L9 and the distal SMS-REP flanking BACs RP11-416I2 and RP11-209J20 are differentially labeled and are detected with red and green colors, respectively. Below the chromosome map, in the left chamber of the slide, two adjacent green and red dots represent the normal chromosome 17, and the presence of a single green signal demonstrates that the deletion breakpoint occurred between clones RP11-209J20 and RP11-416I2, within the distal SMS-REP. Similarly, the absence of the second green signal on the right side indicates that the breakpoint maps between clones RP5-836L9 and RP11-344E13. The red and green signals flanking SMS-REPs do not overlap, because the distance between the clones is greater than the ∼100-kb resolution limit of interphase FISH. The three other hypothetical microscope slides give examples of the FISH results obtained with the same clones. B, A small deletion with the telomeric breakpoint mapping within the distal SMS-REP and the centromeric breakpoint mapping between the proximal and distal SMS-REPs. C, A large deletion with the distal breakpoint mapping telomeric to the distal SMS-REP and the centromeric breakpoint mapping within the proximal SMS-REP. D, A large deletion with the telomeric breakpoint mapping within the distal SMS-REP and the proximal breakpoint mapping centromeric to the proximal SMS-REP.
Genotyping
We determined both (1) the parental origin of the rearranged chromosomes and (2) distinguished inter- and intrachromosomal recombination mechanisms resulting in the deletion, using a combination of microsatellite haplotype reconstruction and the segregation of marker genotypes, on genomic DNA purified from peripheral blood (Gentra), as described by Shaw et al. (2002). Phases of parental haplotypes were defined on the basis of the most parsimonious explanation for observed genotypes in the siblings and under the assumption of no recombination.
Long-Range PCR
For the long-range PCR of the KER cluster, the following ⩾30-bp primers of 50% GC content and melting temperature 65–70°C were designed: F (CCGTGACTACAGCCAGTACTACAGGATAATCG) and R (CTCTGCAGTCTCCAGGACATAGATTTGCTC). The reaction was performed using the TaKaRa LA PCR Kit (Takara Shuzo), following the manufacturer’s recommendations. Initial denaturation at 94°C for 15 min was followed by 40 cycles of denaturation at 98°C for 20 s, extension at 68°C for 10 min, and a final extension at 72°C for 10 min. The 4,134-bp product was extracted from the 1% agarose gel through use of a Gel Extraction Kit (Qiagen).
Somatic Cell Hybrids
We performed polyethylene glycol fusion between the lymphoblastoid cell line from patients 765 (Elsea et al. 1997) and 1153 and from a thymidine kinase–deficient (TK−) hamster cell line, A23 (Chen et al. 1997). For this fusion, 24 independent clones were isolated with cloning rings and were transferred to a 24-well microtiter plate. We obtained cells representing each clone by trypsinization of a confluent well of a 24-well plate, and we then transferred them to a 6-well plate and then to T25 flasks. Two-color FISH with probes mapping within and outside the deleted region was used to analyze the hybrids and identify those with the 17p11.2 deletion chromosome.
PFGE Analysis
High-molecular-weight DNA was isolated in agarose plugs from peripheral blood samples, somatic cell hybrid cell lines, and lymphoblastoid cell lines established from patients and controls (Pentao et al. 1992). For Southern analysis, we used the 1.1-kb HindIII fragment from the cDNA clone 41G7A, which contains the 3′ end of the coding region and part of the 3′ UTR, as the CLP probe; and we used the PCR product (F: ATGTCGGTTTGGGTGTTTGT; R: TTAAGCACTTGGCTCAAGCA) of the PRPSAP2 gene, localized adjacent to the middle SMS-REP on the centromeric side within the BAC clone RP11-28B23, as the PRPSAP2 probe.
DNA Sequence Analysis
The search for additional LCRs was performed using NCBI BLAST analysis against the high-throughput and the nonredundant sequence database, and the sequence was assembled using NCBI BLAST 2 and the Sequencher software (Gene Codes).
Results
A Novel FISH Assay for Distinguishing the Common versus Unusual SMS Deletions
To replace technically challenging and time-consuming PFGE-based screening for the SMS common versus nonrecurrent deletions, we developed a novel FISH assay (fig. 2). Using dual-color interphase FISH with proximal SMS-REP–flanking BAC/PAC clones RP11-344E13 and RP5-836L9 and concurrently using the distal SMS-REP flanking BAC clones RP11-416I2 and RP11-209J20 (fig. 2), we were able to determine simultaneously whether the SMS deletion was of an unusual size and whether it was smaller or larger than the common deletion. The SMS-REPs are ∼200 kb in size and thus larger than the ∼100-kb resolution limit of interphase FISH. Therefore, cohybridized SMS-REP–flanking clones can be visualized as distinct signals by FISH. Direct FISH on uncultured cells can be used, because this approach does not require metaphase chromosomes, thus enabling rapid analyses. Compared with the PFGE analysis (Chen et al. 1997), this novel FISH approach is an easier, faster, much less expensive, and at least equally reliable method for screening for common SMS deletions.
Of note, two of the SMS-REP–flanking BAC clones, RP11-416I2 and RP11-344E13, contain the 33-kb and 23-kb fragments homologous to SMS-REP; however, the size of these segmental duplications did not affect the interpretation of the FISH studies. If a smaller-sized deletion is identified, the middle SMS-REP–flanking BAC clone RP11-28B23 (centromeric to the middle SMS-REP), together with the PAC RP1-178F10 (telomeric to the middle SMS-REP) (fig. 2), can be cohybridized to determine whether the middle SMS-REP is involved in the deletion. Moreover, the same FISH approach can be used for screening of the common duplication, dup(17)(p11.2p11.2) (Potocki et al. 2000). In addition to identifying the common duplication, it will reveal whether the fragment is direct or inverted in orientation.
Uncommon Deletion Breakpoints Map within LCRs
To investigate the recombination products for the unusual-sized deletions, we mapped both breakpoints for each. Surprisingly, we found that 13 of the 14 deletion breakpoints (in patients 147, 572, 765, 1153, 1190, 1456, and 1939) map within the recently identified LCRs: LCR17pA, LCR17pB, and LCR17pD (figs. 1 and 3). Moreover, FISH and PFGE results showed that eight deletions have one breakpoint mapping within proximal (patients 566, 993, 1615, 1774, and 2011), middle (patients 540 and 641), or distal (patient 1195) SMS-REPs (Bi et al. 2002) (figs. 1 and 3). The remaining three unusual deletions (patients 357, 1354, and 1931) do not have a breakpoint in any recognizable LCRs (figs. 1 and 3). These findings indicate that most (21/33) breakpoints of unusual-sized deletions in 17p11.2 may in fact be mediated by genome architectural features such as LCRs.
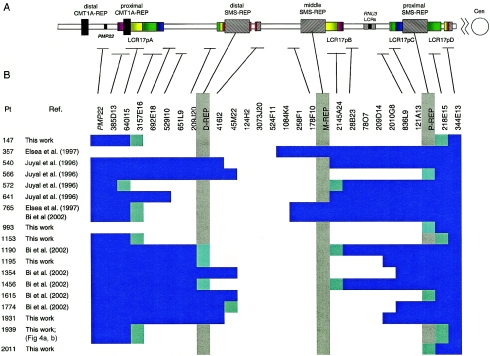
Figure 3.
Summary of deletion breakpoint mapping. A, Schematic representation of LCRs within 17p11.2-p12, with horizontal lines attached to the table showing the position of individual BAC/PAC clones. B, A table with the clones used in FISH studies. PAC/BAC clones that gave positive FISH signals are represented by filled blue bars, and white spaces depict BAC/PAC clones that did not give a hybridization signal from the deleted chromosome (however, each individual clone was not assayed by FISH). Clones, within which the breakpoint was mapped, are depicted by half blue-green shading. The gray vertical shading indicates the three SMS-REPs. Selected clones used for the FISH analysis of uncommon deletions are taken from the complete BAC/PAC contig (Bi et al. 2002) and are labeled in the upper row of the table. Our analysis of the currently available databases reveals that clones RP11-416I2 and RP11-45M22 appear not to overlap and are spanned by the BAC clone RP11-367G9.
Three patients with larger-sized deletions (patients 147, 1153, and 1939) have distal breakpoints within LCR17pA and proximal breakpoints within LCR17pD (figs. 1 and 3). In these patients, we mapped the proximal breakpoint centromeric to the proximal SMS-REP, using FISH (figs. 1, 3, and 4A), and we mapped the distal breakpoint telomeric to the distal SMS-REP (figs. 1, 3, and 4B). PCR on the somatic cell hybrid from patient 1153 indicated that the distal breakpoint occurred within BAC CTD-3157E16, and the proximal breakpoint within BAC RP11-218E15, at the centromeric ends of LCR17pA and LCR17pD, respectively (data not shown). BLAST analysis of the LCRs revealed that LCR17pA, LCR17pC, and LCR17pD are oriented in the same direction, whereas LCR17pB is inverted with respect to the other LCR17p copies. These data suggest that LCR17pA, together with directly repeated LCR17pD, may serve as substrates for NAHR, thus potentially explaining the apparent clustering, or recurrence, of deletion breakpoints in patients 147, 1153, and 1939.
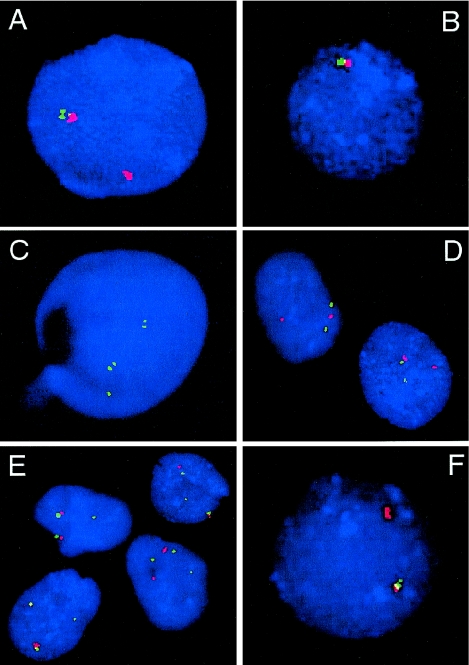
Figure 4.
FISH analyses of interphase nuclei used to map the rearrangement breakpoints. A and B, Interphase nuclei of patient 1939 after FISH with SMS-REPs–flanking clones (fig. 2). In A, the absence of the green signal (RP5-836L9) and presence of the red signal (RP11-344E13) on the del(17) indicates that the proximal breakpoint maps within the proximal SMS-REP (or directly adjacent LCR17pB). In B, the absence of both red (RP11-416I2) and green (RP11-209J20) signals on the del(17) shows that the distal breakpoint maps telomeric to the distal SMS-REP. C, FISH with BAC clone CIT-3157E16 (LCR17pA), enabling mapping the distal breakpoint of the deletion in patient 572 to the distal portion of LCR17pA. The two closely spaced green signals on the normal and deleted chromosomes 17 represent LCR17pC and LCR17pD copies. A single hybridization signal corresponds to the LCR17pA copy on the normal chromosome 17; the LCR17pA on der(17) is deleted. D, FISH with the BAC RP11-344E13 (red) and a centromeric probe (green) on cells from patient GM02836, showing the breakpoint mapped between them. On the normal chromosome 17, the red and green signals are relatively close to each other, whereas the separation of the red and green signals indicates that they are on different chromosomes, der(9) and der(17), respectively. E, The centromeric breakpoint on chromosome 17 in the patient GM03119, identified after the cohybridization of BAC RP11-344E13 (red) and the centromeric probe D17Z1 (green). In addition to the adjacent red and green pair of signals on both chromosomes 17 and the der(9), the single green signal on the der(17) is of weaker intensity when compared with the other two green signals, indicating the localization of this breakpoint to the q11.1 portion of the chromosome 17 centromere. Note the variability of distances between RP11-344E13 (red) and the D17Z1 centromeric probe (green) in D and E, demonstrating different condensation of the pericentromeric heterochromatin. F, FISH with the distal SMS-REP flanking BAC clones RP11-209J20 (green) and RP11-416I2 (red) on an interphase nucleus of patient 1576, a carrier of an unbalanced translocation. The presence of only the red signal on the der(17) chromosome indicates that the breakpoint maps between these two clones.
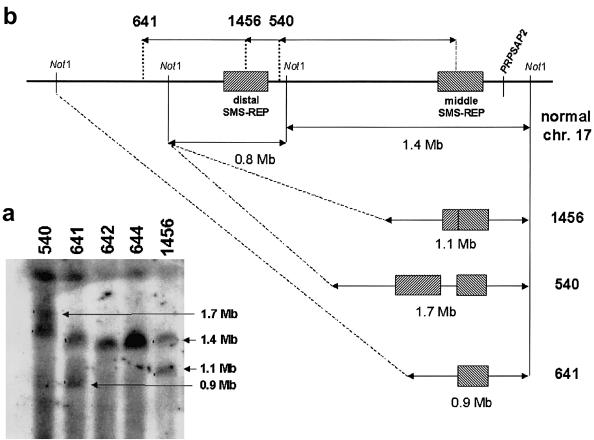
The distal deletion breakpoint in patient 572 mapped within LCR17pA (fig. 4C), and the proximal deletion breakpoint mapped within the inverted LCR17pB copy. Two proximal deletion breakpoints (in patients 540 and 641) involved the middle SMS-REP (which is inverted with respect to other SMS-REPs) (figs. 1 and 3). In patients 1190 and 1456, the proximal breakpoints mapped within the LCR17pB copy, and the distal breakpoints involved the distal SMS-REP (Park et al. 2002). In support of this contention, PFGE experiments and Southern analysis in patients 540, 641, and 1456—through use of PRPSAP2, which maps centromeric and adjacent to the middle SMS-REP and LCR17pB, as a probe—identified junction fragments of 1.7-Mb, 0.9-Mb, and 1.1-Mb, respectively, in addition to the normal 1.4-Mb DNA fragment (fig. 5).
Figure 5.
A, PFGE detection of novel junction fragments in three patients with SMS (patients 540, 641, and 1456) with smaller-sized deletions in which the proximal breakpoint maps to middle SMS-REP. The Southern blot was hybridized with a PRPSAP2 PCR probe, which, in addition to the normal 1.4-Mb NotI fragment (control patients 642 and 644), identified the 0.9-, 1.1-, and 1.7-Mb junction fragments (arrows) spanning the proximal breakpoints within middle SMS-REP or LCR17pB. B, Schematic diagram represents the derivation of novel PFGE junction fragments in patients with SMS with uncommon deletions.
Six deletions have one breakpoint involving the proximal or distal SMS-REPs (in patients 566, 993, 1195, 1615, 1774, and 2011), and the other breakpoint appears to map within unique sequence. However, FISH analysis demonstrates the clustering of the distal breakpoints within one BAC clone in patients 566, 1354, 1774, and 1931 (RP11-45M22) and the clustering of proximal breakpoints in patients 1195 and 1931 (CTD-2010G8), suggesting a potential genome architectural feature stimulating these rearrangements, although analyses of the genomic sequence available at the time of writing (November 2002) failed to identify an obvious LCR or other higher-order sequence structure (figs. 1 and 3).
PCR analysis of a somatic cell hybrid retaining the deleted chromosome from patient 765 showed that the proximal breakpoint maps to a 100-bp interval (nucleotide position 87925–88024 bp) in the finished sequence BAC clone RP11-258F1. The precise mapping of the distal breakpoint within BAC CIT-3157E16 (LCR17pA) was hampered by the presence of the highly homologous and nondeleted LCR17pB sequences in this somatic cell hybrid (figs. 1 and 3). Since this patient did not manifest the typical features of SMS, the mapping of this breakpoint potentially narrowed the SMS critical region to 210 kb (Bi et al. 2002).
Unequal Crossing-Over as a Mechanism for Uncommon Deletions
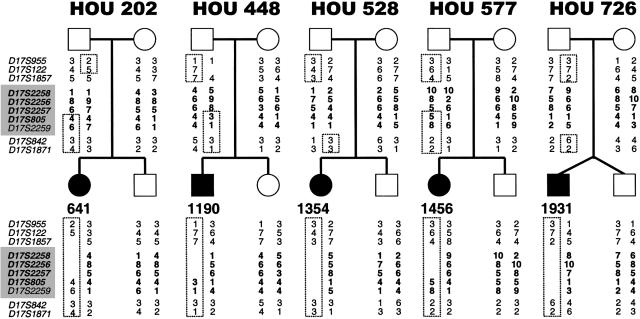
Recently, the common ∼4-Mb SMS deletion has been shown to occur via unequal meiotic crossing-over between the proximal and distal SMS-REP copies (Shaw et al. 2002). However, it is unknown whether deletions with uncommon breakpoints have arisen through the same mechanism. To investigate the genetic mechanism for generating uncommon deletions, we used microsatellite markers in patients and family members to reconstruct the haplotypes of five patients with SMS with unusual-sized deletions (patients 641, 1190, 1354, 1456, and 1931). Surprisingly, three deletions (in patients 641, 1190, and 1354) result from unequal interchromosomal recombination occurring between the genetic markers flanking each breakpoint (fig. 6).
Figure 6.
Haplotypes of five patients with unusual deletions and their families. Standard pedigree symbols are used; a circle denotes a female, a square denotes a male. Blackened circles or squares indicate an affected individual. To the left of each pedigree is a list of microsatellite markers used for genotyping; those within the SMS common deletion region are bold and shaded. The allele numbers are located under each family member. The genotypes of markers within the SMS common deletion region are bold in the patients and the parent of origin. The dotted lines outline alleles inherited by the patient from the parent of origin. In patient 641, recombination occurred between the region flanked by loci D17S122 and D17S1857 and the region between D17S2257 and D17S805 (including the middle SMS-REP), resulting in the deletion. In patient 1190, recombination between the region flanked by markers D17S1857 and D17S2258 (including the distal SMS-REP) and the region flanked by markers D17S2257 and D17S805 (including the middle SMS-REP) resulted in the deletion. Recombination between the region flanked by loci D17S1857 and D17S2258 and the region between D17S2259 and D17S842 (including the proximal SMS-REP) resulted in the deletion in patient 1354. Patients 1456 and 1931 may have deletions resulting from intrachromosomal recombination. (Both of these patients had crossovers on their intact, maternally derived chromosomes 17 [between loci D17S842 and D17S1871 for patient 1456 and between loci D17S955 and D17S122 for patient 1931]). Interestingly, each of the five deletions are paternally derived, as evidenced by the lack of a paternal allele for loci D17S1857, D17S2258, D17S2256, and D17S2257 for patient 641; loci D17S2258, D17S2256, and D17S2257 for patients 1190 and 1456; and loci D17S2258, D17S2256, D17S2257, D17S805, and D17S2259 for patients 1354 and 1931. The locations of markers used in genotyping are shown in figure 1.
The deletions in patients 1190 and 1456 have the same breakpoints within the LCRs, distal SMS-REP, and LCR17pB. It is possible that, similar to the deletion in patient 1190, the deletion in patient 1456 may also result from an unequal crossing-over event but between chromatids and not chromosomes (i.e., it is intrachromosomal). However, because SMS-REP and LCR17pB blocks are not homologous to each other, the unequal crossing-over could result from either minor homology regions (e.g., repetitive sequences within the LCRs) or nonhomologous end-joining (NHEJ). Interestingly, such LCR-stimulated NHEJ events have been described in the very rare group of patients with Pelizaeus-Merzbacher disease, resulting from chromosome deletions (and not duplications) involving genomic segments containing the causative gene PLP1 (Inoue et al. 2002). The same end-joining mechanism following unequal crossing-over events between LCR-free unique-sequence DNA fragments could be responsible for the origin of smaller-sized deletions in patient 1354 (interchromosomal) and possibly 1931 (intrachromosomal).
Interestingly, of 14 analyzed cases, 12 deletions (in patients 572, 641, 1153, 1190, 1195, 1354, 1456, 1615, 1774, 1931, 1939, and 2011) were of paternal origin and two (in patients 540 and 993) were maternal in origin (P⩽.013).
Nonrecurrent Translocation Breakpoints Cluster at the Chromosome 17 Centromere
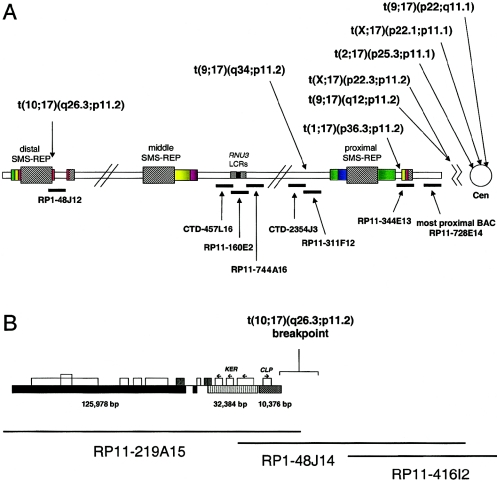
To determine whether nonrecurrent chromosome translocation breakpoints are associated with genome architecture and potential susceptibility to breakage, we investigated the breakpoints of eight nonrecurrent, reciprocal chromosome translocations (table 1). Unexpectedly, we found six breakpoints clustered between the proximal SMS-REP and the chromosome 17 centromeric α-satellite sequence D17Z1; one between proximal and middle SMS-REPs within two overlapping BAC clones, CTD-2354J3 and RP11-311F12; and one at the centromeric end of the distal SMS-REP (fig. 7; table 1). Using BLAST, the Golden Path physical map, and interphase FISH analyses, we estimated the size of this pericentromeric segment as <1 Mb and the distance between the most proximal BAC clone RP11-728E14 and the centromere as <0.5 Mb (fig. 7). Of note, using dual color interphase FISH with the BAC clone RP11-344E and the α-satellite centromeric probe D17Z1, we observed a large variability in the distance between the signals, indicating significant differences in the DNA condensation of this region or polymorphic variation in pericentromeric/centromeric sequence size (fig. 4D and 4E). Interestingly, four of eight partner chromosome breakpoints mapped within the most telomeric subbands: 1p36.3, 2p25.3, 10q26.3, and Xp22.3.
Figure 7.
A, Schematic diagram of identified translocation breakpoints within proximal 17p. FISH experiments on cells from patients harboring translocations with breakpoints in proximal 17p (table 1) showed that five of eight analyzed breakpoints cluster centromeric to the most proximal BAC clone RP11-728E14: one within clone RP11-344E13, one within two overlapping BAC clones (CTD-2354J3 and RP11-311F12), and one at the centromeric end of the distal SMS-REP, within the PAC clone RP1-48J12. B, Schematic representation of the chromosome 17 translocation breakpoint in patient 1576. The breakpoint was mapped between the BAC clones RP11-416I2 and RP11-209J20, indicating that it occurred within or adjacent to the distal SMS-REP on the centromeric side. FISH with the long-range PCR product specific to the KER gene cluster localized within the ∼10–42-kb proximal portion of the distal SMS-REP (Park et al. 2002) showed that it was translocated on the der(10) chromosome. Subsequent FISH mapping with the PAC clone RP1-48J12 that overlaps the distal SMS-REP by ∼20 kb showed that only a small fragment of the clone RP1-48J2 was translocated. Thus, the chromosome 17 breakpoint was mapped at the proximal end of the distal SMS-REP.
LCRs Comprise at Least 23% of Proximal 17p Genomic Sequence
Estimates of the percentage of low-copy repeat sequence in the human genome have varied from 5% to 10% (Mazzarella and Schlessinger 1998; Bailey et al. 2001; Cheung et al. 2001; Eichler 2001; IHGSC 2001; Johnson et al. 2001; Samonte and Eichler 2002) and are based on the virtual analyses of an ever-changing (version 30, August 2002) draft of the human genome. The size and percent identity of LCR necessary and sufficient to mediate genomic disorders remain to be elucidated, but the usual minimal size for an LCR associated with a large genomic segment rearrangement is ∼10 kb with ∼98% sequence identity (Lakich et al. 1993; Lupski 1998; Bailey et al. 2001; Stankiewicz and Lupski 2002a). Extensive studies of proximal 17p rearrangement breakpoints in combination with nearly complete genome sequence enable estimates of the percent genomic sequence contained within LCR, based on experimental observations. Remarkably, in the 7.4 Mb of proximal 17p genomic sequence from the centromere to the distal CMT1A-REP, at least 1.7 Mb (23%) of the genome is contained within LCRs. These LCRs include the proximal and distal CMT1A-REPs (24,011 bp) (Reiter et al. 1997); proximal (∼256 kb), middle (∼241 kb), and distal (∼176 kb) SMS-REPs; LCR17pA (∼383 kb), LCR17pB (∼191 kb), LCR17pC (∼91 kb), LCR17pD (∼118 kb) (Park et al. 2002), LCR17pE (∼31 kb), LCR17pF (∼33 kb), and LCR17pG (∼23 kb); at least two inverted repeats flanking the RNU3 gene (2 × ∼45 kb) (Gao et al. 1997); and three LCRAs in and around the CMT1A region (3 × ∼11 kb) (Inoue et al. 2001) (fig. 1), the majority of which (with the exception of LCRAs, LCR17pC, and RNU3 repeats) have been identified at the breakpoints of the rearranged chromosomes.
Discussion
Genome Architecture and Susceptibility to Nonrecurrent Rearrangements in Proximal Chromosome 17p
In contrast to recurrent common chromosome aberrations, in which the breakpoints are associated with various genomic architectural features such as LCRs, AT-rich palindromes, or fragile sites, the unusual-sized nonrecurrent rearrangements were thought to represent random events. To investigate this hypothesis, we studied 26 nonrecurrent chromosome rearrangements involving proximal 17p. Surprisingly, we found that many of the unusual-sized deletions and chromosome translocations have breakpoints clustering within apparently breakage/recombination–prone genome architectural structures.
Recently, the construction and DNA sequence analysis of the complete BAC/PAC contig covering the CMT1A and SMS common deletion regions within chromosome 17p11.2-p12, in combination with patient breakpoint analysis, have enabled us to identify the novel LCR17p structures (Inoue et al. 2001; Bi et al. 2002; Park et al. 2002). We now show that LCR17pA, LCR17pC, and LCR17pD are in a direct orientation with respect to each other, whereas the fourth copy, LCR17pB (adjacent to the middle SMS-REP on the centromeric side), is inverted. Remarkably, each of these repeats (except LCR17pC), as well as middle SMS-REP, has now been mapped to the breakpoints of rearranged chromosomes.
We have demonstrated that, similar to the recurrent common genomic deletions and duplications in several other contiguous gene syndromes (Emanuel and Shaikh 2001; McDermid and Morrow 2002; Stankiewicz and Lupski 2002a), LCRs may also underlie nonrecurrent rearrangements. We found that the breakpoints of three larger-sized SMS deletions mapped within directly oriented LCRs. Like the SMS-REPs and CMT1A-REPs, these newly identified LCRs in proximal 17p also appear to serve as genomic substrates mediating NAHR, resulting in chromosome rearrangements.
On the basis of the presence of the same junction fragment as shown by PFGE analysis, Chen et al. (1997) reported several patients with SMS with common deletions, and Potocki et al. (2000) described seven unrelated patients with duplication 17p11.2, the reciprocal product of the common SMS deletions. Recently, Shaw et al. (2002) demonstrated that both common SMS deletions, as well as reciprocal duplications flanked by the proximal and distal SMS-REP, result from unequal crossing-over events with no parental origin bias. These genetic data further support the model of NAHR-mediated reciprocal deletion/duplication events and indicate the relevance of genome architectural features such as LCRs (SMS-REPs) in the origin of recurrent DNA rearrangements within proximal 17p. We now show that unusual-sized deletions can also result from unequal crossing-over events, suggesting they may be stimulated by the presence of some yet-unidentified LCRs. Supporting this hypothesis, the LCRs flanking the RNU3 gene (localized between proximal and distal SMS-REPs) (figs. 1 and 3) were identified by molecular methods (Gao et al. 1997) but are yet to be identified in the nearly complete DNA sequence of the SMS region. Interestingly, similar to CMT1A (Palau et al. 1993) and spinal muscular atrophy (Wirth et al. 1997), 12 of 14 unusual-sized deletions analyzed by genotyping (in patients 572, 641, 1153, 1190, 1195, 1354, 1456, 1615, 1774, 1931, 1939, and 2011) were of paternal origin, suggesting a potential increased proclivity to deletions in this region during spermatogenesis.
In addition to the patients with the common duplication dup(17)(p11.2p11.2) (Potocki et al. 2000; Shaw et al. 2002), we have identified several individuals (Roa et al. 1996) with unusual-sized duplications (FISH-positive for duplication, but junction fragment–negative in PFGE screening using the CLP probe) involving this genomic region. We suggest that some of these duplications may also result from LCRs/NAHR-based mechanisms, and this hypothesis is currently under investigation.
Nonrecurrent Translocation Breakpoints Cluster at the Chromosome 17 Centromere
Centromeres and the pericentromeric intervals of the human genome still remain among the greatest DNA sequencing challenges for the Human Genome Project. The border between centromeric heterochromatin and euchromatin is significantly enriched (10-fold) with various repetitive elements (Eichler et al. 1997; Eichler 1999; Bailey et al. 2001; Horwath et al. 2001; IHGSC 2001), thus making both the physical mapping and computational assemblies challenging (Katsanis et al. 2001).
Our complete BAC/PAC contig over the entire chromosome subband 17p11.2 (Bi et al. 2002) apparently ends <0.5–1 Mb from the centromere. Because of this proximity, we were able to identify the cluster of translocation breakpoints between proximal SMS-REP and the chromosome 17 centromere. We found that three of eight translocation breakpoints involving 17p11 were located within the α satellite, three others were located within an ∼1-Mb segment from the centromere, and one was located within the distal SMS-REP. We suggest that the identified (peri)centromeric clustering of constitutional chromosome 17p11 translocation breakpoints may be associated with the observed variability of the DNA condensation (patients GM02836 and GM03119) of this genomic region (fig. 4D and 4E). Such decondensation of the pericentromeric heterochromatin has been proposed as a mechanism leading to the origin of jumping translocations of chromosome 1q in multiple myeloma (Sawyer et al. 1998). The abnormal condensation may in turn be related to the abundance of the LCRs in the hetero-euchromatin transition or histone modification (Horvath et al. 2001; Briggs and Strahl 2002).
Interestingly, in addition to isochromosomes (Mertens et al. 1994) and Robertsonian translocations (Han et al. 1994; Page et al. 1996; Bandyopadhyay et al. 2001), several chromosome breakpoints of different genomic rearrangements have been found to map within the centromere or within pericentromeric regions (Wolff et al. 1996; Tümer et al. 1998; Berger et al. 1999; Beheshti et al. 2000; Fauth et al. 2001). Until now, only a few reciprocal chromosome translocation breakpoints have been shown to be associated with LCRs. AT-rich palindromes within an LCR on 22q11 are responsible for the most common recurrent non-Robertsonian constitutional translocation in humans, resulting in the der(22)t(11;22) syndrome (Kurahashi et al. 2000; Edelmann et al. 2001). Kehrer-Sawatzki et al. (1997) reported a reciprocal t(17;22)(q11;q11) in a family with neurofibromatosis type 1 with a breakpoint mapping to the same AT-rich sequence in the same LCR, and Rhodes et al. (1997) described a translocation, t(1;22), involving LCR22. Recently, Giglio et al. (2002) demonstrated that t(4;8)(p16;p23), probably the second-most-common recurrent reciprocal translocation in humans after the t(11;22) translocation, is also mediated by the LCRs that consist of an olfactory receptor–gene cluster. Similar to our findings, the chromosome breakpoints of 14 different reciprocal translocations involving chromosome 22q11 were reported recently (Morrow et al. 2002). These authors found that five breakpoints mapped within LCR22-3, and an additional four occurred within the vicinity of other LCR22s. Interestingly, all 14 partner chromosome breakpoints mapped within the most telomeric bands.
Similar to constitutive (germline) rearrangements, somatic genomic events (e.g., isochromosome 17q, frequently found in patients with neoplasias such as leukemia and medulloblastoma) also have been proposed to involve genome architectural features such as LCRs (Fioretos et al. 1999; Scheurlen et al. 1999). In support of this notion, Saglio et al. (2002) recently reported a possible involvement of the 76-kb LCR22 in the origin of the Philadelphia chromosome translocation, t(9;22)(q34;q11.2). Mitotic rearrangement events present a challenge for breakpoint analyses because of tissue and cell mosaicism, and thus they likely remain underascertained.
We identified one translocation breakpoint (in patient 1576) mapping within or just adjacent and centromeric to the distal SMS-REP. Interestingly, this breakpoint is localized in the direct vicinity of the evolutionarily unstable portion of the distal SMS-REP. Recently, Park et al. (2002) proposed that the proximal SMS-REP was the progenitor copy that, through several genomic rearrangements 40–65 million years ago, resulted in the middle and the distal SMS-REPs. The evolutionary inversion of the entire proximal SMS-REP, generating the middle SMS-REP copy, was accompanied by the truncation of the terminal ∼14-kb genomic interval including the CLP gene. An interstitial ∼39-kb deletion of the genomic segment between the KER and CLP loci was one of the rearrangements associated with the origin of the distal SMS-REP (fig. 7B). These data, together with the identified localization of the t(10;17)(q26.3;p11.2) breakpoint within or directly adjacent to the KER-CLP portion of the distal SMS-REP, further suggests that this genomic interval containing the SMS-REPs is unstable and prone to rearrangements.
Genome Architecture and Rearrangements
LCRs have been recognized relatively recently, because of their association with DNA rearrangements resulting in disease traits and, in contrast to interspersed repeat sequences (e.g., Alu or LINE), are not identifiable through reassociation kinetics. Nevertheless, the involvement of LCRs in chromosome rearrangements and evolution has received widespread attention (Lupski 1998; Bailey et al. 2001; Emanuel and Shaikh 2001; Johnson et al. 2001; Inoue and Lupski 2002; Samonte and Eichler 2002; Stankiewicz and Lupski 2002a, 2002b). Estimates of the amount of human genomic sequence contained within LCRs have ranged from 5% to 10% (Bailey et al. 2001; Johnson et al. 2001), but, to date, they have been based on the bioinformatic analyses of the draft genome sequences. Assembly of the human genome sequence (IHGSC 2001)—and, in particular, that determined by a shotgun approach—is challenging because of such LCRs (Lupski 1998; Katsanis et al. 2001), and the present genome content of LCRs may be grossly underestimated. Our analysis of the genome sequence in proximal 17p suggests that LCRs may constitute >23% of primary DNA sequence in some parts of the human genome. Furthermore, we demonstrate that each identified LCR within proximal 17p can be involved in a rearrangement event. In fact, 21/33 (64%) deletion breakpoints mapping within 17p11.2 occur in LCRs. This indicates the breakage/recombination–stimulating role of LCRs.
On the basis of our FISH and genotyping findings, we propose three different mechanisms resulting in deletion rearrangements: (i) Similar to common SMS deletions, the unusual-sized deletions with breakpoints mapping within directly oriented copies of LCRs (patients 147, 1153, and 1939; ∼17%) are stimulated by LCRs and mediated by the LCR/NAHR–based inter- or intrachromosomal unequal crossing-over. (ii) In the other group of unusual-sized deletions, represented by those with both breakpoints mapping within nonhomologous copies of LCRs (patients 572, 1190, and 1456; ∼17%) or those with one breakpoint mapping within LCR and the other in LCR-free unique DNA sequence (patients 540, 566, 641, 765, 993, 1195, 1615, 1774, and 2011; 50%), the chromosome deletion is stimulated—but not mediated—by the LCR(s) and may occur via either NAHR utilizing small repeat segments or by NHEJ (Inoue et al. 2002). (iii) Finally, deletions, in which breakpoints do not appear to involve LCRs (patients 357, 1354, and 1931; ∼17%) may occur through NHEJ between repeat-free DNA fragments. It is possible that the completion of the DNA sequence of this region will reveal the presence of yet unknown additional low- or high-copy repeats at the apparently unique sequence breakpoints. Interestingly, the distal breakpoints of the deletions in patients 1354 and 1931 appear to cluster with the breakpoints in patients 566 and 1774, suggesting the presence of a breakage-prone genomic architectural feature.
Our findings in proximal 17p and the recent data from 22q11.2 reported by Morrow et al. (2002) document that genome architecture is important to recurrent chromosomal rearrangements, including interstitial deletions and reciprocal translocations. Further studies are required to determine the extent to which LCRs influence susceptibility to chromosome rearrangements in other regions of the genome. Nevertheless, as in the case of recurrent rearrangements, the findings in proximal 17p (Pentao et al. 1992; Chen et al. 1997) may model what will be found in many other genomic regions.
Recombination-based Disorders and Disease Burden
Unlike conventional monogenic diseases reflecting errors of DNA replication and/or repair, genomic disorders are recombination-based conditions and thus cannot be prevented/repaired by any cellular machinery (Lupski 2003). This has been proposed as a possible explanation for the high frequency and worldwide prevalence of new mutations in genomic disorders (Lupski 1998; Shaffer and Lupski 2000). Depending on the size of the genomic segment involved in the genomic disorder, it can result in a Mendelian disease, a contiguous gene syndrome, or a chromosomal disorder (Stankiewicz and Lupski 2002a). A wide variety of traits have been described as resulting from genomic disorders, including mental retardation, color blindness, hypertension, infertility, panic and phobic disorder, and peripheral neuropathy (Stankiewicz and Lupski 2002b).
In summary, our data demonstrate that genomic architecture involving LCRs plays a significant role in the origin not only of recurrent common chromosome rearrangements (e.g., contiguous gene syndromes) but also of unusual-sized deletions and nonrecurrent translocations.
Acknowledgments
We appreciate the critical reviews of Drs. K. Inoue and B. Morrow. We thank Drs. A. T. Midro (Bialystok, Poland) and R. J. Hastings (Oxford, United Kingdom), for providing us patient samples TIC and UK, respectively; C. Kashork, for excellent technical assistance; and Dr. E. O’Brian Smith, for assistance with statistical analysis. This study was supported, in part, by National Institute of Child Health and Human Development grant PO1 HD39420 and Mental Retardation Research Center grant HD24064.
Electronic-Database Information
The URLs for data presented herein are as follows:
- BACPAC Resources Center Home Page, Children’s Hospital Oakland, http://www.chori.org/bacpac/
- Coriell Cell Repositories, http://locus.umdnj.edu/
- NCBI BLAST Home Page, http://www.ncbi.nlm.nih.gov/blast/
- NCBI Home Page, http://www.ncbi.nlm.nih.gov/
- Online Mendelian Inheritance in Man (OMIM), http://www.ncbi.nlm.nih.gov/Omim/ (for SMS) [PubMed]
- RepeatMasker Web Server, http://repeatmasker.genome.washington.edu/cgi-bin/RepeatMasker
- Research Genetics, http://www.resgen.com/index.php3
- UCSC Genome Bioinformatics, http://genome.ucsc.edu/
References
- Bailey JA, Yavor AM, Massa HF, Trask BJ, Eichler EE (2001) Segmental duplications: organization and impact within the current human genome project assembly. Genome Res 11:1005–1017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bandyopadhyay R, Berend SA, Page SL, Choo KHA, Shaffer LG (2001) Satellite III sequences on 14p and their relevance to Robertsonian translocation formation. Chromosome Res 9:235–242 [DOI] [PubMed] [Google Scholar]
- Beheshti B, Karaskova J, Park PC, Squire JA, Beatty BG (2000) Identification of a high frequency of chromosomal rearrangements in the centromeric regions of prostate cancer cell lines by sequential Giemsa banding and spectral karyotyping. Mol Diagn 5:23–32 [DOI] [PubMed] [Google Scholar]
- Berger R, Busson-Le Coniat M (1999) Centric and pericentric chromosome rearrangements in hematopoietic malignancies. Leukemia 13:671–678 [DOI] [PubMed] [Google Scholar]
- Bi W, Yan J, Stankiewicz P, Park S-S, Walz K, Boerkoel CF, Potocki L, Shaffer LG, Devriendt K, Nowaczyk MJM, Inoue K, Lupski JR (2002) Genes in the Smith-Magenis syndrome critical deletion interval on chromosome 17p11.2 and the syntenic region of the mouse. Genome Res 12:713–728 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brewer C, Holloway S, Zawalnyski P, Schinzel A, FitzPatrick D (1998) A chromosomal deletion map of human malformations. Am J Hum Genet 63:1153–1159 [DOI] [PMC free article] [PubMed] [Google Scholar]
- ——— (1999) A chromosomal duplication map of malformations: regions of suspected haplo- and triplolethality—and tolerance of segmental aneuploidy—in humans. Am J Hum Genet 64:1702–1708 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Briggs SD, Strahl BD (2002) Unraveling heterochromatin. Nat Genet 30:241–242 [DOI] [PubMed] [Google Scholar]
- Chance PF, Abbas N, Lensch MW, Pentao L, Roa BB, Patel PI, Lupski JR (1994) Two autosomal dominant neuropathies result from reciprocal DNA duplication/deletion of a region on chromosome 17. Hum Mol Genet 3:223–228 [DOI] [PubMed] [Google Scholar]
- Chen K-S, Manian P, Koeuth T, Potocki L, Zhao Q, Chinault AC, Lee CC, Lupski JR (1997) Homologous recombination of a flanking repeat gene cluster is a mechanism for a common contiguous gene deletion syndrome. Nat Genet 17:154–163 [DOI] [PubMed] [Google Scholar]
- Cheung VG, Nowak N, Jang W, Kirsch IR, Zhao S, Chen X-N, Furey TS, et al (2001) Integration of cytogenetic landmarks into the draft sequence of the human genome. Nature 409:953–958 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edelmann L, Spiteri E, Koren K, Pulijaal V, Bialer MG, Shanske A, Goldberg R, Morrow BE (2001) AT-rich palindromes mediate the constitutional t(11;22) translocation. Am J Hum Genet 68:1–13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eichler EE (1999) Repetitive conundrums of centromere structure and function. Hum Mol Genet 8:151–155 [DOI] [PubMed] [Google Scholar]
- ——— (2001) Segmental duplications: what’s missing, misassigned, and misassembled—and should we care? Genome Res 11:653–656 [DOI] [PubMed] [Google Scholar]
- Eichler EE, Budarf ML, Rocchi M, Deaven LL, Doggett NA, Baldini A, Nelson DL, Mohrenweiser HW (1997) Interchromosomal duplications of the adrenoleukodystrophy locus: a phenomenon of pericentromeric plasticity. Hum Mol Genet 6:991–1002 [DOI] [PubMed] [Google Scholar]
- Elsea SH, Purandare SM, Adell RA, Juyal RC, Davis JG, Finucane B, Magenis RE, Patel PI (1997) Definition of the critical interval for Smith-Magenis syndrome. Cytogenet Cell Genet 79:276–281 (erratum 81:67) [DOI] [PubMed] [Google Scholar]
- Emanuel BS, Shaikh TH (2001) Segmental duplications: an ‘expanding’ role in genomic instability and disease. Nat Rev Genet 2:791–800 [DOI] [PubMed] [Google Scholar]
- Fauth C, Bartels I, Haaf T, Speicher MR (2001) Additional dark G-band in the p-arm of chromosome 19 due to a paracentric inversion with a breakpoint in the pericentromeric heterochromatin. Am J Med Genet 103:160–162 [DOI] [PubMed] [Google Scholar]
- Fioretos T, Strömbeck B, Sandberg T, Johansson B, Billström R, Borg Å, Nilsson P-G, Van Den Berghe H, Hagemeijer A, Mitelman F, Höglund M (1999) Isochromosome 17q in blast crisis of chronic myeloid leukemia and in other hematologic malignancies is the result of clustered breakpoints in 17p11 and is not associated with coding TP53 mutations. Blood 94:225–232 [PubMed] [Google Scholar]
- Gao L, Frey MR, Matera AG (1997) Human genes encoding U3 snRNA associate with coiled bodies in interphase cells and are clustered on chromosome 17p11.2 in a complex inverted repeat structure. Nucleic Acids Res 25:4740–4747 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giglio S, Calvari V, Gregato G, Gimelli G, Camanini S, Giorda R, Ragusa A, Guerneri S, Selicorni A, Stumm M, Tonnies H, Ventura M, Zollino M, Neri G, Barber J, Wieczorek D, Rocchi M, Zuffardi O (2002) Heterozygous submicroscopic inversions involving olfactory receptor–gene clusters mediate the recurrent t(4;8)(p16;p23) translocation. Am J Hum Genet 71:276–285 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han J-Y Choo KHA, Shaffer LG (1994) Molecular cytogenetic characterization of 17 rob(13q14q) Robertsonian translocations by FISH, narrowing the region containing the breakpoints. Am J Hum Genet 55:960–967 [PMC free article] [PubMed] [Google Scholar]
- Horvath JE, Bailey JA, Locke DP, Eichler EE (2001) Lessons from the human genome: transitions between euchromatin and heterochromatin. Hum Mol Genet 10:2215–2223 [DOI] [PubMed] [Google Scholar]
- Hussain SZ, Evans AL, Ahmed OA, Jones D, McDermot KD, Svennevik EC, Hastings RJ (2000) Non-syndromic mental retardation segregating with an apparently balanced t(1;17) reciprocal translocation through three generations. Am J Med Genet 95:99–104 [DOI] [PubMed] [Google Scholar]
- Inoue K, Dewar K, Katsanis N, Reiter LT, Lander ES, Devon KL, Wyman DW, Lupski JR, Birren B (2001) The 1.4-Mb CMT1A duplication/HNPP deletion genomic region reveals unique genome architectural features and provides insights into the recent evolution of new genes. Genome Res 11:1018–1033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inoue K, Lupski JR (2002) Molecular mechanisms for genomic disorders. Annu Rev Genomics Hum Genet 3:199–242 [DOI] [PubMed] [Google Scholar]
- Inoue K, Osaka H, Thurston VC, Clarke JTR, Yoneyama A, Rosenbarker L, Bird TD, Hodes ME, Shaffer LG, Lupski JR (2002) Genomic rearrangements resulting in PLP1 deletion occur by nonhomologous end joining and cause different dysmyelinating phenotypes in males and females. Am J Hum Genet 71:838–853 [DOI] [PMC free article] [PubMed] [Google Scholar]
- International Human Genome Sequencing Consortium (2001) Initial sequencing and analysis of the human genome. Nature 409:860–921 [DOI] [PubMed] [Google Scholar]
- Johnson ME, Viggiano L, Bailey JA, Abdul-Rauf M, Goodwin G, Rocchi M, Eichler EE (2001) Positive selection of a gene family during the emergence of humans and African apes. Nature 413:514–519 [DOI] [PubMed] [Google Scholar]
- Juyal RC, Figuera LE, Hauge X, Elsea SH, Lupski JR, Greenberg F, Baldini A, Patel PI (1996) Molecular analyses of 17p11.2 deletions in 62 Smith-Magenis syndrome patients. Am J Hum Genet 58:998–1007 [PMC free article] [PubMed] [Google Scholar]
- Katsanis N, Worley KC, Lupski JR (2001) An evaluation of the draft human genome sequence. Nat Genet 29:88–91 [DOI] [PubMed] [Google Scholar]
- Kehrer-Sawatzki H, Häussler J, Krone W, Bode H, Jenne DE, Mehnert KU, Tümmers U, Assum G (1997) The second case of a t(17;22) in a family with neurofibromatosis type 1: sequence analysis of the breakpoint regions. Hum Genet 99:237–247 [DOI] [PubMed] [Google Scholar]
- King PH, Waldrop R, Lupski JR, Shaffer LG (1998) Charcot-Marie-Tooth phenotype produced by a duplicated PMP22 gene as part of a 17p trisomy-translocation to the X chromosome. Clin Genet 54:413–416 [DOI] [PubMed] [Google Scholar]
- Kurahashi H, Shaikh TH, Hu P, Roe BA, Emanuel BS, Budarf ML (2000) Regions of genomic instability on 22q11 and 11q23 as the etiology for the recurrent constitutional t(11;22). Hum Mol Genet 9:1665–1670 [DOI] [PubMed] [Google Scholar]
- Lakich D, Kazazian HH Jr, Antonarakis SE, Gitschier J (1993) Inversions disrupting the factor VIII gene are a common cause of severe haemophilia A. Nat Genet 5:236–241 [DOI] [PubMed] [Google Scholar]
- Lupski JR (1998) Genomic disorders: structural features of the genome can lead to DNA rearrangements and human disease traits. Trends Genet 14:417–422 [DOI] [PubMed] [Google Scholar]
- ——— (2003) Genomic disorders: recombination-based disease resulting from genome architecture. Am J Hum Genet 72:246–252 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mazzarella R, Schlessinger D (1998) Pathological consequences of sequence duplications in the human genome. Genome Res 8:1007–1021 [DOI] [PubMed] [Google Scholar]
- McDermid HE, Morrow BE (2002) Genomic disorders on 22q11. Am J Hum Genet 70:1077–1088 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mertens F, Johansson B, Mitelman F (1994) Isochromosomes in neoplasia. Genes Chromosomes Cancer 10:221–230 [DOI] [PubMed] [Google Scholar]
- Morrow BE, Spiteri E, Wakui K, Pulijaal V, Minoshima S, Shimizu N, Shaffer LG (2002) Frequent rearrangements between low copy repeats on 22q11 (LCR22) and telomeric bands of translocation partner chromosomes. Am J Hum Genet Suppl 71:296 [Google Scholar]
- Page SL, Shin J-C, Han J-Y, Choo KHA, Shaffer LG (1996) Breakpoint diversity illustrates distinct mechanisms for Robertsonian translocation formation. Hum Mol Genet 5:1279–1288 [DOI] [PubMed] [Google Scholar]
- Palau F, Löfgren A, De Jonghe P, Bort S, Nelis E, Sevilla T, Martin J-J, Vilchez J, Prieto F, Van Broeckhoven C (1993) Origin of the de novo duplication in Charcot-Marie-Tooth disease type 1A: unequal nonsister chromatid exchange during spermatogenesis. Hum Mol Genet 2:2031–2035 [DOI] [PubMed] [Google Scholar]
- Park S-S, Stankiewicz P, Bi W, Shaw C, Lehoczky J, Dewar K, Birren B, Lupski JR (2002) Structure and evolution of the Smith-Magenis syndrome repeat gene clusters, SMS-REPs. Genome Res 12:729–738 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pentao L, Wise CA, Chinault AC, Patel PI, Lupski JR (1992) Charcot-Marie-Tooth type 1A duplication appears to arise from recombination at repeat sequences flanking the 1.5 Mb monomer unit. Nat Genet 2:292–300 [DOI] [PubMed] [Google Scholar]
- Potocki L, Chen K-S, Park S-S, Osterholm DE, Withers MA, Kimonis V, Summers AM, Meschino WS, Anyane-Yeboa K, Kashork CD, Shaffer LG, Lupski JR (2000) Molecular mechanism for duplication 17p11.2—the homologous recombination reciprocal of the Smith-Magenis microdeletion. Nat Genet 24:84–87 [DOI] [PubMed] [Google Scholar]
- Reiter LT, Murakami T, Koeuth T, Gibbs RA, Lupski JR (1997) The human COX10 gene is disrupted during homologous recombination between the 24 kb proximal and distal CMT1A-REPs. Hum Mol Genet 6:1595–1603 [DOI] [PubMed] [Google Scholar]
- Reiter LT, Murakami T, Koeuth T, Pentao L, Muzny DM, Gibbs RA, Lupski JR (1996) A recombination hotspot responsible for two inherited peripheral neuropathies is located near a mariner transposon-like element. Nat Genet 12:288-297 (erratum 19:303) [DOI] [PubMed] [Google Scholar]
- Rhodes CH, Call KM, Budarf ML, Barnoski BL, Bell CJ, Emanuel BS, Bigner SH, Park JP, Mohandas TK (1997) Molecular studies of an ependymoma-associated constitutional t(1;22)(p22;q11.2). Cytogenet Cell Genet 78:247–252 [DOI] [PubMed] [Google Scholar]
- Richards RI (2001) Fragile and unstable chromosomes in cancer: causes and consequences. Trends Genet 17:339–345 [DOI] [PubMed] [Google Scholar]
- Roa BB, Greenberg F, Gunaratne P, Sauer CM, Lubinsky MS, Kozma C, Meck JM, Magenis RE, Shaffer LG, Lupski JR (1996) Duplication of the PMP22 gene in 17p partial trisomy patients with Charcot-Marie-Tooth type-1A neuropathy. Hum Genet 97:642–649 [PubMed] [Google Scholar]
- Saglio G, Storlazzi CT, Giugliano E, Surace C, Anelli L, Rege-Cambrin G, Zagaria A, Jimenez Velasco A, Heiniger A, Scaravaglio P, Torres Gomez A, Roman Gomez J, Archidiacono N, Banfi S, Rocchi M (2002) A 76-kb duplicon maps close to the BCR gene on chromosome 22 and the ABL gene on chromosome 9: possible involvement in the genesis of the Philadelphia chromosome translocation. Proc Natl Acad Sci USA 99:9882–9887 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Samonte RV, Eichler EE (2002) Segmental duplications and the evolution of the primate genome. Nat Rev Genet 3:65–72 [DOI] [PubMed] [Google Scholar]
- Sawyer JR, Tricot G, Mattox S, Jagannath S, Barlogie B (1998) Jumping translocations of chromosome 1q in multiple myeloma: evidence for a mechanism involving decondensation of pericentromeric heterochromatin. Blood 91:1732–1741 [PubMed] [Google Scholar]
- Scheurlen WG, Schwabe GC, Seranski P, Joos S, Harbott J, Metzke S, Döhner H, Poustka A, Wilgenbus K, Haas OA (1999) Mapping of the breakpoints on the short arm of chromosome 17 in neoplasms with an i(17q). Genes Chromosomes Cancer 25:230–240 [DOI] [PubMed] [Google Scholar]
- Shaffer LG, Kennedy GM, Spikes AS, Lupski JR (1997) Diagnosis of CMT1A duplications and HNPP deletions by interphase FISH: implications for testing in the cytogenetics laboratory. Am J Med Genet 69:325–331 [PubMed] [Google Scholar]
- Shaffer LG, Lupski JR (2000) Molecular mechanisms for constitutional chromosomal rearrangements in humans. Annu Rev Genet 34:297–329 [DOI] [PubMed] [Google Scholar]
- Shaw CJ, Bi W, Lupski JR (2002) Genetic proof of unequal meiotic crossovers in reciprocal deletion and duplication of 17p11.2. Am J Hum Genet 71:1072–1081 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stankiewicz P, Lupski JR (2002a) Genome architecture, rearrangements and genomic disorders. Trends Genet 18:74–82 [DOI] [PubMed] [Google Scholar]
- ——— (2002b) Molecular-evolutionary mechanisms for genomic disorders. Curr Opin Genet Dev 12:312–319 [DOI] [PubMed] [Google Scholar]
- Stankiewicz P, Park S-S, Inoue K, Lupski JR (2001) The evolutionary chromosome translocation 4;19 in Gorilla gorilla is associated with microduplication of the chromosome fragment syntenic to sequences surrounding the human proximal CMT1A-REP. Genome Res 11:1205–1210 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tümer Z, Wolff D, Silahtaroglu AN, Ørum A, Brøndum-Nielsen K (1998) Characterization of a supernumerary small marker X chromosome in two females with similar phenotypes. Am J Med Genet 76:45–50 [PubMed] [Google Scholar]
- Venter JC, Adams MD, Myers EW, Li PW, Mural RJ, Sutton GG, Smith HO, et al (2001) The sequence of the human genome. Science 291:1304–1351 [DOI] [PubMed] [Google Scholar]
- Wirth B, Schmidt T, Hahnen E, Rudnik-Schöneborn S, Krawczak M, Müller-Myhsok B, Schönling J, Zerres K (1997) De novo rearrangements found in 2% of index patients with spinal muscular atrophy: mutational mechanisms, parental origin, mutation rate, and implications for genetic counseling. Am J Hum Genet 61:1102–1111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolff DJ, Miller AP, Van Dyke DL, Schwartz S, Willard HF (1996) Molecular definition of breakpoints associated with human Xq isochromosomes: implications for mechanisms of formation. Am J Hum Genet 58:154–160 [PMC free article] [PubMed] [Google Scholar]