Abstract
Linkage evidence suggests that chromosome 13 (13q32-33) contains susceptibility genes for both bipolar disorder and schizophrenia. Recently, genes called “G72” and “G30” were identified, and polymorphisms of these overlapping genes were reported to be associated with schizophrenia. We studied two series of pedigrees with bipolar disorder: the Clinical Neurogenetics (CNG) pedigrees (in which linkage to illness had been previously reported at 13q32-33), with 83 samples from 22 multiplex families, and the National Institute of Mental Health (NIMH) Genetics Initiative pedigrees, with 474 samples from 152 families. Sixteen single-nucleotide polymorphisms (SNPs) were genotyped at and around the G72/G30 locus, which covered a 157-kb region encompassing the entire complementary DNA sequences of G72 and G30. We performed transmission/disequilibrium testing (TDT) and haplotype analysis, since a linkage-disequilibrium block was present at this gene locus. In the CNG and NIMH data sets, the results of global TDT of the entire haplotype set were significant and consistent (P=.0004 and P=.008, respectively). In the CNG series, the associated genotypes divided the families into those with linkage and those without linkage (partitioned by the linkage evidence). Analysis of the decay of haplotype sharing gave a location estimate that included G72/G30 in its 95% confidence interval. Although statistically significant association was not detected for individual SNPs in the NIMH data set, the same haplotype was consistently overtransmitted in both series. These data suggest that a susceptibility variant for bipolar illness exists in the vicinity of the G72/G30 genes. Taken together with the earlier report, this is the first demonstration of a novel gene(s), discovered through a positional approach, independently associated with both bipolar illness and schizophrenia.
Introduction
Bipolar disorder is a common disease with a lifetime prevalence rate of 1% and involves episodic disturbances of mood, sleep, appetite, and other physiological functions. Although a single gene may occasionally have a major effect in some families, multiple genes and environmental factors are generally considered to play roles in the development of bipolar disorder (Craddock and Jones 1999). Two chromosomal regions—13q32-33 and 22q11—have been found to confer susceptibility, by meta-analysis of genomewide linkage studies (Badner and Gershon 2002). In that analysis, the same two regions were significantly linked to schizophrenia (MIM 181500). This result suggests the possibility that, in these chromosomal regions, there are shared genes predisposing to these psychiatric illnesses (Berrettini 2000; Gershon 2000).
On 13q32-33, there had been no gene whose association with major psychiatric illnesses was significant until genes called “G72” and “G30” were recently identified in a 65-kb region where there is evidence of association with schizophrenia (Chumakov et al. 2002). These two genes overlap on complementary chromosomal strands. On the basis of the integrated physical map of this region (Christian et al. 2002), these genes are located ∼90 cM from the short-arm telomere of chromosome 13, where there had been no known genes. Significant association with schizophrenia was found in several SNPs and haplotypes at this locus in a case-control study analyzing French-Canadian samples, with the association of two SNPs being replicated in a Russian cohort. They also performed expression and functional studies of these genes. The G72 gene was suggested to interact with d-amino-acid oxidase and to exert an effect on the regulation of d-serine, an agonist for the glycine-binding site of the N-methyl-d-aspartate–type glutamate receptor, whereas the function of the G30 gene remained unclear.
Since linkage to 13q32-33 was the most significant finding across the entire genome in this pedigree series with bipolar disorder in previous studies (Detera-Wadleigh et al. 1999; C. Liu et al. 2001), we examined the G72/G30 gene locus for possible association with bipolar disorder by analysis of the same pedigree series along with an additional independent pedigree series with bipolar disorder. We performed transmission/disequilibrium testing (TDT) (Spielman et al. 1993), for individual SNPs and haplotypes, and two recently developed analyses: partitioning of linkage evidence according to genotype (Horikawa et al. 2000) and decay of haplotype sharing (DHS) (McPeek and Strahs 1999). In addition, the structure of G72 cDNA from human brain and testis was examined.
Subjects and Methods
Bipolar Pedigrees
We studied two pedigree series with bipolar illness. One of them, the Clinical Neurogenetics (CNG) pedigrees, with 371 individuals from 22 extended multiplex families, has been described elsewhere (Berrettini et al. 1991). The initial genome scan and the subsequent fine mapping of 13q32-33 detected the strongest evidence of linkage around D13S779-D13S225 (LOD score 3.25 [P=.000546]), by multipoint linkage analysis with Aspex and Genehunter Plus in nuclear families, whereas linkage was not significant when all 371 individuals were analyzed by Genehunter in extended pedigrees (Detera-Wadleigh et al. 1999; C. Liu et al. 2001). This discrepancy can result from a common susceptibility allele, as discussed elsewhere (Badner et al. 1998). Consequently, in the present study, we selected 83 individuals for genotyping, so that each extended family is represented by one nuclear family with parents and one or two affected offspring. Sib pairs with increased sharing for markers at 13q32 were preferentially selected, when possible, to enhance the likelihood that they would carry the susceptibility locus. In this series, significant linkage disequilibrium (LD) was observed by TDT at marker D13S280 (C. Liu et al. 2001), which is 2.8 Mb proximal to the G72 locus. Another series, the National Institute of Mental Health (NIMH) Genetics Initiative pedigrees, has been described in detail elsewhere (Nurnberger et al. 1997), and evidence of linkage had been nominally significant at 13q in the initial genome scan (Stine et al. 1997) but was no longer significant (although positive) after the number of samples was increased, reaching 474 individuals in 152 families in the present study (data not shown). As in the first series, sib pairs with increased sharing for markers in the 13q32 region were preferentially selected when possible. These two series predominantly consisted of individuals of European descent and included patients with bipolar I disorder, bipolar II disorder, schizoaffective disorder, and recurrent depression.
SNP Selection and Genotyping
SNP-Cruncher software (C. Liu and Gershon 2001) was used to collect SNP data around the G72/G30 gene locus from the public databases, including the National Center for Biotechnology (see the dbSNP Home Page), the SNP Consortium, and the Human Genome Browser Gateway (see the UCSC Genome Bioinformatics Web site). With the tools provided in SNP-Cruncher, we selected SNPs suitable for genotyping by filtering out repeat sequences, removing possible duplications of SNPs in the databases, checking the validation status, and searching for restriction enzymes that recognize each polymorphism. All discovered SNPs, STSs, and ESTs and the G72/G30 genes were mapped to a reference genomic DNA sequence by DNannotator. This sequence was assembled and evaluated locally and is nearly identical to part of the sequence in GenBank (accession numbers NT_009952.10, AE014312, and AE014313). SNPs were then selected for genotyping, considering factors such as minor-allele frequency (if available), distance from the G72/G30 genes, marker intervals, and availability of inexpensive restriction enzymes for genotyping.
Initially, six SNPs (rs1935058, rs1341402, rs2391191, rs1935062, rs947267, and rs954581) close to or at the G72/G30 gene locus were genotyped and tested for association by analysis of the CNG pedigree series. After finding significant association between five of these SNPs and illness either by TDT or by partitioning of linkage evidence (as described below, in the “Results” section), we also genotyped these six markers for analysis in the NIMH pedigree series. Also, to delimit the range of association by fine mapping of a 157-kb region surrounding the G72/G30 genes, we added eight SNPs (rs1998654, rs2181953, rs978714, rs1359387, rs1815686, rs778294, rs778334, and rs2012887) from the public databases and two SNPs (M-13 and M-23) described elsewhere (Chumakov et al. 2002). We typed all of these additional 10 markers in the CNG series but only 1 (rs778294) in the NIMH series, because it did not seem to be suitable for the purpose of delimiting the region of association, as described in the “Discussion” section.
Fragments containing the selected SNPs were amplified by PCR with primers designed by Primer3 software (Rozen and Skaletsky 2000). Of all 16 SNPs, 13 were genotyped by RFLPs, whereas SNPs rs2391191, M-13, and M-23 were typed by direct sequencing of PCR products with an automated sequencer (ABI 3700 DNA analyzer; Applied Biosystems). The SNPs, along with their positions, are given in table 1.
Table 1.
TDT by Locus and Partitioning of Linkage Evidence According to Genotype
| TDT |
Partitioning of LinkageEvidence, for Genotype |
||||||||||
| 1/1 |
1/2 |
2/2 |
|||||||||
| Series and SNPa | Variant | Allele 1 | Distanceb(kb) | P | TransmissionRatioc | nd | LODe | nd | LOD (P)e | nd | LODe |
| CNG pedigrees: | |||||||||||
| rs1998654 | CT | T | .0 | 1 | .38 | 18 | 2.87 | 9 | .1 | 0 | 0 |
| rs2181953 | AT | T | 27.8 | .39 | .47 | 11 | 2.59 | 9 | .5 | 7 | .07 |
| rs978714 | AG | G | 38.3 | .62 | .67 | 9 | .73 | 15 | .7 | 3 | 2.44 |
| rs1359387 | AG | A | 43.3 | .53 | .65 | 10 | .36 | 12 | 1.46 | 5 | .92 |
| rs1815686 | CG | C | 80.6 | .041 | .93 | 12 | .11 | 9 | 2.62 (.016) | 6 | .6 |
| M-13 | AC | C | 82.1 | .11 | .81 | 11 | .01 | 8 | 2.48 (.02) | 8 | 1.32 |
| rs1935058 | CT | C | 82.5 | .00077 | 1.00 | 14 | .39 | 7 | 2.01 (.042) | 6 | .6 |
| rs1341402 | CT | T | 86.7 | .0075 | .80 | 19 | .85 | 8 | 2.08 (.046) | 0 | 0 |
| rs2391191f | AG | A | 90.6 | .08 | .84 | 11 | .01 | 8 | 2.48 (.02) | 8 | 1.32 |
| rs1935062 | AC | A | 99.3 | .0078 | .11 | 7 | .4 | 12 | 2.8 (.027) | 8 | .07 |
| rs947267f | AC | A | 110.8 | .17 | .70 | 13 | .05 | 11 | 3.63 (.0042) | 3 | .42 |
| rs778294f | AG | G | 113.4 | .018 | .83 | 20 | 1.03 | 7 | 1.9 (.05) | 0 | 0 |
| rs954581 | CT | T | 123.4 | .62 | .57 | 19 | 2.99 | 7 | .04 | 1 | 0 |
| rs778334 | TC | T | 131.1 | .8 | .50 | 2 | .61 | 5 | .03 | 20 | 2.22 |
| rs2012887 | CT | C | 141.1 | .17 | .58 | 10 | 1.17 | 12 | 1.05 | 5 | .33 |
| M-23 | CT | C | 156.8 | .074 | .38 | 5 | .33 | 11 | .54 | 11 | 1.87 |
| NIMH pedigrees: | |||||||||||
| rs1935058 | CT | C | 82.5 | .055 | .63 | 38 | .06 | 69 | −.14 | 42 | .00 |
| rs1341402 | CT | T | 86.7 | .58 | .56 | 87 | −.01 | 54 | .00 | 8 | -0.01 |
| rs2391191f | AG | A | 90.6 | .39 | .57 | 29 | .00 | 68 | −.06 | 52 | .00 |
| rs1935062 | AC | A | 99.3 | .38 | .43 | 60 | −.05 | 66 | .00 | 23 | .00 |
| rs947267f | AC | A | 110.8 | .58 | .48 | 50 | .00 | 68 | −.11 | 31 | .05 |
| rs778294f | AG | G | 113.4 | .24 | .61 | 80 | −.11 | 53 | .14 | 15 | −.11 |
| rs954581 | CT | T | 123.4 | .11 | .65 | 114 | .06 | 33 | −.47 | 2 | −.10 |
SNPs are ordered from the centromere to the telomere of 13q.
Positions of SNPs are shown as distances from rs1998654, where ranges of G72 and G30 are 89.4–114.5 kb and 82.6–129.2 kb, respectively.
Frequency with which allele 1 is transmitted from a heterozygous parent to an affected offspring.
Number of probands for each genotype.
Calculated for a group of families with probands that have each specific genotype, testing for difference of linkage scores among the separate groups of families (Horikawa et al. 2000). P values <.05 are omitted.
SNPs rs2391191, rs947267, and rs778294 correspond to M-15, M-18, and M-19, respectively, of Chumakov et al. (2002).
Genotyping results were examined for incompatibilities within families, by PedCheck (O’Connell and Weeks 1998), and also for unlikely recombinations, by Merlin (Abecasis et al. 2002). Detection of incompatibilities or unlikely recombinants led to rechecking of genotypes and subsequent correction as necessary. None of the SNPs deviated from Hardy-Weinberg equilibrium in either of the pedigree series.
Statistical Analyses
TDT by individual SNP locus was performed using the Aspex/SibTDT program, which tests for LD in sibships, controlling for evidence of linkage. Empirical probabilities for χ2 statistics are calculated, which will accurately reflect association independently of linkage within families. This calculation is done by permuting the parent alleles while fixing the IBD status of sibs within a family. Transmission and frequencies of haplotypes were analyzed by TDTPhase (Dudbridge et al. 2000), using the “tsu” correction for sib pairs, which controls for the presence of linkage. This correction includes only parents transmitting the same haplotype to all sibs. When one or more sibs are ambiguous, only the first sib is included in the analysis. In the calculation of the global tests, haplotypes with counts <5 were dropped. In TDT, the transmission ratio for one randomly chosen individual per sibship was also calculated for each allele and haplotype. The transmission ratio was estimated as the frequency with which an allele or haplotype is transmitted from a heterozygous parent to an affected offspring.
We also tested partitioning evidence of linkage according to genotype, as described elsewhere (Horikawa et al. 2000). Positive results obtained using this test are regarded as further evidence of association. In the present analysis, 22 and 152 probands were randomly selected from the CNG and NIMH series, respectively, and families were separated into groups according to the genotypes of probands for each SNP. If there was no association between the SNP and the evidence for linkage, the resulting LOD score within each genotype group should be proportional to the number of families within that group. If there is association, the LOD score should be disproportionately high within one or two of the genotype groups. Linkage score for each group was calculated on the basis of the original linkage evidence at 13q32-33 in the CNG series, as demonstrated by the Genehunter Plus (Kong and Cox 1997) analysis in nuclear families by use of the narrow affection model (Detera-Wadleigh et al. 1999; C. Liu et al. 2001), and in the NIMH series (data not shown). Families in which the proband contained the SNP genotype being tested were weighted 1, whereas other families were weighted 0. If there was more than one nuclear family within a pedigree, then all were weighted the same on the basis of the proband’s genotype. Empirical P values, for the assessment of the significance of the increase in the LOD score for the group defined by SNP genotype, were obtained by randomly choosing the same number of families included in the group generating the increased LOD score by use of Weight (N. J. Cox, personal communication). Enough replicates were simulated until ⩾20 replicates achieving a LOD score at least as high as that observed in the genotype group were generated.
DHS analysis as implemented in DHSMap (McPeek and Strahs 1999) was also performed, to plot the likelihood of association for the entire 157-kb region and to delimit the 95% CI of association. Haplotypes from a single randomly selected proband in each pedigree from the CNG data set were used. The transmitted haplotypes were considered as the “cases,” and the nontransmitted haplotypes were considered as the “controls.” The conditional-coalescent model was used. All analyses were based on one affection status model, wherein the definition of affected category was restricted to bipolar I, bipolar II with major depression, and schizoaffective disorder.
Results
Pairwise LD
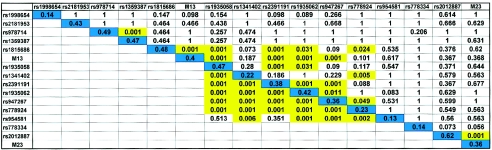
To calculate the P value for LD between pairs of loci, we used genotypes from one individual per pedigree. Haplotypes that could be unambiguously inferred from transmitted and nontransmitted genotypes from the parents were estimated using software written by our group (J. A. Badner, unpublished data). P values of LD were calculated using Clump (Sham and Curtis 1995). We present P values, rather than D′ values, because D′ may be high but nonsignificant owing to the absence of a low-frequency haplotype in our small sample, thereby leading to spurious evidence of LD. Although rs1341402 was not in significant LD with adjacent markers in the CNG series, the number of samples in this series did not seem to be large enough to detect weak-to-moderate LD between loci. The LD analysis in the NIMH series, with 152 samples, revealed that all seven SNPs between rs1935058 and rs778924 were in an LD block (fig. 1).
Figure 1.
Frequencies for each SNP—and LD between pairs of loci. The diagonal pattern composed of blue boxes represents the minor-allele frequencies of each SNP in the CNG pedigrees. To either side of the diagonal are the P values for LD between pairs of loci in the CNG and NIMH pedigrees, respectively; significant LD (P<.05) is shown in yellow. In the NIMH series, an LD block extended across the entire region examined between rs1935058 and rs954581.
TDT
In the CNG series, significant LD with illness was detected in three of the six SNPs that were initially chosen because of their positions' being close to or at G72/G30 (table 1). The strongest association was detected in rs1935058 (P=.00077) (table 1). This SNP is 6.9 kb upstream of G72 exon 1 and 54 bp downstream of the last G30 exon on the opposite strand. This SNP also showed near-significant association in the NIMH series (P=.055). Although TDT gave no P values <.05 for any SNPs in the NIMH series, transmission ratios showed that the same alleles were consistently over- or undertransmitted to affected offspring in both series. When we looked at the subset of families with increased evidence for linkage in the NIMH series (Genehunter Plus nonparametric linkage score >0.7 for the family), transmission ratios, in general, changed in the direction expected (i.e., increased when >0.5 in the total sample and decreased when <0.5 in the total sample). However, TDT was not significant, possibly owing to the small sample size. SNP rs2391191 was the only marker located in exons of either gene, and this variant alters the amino acid sequence of the G72 protein as presented by Chumakov et al. (2002) (although it would be in the 5′ UTR of other transcripts, as described below, in the “G72 and G30 cDNA Sequences” subsection). However, no association was found between this marker and bipolar disorder.
We then added 10 SNPs for genotyping in the CNG series, to delineate the boundaries of association with bipolar disorder. Among these, two showed significant LD with illness (table 1). In all, 16 SNPs were genotyped in the CNG pedigree series, and five of them were significantly associated with illness. All five SNPs were close to or at the G72/G30 genes.
We also analyzed the frequency of the haplotypes of seven SNPs that are in disequilibrium with each other, and we tested the transmission of the haplotypes in relation to illness (table 2). The results showed that, in the CNG series, the most common haplotype, 1112111, was significantly overtransmitted (P=.0003) and the second most common haplotype, 2221221, which is almost the mirror image of the first haplotype, was significantly undertransmitted (P=.008) (table 2). The global TDT for haplotype transmission gave significant results in both the CNG series and the NIMH series (P=.0004 and P=.008, respectively), with the most common haplotype being overtransmitted to affected individuals in both series.
Table 2.
TDT by Haplotype
|
CNG Pedigrees |
NIMH GeneticsInitiative Pedigrees |
|||||
| Haplotypea | Frequency | P | TransmissionRatiob | Frequency | P | TransmissionRatiob |
| 1112111 | .25 | .0003 | .94 | .30 | .06 | .60 |
| 2221221 | .25 | .008 | .18 | .25 | .3 | .44 |
| 2121111 | .12 | .1 | .17 | |||
| 1121211 | .09 | .1 | .83 | |||
| 2121112 | .07 | .7 | .40 | .08 | .06 | .32 |
| 2121211 | .10 | .1 | .62 | |||
| 1111112 | .06 | .8 | .43 | |||
| 2121221 | .06 | .04 | .29 | |||
| Global | .0004 | .008 | ||||
For each haplotype, alleles are concatenated according to the order rs1935058, rs1341402, rs2391191, rs1935062, rs947267, rs778294, and rs954581. Haplotypes with a frequency <.05 are omitted.
Frequency with which the haplotype is transmitted from a heterozygous parent to an affected offspring.
Partitioning of Linkage Evidence
To obtain more evidence of association, we performed another recently developed independent test: partitioning of linkage evidence (Horikawa et al. 2000). In the CNG series, this test gave significant results in families with heterozygous genotypes of eight consecutive SNPs encompassing G72/G30, with the most significant P value <.0042 for rs947267 heterozygotes (table 1). All five SNPs giving significant results in TDT were also significant in this test. In the NIMH series, we found no significant partitioning of linkage evidence.
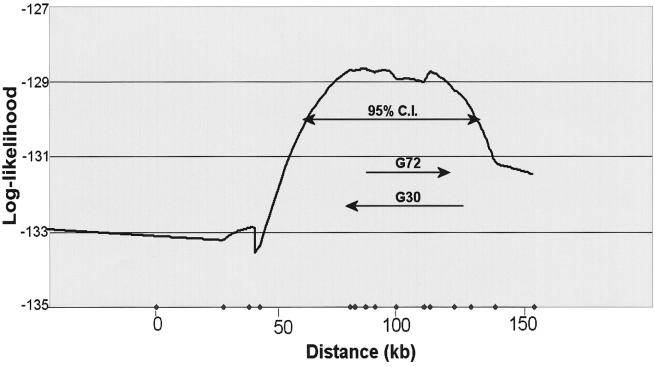
DHS
In this analysis, the likelihood of association reached peak at the G72/G30 gene locus, giving a 95% CI of association in a 78-kb region surrounding it (fig. 2). No other genes are known in this interval. The haplotype associated with bipolar disorder in this analysis is the same as that in the TDTPhase analysis.
Figure 2.
Results from DHS analysis. Log-likelihood of association for each locus in the CNG pedigrees was plotted by the DHSMap program. The horizontal axis shows the distance from rs1998654, the most proximal marker in the present study. Dots along the X-axis denote the positions of SNPs whose genotypic data were used for this analysis. The ranges of all exons for G72/G30 are also shown with their 5′ to 3′ directions. This analysis gave a 95% CI of association in a 78-kb region encompassing the G72/G30 genes.
G72 and G30 cDNA Sequences
We have characterized cDNAs of G72 and G30, to define the exons of these two genes so that one can assess the functional significance of polymorphisms in the gene region and to define potentially functional targets for further polymorphism screening. The cDNAs have been amplified, by PCR methods, from human testis and brain cDNAs (Marathon Ready cDNA; Clontech) and occasionally from fetal brain or amygdala cDNA. The two genes appear to be expressed at quite low levels, since it typically requires ⩾40 cycles of PCR using 0.5–1.0 ng of template to generate significant amounts of product, whereas 30–35 cycles suffices for most other genes. This low level of expression may contribute to the variability as to which splice variants are observed in different PCRs using the same template.
The G72 cDNA from human testis was initially amplified by rapid amplification of cDNA ends (RACE) PCR and, subsequently, by PCR with gene-specific primers from both ends. Different splice variants (GenBank accession numbers AY170469–AY170471) defined a total of nine exons, implying that the exon structure is more complicated than had been indicated by the GenBank accession numbers (AY138546 and AY138547) submitted by Chumakov et al. (2002). Although our attempts at RACE on brain cDNA were unsuccessful, we did succeed in the amplification of products from adult brain and fetal brain and amygdala (GenBank accession number AY223901) by using primers in exons 1 (TGCTGAATGGAAAGCCAGAAAGTAGAGTG) and 9 (GATTCTCCCAGTCACACAGGC). On the basis of the products we have seen, transcripts typically contain exons 1–3, 5, 8, and 9, with the variable inclusion of exons 4, 6, and 7. In addition, intron 1 is frequently retained, and exon 3 (exon N/N2 of Chumkov et al.) and exon 8 use two alternative splice acceptors to produce long and short forms of the exon. The predicted protein products are numerous, depending on which exons are included—and, especially, on which splice acceptor is used for exon 3, since the short form of exon 3 permits the use of an initiation codon in exon 2 while the long form indicates the use of an initiation codon in exon 5 (exon O of Chumkov et al.). In light of both the variability we have seen as to which splice variants are present in PCRs using different primer pairs and the biases inherent in PCRs that select only transcripts containing both primers, it is difficult to assess which transcripts are the most significant. If the gene encodes a protein of conserved function, then the reported existence of frameshifts in exons 3 and 5 in rhesus monkey, chimpanzee, and gorilla favors the use of the initiation codon in exon 5.
The G30 cDNA as described in GenBank (accession number AY138548) defines seven exons. Our preliminary characterization of the G30 gene indicates that it is expressed at levels as low as G72 is and that it contains at least nine exons. PCRs using primers in exons 1 and 7 have given product with testis but not brain template. These include transcripts that lack exon 4 and others that contain an alternative exon 4. Furthermore, 5′ RACE reactions identify an alternative exon 5. Note that parts of exons 4 and 5 are complementary to parts of G72 exons 8 and 2, respectively, which could lead to RNA interference in cells expressing both transcripts.
Discussion
In the present study, we have examined the association between the G72/G30 gene locus on 13q33 and bipolar illness in two data sets, by two different tests for association—TDT, of individual SNPs and the global haplotype of six SNPs, and partitioning of linkage evidence. This is a family-based study and has the advantage of avoiding false-positive results due to population stratification, which could not be ruled out in the case-control study of Chumakov et al. (2002). In the CNG series, the latter gave eight consecutive SNPs at the G72/G30 locus, with five of them being significant also in TDT. In the NIMH series, the individual SNPs were not statistically significant in either test. Bipolar disorder is thought to be a heterogeneous illness, in which one susceptibility gene could not account for the entire population of affected subjects. The NIMH series, on the basis of weak evidence of original linkage to the 13q32-33 region, may contain a smaller proportion of families in which a variant at or around G72/G30 is responsible for susceptibility to illness, as compared to the CNG pedigrees. This is also the reason why we included only the CNG series for the purpose of delimiting the region of association by DHS analysis. Still, the global TDT for haplotypes was significant in both series, with the most common haplotype being consistently overtransmitted to affected offspring. Therefore, the two data sets are consistent with each other, supporting association between this locus and bipolar disorder.
Power analyses were performed to estimate the smallest-effect gene that could be detected in each of the pedigree series. Slink (Ott 1989; Weeks et al. 1990; Cottingham et al. 1993) was used for the simulations. For the simulations, data on pedigree structure, affection status, and genotype at a single 13q32 marker were kept the same as in the original series. A disease gene, with allele frequency of 0.25 and varying degrees of multiplicative genotypic relative risks (MGRRs), and a SNP marker, with allele frequency of 0.50, in complete LD with the disease locus, and linked to the 13q32 marker, were simulated. In the CNG series, for P<.05, there was 80% power to detect a locus with MGRR 5, and there was 50% power to detect a locus with MGRR 3. In the NIMH series, for P<.05, there was 80% power to detect a locus with MGRR 1.75, and there was 50% power to detect a locus with MGRR 1.6. This is consistent with the expectation that the effect of the locus in the CNG series, a sample showing strong evidence of linkage, would be larger than that in the NIMH series, which does not show evidence of linkage (Göring et al. 2001).
Partitioning of linkage evidence gave significant results in heterozygotes, rather than homozygotes, in eight SNPs. There are several possible explanations for this finding. For example, the number of homozygous probands was not large enough to detect significant partitioning of linkage evidence. Alternatively, there may be an unknown biological mechanism for “heterosis,” wherein heterozygotes are more likely to develop bipolar disorder, as in the study of the calpain 10 gene and type 2 diabetes (Horikawa et al. 2000). The significant result in this test supports the possibility that the original linkage to 13q32-33 may be attributed to either G72 or G30. However, since this locus is actually ∼3 cM away from the distal edge of the 95% CI of linkage (Detera-Wadleigh et al. 1999; C. Liu et al. 2001), the existence of another susceptibility gene within the limits of this CI is still plausible.
There has been considerable progress in the identification of possible susceptibility genes for major psychiatric illnesses in chromosomal regions where linkage with them was previously reported. Dysbindin, on 6p (Straub et al. 2002), and neuregulin 1, on 8p (Stefansson et al. 2002), have been reported to be associated with schizophrenia. On 22q11, a few genes have been suggested for susceptibility to major psychiatric illnesses. These genes include cathecol-O-methyltransferase (COMT), PRODH2, and G-protein–coupled receptor kinase 3 (GRK3) and are located within a range of a microdeletion, which occurs with increased frequency in schizophrenia. COMT has a long history of association reports in major psychiatric disorders, starting with reports of different enzyme activities in patients (Cohn et al. 1970). Although results from multiple studies of association between a functional polymorphism at codon 108/158 (Val108/158 Met) of COMT and major psychiatric illnesses are inconsistent, this polymorphism has been suggested both to play a role in prefrontal cognition impairment in schizophrenia (Egan et al. 2001) and to modify the course of rapid-cycling–type bipolar disorder (Craddock et al. 2001). Also, a large-scale case-control study showed association between polymorphisms within the genomic sequence of the COMT gene and schizophrenia, suggesting other susceptibility loci within this gene (Shifman et al. 2002). Association between the PRODH2 gene and schizophrenia has also been reported (H. Liu et al. 2002). Expression level of GRK3 has been correlated with severity of bipolar disorder (Niculescu et al. 2000). The different susceptibility associations on chromosome 22, particularly for bipolar illness, remain to be resolved.
The G72/G30 gene locus, on 13q33, is one of the few loci reported to be associated with both major psychiatric illnesses that are present in regions where linkage to both disorders has been consistently reported (Badner and Gershon 2002). COMT is also associated with both disorders, as noted above. These findings are consistent with the speculation from the linkage evidence that the same gene may be implicated in both disorders (Berrettini 2000; Gershon 2000).
Do schizophrenia and bipolar disorder share a common disease-susceptibility variant at the G72/G30 locus? Since the range of the 95% CI of association given by DHS in the CNG series covered some of the SNPs that Chumakov et al. (2002) had associated with schizophrenia, this remains possible; however, this range did not include two SNPs that were associated with schizophrenia in both French-Canadian and Russian data sets in their study. Obviously, linkage or LD data do not tell which gene, G72 or G30, confers susceptibility to each major psychiatric illness. Chumakov et al. (2002), noting the absence of a detectable in vitro translation product of the antisense gene G30, have suggested transcriptional regulation of G72 by G30. The existence of multiple splice variants of both genes, together with the presence of complementary segments in at least some of these transcripts, provides the basis for a complex transcriptional relationship between the two genes, but it is unclear which gene regulates which. The present evidence shows a bipolar association with the same gene locus that had been associated with schizophrenia (Chumakov et al. 2002) and may be considered as supportive of a shared susceptibility-gene complex, although possibly not of a shared variant or even a shared gene.
In the haplotype analysis, two common haplotypes were significant in the CNG series. This result implies several possibilities. One of them is that the most common haplotype, 1112111, which is overtransmitted to affected offspring, carries one or more predisposing variants. Conversely, it is also possible that the undertransmitted haplotype, 2221221, contains a variant that is protective against the illness. In any case, the susceptibility variant is expected to be frequent, on the basis of associated haplotype frequencies and the previous finding, in the CNG series, that linkage to 13q32-33 was more significant in the nuclear families than in all extended families (Detera-Wadleigh et al. 1999; C. Liu et al. 2001), given that power to detect evidence of linkage in extended pedigrees can be less when a causal variant is common (Badner et al. 1998). Therefore, we could reasonably expect that intensive mutational screening of the associated genomic region can lead to detection of a susceptibility allele of bipolar disorder. DHS analysis, although based on limited data of randomly chosen probands from each CNG family in the present study, provides a rational range in which to search for such a susceptibility allele.
Acknowledgments
We thank all the patients and families included in this study. Data and biomaterial were partly collected in four projects as part of the NIMH Bipolar Disorder Genetics Initiative. From 1991 to 1998, the Principal Investigators and Co-Investigators were as follows: Indiana University, Indianapolis.—John Nurnberger, M.D., Ph.D., Marvin Miller, M.D., and Elizabeth Bowman, M.D. (supported by grant U01 H46282); Washington University, St. Louis.—Theodore Reich, M.D., Allison Goate, Ph.D., and John Rice, Ph.D. (supported by grant U01 MH46280); Johns Hopkins University, Baltimore.—J. Raymond DePaulo, Jr., M.D., Sylvia Simpson, M.D., M.P.H., and Colin Stine, Ph.D. (supported by U01 H46274); NIMH Intramural Research Program, Clinical Neurogenetics Branch, Bethesda.—Elliot Gershon, M.D., Diane Kazuba, B.A., and Elizabeth Maxwell, M.S.W. This study was supported, in part, by National Institutes of Health grants R01 MH65560-01 and R01 MH59535 (both to E.S.G.), a National Alliance for Research on Schizophrenia and Depression (NARSAD) Young Investigator Award (to C.L.), a NARSAD Independent Investigator Award (to S.L.C.), a NARSAD Distinguished Investigator Award (to E.S.G.), and a grant from Japan Foundation for Aging and Health (to E.H.). Support from the Geraldi Norton Memorial Corporation, the Eklund family, and Anita Kaskel Roe is gratefully acknowledged. We also are grateful to Dr. Hongwei Zou and Ms. Jennifer Lyons, both of whose work on genotyping and sample management was invaluable, and Dr. Takeo Yoshikawa, for critical advice on experiments.
Footnotes
Presented, in part, in a slide show at the 52nd annual meeting of The American Society of Human Genetics, in Baltimore, on October 15–19, 2002.
Electronic-Database Information
The accession numbers and URLs for data presented herein are as follows:
- dbSNP Home Page, http://www.ncbi.nlm.nih.gov/SNP/
- DNannotator, http://sky.bsd.uchicago.edu/DNannotator.htm (for reference genomic DNA sequence)
- GenBank, http://www.ncbi.nlm.nih.gov/Genbank/ (for our G72 cDNA sequences [accession numbers AY170469–AY170471 and AY223901], cDNA G72/G30 sequences of Chumakov et al. [accession numbers AY138546–AY138548], chromosome 13 reference genomic contig [accession number NT_009952.10], and G72/G30 genomic sequences [accession numbers AE14312 and AE14313])
- Online Mendelian Inheritance in Man (OMIM), http://www.ncbi.nlm.nih.gov/Omim/ (for schizophrenia)
- SNP Consortium Ltd, The, http://snp.cshl.org/
- UCSC Genome Bioinformatics, http://genome.ucsc.edu/ (for Human Genome Browser Gateway)
References
- Abecasis GR, Cherny SS, Cookson WO, Cardon LR (2002) Merlin—rapid analysis of dense genetic maps using sparse gene flow trees. Nat Genet 30:97–101 [DOI] [PubMed] [Google Scholar]
- Badner JA, Gershon ES (2002) Meta-analysis of whole-genome linkage scans of bipolar disorder and schizophrenia. Mol Psychiatry 7:405–411 [DOI] [PubMed] [Google Scholar]
- Badner JA, Gershon ES, Goldin LR (1998) Optimal ascertainment strategies to detect linkage to common disease alleles. Am J Hum Genet 63:880–888 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berrettini WH (2000) Are schizophrenic and bipolar disorders related? A review of family and molecular studies. Biol Psychiatry 48:531–538 [DOI] [PubMed] [Google Scholar]
- Berrettini WH, Goldin LR, Martinez MM, Maxwell ME, Smith AL, Guroff JJ, Kazuba DM, Nurnberger JI Jr, Hamovit J, Simmons-Allings S, Muniec D, Choi H, York C, Robb AS, Gershon ES (1991) A bipolar pedigree series for genomic mapping of disease genes: diagnostic and analytic considerations. Psychiatr Genet 2:125–160 [Google Scholar]
- Christian SL, McDonough J, Liu CY, Shaikh S, Vlamakis V, Badner JA, Chakravarti A, Gershon ES (2002) An evaluation of the assembly of an approximately 15-Mb region on human chromosome 13q32-q33 linked to bipolar disorder and schizophrenia. Genomics 79:635–656 [DOI] [PubMed] [Google Scholar]
- Chumakov I, Blumenfeld M, Guerassimenko O, Cavarec L, Palicio M, Abderrahim H, Bougueleret L, et al (2002) Genetic and physiological data implicating the new human gene G72 and the gene for d-amino acid oxidase in schizophrenia. Proc Natl Acad Sci USA 99:13675–13680 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cohn CK, Dunner DL, Axelrod J (1970) Reduced catechol-O-methyltransferase activity in red blood cells of women with primary affective disorder. Science 170:1323–1324 [DOI] [PubMed] [Google Scholar]
- Cottingham RW Jr, Idury RM, Schäffer AA (1993) Faster sequential genetic linkage computations. Am J Hum Genet 53:252–263 [PMC free article] [PubMed] [Google Scholar]
- Craddock N, Dave S, Greening J (2001) Association studies of bipolar disorder. Bipolar Disord 3:284–298 [DOI] [PubMed] [Google Scholar]
- Craddock N, Jones I (1999) Genetics of bipolar disorder. J Med Genet 36:585–594 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Detera-Wadleigh SD, Badner JA, Berrettini WH, Yoshikawa T, Goldin LR, Turner G, Rollins DY, Moses T, Sanders AR, Karkera JD, Esterling LE, Zeng J, Ferraro TN, Guroff JJ, Kazuba D, Maxwell ME, Nurnberger JI Jr, Gershon ES (1999) A high-density genome scan detects evidence for a bipolar-disorder susceptibility locus on 13q32 and other potential loci on 1q32 and 18p11.2. Proc Natl Acad Sci USA 96:5604–5609 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dudbridge F, Koeleman BP, Todd JA, Clayton DG (2000) Unbiased application of the transmission/disequilibrium test to multilocus haplotypes. Am J Hum Genet 66:2009–2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Egan MF, Goldberg TE, Kolachana BS, Callicott JH, Mazzanti CM, Straub RE, Goldman D, Weinberger DR (2001) Effect of COMT Val108/158 Met genotype on frontal lobe function and risk for schizophrenia. Proc Natl Acad Sci USA 98:6917–6922 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gershon ES (2000) Bipolar illness and schizophrenia as oligogenic diseases: implications for the future. Biol Psychiatry 47:240–244 [DOI] [PubMed] [Google Scholar]
- Göring HH, Terwilliger JD, Blangero J (2001) Large upward bias in estimation of locus-specific effects from genomewide scans. Am J Hum Genet 69:1357–1369 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horikawa Y, Oda N, Cox NJ, Li X, Orho-Melander M, Hara M, Hinokio Y, et al (2000) Genetic variation in the gene encoding calpain-10 is associated with type 2 diabetes mellitus. Nat Genet 26:163–175 [DOI] [PubMed] [Google Scholar]
- Kong A, Cox NJ (1997) Allele-sharing models: LOD scores and accurate linkage tests. Am J Hum Genet 61:1179–1188 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu C, Badner JA, Christian SL, Guroff JJ, Detera-Wadleigh SD, Gershon ES (2001) Fine mapping supports previous linkage evidence for a bipolar disorder susceptibility locus on 13q32. Am J Med Genet 105:375–380 [DOI] [PubMed] [Google Scholar]
- Liu C, Gershon ES (2001) “SNP-Cruncher”: a Perl script toolbox for large-scale SNP data mining and its application on 13q32. Am J Hum Genet Suppl 69:449 [Google Scholar]
- Liu H, Heath SC, Sobin C, Roos JL, Galke BL, Blundell ML, Lenane M, Robertson B, Wijsman EM, Rapoport JL, Gogos JA, Karayiorgou M (2002) Genetic variation at the 22q11 PRODH2/DGCR6 locus presents an unusual pattern and increases susceptibility to schizophrenia. Proc Natl Acad Sci USA 99:3717–3722 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McPeek MS, Strahs A (1999) Assessment of linkage disequilibrium by the decay of haplotype sharing, with application to fine-scale genetic mapping. Am J Hum Genet 65:858–875 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niculescu AB III, Segal DS, Kuczenski R, Barrett T, Hauger RL, Kelsoe JR (2000) Identifying a series of candidate genes for mania and psychosis: a convergent functional genomics approach. Physiol Genomics 4:83–91 [DOI] [PubMed] [Google Scholar]
- Nurnberger JI Jr, DePaulo JR, Gershon ES, Reich T, Blehar MC, Edenberg HJ, Foroud T, et al (1997) Genomic survey of bipolar illness in the NIMH Genetics Initiative pedigrees: a preliminary report. Am J Med Genet 74:227–237 [DOI] [PubMed] [Google Scholar]
- O’Connell JR, Weeks DE (1998) PedCheck: a program for identification of genotype incompatibilities in linkage analysis. Am J Hum Genet 63:259–266 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ott J (1989) Computer-simulation methods in human linkage analysis. Proc Natl Acad Sci USA 86:4175–4178 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rozen S, Skaletsky HJ (2000) Primer3 on the WWW for general users and for biologist programmers. In: Krawetz S, Misener S (eds) Bioinformatics methods and protocols: methods in molecular biology. Humana Press, Totowa, NJ, pp 365–386 [DOI] [PubMed] [Google Scholar]
- Sham PC, Curtis D (1995) Monte Carlo tests for associations between disease and alleles at highly polymorphic loci. Ann Hum Genet 59 Part 1:97–105 [DOI] [PubMed] [Google Scholar]
- Shifman S, Bronstein M, Sternfeld M, Pisanté-Shalom A, Lev-Lehman E, Weizman A, Reznik I, Spivak B, Grisaru N, Karp L, Schiffer R, Kotler M, Strous RD, Swartz-Vanetik M, Knobler HY, Shinar E, Beckmann JS, Yakir B, Risch N, Zak NB, Darvasi A (2002) A highly significant association between a COMT haplotype and schizophrenia. Am J Hum Genet 71:1296–1302 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spielman RS, McGinnis RE, Ewens WJ (1993) Transmission test for linkage disequilibrium: the insulin gene region and insulin-dependent diabetes mellitus (IDDM). Am J Hum Genet 52:506–516 [PMC free article] [PubMed] [Google Scholar]
- Stefansson H, Sigurdsson E, Steinthorsdottir V, Bjornsdottir S, Sigmundsson T, Ghosh S, Brynjolfsson J, et al (2002) Neuregulin 1 and susceptibility to schizophrenia. Am J Hum Genet 71:877–892 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stine OC, McMahon FJ, Chen L, Xu J, Meyers DA, MacKinnon DF, Simpson S, McInnis MG, Rice JP, Goate A, Reich T, Edenberg HJ, Foroud T, Nurnberger JI Jr, Detera-Wadleigh SD, Goldin LR, Guroff J, Gershon ES, Blehar MC, DePaulo JR (1997) Initial genome screen for bipolar disorder in the NIMH Genetics Initiative pedigrees: chromosomes 2, 11, 13, 14, and X. Am J Med Genet 74:263–269 [PubMed] [Google Scholar]
- Straub RE, Jiang Y, MacLean CJ, Ma Y, Webb BT, Myakishev MV, Harris-Kerr C, Wormley B, Sadek H, Kadambi B, Cesare AJ, Gibberman A, Wang X, O’Neill FA, Walsh D, Kendler KS (2002) Genetic variation in the 6p22.3 gene DTNBP1, the human ortholog of the mouse dysbindin gene, is associated with schizophrenia. Am J Hum Genet 71:337–348 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weeks DE, Ott J, Lathrop GM (1990) SLINK: a general simulation program for linkage analysis. Am J Hum Genet Suppl 47:A204 [Google Scholar]