Abstract
Autism spectrum disorder (ASD) is a neurodevelopmental disability condition arising from a combination of genetic and environmental factors. Despite the blood-brain barrier (BBB) serving as a crucial gatekeeper, conveying environmental influences into the brain parenchyma, the contributions of BBB in ASD pathogenesis remain largely uncharted. Here we report that SHANK3, an ASD-risk gene, expresses in the BBB-forming brain endothelial cells (BECs) and regulates tight junctional (TJ) integrity essential for BBB’s barrier function. Endothelium-specific Shank3 (eShank3) knockout (KO) neonatal mice exhibit male-specific BBB-hyperpermeability, reduced neuronal excitability, and impaired ultra-sonic communications. Although BBB permeability is restored during adult age, the male mutant mice display reduced neuronal excitability and impaired sociability. Further analysis reveals that the BBB-hyperpermeability is attributed to the β-Catenin imbalance triggered by eShank3-KO. These findings highlight a pathogenic mechanism stemming from the ASD-risk Shank3, emphasizing the significance of neonatal BECs in the BBB as a potential therapeutic target for ASD.
Subject terms: Autism spectrum disorders, Blood-brain barrier, Social behaviour
This study reveals that SHANK3 deficiency in brain endothelial cells disrupts BBB function during the neonatal period through β-Catenin imbalance, leading to ASD-related neuronal and behavioral abnormalities.
Introduction
Despite decades of efforts investigating intrinsic functions of genes associated with autism spectrum disorder (ASD) in neurons and neuroglia, we still have limited insights into the pathogenic mechanisms of ASD due to an insufficient understanding of the etiological trajectory leading to ASD, resulting in a paucity of therapeutic options for this common but devastating mental illness. In the present study, we shift our focus to the blood-brain barrier (BBB), the most front-line interface between environmental factors and brain parenchyma, as one of the potential origins of ASD pathogenesis. While studies suggest that ASD arises from a complex interplay of genetic predisposition and environmental influences1, the pathogenic contribution of the BBB to this condition remains poorly understood.
The capillary wall of BBB is formed by adjoined brain endothelial cells (BECs) with additional support by astrocytes and pericytes2. BECs have specialized tight junctions (TJs) in which adhesion molecules, such as Zonula Occludens (ZO), Claudins, and Occludin play critical roles in preventing the free paracellular passage of molecules through the BBB by maintaining the intercellular integrity through complex connections between BECs3. Current hypotheses posit that the barrier system of BBB could be associated with the pathogenesis of ASD4–7. Until now, however, there is a lack of understanding of the precise mechanisms of how the genetic risk of ASD alters TJ integrity of BECs and leads to BBB breakdown, resulting in neuronal dysfunction and ASD-like behaviors.
SHANK3 is known as a critical synaptic scaffolding protein regulating the morphogenesis and function of synapses8. Deficiency in the SHANK3 gene has been reported as a monogenic cause of Phelan-McDermid syndrome (PMS), a neurodevelopmental disorder, characterized by developmental delay, intellectual disability, delayed or absent speech, and lack of sociability9–11. Recent human genetic studies have found that SHANK3 is one of the most frequent and penetrant genetic causes of ASD12,13. Thus, targeting Shank3 in rodents has been an important model system in studying ASD14–16. Although the role of Shank3 in neurons has been extensively investigated, its role in BECs remains unexplored.
In this study, we uncover the critical role of Shank3 in maintaining the integrity of BBB. We show that the Shank3 deficiency in BECs disrupts the BBB function in male neonatal mice by interfering with β-Catenin signaling in BECs, leading to persistent neuronal and behavioral abnormalities associated with ASD. Finally, we demonstrate that normalizing β-Catenin signaling in neonatal BECs significantly alleviates BBB dysfunction, neuronal abnormalities, and impaired social behaviors in eShank3-KO mice.
Results
A novel form of Shank3 is expressed in BECs
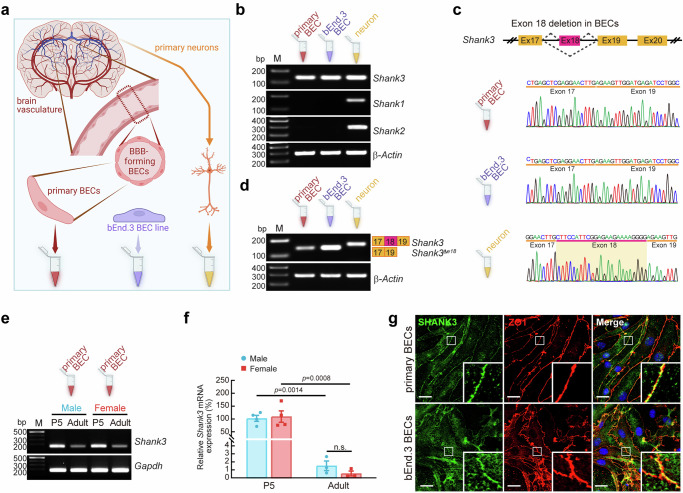
To examine the potential role of Shank3 in the BECs of BBB, we profiled Shank3 expression in BECs. PCR analysis with the mouse and human primary BECs, BEC lines [bEnd.3 (mouse), hCMEC/D3 (human)], and mouse primary neurons (Fig. 1a) revealed that Shank3 is the only Shank member expressed in BECs, while neurons express all three types of Shank genes (Fig. 1b, Supplementary Fig. 1a). Notably, the brain endothelial Shank3 lacks exon 18 (Shank3Δe18) in mouse and human primary BECs and BEC lines, as verified by sequencing and PCR analyses (Fig. 1c, d, Supplementary Fig. 1b, c). The histological confirmation of endothelial Shank3Δe18 (eShank3 hereafter) expression in BECs was conducted using in situ hybridization histochemistry (ISH) analysis with a probe specific for the exon 18 deletion. This analysis demonstrated that eShank3 mRNA is expressed in BECs in the brain vasculature (Supplementary Fig. 2).
Fig. 1. A novel Shank3 variant is expressed in BECs.
a Collection of cDNAs from RNA extracts of mouse primary BECs, bEnd.3 BECs, and primary neurons. Created in BioRender. Kim, S. (2025) https://BioRender.com/q10o614. b PCR analysis revealed that mouse primary BECs and bEnd.3 BECs express only Shank3 mRNA, while neurons express all Shank family. β-Actin: input control. Created in BioRender. Kim, S. (2025) https://BioRender.com/c22u541. c Sequencing chromatograms showing the lack of exon 18 in Shank3 mRNA in mouse primary BECs and bEnd.3 BECs, whereas neuronal Shank3 has exon 18. d PCR analysis confirmed exon 18-deleted Shank3 in the mouse primary BECs and bEnd.3 BECs. Shank3Δe18: exon 18 deletion form of Shank3. PCR (e) and qRT-PCR (f) analyses demonstrated that neonatal Shank3 mRNA expression at P5 (n = 4) was significantly higher than that in adult mice (n = 3), in both males and females. The graph represents the relative expression of Shank3 mRNA normalized to its levels in P5 male mice. Gapdh (Glyceraldehyde-3-phosphate dehydrogenase): input control. g ICC analysis revealed that eSHANK3 is mainly expressed in ZO1-positive cell-cell junctional area of mouse primary BECs and bEnd.3 BECs. Insets showing magnified views of the junctional areas. DAPI: nuclear staining. Scale bar, 20 μm. Each experiment b, d, g was independently repeated three times, yielding similar results. Data are mean ± SEM. Statistical analysis was performed using two-way ANOVA followed by Šídák’s multiple comparisons test (f). Detailed statistical methods and results are described in Supplementary Table 1. Source data are provided as Source Data File.
Next, we analyzed the expression pattern of eShank3 across sexes during postnatal development. We purified the primary BECs from the postnatal day 5 (P5) and 12-week-old male and female mice. PCR (Fig. 1e) and quantitative reverse transcription (qRT)-PCR (Fig. 1f) analyses demonstrated that the neonatal eShank3 mRNA expression in P5 BECs is significantly higher than in adult mice, with no differences between male and female (Fig. 1e, f). The robust expression of eShank3 in BECs during the neonatal period suggests a potential role of eShank3 in vasculature and BBB early in life.
Since various Shank3 isoforms arise via multiple intragenic promoters17, we measured the amount of each isoform’s transcript using a qRT-PCR assay and found that ‘a’ and ‘b’ forms are the predominant Shank3 isoforms in mouse primary and bEnd.3 BECs (Supplementary Fig. 1d, e). In comparison, primary neurons express other isoforms that account for ~40% (Supplementary Fig. 1e). Importantly, immunocytochemical (ICC) analyses using both primary BECs purified from mouse brain and bEnd.3 BEC line demonstrated the predominant expression of eSHANK3 protein within the ZO1-positive cell-to-cell junctional area (Fig. 1g), suggesting the role of eSHANK3 in regulating the junctional characteristics of BECs and, consequently, its potential impact on the barrier function of the BBB.
Increased BBB permeability in male eShank3-KO neonates
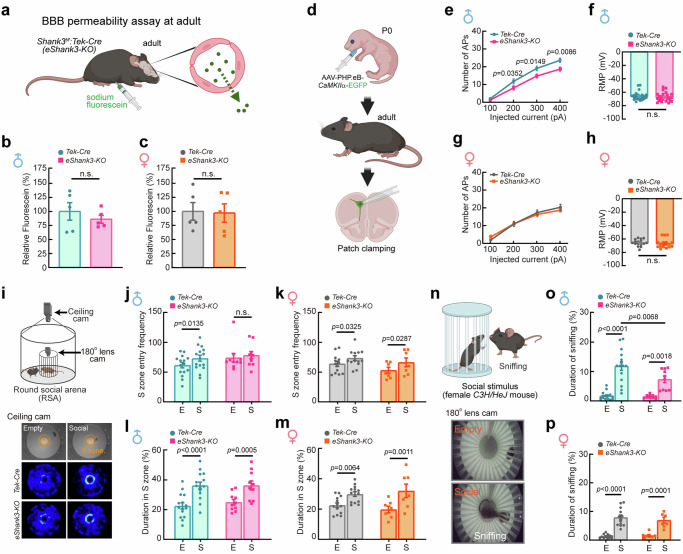
The expression of eSHANK3 in the junctions between BECs led us to promptly test whether the eShank3 deletion could affect the BBB permeability in vivo. We developed endothelium-specific Shank3 a/b knockout (KO) mice [Shank3Δe4-9 (deletion of a and b forms of Shank3)16:Tek-Cre18; eShank3-KO mice hereafter]. The specific Cre expression in endothelium and the depletion of eSHANK3 proteins in BECs were validated through whole brain 3-D imaging and Western blot (WB) analysis, respectively (Supplementary Fig. 3, Supplementary Movie 1). Given the robust neonatal expression of eSHANK3 at the junctions between BECs (Fig. 1e–g) and the fact that ASD commonly occurs in infancy, we examined the paracellular BBB permeability19,20 of eShank3-KO mice at neonatal age. Sodium fluorescein (376.27 Da) was administered via intraperitoneal (IP) injection to P5 mice, and the leakage of fluorescein into the brain parenchyma was measured by fluorescence spectrophotometry (Fig. 2a). This assay revealed a significant increase in paracellular BBB permeability in P5 male eShank3-KO mice compared to that of littermate Tek-Cre controls (Fig. 2b). However, the increased BBB leakage was not observed in female eShank3-KO neonates (Fig. 2c). Further analyses with various brain regions revealed that the increased BBB permeability occurred in several brain areas including the prefrontal cortex (PFC), cerebral cortex, and cerebellum of the male eShank3-KO neonates (Supplementary Fig. 4a–d), whereas no hyperpermeability was observed in any brain regions of female neonates (Supplementary Fig. 4e–h).
Fig. 2. Endothelial SHANK3 deficiency leads to defects in BBB permeability, neuronal function, and ultrasonic communication in neonatal mice.
a Schematic illustration of BBB permeability assay at P5. Created in BioRender. Kim, S. (2025) https://BioRender.com/g37y692. Relative amount of sodium fluorescein leaked into brain parenchyma of male (b) and female (c) Tek-Cre control (n = 17 for male, n = 16 for female) and eShank3-KO (n = 12 for male, n = 14 for female) mice at P5. d Strategy to label the neonatal pyramidal neurons with high probability of being impacted by BBB dysfunction. AAV-PHP.eB-CaMKIIα-EGFP injected into the STV of a P0 pup marked PFC neurons adjacent to CD31-positive endothelium at P5. Created in BioRender. Kim, S. (2025) https://BioRender.com/b65v147. e Current-clamp recordings of GFP-positive mPFC pyramidal neurons in the prelimbic region at P5. Analysis of neuronal excitability in mPFC of eShank3-KO male (f–h) and female (i–k) at P5. Representative traces of APs responded to 100, 200, 300 and 400 pA current injections, respectively (f, i). Graphs represent the number of evoked APs at the indicated current injection (g, j) and the RMP (h, k) in eShank3-KO (n = 14 for male, n = 10 for female from 3 mice per group) and Tek-Cre control (n = 14 for male, n = 8 for female from 3 mice per group) groups. l–p Ultrasonic communication testing at P5. l Spectrograms show representative USVs emitted by P5 male eShank3-KO and Tek-Cre control pups separated from their dams. Graphs show the number of USVs calls per min (m, o) and mean call duration (n, p) from male (m-n) and female (o, p) eShank3-KO (n = 10 for male, n = 7 for female) and Tek-Cre control (n = 13 for male, n = 13 for female) pups. Created in BioRender. Kim, S. (2025) https://BioRender.com/v30v894. Data are mean ± SEM. Statistical analysis was performed using two-tailed unpaired t-tests (b, c, h, k–p) and two-way ANOVA with repeated measures followed by two-stage linear step-up procedure of Benjamini, Kreiger and Yekutieli (false discovery rate) (g, j). Detailed statistical methods and results are described in Supplementary Table 1. Source data are provided as Source Data File.
We next examined whether the hyperpermeability of male eShank3-KO mice could be mediated by an increased transcellular transport across the BBB. However, IgG leakage assay revealed that all four groups of mice exhibited similar levels of transcellular permeability (Supplementary Fig. 5), suggesting that the increased BBB permeability is not due to alterations in transcellular transport.
Endothelial Shank3-KO reduces neuronal excitability and ultrasonic communication in male neonates
Could the BBB dysfunction due to eShank3 deletion contribute to the neuronal and behavioral deficits associated with ASD? We hypothesized that increased BBB permeability during neonatal period may provoke brain parenchyma damage, leading to neuronal and behavioral abnormalities. To test whether the eShank3 deletion could affect the neonatal brain parenchyma, we monitored the expression levels of glial fibrillary acidic protein (GFAP), a marker of reactive astrocytes, in the brains of eShank3-KO pups. Immunohistochemical (IHC) analysis with P5 brains revealed the significantly increased expression of GFAP in the medial prefrontal cortex (mPFC) and somatosensory cortex (S1) regions only in male eShank3-KO pups (Supplementary Fig. 6a–j), suggesting the potential male-specific neuronal dysfunction in the eShank3-KO neonates. To test this possibility, we examined the excitability of neurons in the eShank3-KO mice at P5. To label the excitatory neurons with a high probability of receiving influences from the BBB, we transduced AAV-PHP.eB-CaMKIIα-EGFP21 through the superficial temporal vein (STV) at P0 and found GFP-positive neurons in close proximity to CD31-positive blood vessels at P5 (Fig. 2d). Current-clamp recording was conducted by targeting the GFP-positive pyramidal neurons in the mPFC (Fig. 2e). The mPFC is a critical brain region exhibiting BBB hyperpermeability in eShank3-KO pups (Supplementary Fig. 4a) and is well known for its strong association with ASD-related behaviors across species22–24. The frequency of action potentials (APs) evoked by current injections was significantly decreased in P5 male eShank3-KO pups compared to Tek-Cre controls (Fig. 2f, g). However, the AP frequency in P5 female eShank3-KO pups did not change (Fig. 2i, j). There was no significant alteration in resting membrane potential (RMP) in eShank3-KO pups for both males and females (Fig. 2h, k). Collectively, these data suggest that the neonatal BBB dysfunction arising from the eShank3 deletion in BECs reduces the excitability of the genetically intact neurons in male neonates, which may suppress the output function of these neurons.
To evaluate the behaviors of eShank3-KO neonates, we employed the ultrasonic vocalization (USV) test, which assesses communication deficits frequently observed in children with ASD25–27. P5 eShank3-KO or Tek-Cre littermate control pups were separated from their dam, and the USV calls emitted by the pups were recorded (Fig. 2l). The number and mean duration of USV calls were significantly decreased in P5 male eShank3-KO pups compared to male Tek-Cre controls (Fig. 2m, n, Supplementary Movie 2). This neonatal communication impairment was not observed in female eShank3-KO pups (Fig. 2o, p).
Normal BBB permeability while sustained neuronal and behavioral abnormalities in adult male eShank3-KO mice
Autistic behaviors typically emerge in infancy and mostly persist across the lifespan. Thus, we asked whether the BBB, neuronal, and behavioral deficits in eShank3-KO neonates would endure into adulthood. To address this question, our initial focus was on assessing BBB permeability in adult eShank3-KO mice. Interestingly, in contrast to neonates, adult eShank3-KO mice exhibited intact BBB permeability in both males and females (Fig. 3a–c). Moreover, there were no differences in GFAP expression between the eShank3-KO and control Tek-Cre adult mice (Supplementary Fig. 6k–r). These results suggest that, through a compensatory mechanism yet to be determined, the impaired BBB might be restored as these mice reach adulthood.
Fig. 3. Persistent effects of eSHANK3 deficiency on neuronal function and social behavior in adult mice.
a Schematic illustration of BBB permeability assay at adult (17–19 week-old). Created in BioRender. Kim, S. (2025) https://BioRender.com/g37y692. Relative amount of NaF leaked into brain parenchyma of male (b) and female (c) Tek-Cre control and eShank3-KO (n = 5 per group) mice. d Labeling of the pyramidal neurons with high probability of being affected by neonatal BBB dysfunction by AAV-PHP.eB-CaMKIIa-EGFP injection into the STV at P0. Patch clamp analyses for the GFP-labeled mPFC neurons were conducted at adult age (17–19 week-old). Created in BioRender. Kim, S. (2025) https://BioRender.com/b65v147. e–h Analysis of neuronal excitability in the mPFC of adult male (e) and female (g) eShank3-KO and Tek-Cre control mice. Graphs represent the number of evoked APs at the indicated current injection (e, g) and the RMP (f, h) in male and female eShank3-KO (n = 25 for male, n = 18 for female from 5 mice) and Tek-Cre control (n = 24 for male from 5 mice, n = 16 for female from 4 mice) mice. i–p Sociability testing at adult age (17-19 week-old) using RSA assay. i A representative heatmaps show the movement traces of eShank3-KO and Tek-Cre control mice. Graphs display the S zone (3 cm radius from the inner cage) entry frequency (j, k), total duration spent in S zone (l, m) of male and female eShank3-KO (n = 10 for male, n = 7 for female) and Tek-Cre control (n = 15 for male, n = 13 for female) mice. n Representative snapshots of sniffing behavior captured by a 180° fish-eye camera installed in the inner cage. Total duration of sniffing behaviors of male (o) and female (p) eShank3-KO and Tek-Cre control mice. Created in BioRender. Kim, S. (2025) https://BioRender.com/v30v894. Data are mean ± SEM. Statistical analysis was performed using two-tailed unpaired t-tests (b, c, f, h, k–p), two-way ANOVA with repeated measures followed by two-stage linear step-up procedure of Benjamini, Kreiger and Yekutieli (false discovery rate) (e, g), and two-way ANOVA followed by Šídák’s multiple comparisons test (j–p). Detailed statistical methods and results are described in Supplementary Table 1. Source data are provided as Source Data File.
We then assessed neuronal excitability in adult mice (17–19 week-old) by conducting current-clamp recordings targeting the GFP-positive mPFC pyramidal neurons that were labeled by AAV-PHP.eB-CaMKIIα-EGFP at P0 (Fig. 3d). The AP frequency of the neurons in adult male eShank3-KO brains was significantly lower than that of male Tek-Cre controls, as observed in neonates (Fig. 3e). However, this hypo-excitability was not found in female eShank3-KO mice (Fig. 3g). There were no differences in RMP between each group of mice (Fig. 3f, h). These results suggest that the neuronal impairment driven by neonatal BBB damage persists into adult age despite restoration of BBB permeability.
Next, we investigated social and repetitive behaviors, the core behavioral domains commonly affected by ASD28. To precisely monitor detailed social behaviors, we used a Round Social Arena (RSA) paradigm as described in our previous studies23,29 (Fig. 3i). In the first phase, the adult test mouse (17–19 week-old) explores the arena with an empty inner cage (E). In the second phase, the mouse is allowed to interact with a social stimulus (P30-P50 C3H/HeJ mouse) placed in the inner cage (S). We monitored the frequency of entry into the social zone (S zone) as well as the duration in the S zone. Quantification revealed that male and female Tek-Cre control and female eShank3-KO mice exhibited an increased frequency of entries into the S zone when a social stimulant was present in the inner cage (Fig. 3j, k). In contrast, male eShank3-KO mice showed a consistent entry frequency into the S zone, irrespective of the presence or absence of a social stimulus (Fig. 3j), indicating a reduced preference for the social stimulus in male eShank3-KO mice compared to male Tek-Cre control mice. However, the total duration in the S zone was not different between the eShank3-KO and Tek-Cre controls (Fig. 3l, m). To examine the quality of sociability in the S zone, we monitored direct social interaction (sniffing) via a fish-eye camera installed in the inner cage (Fig. 3n, Supplementary Movie 3). Quantification demonstrated that all four groups of mice spent more time sniffing the social stimulus than the empty inner cage (Fig. 3n–p). Notably, male eShank3-KO mice sniffed significantly less with the social stimulus compared to male Tek-Cre controls (Fig. 3o). However, this difference was not significant in female mice (Fig. 3p), indicating male-specific reduction in sociability in eShank3-KO mice. Note that this social behavioral abnormality in male eShank3-KO mice was consistently observed during interactions with both same-sex (male-male) and opposite-sex (male-female) social stimuli (Fig. 3j, o, Supplementary Fig. 7).
We also examined the repetitive response, motor performance, and anxiety-like behaviors of each group of mice. In the grooming and marble-burying assays, male eShank3-KO mice displayed significantly increased repetitive behaviors compared to other groups of animals (Supplementary Fig. 8a–d). However, open field and light-dark box tests revealed normal motor function and anxiety level (Supplementary Fig. 8e–r), suggesting that the effects of eShank3-KO in BECs are confined to the social and repetitive behaviors of male mice. Overall, while the impact of eShank3 deletion on BBB damage is limited to neonatal stages, the neuronal and specific behavioral deficits persist until adult age, particularly in male mice.
eSHANK3 interacts with tight junction proteins in BECs
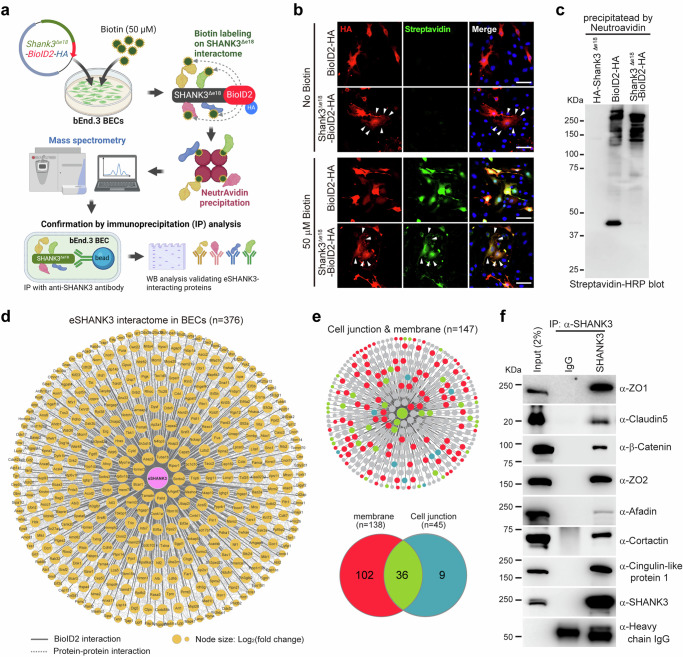
Although numerous SHANK3 interacting proteins in neurons have been identified8,30,31, the specific eSHANK3 interactome in BECs remains unexplored. We anticipated that mapping the eSHANK3 interactome in BECs could provide fundamental insight into the unique role of eSHANK3 in BBB integrity. To profile the eSHANK3 interactome in BECs, we created a specific probe by fusing Shank3Δe18 and a biotin ligase (Shank3Δe18-BioID2-HA) to label biotin onto the eSHANK3-interacting proteins (Fig. 4a), as previously described32. For negative controls, BioID2-HA and Shank3Δe18-HA were used. After validating the successful biotinylation (Fig. 4b, Supplementary Fig. 9a–c), eSHANK3-interacting proteins were purified using NeutrAvidin beads (Fig. 4c) and subjected to proteomic analysis. Mass spectrometry analysis, with four replicates for each group, identified 376 proteins that are significantly enriched in the Shank3Δe18-BioID2 group compared to BioID2 or Shank3Δe18 controls (Fig. 4d, Supplementary Data 1). As expected from our ICC results showing eSHANK3 expression in the junctional area (Fig. 1g), Gene Ontology (GO) analysis revealed a prominent and statistically significant enrichment of eSHANK3-interacting proteins associated with membrane and cell junction functions (Fig. 4e, Supplementary Fig. 9d, e). The interactions between eSHANK3 and some candidate proteins (e.g., ZO1, Claudin5, and β-Catenin) were confirmed by immunoprecipitation (IP) analysis using bEnd.3 BECs (Fig. 4f). Given that eSHANK3 interacts with critical TJ proteins, we reasoned that the loss of eSHANK3 could affect the TJ integrity in BECs.
Fig. 4. TJ and membrane proteins are major eSHANK3 interactome in BECs.
a Workflow of BioID2 approach illustrating biotinylation of eSHANK3-interacting proteins, purification of the biotinylated proteins by NeutrAvidin agarose beads, mass spectrometry analysis, and validation through IP analysis. Created in BioRender. Kim, S. (2025) https://BioRender.com/q75n212. b ICC analysis using anti-HA antibody and Alexa Fluor® 488-Streptavidin shows colocalization of eSHANK3-BioID2-HA (or BioID2-HA) and biotin labeled proteins only in the biotin-treated bEnd.3 BECs. Arrowheads indicate the co-expression of SHANK3Δe18-BioID2 and biotinylated proteins in the cell membrane. Scale bars, 50 μm. Blue: DAPI. c Streptavidin blot analysis, using purified biotinylated proteins, showed the protein bands of various sizes that were biotinylated by BioID2-HA or Shank3Δe18-BioID2-HA. Four biological replicates were analyzed, all yielding consistent results. 5% of purified proteins were loaded for the blotting analysis. 95% of purified proteins were used for subsequent mass spectrometry analysis. d Mass spectrometry analysis identified 376 candidate eSHANK3 interacting proteins. Node size represents protein abundance [log2 (fold change)] over eSHANK3Δe18-only (no BioID2) and BioID2-only control groups. Solid gray edges delineate interactions between the eSHANK3 and the identified interactome. Dashed edges indicate known interactions. e GO analysis revealed membrane and junctional proteins are the primary eSHANK3 interactome in the bEnd.3 BECs. f IP analysis confirmed the interactions between endogenous eSHANK3 and some of the identified interactome in the bEnd.3 BECs. Three times independent experiments yielded similar results. IgG: control for non-specific antibody binding. Heavy chain IgG: loading control. The detailed information of the identified eSHANK3 interactome and GO analysis are provided in Supplementary Data 1. Source data are provided as Source Data File.
eSHANK3 deficiency disrupts tight junctions in BECs
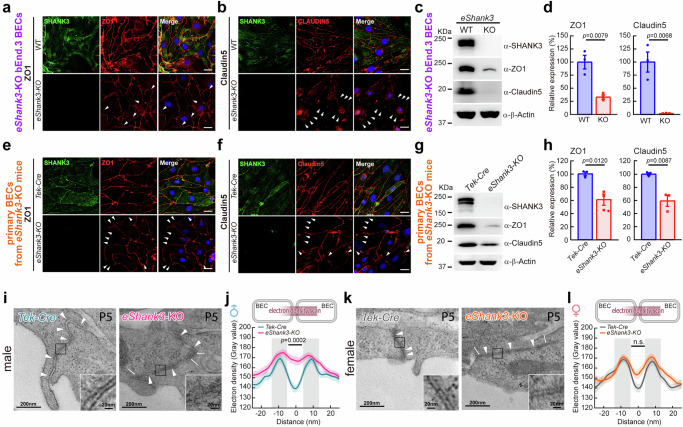
Due to the extremely low yield of primary BECs purified from mouse brains, we generated a stable eShank3-KO bEnd.3 BEC line using CRISPR strategy to expedite the investigations of eSHANK3 function in BECs (Supplementary Fig. 10). Because eSHANK3 turned out to be a part of the TJ complex in BECs, we speculated that the loss of eSHANK3 could destabilize the eSHANK3-interacting TJ proteins32. Indeed, ICC analysis revealed reduced, discontinuous, and patchy expression of ZO1 and Claudin5, the key components of TJ (Fig. 5a, b), and WB analysis confirmed reduced levels of ZO1 and Claudin5 in the eShank3-KO BECs (Fig. 5c, d), while other TJ-related proteins were unchanged or slightly upregulated (Supplementary Fig. 11a, b).
Fig. 5. eSHANK3 depletion leads to aberrant ZO1/Claudin5 expressions and structural abnormalities in the junctions between BECs.
a–h eShank3 deletion effects on ZO1 and Claudin5 in BECs. ICC analysis showed discontinuous and patched (arrowheads) expressions of ZO1 and Claudin5 in both eShank3-KO bEnd.3 BEC line (a, b) and primary BECs from eShank3-KO mice (e, f) compared to controls. Blue: DAPI. Scale bars, 20 μm. WB analysis showing reduced expressions of ZO1 and Claudin5 in the eShank3-KO bEnd.3 BEC line (c, d) and primary BECs from eShank3-KO mice (g, h) compared to WT controls (n = 3 per group). Band intensities were normalized with β-Actin. i–l Ultrastructure of junctional area between BECs in the PFC. Representative electron micrographs of BEC junctions observed in the brain capillaries of P5 male (i) and female (k) eShank3-KO mice and control Tek-Cre mice (n = 3 mice per group). Arrowheads indicate cell-cell junctions along the adjacent BECs. Arrows indicate abnormal high electron densities in the cleft between BEC membranes. Insets are magnified views of the junctional areas. j Electron density scanning across the junction between BECs revealed elevated electron densities in the cleft of P5 male eShank3-KO mice. l However, this phenotype was not significant in female group. Data are mean ± SEM. Statistical analysis was performed using two-tailed unpaired t-tests (d, h) and mixed-effects analysis followed by Šídák’s multiple comparisons test (j, l). Detailed statistical methods and results are described in Supplementary Table 1. Source data are provided as Source Data File.
Alterations in ZO1 and Claudin5 were also observed in primary BECs purified from the brains of eShank3-KO mice. First of all, we verified the loss of eShank3 gene in the eShank3-KO primary BECs (Supplementary Fig. 12). ICC and WB analyses showed the diminished and discontinuous expression of ZO1 and Claudin5 in the eShank3-KO primary BECs compared to those of control Tek-Cre mice (Fig. 5e–h, Supplementary Fig. 11c, d). Additionally, IHC analysis revealed that the expression of ZO1 and Claudin5 in the brain vasculature was significantly reduced in eShank3-KO pups compared to Tek-Cre pups at P5 (Supplementary Fig. 11e–h).
Studies have shown that Claudin5, an integral membrane protein, is a critical TJ protein that forms a complex with the intracellular assemblers (e.g., ZO1/2) to complete a TJ structure33–35 which is essential for BBB integrity36. Our additional IP analysis demonstrated that Claudin5 interacts with eSHANK3 via the PDZ domain of eSHANK3 (Supplementary Fig. 13). We reasoned that the Claudin5-eSHANK3 interaction may contribute to the stabilization of the TJ in BECs. Consequently, the deletion of eShank3 could disrupt TJ structure in the BECs, leading to BBB dysfunction.
To further explore this idea, we examined the ultrastructure of TJs in eShank3-KO neonatal brains using transmission electron microscopy. P5 Tek-Cre mice (controls) exhibited tightly connected TJs between adjacent BECs and maintained a constant cleft width of ~10 nm, whereas male P5 eShank3-KO mice frequently displayed indistinct and discontinuous BEC membrane layers and irregular cleft widths between BECs (Fig. 5i). Moreover, density-scanning analysis conducted across the cleft revealed that the male mutant pups exhibit atypical electron densities dispersed along the intercellular cleft between BECs compared to Tek-Cre control (Fig. 5j), indicating altered junctional structure in eShank3-KO BECs. Again, this structural abnormality was not significant in female eShank3-KO mice (Fig. 5k, l).
eSHANK3 regulates ZO1 and Claudin5 via β-Catenin signaling
We proceeded to investigate the mechanisms underlying the downregulation of ZO1 and Claudin5 due to Shank3 deletion in BECs. SHANK3 is known to interact with β-Catenin in the brain37, and our proteomic and IP analyses demonstrated that this interaction also occurs in BECs (Fig. 4d–f). Further co-IP analysis showed the interactions between eSHANK3 and β-Catenin destruction complex proteins such as β-CateninS33Y (stable form), Casein kinase 1α (CK1α), and Glycogen synthase kinase 3β (GSK3β) (Fig. 6a), suggesting a potential role of eSHANK3 in the β-Catenin destruction machinery in the BECs. β-Catenin is located at the peripheral membrane via interaction with E-cadherin38 and is released to the cytoplasm and degraded by the β-Catenin destruction complex. However, in the presence of a Wnt stimulus, β-Catenin is translocated to the nucleus and regulates the transcription of Wnt/β-Catenin target genes39,40. The activation of β-Catenin is known to downregulate ZO1 and Claudin5 expression41,42. Thus, we tested whether eShank3-KO could alter the β-Catenin level in BECs. WB analysis using fractionated BECs showed a significant accumulation of total β-Catenin as well as active phospho-β-CateninS552 41 in both cytoplasm and nucleus of eShank3-KO bEnd.3 BECs compared to WT controls (Fig. 6b–d).
Fig. 6. eSHANK3 regulates the TJ integrity and barrier function via β-Catenin destruction pathway.
a Co-IP analysis showing the interaction between eSHANK3 (HA-SHANK3Δe18) and β-Catenin destruction complex including β-CateninS33Y-FLAG, V5-CK1α, and GSK3β-Myc. β-Actin: loading control. b Subcellular fractionation followed by WB analysis using eShank3-KO and WT bEnd.3 BECs (n = 3). c Phospho-β-CateninS552 in nucleus and cytoplasm were increased in bEnd.3 BECs by eShank3 disruption. α-Tubulin: cytoplasmic marker. Histone H3: nuclear marker. d Hypothetical model illustrating enhanced nuclear translocation of β-CateninS552 by eSHANK3 depletion in BECs. Created in BioRender. Kim, S. (2025) https://BioRender.com/l10t237. e–j, IWR-1-endo (1 μM) effects on eShank3-KO BECs. e WB analysis with IWR-1-endo-treated fractionated eShank3-KO bEnd.3 BECs. f Enhanced phospho-β-CateninS552 level was normalized upon IWR-1-endo treatment in cytoplasm and nucleus (n = 3). g WB analysis for ZO1 and Cladudin5 levels in eShank3-KO bEnd.3 BECs following IWR-1-endo treatment. h Reduced ZO1 and Cladudin5 in eShank3-KO bEnd.3 BECs were restored by IWR-1-endo treatment (n = 3). TWP (n = 4) (i) and TEER (n = 12) (j) assays revealed that IWR-1-endo treatment rescues hyperpermeability of eShank3-KO bEnd.3 BECs. Arrow: time point of IWR-1-endo treatment. Bar graph (j; right panel) showed impedance at 95 h time point. k–p GSK3β activation effects on eShank3-KO bEnd.3 BECs. k WB analysis measuring total β-Catenin and phospho-β-CateninS552 levels in GSK3β S9A/eShank3-KO double mutant bEnd.3 BECs. l The increased β-CateninS552 level in eShank3-KO bEnd.3 BECs was normalized by GSK3βS9A overexpression both in cytoplasm and nucleus (n = 3). m WB analysis for ZO1 and Claudin5 levels in GSK3βS9A/eShank3-KO bEnd.3 BECs. The HA band showed the overexpression of GSK3βS9A in double mutant bEnd.3 BECs. n The reduced expressions of ZO1 and Claudin5 by eSHANK3 depletion were significantly restored by GSK3βS9A overexpression (n = 3). TWP (n = 4) (o) and TEER (n = 12) (p) assays revealed that GSK3βS9A overexpression rescues hyperpermeability of eShank3-KO bEnd.3 BECs. Bar graph (p; right panel) showed impedance at 95 h time point. Data are mean ± SEM. Statistical analysis was performed using a two-tailed unpaired t-test (c) and one-way ANOVA followed by Dunnett’s multiple comparisons test (f, h–j, l, n–p). Detailed statistical methods and results are described in Supplementary Table 1. Source data are provided as Source Data File.
To examine whether β-Catenin is involved in TJ disruption in eShank3-KO BECs, we treated eShank3-KO bEnd.3 BECs with IWR-1-endo, a Wnt/β-Catenin inhibitor43, resulting in the reduction of cytoplasmic and nuclear β-Catenin to the level of WT BECs (Fig. 6e, f). Moreover, the reduced ZO1 and Claudin5 proteins and mRNAs were significantly restored by IWR-1-endo treatment (Fig. 6g, h, Supplementary Fig. 14a, b). IWR-1-endo effects on permeability in the eShank3-KO bEnd.3 BECs were measured by both trans-well permeability (TWP) and trans-endothelial electrical resistance (TEER) assays. Quantification of the TWP revealed increased passage of FITC-Dextran through the mutant BEC monolayer, which was normalized by IWR-1-endo treatment (Fig. 6i). The TEER assay demonstrated reduced impedance in eShank3-KO bEnd.3 BECs compared to the WT BEC control. However, IWR-1-endo treatment significantly elevated impedance levels in eShank3-KO bEnd.3 BECs (Fig. 6j). These results suggest that IWR-1-endo has a rescuing effect on the barrier dysfunction observed in eShank3-KO bEnd.3 BECs.
To validate the rescue effects of the IWR-1-endo treatment on BECs, we generated stable eShank3-KO BEC line that express constitutively active GSK3βS9A (3X HA-GSK3βS9A / eShank3-KO double mutant bEnd.3 BECs) to facilitate the β-Catenin destruction pathway44 (Supplementary Fig. 14c). Note that the active form of GSK3β phosphorylates β-Catenin, leading to enhanced β-Catenin degradation by the proteasome45–47. WB analysis confirmed normalized β-Catenin and phospho-β-CateninS552 levels in both the cytoplasm and nucleus of the 3X HA-GSK3βS9A / eShank3-KO double mutant bEnd.3 BECs (Fig. 6k, l). In addition, we observed significantly restored ZO1 and Claudin5 protein and mRNA levels in the 3X HA-GSK3βS9A / eShank3-KO double mutant bEnd.3 BECs (Fig. 6m, n, Supplementary Fig. 14d, e). TWP and TEER assays revealed that 3X HA-GSK3βS9A expression substantially restored the impaired barrier functions observed in eShank3-KO bEnd.3 BECs (Fig. 6o, p). These results indicate that eShank3-KO-mediated β-Catenin accumulation inhibits the expressions of ZO1 and Claudin5, leading to defects in TJ integrity and paracellular permeability.
The transcription of Claudin5 is known to be regulated by β-Catenin via transcription factor C/EBPα41. Inhibition of β-Catenin’s transcriptional activity upregulates C/EBPα, subsequently promoting the transcription of Claudin541. In our investigation, we observed a significant downregulation of both C/EBPα mRNA and protein levels in the eShank3-KO bEnd.3 BECs compared to WT bEnd.3 BECs (Supplementary Fig. 14f–h). Thus, we hypothesized that overexpressing C/EBPα could increase the Claudin5 mRNA expression in eShank3-KO bEnd.3 BECs, restoring the impaired barrier function. To test this hypothesis, we generated a stable 3X HA-C/EBPα (overexpression) / eShank3-KO double mutant bEnd.3 BEC line (Supplementary Fig. 14i). qRT-PCR analysis revealed a normalized Claudin5 mRNA level in the double mutant bEnd.3 BECs (Supplementary Fig. 14j, k), elucidating a Claudin5 disruption mechanism caused by eSHANK3 depletion through the β-Catenin-C/EBPα transcriptional pathway. The permeability of the double mutant bEnd.3 BECs was assessed by TEER assay, showing a significantly restored impedance level compared to that of eShank3-KO bEnd.3 BECs (Supplementary Fig. 14l, m).
These results suggest that eSHANK3 plays a crucial role in regulating TJ and BBB functions by interacting with the β-Catenin destruction complex to ensure the maintenance of optimal levels of β-Catenin, C/EBPα, and TJ proteins in BECs.
GSK3β activation in BECs restores BBB and neuronal dysfunctions in eShank3-KO mice
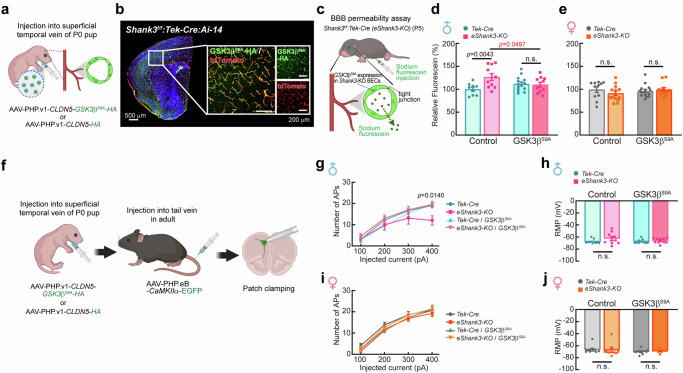
Our next question is whether the normalization of β-Catenin signaling specifically in BECs could improve the BBB dysfunction of P5 eShank3-KO mice in vivo. To address this question, we expressed GSK3βS9A exclusively in the BECs of eShank3-KO mice by administering AAV-PHP.v1 (brain endothelium-specific serotype)48,49-CLDN5 (Claudin5 promoter)-GSK3βS9A-HA or control AAV-PHP.v1-CLDN5-HA through STV at P0 (Fig. 7a). After confirming the specific expression of GSK3βS9A-HA in brain endothelium at P5 (Fig. 7b), we conducted a BBB permeability assay at the same stage (Fig. 7c). The results revealed that the elevated fluorescein leakage observed in male eShank3-KO pups was restored to normal level by the targeted overexpression of active GSK3βS9A-HA within BECs (Fig. 7d, e), indicating the rescuing effect of GSK3βS9A on the neonatal BBB hyperpermeability induced by eSHANK3 disruption in vivo.
Fig. 7. Restoration of neonatal BBB function through GSK3β activation in BECs normalizes neurological function in adult mice.
a–e GSK3βS9A overexpression effect on the BBB permeability in vivo. a Injection of AAV-PHP.v1-CLDN5-GSK3βS9A-HA through STV at P0. b IHC analysis using Shank3f/f:Tek-Cre:Ai-14 triple mutant mice shows specific expression of GSK3βS9A-HA (green) in the tdTomato-positive BECs of P5 brain. c Schematic illustration of BBB permeability assay using AAV-PHP.v1-CLDN5-GSK3βS9A-HA-injected eShank3-KO mice at P5. In vivo BBB permeability was assessed with P5 male (d) and female (e) eShank3-KO and control Tek-Cre mice injected by AAV-PHP.v1-CLDN5-GSK3βS9A-HA (n = 10 for male eShank3-KO, n = 13 for male Tek-Cre, n = 11 for female eShank3-KO, n = 14 for female Tek-Cre) or control AAV-PHP.v1-CLDN5-HA (n = 10 for male eShank3-KO, n = 9 for male Tek-Cre, n = 13 for female eShank3-KO, n = 11 for female Tek-Cre) at P0. The increased BBB permeability in male eShank3-KO pups was normalized by overexpression of GSK3βS9A in the BECs. Created in BioRender. Kim, S. (2025) https://BioRender.com/z54o523. f GSK3βS9A overexpression effect on the neuronal excitability. AAV-PHP.v1-CLDN5-GSK3βS9A-HA or control AAV-PHP.v1-CLDN5-HA) was injected into STV at P0 to express GSK3βS9A in the BECs from the neonatal period. To label the excitatory neurons, AAV-PHP.eB-CaMKIIα-EGFP was injected into the tail vein of adult mice (13–15 weeks old). Patch clamp analysis was then performed on GFP-labeled mPFC neurons of 17–19 week-old mice. Created in BioRender. Kim, S. (2025) https://BioRender.com/r42u905. g–j Analysis of neuronal excitability in adult male (n = 9 for eShank3-KO with HA expression, n = 11 for Tek-Cre with HA expression, n = 10 for eShank3-KO with GSK3βS9A expression, n = 9 for Tek-Cre with GSK3βS9A expression) (g) and female (n = 9 for each group) (i) eShank3-KO and Tek-Cre control mice, with or without GSK3βS9A expression in BECs. Graphs represent the number of evoked APs at the indicated current injection (g, i) and the RMP (h, j) of each group of mice. Data are mean ± SEM. Statistical analysis was performed using two-way ANOVA followed by Šídák’s multiple comparisons tests (d, e, h, j) and two-way ANOVA with repeated measures followed by two-stage linear step-up procedure of Benjamini, Kreiger and Yekutieli (false discovery rate) (g, i). Detailed statistical methods and results are described in Supplementary Table 1. Source data are provided as Source Data File.
We also examined whether the GSK3β activation in the BECs could normalize the neuronal dysfunction of eShank3-KO mice. AAV-PHP.v1-CLDN5-GSK3βS9A-HA was injected into the STV of male and female eShank3-KO (or Tek-Cre control) pups at P0 to activate GSK3β in the neonatal BECs (Fig. 7f). To label the excitatory neurons with a high probability of receiving influences from the BBB, AAV-PHP.eB-CaMKIIα-EGFP was administered via the tail vein to each group of mice one month prior to current-clamp recordings (Fig. 7f). The AP frequency of the GFP-positive pyramidal neurons in the mPFC was analyzed at adult age (17–19 week-old). As shown in Fig. 3e, without GSK3βS9A overexpression (control AAV-PHP.v1-CLDN5-HA expression), the neurons of male eShank3-KO mice exhibited significantly decreased AP frequency compared to male Tek-Cre controls (Fig. 7g). Importantly, this disrupted AP frequency of male eShank3-KO mice was normalized by the overexpression of GSK3βS9A in the BECs (AAV-PHP.v1-CLDN5-GSK3βS9A-HA) (Fig. 7g), demonstrating a rescue effect of the BEC-specific GSK3βS9A expression on the neuronal hypo-excitability observed in male eShank3-KO mice. However, in the female group, there was no difference in AP frequency between eShank3-KO mice and Tek-Cre controls, regardless of GSK3βS9A overexpression in BECs (Fig. 7i). Note, GSK3βS9A overexpression in BECs did not change the RMP of the excitatory neurons in both male and female eShank3-KO mice and Tek-Cre mice (Fig. 7h, j).
GSK3β activation in BECs restores social behavioral deficits in eShank3-KO mice
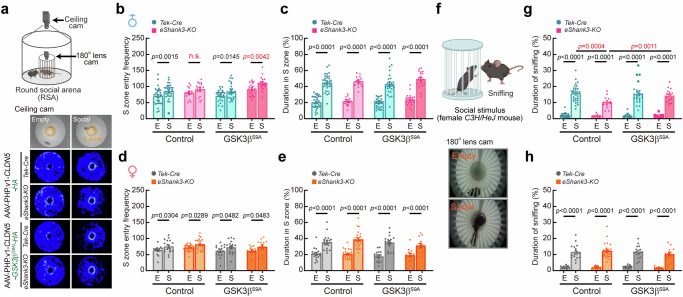
We also investigated whether activating GSK3β in BECs could rescue the social behavior deficits in eShank3-KO mice. Using the RSA paradigm, we analyzed the sociability of the mice by measuring their entry frequency into the S zone, both with and without a social stimulus (Fig. 8a). As shown in Fig. 3j, male eShank3-KO mice (injected by control AAV-PHP.v1-CLDN5-HA) entered similar times to both S zone (Fig. 8b-left), indicating social deficit in this group of mice. However, expression of AAV-PHP.v1-CLDN5-GSK3βS9A-HA in BECs of male eShank3-KO mice significantly increased the number of entries into the S zone in the presence of a social stimulus (Fig. 8b-right), indicating the rescue effect of GSK3βS9A expression in BECs on social behavior. Note that female eShank3-KO mice did not exhibit social deficits in this test (Fig. 8d).
Fig. 8. Restoration of neonatal BBB function through GSK3β activation in BECs normalizes social behavior in adult mice.
a–h Social behavioral tests at adult age (17-19 week-old) using RSA assay. a A representative heatmaps show the movement traces of male eShank3-KO and Tek-Cre control mice injected with AAV-PHP.v1-CLDN5-GSK3βS9A-HA or AAV-PHP.v1-CLDN5-HA. Graphs display the S zone (3 cm radius from the inner cage) entry frequency (b, d), total duration spent in S zone (c, e) of all groups of mice. f Representative snapshots of sniffing behavior captured by a 180° fish-eye camera installed in the inner cage. Total duration of sniffing behaviors of male (g) and female (h) eShank3-KO and Tek-Cre control mice injected with AAV-PHP.v1-CLDN5-GSK3βS9A-HA (n = 17 for male eShank3-KO, n = 24 for male Tek-Cre, n = 13 for female eShank3-KO, n = 17 for female Tek-Cre) or AAV-PHP.v1-CLDN5-HA (n = 13 for male eShank3-KO, n = 29 for male Tek-Cre, n = 24 for female eShank3-KO, n = 17 for female Tek-Cre). Created in BioRender. Kim, S. (2025) https://BioRender.com/v30v894. Data are mean ± SEM. Statistical analyses were performed using two-way ANOVA followed by Šídák’s multiple comparisons tests (b–e, g, h). Detailed statistical methods and results are described in Supplementary Table 1. Source data are provided as Source Data File.
We also analyzed the total duration spent in the S zone and found that all groups (eShank3-KO and Tek-Cre control; GSK3βS9A-HA and HA control; male and female) spent similar time in the S zone with a social stimulus (Fig. 8c, e). Thus, we monitored direct social interaction by analyzing the sniffing behavior with social stimulus in the S zone (Fig. 8f). This analysis revealed that male eShank3-KO mice exhibited significantly reduced sniffing behavior compared to Tek-Cre controls (Fig. 8g-left). Notably, the reduction of sniffing behavior in male eShank3-KO mice was restored by AAV-PHP.v1-CLDN5-GSK3βS9A-HA expression in BECs (Fig. 8g-right). However, female eShank3-KO mice exhibited normal sniffing behavior with social stimulus, regardless of GSK3βS9A expression (Fig. 8h).
We next assessed motor performance and anxiety-like behaviors across the groups. The open field test revealed that all groups of mice traveled similar distances in the field at comparable velocities (Supplementary Fig. 15a–d), indicating normal motor performance. Additionally, the time spent in the center and marginal areas of the open field was similar across groups (Supplementary Fig. 15e–h), suggesting normal anxiety levels in all groups of mice. The light-dark box test further confirmed that all groups exhibited similar levels of anxiety (Supplementary Fig. 15i–l). These results indicate that neither eShank3 deletion nor GSK3βS9A overexpression in BECs affects motor performance or anxiety-like behaviors.
Taken together, we demonstrated that rescuing TJ and BBB function in eShank3-KO mice through normalization of β-Catenin during the neonatal stage restores normal neuronal excitability and social behavior in adulthood. These findings highlight the β-Catenin pathway in BECs as a potential intervention target to alleviate TJ and BBB abnormalities in eShank3-related ASD.
Discussion
The present study provides novel insights into ASD research by emphasizing the BECs of BBB as one of the potential origins of ASD pathogenesis. We have demonstrated the mechanism through which the loss of ASD-risk gene Shank3 leads to TJ disruption in the BECs via perturbation of the β-Catenin signaling, resulting in neonatal BBB breakdown, deterioration of the excitability of genetically intact neurons, and behavioral abnormalities associated with ASD. Further studies are needed to investigate the yet undiscovered functions of other ASD-associated genes in the BECs to understand their role in regulating BBB integrity, neuronal function, and behavior. Indeed, we identified various ASD-associated genes (e.g., FMR1, MECP2, UBE3A, OXTR, NRGN3, ANK2, CDH9, CHD8, CYFIP2, HOXB1, KCNQ5, NLGN4, PTEN, and TSC1/2) that show high expression in human and mouse primary BECs (Supplementary Fig. 16). Therefore, there is a high likelihood that these ASD-risk genes may play their distinctive roles in maintaining normal BEC/BBB function. Future studies that concentrate on these ASD-associated genes within the context of BECs have the potential to broaden the ASD research landscape. This expansion could significantly contribute to unraveling the mechanisms underlying the pathogenesis of ASD, particularly those stemming from neonatal BBB dysfunction.
We unexpectedly but constantly observed male-predominant phenotypes in eShank3-KO mice, including abnormal TJ structure, increased neonatal BBB permeability, impaired neuronal excitability, and autistic-like behaviors. Although accumulated studies indicate a higher prevalence of ASD in males than in females, both in human patients and animal models50–52, the underlying mechanisms remain undetermined. Further comprehensive studies using the BBB-oriented model system could contribute to elucidating the mechanisms of why the female is more protected from the genetic risk of ASD. This insight may offer clues for developing new medical interventions or protective strategies to mitigate ASD development.
Another striking finding was that eShank3 deletion drove BBB dysfunction in a focal time point during neonatal age. This early BBB hyperpermeability continued to affect neuronal excitability, social behavior, and repetitive responses of adult male eShank3-KO mice. Accumulated studies have found that BBB disruption enhances abnormal influx of ions, toxins, and inflammatory cytokines into the parenchyma, resulting in neuronal dysfunction and behavioral changes53–55. In addition to BBB dysfunction, the male eShank3-KO mice exhibited increased GFAP expression across various brain regions. Elevated GFAP levels in the brain are known to disrupt neurotransmitter regulation56–58 and increase inflammatory responses59,60, both of which contribute to neuronal damage. The detailed pathogenic mechanisms and precise time frame through which neonatal BBB dysfunction leads to neuronal and behavioral defects remain to be fully elucidated in future studies.
The haploinsufficiency of ASD-risk genes often drives biological and behavioral deficits associated with ASD. Thus, we tested the BBB permeability and various behaviors using eShank3 heterozygous mice [Shank3Δe4-9/wild (heterozygous deletion of a and b forms of Shank3):Tek-Cre; eShank3-Het mice hereafter]. However, the eShank3-Het mice did not exhibit any deficits in BBB integrity, USV communication, social interaction, repetitive behaviors, motor performance, and anxiety-like behavior (Supplementary Figs. 17 and 18). These results suggest that haploinsufficiency of eShank3 in the endothelial cells may not be sufficient to affect BBB function or alter the behaviors of the mouse model.
We expect that our findings and unique approach with an established experimental platform will serve as a foundation for the growth of research in the realm of ASD pathogenesis originating from neonatal BBB disruption. Future studies, based on this groundwork, have the potential to yield valuable insights into elucidating detailed pathogenic mechanisms and developing therapeutic strategies for ASD, specifically those targeting the BECs in the BBB.
Methods
Animals
Shank3f/f(e4-9) mouse line was developed in our previous study16. Tek-Cre (B6.Cg-Tg(Tek-cre)1Ywa/J, stock no. 008863), Ai-14 (B6;129S6-Gt(ROSA)26Sortm14(CAG-tdTomato)Hze/J, stock no. 007908), C3H/HeJ (stock no. 000659), and C57BL/6 J (stock no. 000664) mice were purchased from Jackson Laboratory. Tek-Cre males were crossed with Shank3f/f females to obtain heterozygous double mutant Shank3f/wt:Tek-Cre offspring. Conditional Shank3f/f:Tek-Cre (eShank3-KO) or Shank3f/wt:Tek-Cre (eShank3-Het) mutants were obtained from breeding between Shank3f/wt:Tek-Cre males with Shank3f/wt females. eShank3-KO mutants and littermate control Tek-Cre were obtained from breeding between Shank3f/wt:Tek-Cre males and Shank3f/wt females. Ai-14 Cre reporter mice were crossed with Tek-Cre mice and used to visualize endothelial cells. The eShank3-KO:Ai-14f/wt (eShank3-KO:Ai-14) triple mutants were obtained from breeding between eShank3-KO male and Shank3 f/f:Ai-14f/f females. Tek-Cre males were crossed with Ai-14f/f females to obtain Tek-Cre:Ai-14f/wt (Tek-Cre: Ai-14) offspring as a control. Genotyping was conducted using PhireTM Tissue Direct PCR Master Mix (ThermoFisher Scientific, Waltham, MA, USA) according to the manufacturer’s instructions. To determine the sex of P5 pups, Rbm31x/y genes were examined61. All the primers used for genotyping were described in Supplementary Table 2. All mice were housed together with their littermates at a maximum of five per cage. The mice were kept on a regular 12 h light/dark cycle and were given ad libitum access to food and water. All tests were performed during the light cycle. After completing the experiments, mice were euthanized in CO2 chamber, followed by cervical dislocation. All experimental procedures were performed with protocols approved by the Institutional Animal Care and Use Committee of both University of Tennessee Health Science Center and Louisiana State University Health Science Center in accordance with U.S. National Institutes of Health guidelines.
Primary BEC isolation and enrichment
Mouse BEC isolation was performed using Adult Brain Dissociation Kit (Milterny Biotec, Gaithersburg, MD, USA, #130-107-677), CD45 MicroBeads (Milterny Biotec, #130-052-301), and CD31 MicroBeads (Milterny Biotec, #130-097-418) according to the manufacturer’s instructions. Brains dissected from 8 to 12 weeks old mice or postnatal day 5 pups were washed with cold D-PBS containing calcium, magnesium, glucose, and pyruvate (ThermoFisher Science). The brains were cut into eight sagittal slices using a scalpel and transferred into 15 ml tube containing enzyme mixture 1 and 2 supplied in Adult Brain Dissociation Kit. Minced brain slices were placed into a C tube (Milteny Biotec, #130-093-237). The C tube was then securely sealed and, following the manufacturer’s instructions, mounted upside down onto the sleeve of the gentleMACS Octo Dissociator with Heaters. Subsequently, the gentleMACS (Milteny Biotec, #130-096-427) Program 37C_ABDK_01 was run to process the tissue. Alternatively, the minced brain slices were incubated at 37 °C for 1 h, shaking gently every 5 min. The dissociated cell suspension was strained through a 70 μm cell strainer placed on top of a 15 ml centrifuge tube. The collected cell suspension was centrifuged at 300 × g for 10 min at 4 °C. After completely aspirating the supernatant, the cell pellet was resuspended with 8 ml of debris removal solution. The 4 ml of cold D-PBS was gently overlaid on top of the debris removal solution and cell mixture in 15 ml centrifuge tube, forming a clear layer on top. The 15 ml centrifuge tube was centrifuged at 3000 × g for 10 min at 4 °C. The clear top phase and debris middle layers were aspirated, leaving a milky mixture beneath the debris. After washing with cold D-PBS, cell pellet was resuspended with 1 ml of cold 1x red blood cell removal solution and incubated for 10 min at 4 °C. Then, 10 ml of 0.5% bovine serum albumin diluted in D-PBS (D-PBS/BSA buffer) added and the mixture was centrifuged at 300 × g for 10 min at 4 °C. The supernatant was completely aspirated, and the pellet was resuspended in 80 μl of D-PBS/BSA buffer. For depletion of leukocytes, the resuspended cells were incubated with 20 μl of CD45 MicroBead for 15 min in the dark at 4 °C by gently inverting on a mini-tube rotator. The cells were washed by adding 1 ml of D-PBS/BSA buffer and centrifuging at 300 × g for 5 min. The pellet was resuspended 500 μl of D-PBS/BSA buffer and applied on the pre-wet LD column (Miltenyi Biotech, #130-042-901) placed at QuadroMACS™ Separator. The flow-through containing unlabeled cells was collected and centrifuged at 300 × g for 10 min. Subsequently, cell pellet was resuspended in 90 μl of D-PBS/BSA buffer and incubated with 10 μl of CD31 MicroBead for 15 min in the dark at 4 °C by gently inverting on a mini-tube rotator to magnetically label the CD31+ endothelial cells. The cells were washed by adding 1 ml of D-PBS/BSA buffer and centrifuging at 300 × g for 5 min. Then, the pellet resuspended with 500 μl of D-PBS/BSA buffer was applied on the pre-wet MS column (Miltenyi Biotech, #130-042-201) placed at OctoMACS™ Separator. The CD31+ cells bound MS column was washed three times with 500 μl of D-PBS/BSA buffer. After removing MS column from the separator, magnetically labeled CD31+ endothelial cells were collected by adding 1 ml of D-PBS/BSA buffer. The CD31+ endothelial cells were subjected to RNA extraction/cDNA synthesis and western blotting or cultured in Endothelial Cell Basal Medium MV2 supplemented with Supplement Pack, 100 μ/ml penicillin, 100 μg/ml streptomycin on plates coated with rat tail collagen I.
To obtain the pure CD31+/CD45– endothelial cells from mice brains for genomic DNA or RNA extractions, FACS sorting was performed after removing debris and red blood cells using the same method as described above. Dissociated cells were stained with CD31-PE (BioLegend, San Diego, CA, USA, #102508) and CD45-APC (BioLegend, #103112) antibodies, diluted 1:150 in 4% calf serum, for 20 min in the dark at RT. The stained cells were washed twice, each time adding 1 mL of 4% calf serum, followed by three inversions and centrifugation at 400 × g for 5 min at 4 °C. Cells were then resuspended at a concentration of 3 × 106 per mL and sorted out of a CD31+/CD45- gate on low speed to ensure maximum efficiency and purity using the 5 laser Invitrogen Bigfoot spectral cell sorter. Data analysis was performed using Sasquatch Software (SQS).
Transmission electron microscopy
Pups at P5 were anesthetized by hypothermia and intracardially perfused with PBS containing 25 μ/ml heparin for 5 min, followed by perfusion with 10 ml of fixative solution [4% PFA and 2% glutaraldehyde in 0.13 M sodium cacodylate buffer (pH7.2)]. Perfusion was performed by gravity-fed method to minimize the rupture of cerebral vasculature. After dissecting the brains, the PFC region was cut at a thickness of 2 mm coronally, and then fixed overnight at 4 °C in fixative solution. Tissues were washed in 0.13 M sodium cacodylate buffer, and post-fixed with 1% osmium tetroxide in 0.13 M sodium cacodylate buffer for 2 h. Tissues were washed in 0.13 M sodium cacodylate buffer and distilled water followed by dehydration in a series of ethanol dilutions. Infiltration was performed with 1:1 ratio mixture of Embed812 resin and Acetone overnight at RT followed by incubation with 100% Embed812 resin three times for 2 h. Tissues were embedded in resin at 65 °C overnight. Subsequently, tissue blocks were trimmed and sectioned in a Leica EM UC 7 ultramicrotome at 60–65 nm thick and mounted on copper 200 mesh grids. Sections were stained using uranyless and lead citrate and visualized at 60 kV in JEM 2000EX II Transmission Electron Microscope (JEOL Co, Tokyo, Japan). For the quantification of the electron densities between BECs, the images of 15~30 cortical vessels, ranging in diameter between 3 and 6 μm, were randomly obtained from 3 mice per group. The electron density scan across adjoined BEC membranes was performed using ImageJ software (NIH).
Proteomics
BioID2 method for eSHANK3 interactome analysis
To identify eSHANK3 interactome, we used BioID2 method32,62. bEnd.3 BECs were transfected with Shank3Δe18-BioID2-HA, BioID2-HA, or HA-Shank3Δe18 plasmid. After 48 h transfection, the media were replaced by fresh media containing 50 μM biotin (Millipore sigma) and further incubated for 24 h. After three times washing with PBS for removal of biotin remaining in media, cells were extracted with high-salt RIPA buffer [25 mM Tris-HCl (pH 7.4), 500 mM NaCl, 1% NP-40, 1% sodium deoxycholate, 0.1% SDS, 1 mM EDTA] containing protease inhibitor cocktail, 1 mM PMSF, and 1 mM Na3VO4. The cell lysates were incubated for 30 min on ice and sonicated three times for 10 s, followed by centrifugation at 16,000 × g for 20 min at 4 °C. The supernatants were subjected to filtration using Amicon® Ultra-4 Centrifugal Filter Units (Millipore sigma) with 10 kDa molecular weight cutoff to prevent the binding of any residual free biotin in the cell lysate to NeutrAvidin agarose. Biotin-free and concentrated cell lysates obtained through filtration were incubated with 15 μl of NeutrAvidin agarose (ThermoFisher Scientific) overnight at 4 °C by gently inverting on a mini-tube rotator. NeutrAvidin agarose beads were collected by centrifugation at 800 × g for 1 min at 4 °C and washed sequentially with the following washing buffers; twice with high-salt RIPA buffer, twice with wash buffer A (2% SDS in D.W), once with wash buffer B [50 mM HEPES (pH 7.5), 500 mM NaCl, 1 mM EDTA, 0.1% sodium deoxycholate, and 1% Triton X-100], once with wash buffer C [10 mM Tris-HCl (pH 8.1), 1 mM EDTA, 250 mM LiCl, 0.5% NP-40, and 0.5% sodium deoxycholate], twice with wash buffer D [50 mM Tris-HCl (pH 7.4) and 50 mM NaCl], and once with wash buffer E [50 mM ammonium bicarbonate (pH8.3)] at 4 °C. After wash, the NeutrAvidin agarose beads were resuspended with 100 μl of 100 mM ammonium bicarbonate. 5% of the samples were used for WB analysis and 95% of the samples were analyzed by liquid chromatography-tandem mass spectrometry (LC-MS/MS).
LC-MS/MS sample preparation and analysis
Sample processing, LC-MS/MS analysis, and data acquisition for proteins identification were performed at Proteomics and Metabolomics Core, the mass spectrometry facility at UTHSC. The samples, bound to NeutrAvidin agarose beads, were reduced with 1 mM dithiothreitol (DTT) for 45 min at 4 °C and alkylated with 5 mM iodoacetamide for 20 min at room temperature, and further reduced with 5 mM DTT for 15 min at RT. Subsequently, LysC/Trypsin (Promega, Madison, WI, USA) was added to the samples and the digests were incubated at 37 °C overnight with shaking at 500 rpm using an Eppendorf ThermoMixer F1.5. Supernatant containing peptide digest was collected, and NeutrAvidin agarose beads were extracted with 100 μl of 50% acetonitrile/0.5% trifluoroacetic acid (TFA) for 10 min at 4 °C. The extracts were combined with their corresponding peptide digests and subjected to vacuum drying using a SpeedVac concentrator for 3 h. The peptide digests were desalted using Pierce C-18 spin tips (ThermoFisher Scientific) according to the manufacturer’s instructions, and then subjected to vacuum drying. Each sample was dissolved in 100 μl of 3% acetonitrile/0.05% TFA.
Quantitative LC-MS/MS was performed with 5 μl of each sample using an Ultimate 3000RSLCnano HPLC system (ThermoFisher Scientific) coupled with a Orbitrap Fusion Lumos Mass Spectrometer (ThermoFisher Scientific) via a nanoelectrospray ionization source. The sample was first trapped on C18 Acclaim PepMap 100 (75 μl × 20 mm) column (ThermoFisher Scientific), and then the analytical separation was performed using C18 Acclaim PepMap RSLC (75 μl × 500 mm) column (ThermoFisher Scientific). The mobile phase A consisted of 0.1% formic acid in water, and mobile phase B consisted of 0.1% formic acid in acetonitrile. Peptides were separated at a flow rate of 300 nl/min with a column temperature of 40 °C using the following linear gradient parameters: 3% B over 4 min, 3% B to 5% B over 1 min, 5% B to 25% B over 50 min, 25% B to 30% B over 5 min, 30% B to 90% B over 3 min, held at 90% B for 10 min, 90% B to 3% B over 3 min, and then 3% B was held for 24 min. Data collection was performed in a data-dependent acquisition mode. The full MS scan (full) was performed using an Orbitrap analyzer with a resolution of 120,000 (FWHM, at m/z = 200). The MS2 scan were performed by 0.7 m/z isolation with the quadrupole, normalized HCD collision energy of 30%, and analyzed with Orbitrap with a resolution of 30,000 (FWHM, at m/z = 200). Dynamic exclusion was set to 30 s, and Monoisotopic Precursor Selection (MIPS) was set to Peptide. An intensity threshold of ≥10,000 was applied, and charge states was set to between 2 and 6. The raw MS data was searched on Proteome Discoverer 2.4 (ThermoFisher Scientific) software using SequestHT as a search engine with following parameters: digestion reagent, trypsin; dynamic modification, oxidation of Met, acetylation of the protein N-terminus; static modification, carbamidomethylation of Cys; precursor ion mass tolerance, 10 ppm; fragment ion mass tolerance; 0.02 Da. The database for protein identification used SwissProt ver.2017-10-25 (25,097 entries) with TaxID 10090 (Mus musculus). For each sample, four biological replicates were analyzed.
The data generated by the mass spectrometry analysis was transferred to the UTHSC Molecular Bioinformatics Core using SFTP. The data was normalized using normalizeCylclicLoess function of R/Bioconductor-package limma63 after log2 transformation. The normalized data matrix was loaded into R to gather statistics and determine differential expression. The mean, variance, and standard deviation were calculated for each protein across each condition. The fold change was then calculated for all proteins. One-way ANOVA was implemented to determine significance for each protein. The p values were then adjusted for multiplicity using the Benjamini Hochberg method64. Only proteins with an adjusted p value < 0.05 were considered differentially expressed. Analysis for the protein-protein interaction mapping and the bioinformatic Gene ontology (GO) was performed using StringDB65 and Gorilla66,67, respectively. Protein networks figure was visualized using Cytoscape (v3.10), with nodes representing the gene names of proteins identified in the proteomic analysis. The node positions were manually adjusted to visualize the module more concisely. The node size is proportional to log2 (fold change) over control groups. From the GO analysis, cell component result was visualized in a tree and a bar plots.
AAV injection into the superficial temporal vein (STV) of pups at postnatal day 0
Pups at P0 were anesthetized by placing them on the wet ice for 30 s. For the electrophysiological experiments, 20 μl of the mixture containing AAV-PHP.eB-CaMKIIa-EGFP (5 × 1010 vg) and 0.05% trypan blue dye in PBS was injected into the superficial temporal vein using a syringe with a 30-gauge needle under the surgical stereoscope. Similarly, to induce the expression of HA-GSK3βS9A in the brain endothelium, pups at P0 were anesthetized by placing them on the wet ice for 30 s, and then injected 20 μl of the mixture of AAV-PHP.v1-CLDN5-GSK3βS9A-HA (1 × 1011 vg) and 0.05% trypan blue dye in PBS into the superficial temporal vein. As a control, AAV-PHP.v1-CLDN5-HA was injected into the same way instead of AAV-PHP.v1-CLDN5-GSK3βS9A-HA. GSK3βS9A-induced mice were subsequently assessed for in vivo permeability, electrophysiology, and behaviors.
In vivo BBB permeability assay
BBB permeability assay was performed to assess BBB leakage. For P5 pups, 30 μl of sodium fluorescein (Millipore sigma) dissolved in PBS at a concentration of 50 mg/ml was intraperitoneally injected. After 1 h of circulation, pups were anesthetized by hypothermia, and blood was collected by right heart needle aspiration, followed by transcardial perfusion with 10 ml of PBS containing 25 μ/ml heparin at a rate of 1 ml/min to remove blood from the intravascular compartment. In vivo BBB permeability of GSK3βS9A-induced pups at P5 was assessed using the same method with sodium fluorescein.
For adults, 100 μl sodium fluorescein dissolved in PBS at a concentration of 200 mg/ml was intraperitoneally injected. After 1 h circulation, mice were anesthetized with isoflurane, and blood was collected from right heart. And then these mice were transcardially perfused with 50 ml of PBS/ heparin at a rate of 5 ml /min. Brains were collected and individually weighed. Brains homogenized with 200 μl of PBS, and the lysate was centrifuged at 16,000 × g for 10 min at 4 °C. Two hundred microliters of the supernatant was mixed with 200 μl of 15% trichloroacetic acid (TCA) and centrifuged at 16,000 × g for 10 min at 4 °C. Three hundred microliters of the supernatant was mixed with 75 μl of 5 N NaOH, and fluorescence was measured in 100 μl of the mixture using SpectraMax M2e fluorescence spectrophotometer (Molecular Devices, San Jose, CA) at ex/em = 485 nm/ 535 nm. Serum was obtained by centrifugation at 16,000 × g for 10 min at 4 °C and then mixed with 15 % TCA of ten times volume followed by centrifugation. The supernatants were neutralized with 5 N NaOH at a 4:1 ratio and diluted with PBS, and then fluorescence was measured. The amount of fluorescein for each sample was determined using standards ranging from 0.1 to 3 μg/ml. The BBB leakage of sodium fluorescein was normalized with the amount measured in serum.
Electrophysiology
AAV-PHP.eB-CaMKIIa-EGFP with the ability to cross the BBB21 was used to select pyramidal neurons adjacent to the BBB in PFC. After injecting AAV-PHP.eB-CaMKIIa-EGFP (5 × 1010 vg) into the superficial temporal vein of pups at P0, mouse at P5 or adult age (17–19 weeks) was anesthetized and rapidly decapitated. To verify the effects by GSK3βS9A expression in the brain endothelium, AAV-PHP.v1-CLDN5-GSK3βS9A-HA was injected into the superficial temporal vein of P0 pups. Subsequently, 100 μl of AAV-PHP.eB-CaMKIIa-EGFP (3 × 1011 vg) in PBS was injected through the tail vein of adult mice (13–15 weeks) to label pyramidal neurons.
Brain was quickly removed and submerged in N-methly-D-glutamine (NMDG)-artificial CSF containing 92 mM NMDG, 2.5 mM KCl, 1.25 mM NaH2PO4, 30 mM NaHCO3, 20 mM HEPES, 25 mM D-glucose, 2 mM thiourea, 5 mM sodium ascorbate, 3 mM sodium pyruvate, 0.5 mM CaCl2, and 10 mM MgSO4 with pH 7.4 and osmolarity ~315 mOsm at RT for 1 min. All solutions used for electrophysiological experiment were bubbled with 95% O2 and 5% CO2. Brain was cut with thickness of 400 µm in oxygenated NMDG-aCSF solution using Leica VT1200S vibratome. The slices were placed in oxygenated NMDG-aCSF at 30–32 °C for 12 min, then allowed to recover at RT in oxygenated HEPES-aCSF containing 92 mM NaCl, 2.5 mM KCl, 1.25 mM NaH2PO4, 30 mM NaHCO3, 20 mM HEPES, 25 mM D-glucose, 2 mM thiourea, 5 mM sodium ascorbate, 3 mM sodium pyruvate, 2 mM CaCl2, and 2 mM MgSO4 with pH 7.4 and osmolarity ~315 mOsm for a minimum of 50 min. During the recording, slices were maintained in oxygenated external bath solution containing 119 mM NaCl, 2.5 mM KCl, 1.25 mM NaH2PO4, 24 mM NaHCO3, 12.5 mM D-glucose, 2 mM CaCl2, and 2 mM MgSO4 with pH 7.4 and osmolarity ~310 mOsm at 28–30 °C. Current-clamp recordings of pyramidal neurons in PFC region were performed using patch pipettes (3.5–5.5 MΩ resistance) filled with the internal solution containing 150 mM potassium gluconate, 2 mM MgCl2, 1.1 mM EGTA, 10 mM HEPES, 3 mM sodium ATP, and 0.2 mM sodium GTP with pH 7.2 and osmolarity ~290 mOsm. Pyramidal neurons were identified with EGFP signals. The synaptic responses were amplified with Multiclamp 700B Microelectrode Amplifier (Molecular Devices) and digitized with Digidata 1440A Digitizer (Molecular Devices). To elicit action potentials, depolarizing currents (–300 pA to 400 pA, 100 pA/step, current injection time; 1 s, inter-injection interval; 9 s) were injected in a stepwise manner. During recording, resting membrane potentials were simultaneously measured. The action potentials were analyzed using pCLAMP10 software (Molecular Devices).
Behavioral tests
Ultrasonic vocalization test
The Ultrasonic vocalization test in pups at P5 was performed in isolation container placed in a sound-proof box68. The microphone (M500-384 USB Ultrasound Microphone, Pettersson Elektronik, Uppsala, Sweden) was placed on the top through a hole in the top of the box and adjusted the 12–15 cm from the bottom of the recording box. Pups were isolated from their mother and placed in the recording box, followed by recording for 5 min. After recording, pup was returned to the mother. Ultrasonic vocalization was recorded using the BatSound® Touch Lite (Pettersson Elektronik) software. Acoustic frequencies below 40 kHz and above 150 kHz were filtered away to reduce background noise interference. The number and duration of calls were analyzed using Adobe Audition (Audio recording and editing software) software.
Round social arena (RSA) test
RSA paradigm was developed in our previous study for precise analysis of social behaviors23,29. This test was performed in a round-shaped chamber (49 (D) X 45 (H) cm) under 35 lx illumination. The transparent inner cage (8 (D) X 10.5 (H) cm) was placed in the center of the arena. A camera was placed on the ceiling to track the test animal’s overall behaviors, while wide-angle (180°) fish-eye lens camera installed on the top of the inner cage monitored the detailed social interactions between the test mouse and social stimulus. First, mouse was given full access to empty inner cage for 10 min. After returning the test mouse to its home cage, juvenile female C3H/HeJ (P30-P50) social stimulus was placed into empty cage. After the test mouse was brought back into the chamber, test mouse was allowed free access to social stimulus for 10 min. Behaviors including times spent in S zone, and frequency of entry into S zone were analyzed using Ethovision XT (Noldus, Wageningen, Netherlands) software. A circular zone of 3 cm around the inner cage was named as a S zone. The duration in the S zone was measured as the time the mouse’s nose stayed within the S zone. The duration of sniffing was measured by analyzing video recorded with a fish-eye lens camera using Ethovision XT software. Sniffing was defined as the test mouse actively exploring the inner cage with its nose directed towards it.
Grooming behavior test
Grooming behavior test was performed in a standard mouse cage filled with 1–2 cm thick beddings under 35 lx illumination. The mouse was placed in the cage and the video was recorded for 30 min. The bottom of the cage was replaced with new bedding for the next mouse recording. The total duration of grooming behavior was manually scored by the experimenters in a double-blinded manner.
Marble-burying test
The marble-burying test was performed in a standard rat cage (48 × 27 × 20 cm) filled with 5 cm thick beddings. Twenty glass marbles were placed on the surface of the bedding in 5 rows of 4 marbles. Each mouse was placed into a corner of the cage. During the test, the cage was covered with a filter-top, and the mouse was allowed to explore for 30 min. The number of buried marbles was counted both fully buried and at least two-thirds covered by bedding in a double-blinded manner.
Open field test
Since some of the global Shank3-KO mice have shown various other behavioral abnormalities, including reduced motor performance and anxiety-like behaviors16,51,69,70, we assessed the behaviors using open field and light-dark box tests at adult age. Exploratory behavior in a novel environment was performed in an open field square chamber (37 × 37 × 36 cm) for 15 min under 180 lx illumination. Mouse was placed in the arena and recorded the distance traveled, mean velocity, and the time spent in the center (23 × 23 cm) and the margin of the arena using Ethovision XT software.
Light dark box test
The box for the light dark box test was divided into two compartments having a small removable door (6 × 6 cm) for entry of mouse. The illumination of light on the bottom of the center of the light-side and dark-side was set at 600 lx and 5 lx, respectively. The mouse was placed in the dark-side for 1 min with the door closed. After removing the door to allow access to the other side, the mouse was placed in the center of the dark-side of the box and the video was recorded for 5 min. The time spent in light-side and the number of entries into the light-side was analyzed with Ethovision XT software.
Statistical analysis
All statistical analyses were performed using GraphPad Prism software version 9.1 (GraphPad Software, Inc., CA, US). Two-tailed unpaired t-test was used for comparisons between two group: bEnd.3 BECs and eShank3-KO bEnd.3 BECs, eShank3-KO and Tek-Cre control mice, or eShank3-Het and Tek-Cre control mice. One-way ANOVA followed by Dunnett’s multiple comparisons test was used for three group comparisons of bEnd.3 BECs, eShank3-KO BECs, and IWR-1-endo treated or GSK3βS9A and C/EBP1α induced eShank3-KO BECs. Two-way ANOVA with repeated measure and following Šídák’s multiple comparisons test was used for the time (or stage)-dependent behavior tests. Two-way ANOVA followed by Šídák’s multiple comparisons test was used for the Shank3 mRNA expression test in primary BECs of P5 and adult male and female mice, and for in vivo permeability and behavior tests in the GSK3βS9A-induced mice. Two-way ANOVA with repeated measures followed by two-stage linear step-up procedure of Benjamini, Krieger and Yekutieli was used for current-clamp recordings. Mixed-effects analysis and Šídák’s multiple comparisons test were used for analyzing electron density of TJs. All the detailed statistical methods and results are described in Supplementary Table 1. Data are represented as mean ± SEM. p-value < 0.05 was considered statistically significant.
Reporting summary
Further information on research design is available in the Nature Portfolio Reporting Summary linked to this article.
Supplementary information
Description of Additional Supplementary Files
Source data
Acknowledgements
This work was supported by NIH MH117429 and AG075000 for I.H.K., and HL119291 for C.P. We thank Dr. Kihoon Han for providing Shank3 DNA, Dr. David Kakhniashvili for supporting proteomic analysis, Dr. Eun Bee Cho for assisting in the primary BEC purification, Drs. Daniel Johnson and Hyo Young Choi for supporting bioinformatic analysis, Drs. Matthew Ennis, Lynn Dobrunz, and John Boughter for their critical reading and comments, and LifeCanvas Technologies for their assistance with light-sheet imaging. A subset of the illustrations was created with BioRender.com.
Author contributions
Y.K., C.P., and I.H.K. conceived the design of this study. Y.K., M.K., R.L., C.P., and I.H.K. performed the cloning and generation of stable cell lines. Y.K., M.K., and R.L. performed WB, PCR, qRT-PCR, IP, IHC, and ICC. Y.K. and M.K. performed BBB and cell permeability assays. Y.K., S.K., and Y.U. performed behavioral tests and analyses. S.K. performed electrophysiology and analyses. Y.K. and E.M.M.W. performed TEM analysis. E.Y. and H.K. performed in situ hybridization. Y.K. performed viral production and purification, viral injection, proteomics, and primary cell isolation. Y.K. and C.L. performed bioinformatic analysis. Y.K., M.K., S.K., R.L., Y.U., Y.J., C.L., C.P., and I.H.K. analyzed the data and statistics. This paper was written by Y.K., C.P., and I. H. K. and was edited by other authors.
Peer review
Peer review information
Nature Communications thanks Chris Greene, Zhen Yan and the other, anonymous, reviewer(s) for their contribution to the peer review of this work. A peer review file is available.
Data availability
The protein mass spectrometry (LC-MS/MS) data produced in this study are available in the ProteomeXchange under accession code PXD059466. All data generated or analyzed in this study are included in this published article and its supplementary information files. Source data are provided with this paper.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
These authors contributed equally: Minseong Kim, Sunwhi Kim, Raham Lee.
Contributor Information
Changwon Park, Email: changwon.park@lsuhs.edu.
Il Hwan Kim, Email: ikim9@uthsc.edu.
Supplementary information
The online version contains supplementary material available at 10.1038/s41467-025-56720-1.
References
- 1.Chaste, P. & Leboyer, M. Autism risk factors: genes, environment, and gene-environment interactions. Dialogues Clin. Neurosci.14, 281–292 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Obermeier, B., Daneman, R. & Ransohoff, R. M. Development, maintenance and disruption of the blood-brain barrier. Nat. Med.19, 1584–1596 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Huber, J. D., Egleton, R. D. & Davis, T. P. Molecular physiology and pathophysiology of tight junctions in the blood-brain barrier. Trends Neurosci.24, 719–725 (2001). [DOI] [PubMed] [Google Scholar]
- 4.Ouellette, J. et al. Vascular contributions to 16p11.2 deletion autism syndrome modeled in mice. Nat. Neurosci.23, 1090–1101 (2020). [DOI] [PubMed] [Google Scholar]
- 5.Tarlungeanu, D. C. et al. Impaired amino acid transport at the blood brain barrier is a cause of autism spectrum disorder. Cell167, 1481–1494.e1418 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Memis, I. et al. Altered blood brain barrier permeability and oxidative stress in Cntnap2 knockout rat model. J. Clin. Med.11, 2725 (2022). [DOI] [PMC free article] [PubMed]
- 7.Pepe, G. et al. Blood-brain barrier integrity is perturbed in a Mecp2-null mouse model of Rett syndrome. Biomolecules13, 606 (2023). [DOI] [PMC free article] [PubMed]
- 8.Sheng, M. & Kim, E. The Shank family of scaffold proteins. J. Cell Sci.113, 1851–1856 (2000). [DOI] [PubMed] [Google Scholar]
- 9.Durand, C. M. et al. Mutations in the gene encoding the synaptic scaffolding protein SHANK3 are associated with autism spectrum disorders. Nat. Genet, 39, 25–27 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bonaglia, M. C. et al. Molecular mechanisms generating and stabilizing terminal 22q13 deletions in 44 subjects with Phelan/McDermid syndrome. PLoS Genet, 7, e1002173 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Vyas, Y. et al. Shankopathies in the Developing Brain in Autism Spectrum Disorders. Front Neurosci.15, 775431 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Leblond, C. S. et al. Meta-analysis of SHANK mutations in autism spectrum disorders: a gradient of severity in cognitive impairments. PLoS Genet.10, e1004580 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Moessner, R. et al. Contribution of SHANK3 mutations to autism spectrum disorder. Am. J. Hum. Genet.81, 1289–1297 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Delling, J. P. & Boeckers, T. M. Comparison of SHANK3 deficiency in animal models: phenotypes, treatment strategies, and translational implications. J. Neurodev. Disord.13, 55 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Peca, J. et al. Shank3 mutant mice display autistic-like behaviours and striatal dysfunction. Nature472, 437–442 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wang, X. et al. Synaptic dysfunction and abnormal behaviors in mice lacking major isoforms of Shank3. Hum. Mol. Genet.20, 3093–3108 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Monteiro, P. & Feng, G. SHANK proteins: roles at the synapse and in autism spectrum disorder. Nat. Rev. Neurosci.18, 147–157 (2017). [DOI] [PubMed] [Google Scholar]
- 18.Kisanuki, Y. Y. et al. Tie2-Cre transgenic mice: a new model for endothelial cell-lineage analysis in vivo. Dev. Biol.230, 230–242 (2001). [DOI] [PubMed] [Google Scholar]
- 19.He, J. et al. Sleep restriction impairs blood-brain barrier function. J. Neurosci.34, 14697–14706 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Mathiesen Janiurek, M., Soylu-Kucharz, R., Christoffersen, C., Kucharz, K., & Lauritzen, M. Apolipoprotein M-bound sphingosine-1-phosphate regulates blood-brain barrier paracellular permeability and transcytosis. eLife8, e49405 (2019). [DOI] [PMC free article] [PubMed]
- 21.Chan, K. Y. et al. Engineered AAVs for efficient noninvasive gene delivery to the central and peripheral nervous systems. Nat. Neurosci.20, 1172–1179 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kim, I. H. et al. Dysregulation of the synaptic cytoskeleton in the PFC drives neural circuit pathology, leading to social dysfunction. Cell Rep.32, 107965 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kim, S. et al. Neural circuit pathology driven by Shank3 mutation disrupts social behaviors. Cell Rep.39, 110906 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Mohapatra, A. N. & Wagner, S. The role of the prefrontal cortex in social interactions of animal models and the implications for autism spectrum disorder. Front. Psychiatry14, 1205199 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Silverman, J. L., Yang, M., Lord, C. & Crawley, J. N. Behavioural phenotyping assays for mouse models of autism. Nat. Rev. Neurosci.11, 490–502 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Rapin, I. & Dunn, M. Update on the language disorders of individuals on the autistic spectrum. Brain Dev.25, 166–172 (2003). [DOI] [PubMed] [Google Scholar]
- 27.Lord, C. et al. Autism from 2 to 9 years of age. Arch. Gen. Psychiatry63, 694–701 (2006). [DOI] [PubMed] [Google Scholar]
- 28.Regier, D. A., Kuhl, E. A. & Kupfer, D. J. The DSM-5: classification and criteria changes. World Psychiatry12, 92–98 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kim, S., Kim, Y. E. & Kim, I. H. Simultaneous analysis of social behaviors and neural responses in mice using round social arena system. STAR Protoc.3, 101722 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Han, K. et al. SHANK3 overexpression causes manic-like behaviour with unique pharmacogenetic properties. Nature503, 72–77 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sakai, Y. et al. Protein interactome reveals converging molecular pathways among autism disorders. Sci. Transl. Med.3, 86ra49 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Courtland, J. L. et al. Genetic disruption of WASHC4 drives endo-lysosomal dysfunction and cognitive-movement impairments in mice and humans. eLife10, e61590 (2021). [DOI] [PMC free article] [PubMed]
- 33.Poliak, S., Matlis, S., Ullmer, C., Scherer, S. S. & Peles, E. Distinct claudins and associated PDZ proteins form different autotypic tight junctions in myelinating Schwann cells. J. Cell Biol.159, 361–372 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Umeda, K. et al. ZO-1 and ZO-2 independently determine where claudins are polymerized in tight-junction strand formation. Cell126, 741–754 (2006). [DOI] [PubMed] [Google Scholar]
- 35.Zihni, C., Mills, C., Matter, K. & Balda, M. S. Tight junctions: from simple barriers to multifunctional molecular gates. Nat. Rev. Mol. Cell Biol.17, 564–580 (2016). [DOI] [PubMed] [Google Scholar]
- 36.Haseloff, R. F., Dithmer, S., Winkler, L., Wolburg, H. & Blasig, I. E. Transmembrane proteins of the tight junctions at the blood-brain barrier: structural and functional aspects. Semin Cell Dev. Biol.38, 16–25 (2015). [DOI] [PubMed] [Google Scholar]
- 37.Qin, L. et al. Social deficits in Shank3-deficient mouse models of autism are rescued by histone deacetylase (HDAC) inhibition. Nat. Neurosci.21, 564–575 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ozawa, M., Baribault, H. & Kemler, R. The cytoplasmic domain of the cell adhesion molecule uvomorulin associates with three independent proteins structurally related in different species. EMBO J.8, 1711–1717 (1989). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Valenta, T., Hausmann, G. & Basler, K. The many faces and functions of beta-catenin. EMBO J.31, 2714–2736 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Liu, J. et al. Wnt/beta-catenin signalling: function, biological mechanisms, and therapeutic opportunities. Signal Transduct. Target Ther.7, 3 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kakogiannos, N. et al. JAM-A Acts via C/EBP-alpha to promote claudin-5 expression and enhance endothelial barrier function. Circ. Res.127, 1056–1073 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Mann, B. et al. Target genes of beta-catenin-T cell-factor/lymphoid-enhancer-factor signaling in human colorectal carcinomas. Proc. Natl Acad. Sci. USA96, 1603–1608 (1999). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Aberle, H., Bauer, A., Stappert, J., Kispert, A. & Kemler, R. beta-catenin is a target for the ubiquitin-proteasome pathway. EMBO J.16, 3797–3804 (1997). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Skurk, C. et al. Glycogen-Synthase Kinase3beta/beta-catenin axis promotes angiogenesis through activation of vascular endothelial growth factor signaling in endothelial cells. Circ. Res.96, 308–318 (2005). [DOI] [PubMed] [Google Scholar]
- 45.Behrens, J. et al. Functional interaction of an axin homolog, conductin, with beta-catenin, APC, and GSK3beta. Science280, 596–599 (1998). [DOI] [PubMed] [Google Scholar]
- 46.Hart, M. J., de los Santos, R., Albert, I. N., Rubinfeld, B. & Polakis, P. Downregulation of beta-catenin by human Axin and its association with the APC tumor suppressor, beta-catenin and GSK3 beta. Curr. Biol.8, 573–581 (1998). [DOI] [PubMed] [Google Scholar]
- 47.Liu, C. et al. Control of beta-catenin phosphorylation/degradation by a dual-kinase mechanism. Cell108, 837–847 (2002). [DOI] [PubMed] [Google Scholar]
- 48.Ravindra Kumar, S. et al. Multiplexed cre-dependent selection yields systemic AAVs for targeting distinct brain cell types. Nat. Methods17, 541–550 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Baumann, B. H. et al. Ferroportin-mediated iron export from vascular endothelial cells in retina and brain. Exp. Eye Res.187, 107728 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Fombonne, E. Epidemiology of pervasive developmental disorders. Pediatr. Res.65, 591–598 (2009). [DOI] [PubMed] [Google Scholar]
- 51.Bauer, H. F., Delling, J. P., Bockmann, J., Boeckers, T. M. & Schon, M. Development of sex- and genotype-specific behavioral phenotypes in a Shank3 mouse model for neurodevelopmental disorders. Front. Behav. Neurosci.16, 1051175 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Loomes, R., Hull, L. & Mandy, W. P. L. What Is the male-to-female ratio in autism spectrum disorder? A systematic review and meta-analysis. J. Am. Acad. Child Adolesc. Psychiatry56, 466–474 (2017). [DOI] [PubMed] [Google Scholar]
- 53.Senatorov, V. V. Jr., et al. Blood-brain barrier dysfunction in aging induces hyperactivation of TGFbeta signaling and chronic yet reversible neural dysfunction. Sci. Transl. Med.11, eaaw8283 (2019). [DOI] [PubMed]
- 54.Milikovsky, D. Z. et al. Paroxysmal slow cortical activity in Alzheimer’s disease and epilepsy is associated with blood-brain barrier dysfunction. Sci. Transl. Med.11, eaaw8954 (2019). [DOI] [PubMed]
- 55.Menard, C. et al. Social stress induces neurovascular pathology promoting depression. Nat. Neurosci.20, 1752–1760 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Halassa, M. M., Fellin, T. & Haydon, P. G. The tripartite synapse: roles for gliotransmission in health and disease. Trends Mol. Med.13, 54–63 (2007). [DOI] [PubMed] [Google Scholar]
- 57.Nedergaard, M., Ransom, B. & Goldman, S. A. New roles for astrocytes: redefining the functional architecture of the brain. Trends Neurosci.26, 523–530 (2003). [DOI] [PubMed] [Google Scholar]
- 58.Perea, G., Navarrete, M. & Araque, A. Tripartite synapses: astrocytes process and control synaptic information. Trends Neurosci.32, 421–431 (2009). [DOI] [PubMed] [Google Scholar]
- 59.Linnerbauer, M., Wheeler, M. A. & Quintana, F. J. Astrocyte crosstalk in CNS inflammation. Neuron108, 608–622 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Abdelhak, A. et al. Blood GFAP as an emerging biomarker in brain and spinal cord disorders. Nat. Rev. Neurol.18, 158–172 (2022). [DOI] [PubMed] [Google Scholar]
- 61.Tunster, S. J. Genetic sex determination of mice by simplex PCR. Biol. Sex. Differ.8, 31 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Roux, K. J., Kim, D. I., Raida, M. & Burke, B. A promiscuous biotin ligase fusion protein identifies proximal and interacting proteins in mammalian cells. J. Cell Biol.196, 801–810 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Ritchie, M. E. et al. limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res.43, e47 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Benjamini, Y., Drai, D., Elmer, G., Kafkafi, N. & Golani, I. Controlling the false discovery rate in behavior genetics research. Behav. Brain Res.125, 279–284 (2001). [DOI] [PubMed] [Google Scholar]
- 65.Szklarczyk, D. et al. STRING v10: protein-protein interaction networks, integrated over the tree of life. Nucleic Acids Res.43, D447–D452 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Eden, E., Navon, R., Steinfeld, I., Lipson, D. & Yakhini, Z. GOrilla: a tool for discovery and visualization of enriched GO terms in ranked gene lists. BMC Bioinform.10, 48 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Eden, E., Lipson, D., Yogev, S. & Yakhini, Z. Discovering motifs in ranked lists of DNA sequences. PLoS Comput. Biol.3, e39 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Yin, X. et al. Maternal deprivation influences pup ultrasonic vocalizations of C57BL/6J mice. PLoS ONE11, e0160409 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Pillerova, M. et al. Neuromotor development in the Shank3 mouse model of autism spectrum disorder. Brain Sci.12, 872 (2022). [DOI] [PMC free article] [PubMed]
- 70.Liu, H. et al. Dissection of the relationship between anxiety and stereotyped self-grooming using the Shank3B mutant autistic model, acute stress model and chronic pain model. Neurobiol. Stress15, 100417 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Description of Additional Supplementary Files
Data Availability Statement
The protein mass spectrometry (LC-MS/MS) data produced in this study are available in the ProteomeXchange under accession code PXD059466. All data generated or analyzed in this study are included in this published article and its supplementary information files. Source data are provided with this paper.