Abstract
The putative locus for hereditary mixed polyposis syndrome (HMPS) in a large family of Ashkenazi descent (SM96) was previously reported to map to chromosome sub-bands 6q16–q21. However, new clinical data, together with molecular data from additional family members, have shown 6q linkage to be incorrect. A high-density genomewide screen for the HMPS gene was therefore performed on SM96, using stringent criteria for assignment of affection status to minimize phenocopy rates. Significant evidence of linkage was found only on a region on chromosome 15q13-q14. Since this region encompassed CRAC1, a locus involved in inherited susceptibility to colorectal adenomas and carcinomas in another Ashkenazi family (SM1311), we determined whether HMPS and CRAC1 might be the same. We found that affected individuals from both families shared a haplotype between D15S1031 and D15S118; the haplotype was rare in the general Ashkenazi population. A third informative family, SM2952, showed linkage of disease to HMPS/CRAC1 and shared the putative ancestral haplotype, as did a further two families, SMU and RF. Although there are probably multiple causes of the multiple colorectal adenoma and cancer phenotype in Ashkenazim, an important one is the HMPS/CRAC1 locus on 15q13–q14.
Introduction
Hereditary mixed polyposis syndrome (HMPS [MIM 601228]) is characterized by the development of a variety of different colorectal tumors (Whitelaw et al. 1997), including atypical juvenile polyps, hyperplastic polyps with areas of dysplasia (serrated adenomas), classical adenomas, and carcinoma. In contrast to familial adenomatous polyposis, disease in the only known HMPS kindred (SM96) appears to be confined to the large bowel. Most older individuals in SM96 presented with colorectal carcinoma, whereas younger individuals presented with polyps of either the atypical juvenile or hyperplastic type, suggesting progression from hyperplastic polyp to serrated adenoma to carcinoma. A similar natural history has been shown in other disorders characterized by the presence of polyps and increased risk of malignancy, including juvenile polyposis syndrome (Woodford-Richens et al. 2000), Cowden syndrome (Marsh et al. 1998), and Peutz-Jeghers syndrome (Gruber et al. 1998; Wang et al. 1999).
In family SM96, disease is inherited as an autosomal dominant trait, and a previous study mapped the HMPS locus to chromosome bands 6q16–q21 (Thomas et al. 1996). The HMPS gene was not identified. Over the 6 years since the HMPS locus was mapped to chromosome 6 (Thomas et al. 1996), one individual from SM96 without the putative disease-associated haplotype (patient 4.30) has developed multiple colorectal adenomas, before the age of 40 years. These data strongly suggested that the reported location of the putative HMPS gene on chromosome 6 was incorrect. We have therefore retested SM96 for linkage of disease to 6q16–q21, using updated affection status and genetic markers from recent genetic maps. We then undertook a new genomewide linkage screen in family SM96 with a high-density set of markers and a conservative assignment of affection status.
Subjects and Methods
Pedigree Collection and Assignment of Affection Status
The development of multiple adenomas by individual 4.30 led us to perform a systematic reassessment of the affection statuses in family SM96. In such a large family, a small number of phenocopies would be expected for relatively common conditions such as a single colorectal adenoma or carcinoma, or multiple hyperplastic polyps of the rectum. We therefore used a relatively uncommon and specific phenotype—multiple (⩾3) colorectal adenomas (including polyps with adenomatous areas)—as the basis of our affection criteria. Updated pedigree information was obtained from members of SM96, and patient information, histology reports, and samples were obtained with full informed consent and ethical review board approval. Importantly, all clinicopathological data were reverified from histology reports, and unverified data were excluded. Besides those classed as “affected,” spouses marrying into the family were classed as “unaffected,” and all other individuals, including those with unverified clinicopathological data, were classed as of “unknown” status. Furthermore, all individuals were typed for the Ashkenazi-specific APCI1307K variant, and any carriers of this were classed as having unknown affection status, since their disease might result from I1307K, from the unknown gene, or from both. (In addition to some extra individuals included in this study, the differences between the new and old affection statuses can generally be assessed by comparing fig. 1 of Thomas et al. [1996] with the results in this article.) In addition to family SM96, we also ascertained and analyzed members of family SM1311 and members of three other new families, SM2592, SMU, and RF.
Exclusion of Chromosome 6q
Blood samples were obtained from 57 members of SM96, who provided useful information for linkage analysis. DNA was extracted from either established cell lines or blood, by use of standard methods. Polymorphic microsatellite markers D6S1716, D6S1580, and D6S1592, spanning an interval of ∼8.5 cM and mapping to 6q16–q21, were chosen from The Genetic Location Database. PCR amplification of these plus the original markers (Thomas et al. 1996) comprising the putative disease-associated haplotype (D6S283, D6S434, and D6S301), was performed. Reactions comprised 25 ng of genomic DNA in 25-μl volumes, containing 1× standard PCR buffer, 1.5 mM Mg2+, 0.4 mM dNTPs, 0.25 U Taq polymerase, and 0.5 mM of each oligonucleotide primer. Each forward primer was labeled with HEX, FAM, or TET. Cycling conditions consisted of an initial denaturation of 94°C for 5 min, followed by 35 cycles of 94°C for 30 s, 55°C for 30 s, and 72°C for 30 s, with a final extension step of 72°C for 7 min. 0.2 μl of each PCR product was combined with 0.2 μl of Tamra350 or Tamra500 size standard (PE Applied Biosystems) and 3 μl of formamide loading buffer. After denaturation at 94°C for 5 min, products were electrophoresed on 5% denaturing polyacrylamide gels on an ABI377 semiautomated sequencer for 2 h. Results were analyzed by use of Genescan and Genotyper software (Applied Biosystems).
Genomewide Linkage Analysis
PCR amplification of 387 microsatellite markers, spaced at ∼10 cM intervals across the genome, was performed on 57 individuals from SM96 by use of the Weber 9 set (Research Genetics). PCR and electrophoretic conditions were as described for the chromosome 6 markers. For linkage analysis, HMPS was modeled as a dominant trait (q=0.001), with penetrances AA=0.75, Aa=0.75, and aa=0.001. Two-point LOD scores were calculated for each marker using the subprogram MLINK (version 5.1) of the LINKAGE program package (Lathrop et al. 1984), as implemented in FASTLINK (version 4.1) (Cottingham et al. 1993). Multipoint analyses were undertaken using the VITESSE program (O’Connell and Weeks 1995). Marker allele frequencies were taken from The Genome Database or from the genotyping of pedigree founders.
Typing of Markers on Chromosome 15q13–q14
In addition to marker ACTC from the genome screen, the following additional markers close to ACTC were typed in all five families, to refine the location of the HMPS gene: (cen→tel) D15S1031, D15S1010, D15S144, D15S995, D15S1007, D15S1040, ACTC, D15S971, and D15S118. Two-point and multipoint linkage analyses were undertaken by use of the same affection criteria and model as for the SM96 genome screen.
Loss of Heterozygosity (LOH) Analysis
DNA was extracted from archival paraffin-embedded tumor tissue. The appropriate tissue was microdissected by use of a sterile needle and was digested in 400 μg proteinase K/ml and 1× Perkin Elmer PCR buffer (no Mg2+) at 55°C. After digestion, the proteinase K was heat denatured at 95°C for 10 min. Samples were then centrifuged at 13,000 rpm for 10 min, the supernatant removed, and 2 μl used per PCR, using markers D15S1010, D15S144, D15S995, D15S1007, ACTC, and D15S1040. Allelic loss was only considered present if the ratio of peak areas was either <0.5 or >2.0, after correction for the relative areas in constitutional DNA.
Results
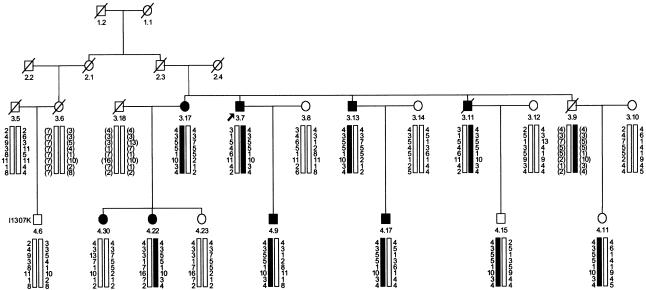
SM96 family members were genotyped at the three original polymorphic markers (Thomas et al. 1996) and three new markers close to the reported site of the HMPS locus at 6q16–q21. These data were used for two-point and multipoint LOD-score analysis using the revised affection criteria. LOD scores throughout the region were uniformly negative and were sufficient to exclude the putative HMPS susceptibility gene from this region (data not shown). Haplotype construction confirmed that disease and 6q16–q21 alleles did not cosegregate (fig. 1). The previously presumed phenocopy (individual 4.30)—who had a single adenoma but has since developed four adenomas—was confirmed as not carrying the putative “linked” haplotype. Moreover, typing of additional markers revealed that one other individual (4.6) had developed adenomas without carrying the linked haplotype, although this individual was found to carry the APCI1307K variant.
Figure 1.
Pedigree of selected part of family SM96, showing haplotypes for the following chromosome 6 markers: D6S1716, D6S468, D6S283, D6S434, D6S1580, D6S301, D6S1592, and D6S447. Black bars denote the haplotype that was previously thought to segregate with disease. Known affected individuals are indicated by a blackened symbol. Inferred haplotypes are bracketed.
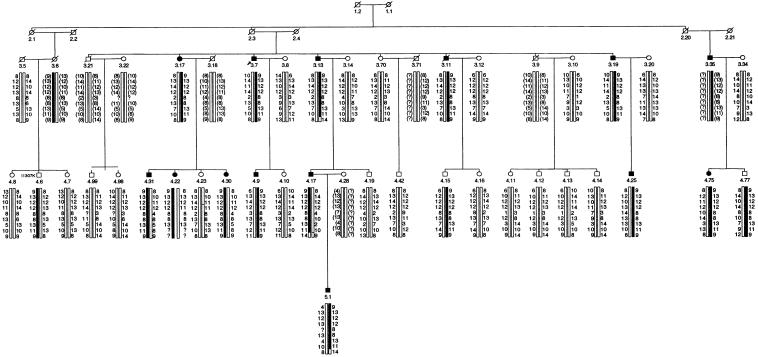
The new genomewide screen on family SM96 revealed only one site in the genome with good evidence of linkage to HMPS. This region was on chromosome 15q13–q21, close to marker ACTC. A maximum two-point LOD score of 3.98 was found at ACTC and a maximum multipoint LOD score of 4.67 was also found at ACTC. Further markers mapping to this region were chosen to give a dense haplotype in an attempt to define the minimal region containing the disease gene. Haplotype construction showed the minimal region containing the HMPS gene to lie between D15S1031 and D15S118, a 10-cM interval (fig. 2). The HMPS gene was shown to be highly penetrant: of 20 individuals in SM96 sharing the haplotype, 18 were affected.
Figure 2.
Pedigree of selected members of family SM96, showing haplotypes for the following chromosome 15 markers: D15S1031, D15S1010, D15S144, D15S995, D15S1007, D15S1040, ACTC, D15S971, and D15S118. Symbols are the same as in figure 1.
There are two main explanations as to why 15q linkage was not found in SM96 by the initial genome screen. First, some individuals whom we previously classed as affected were reclassified as “unknown,” because the original data provided by the patient could not be confirmed from histopathological records (for example, patients 3.70, 3.80, 3.90, and the entire nuclear family of patient 3.1 [see fig. 1 from Thomas et al. 1996]). Second, some individuals were reclassified as “unknown” because they had colorectal tumor(s) but not multiple adenomas. Patient 3.9 and his nuclear family are good examples. He was screened for colorectal tumors in his 50s and was reassured that he had not inherited the family’s predisposition to polyps. He subsequently developed an isolated colorectal carcinoma at age 63 years, but there was still no evidence of the specific HMPS phenotype. To date, none of his four children has developed any adenoma, although his daughter (4.11) had previously reported that she had had multiple polyps and had been classed as affected; on subsequent investigation, the “polyps” turned out to be ulcerative colitis. It is most likely, therefore, that patient 3.9's cancer was “sporadic” and unrelated to an inherited susceptibility. Although allowance was made for such phenocopies in the calculation of LOD scores in the previous genome screen, this did not prevent incorrect assignment of the HMPS locus to chromosome 6. Our revised strategy of relying on individuals with a distinct phenotype to provide linkage information proved to be more prudent.
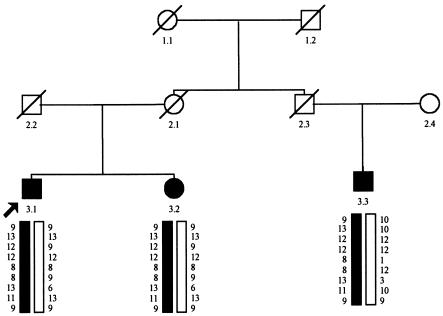
A genomewide screen previously performed on another Ashkenazi family (SM1311) had mapped a new colorectal tumor susceptibility gene, CRAC1, to 15q14–q22 (Tomlinson et al. 1999). The linked haplotype had spanned a 40-cM interval defined by D15S1031 and D15S153, a larger region than that found for SM96. We wondered whether HMPS and CRAC1 might be the same locus. When the CRAC1 and SM96 disease-associated haplotypes were compared, we found that they were identical for markers shared within the HMPS region (D15S1031–D15S118). Following this discovery, we examined an additional Ashkenazi family (SM2952) with multiple colorectal adenomas. No serrated adenomas or dysplastic hyperplastic polyps had been diagnosed in this family, although not all histopathologists use this classification. Typing of markers D15S1031 to D15S118 at 15q13–q14 in this family showed that all affected members shared the minimal HMPS/CRAC1 region haplotype (fig. 3).
Figure 3.
Pedigree of family SM2592, showing haplotypes for the same chromosome 15 markers as are shown in figure 2. Symbols are the same as in figure 1.
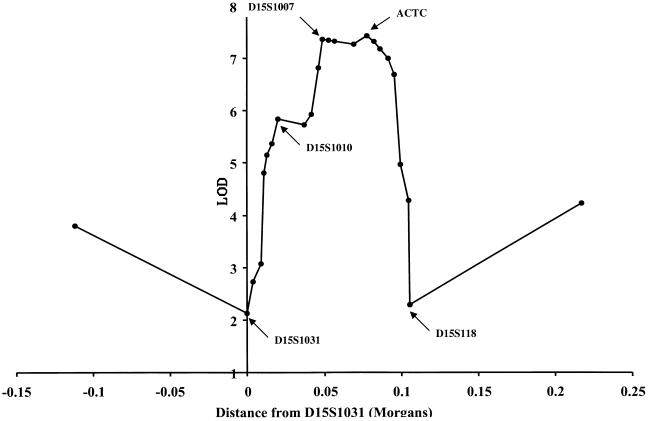
By use of combined genotyping data for markers D15S1031 to D15S118 from families SM96, SM1311, and SM2952, further two-point and multipoint LOD score analyses were performed. By use of the affection criteria adopted for this study (and incorporating a small amount of updated clinical information for SM1311), maximum two-point and multipoint LOD scores of 5.31 and 7.19, respectively, were obtained at marker ACTC (table 1 and fig. 4) for the three families combined.
Table 1.
Two-Point LOD Scores for 15q13–14 Markers, Using Combined Genotyping Data from Families SM96, SM1311, and SM2952[Note]
|
LOD at Recombination Fraction |
||||||
| MarkerandFamily | .001 | .101 | .201 | .301 | .401 | Approximate Chromosome 15 Map Position(cM) |
| D15S1031: | 1.28 | 2.04 | 1.61 | 1.02 | .41 | 26.0 |
| SM96 | 2.54 | 2.11 | 1.59 | 1.00 | .041 | |
| SM1311 | −1.50 | −.22 | −.06 | −.01 | −.01 | |
| SM2592 | .23 | .15 | .08 | .03 | .01 | |
| D15S1010: | 3.48 | 2.65 | 1.83 | 1.05 | .37 | 28.2 |
| SM96 | 2.29 | 1.79 | 1.26 | .71 | .22 | |
| SM1311 | 1.01 | .75 | .51 | .31 | .14 | |
| SM2592 | .18 | .11 | .06 | .02 | .01 | |
| D15S144: | 1.83 | 1.49 | 1.15 | .81 | .42 | 28.7 |
| SM96 | 1.91 | 1.56 | 1.21 | .83 | .42 | |
| SM1311 | −.09 | −.08 | −.06 | −.03 | .00 | |
| SM2592 | .02 | .01 | .00 | .00 | .00 | |
| D15S995: | 3.08 | 2.29 | 1.49 | .73 | .16 | 28.8 |
| SM96 | 2.32 | 1.76 | 1.17 | .58 | .12 | |
| SM1311 | .42 | .30 | .19 | .10 | .03 | |
| SM2592 | .34 | .22 | .12 | .05 | .01 | |
| D15S1007: | 1.51 | 1.07 | 1.15 | .86 | .37 | 28.9 |
| SM96 | 3.35 | 2.73 | 2.05 | 1.31 | .55 | |
| SM1311 | −2.08 | −1.80 | −.98 | −.49 | −.19 | |
| SM2592 | .24 | .15 | .08 | .03 | .01 | |
| D15S1040: | 3.09 | 2.37 | 1.71 | 1.12 | .56 | 29.3 |
| SM96 | 2.35 | 1.93 | 1.49 | 1.02 | .52 | |
| SM1311 | .68 | .41 | .21 | .09 | .04 | |
| SM2592 | .06 | .03 | .02 | .01 | .00 | |
| ACTC: | 5.31 | 4.22 | 3.10 | 1.97 | .87 | 30.4 |
| SM96 | 3.98 | 3.25 | 2.47 | 1.63 | .74 | |
| SM1311 | 1.19 | .88 | .58 | .32 | .13 | |
| SM2592 | .14 | .08 | .04 | .02 | .00 | |
| D15S971: | 1.45 | 2.21 | 1.68 | 1.01 | .33 | 30.7 |
| SM96 | .62 | 1.61 | 1.29 | .79 | .24 | |
| SM1311 | .77 | .57 | .38 | .21 | .08 | |
| SM2592 | .06 | .04 | .02 | .01 | .00 | |
| D15S118: | −.85 | 1.55 | 1.33 | .87 | .36 | 31.0 |
| SM96 | −1.21 | 1.32 | 1.20 | .82 | .35 | |
| SM1311 | .00 | .00 | .00 | .00 | .00 | |
| SM2592 | .36 | .23 | .13 | .05 | .01 | |
Note.— The maximum LOD score obtained was 5.31, for ACTC.
Figure 4.
A sliding map of three markers was used to calculate multipoint linkage analysis between markers D15S1031 and D15S118. Combined genotyping data from families SM96, SM1311, and SM2592 were used.
We then ascertained a further two Ashkenazi kindreds, one with a history of multiple colorectal adenomas and cancer (SMU) and one with mixed hyperplastic/adenomatous polyps (RF). Although these families were not informative for linkage analysis, all four affected individuals typed carried the minimal HMPS haplotype between D15S1031 and D15S118. We subsequently found that the disease-associated haplotype was not present in any spouse marrying into the families studied (figs. 2 and 3) and was present in, at most, 1 of 95 random Ashkenazi controls (although haplotypes could not be assigned for the latter).
To investigate the possibility that the HMPS/CRAC1 gene might act as a tumor suppressor, LOH analysis was performed on 22 colorectal tumors from two SM96 and five SM1311 family members, by use of a selection of microsatellite markers. Only two tumors (from the same SM1311 member) showed loss of the wild-type allele, each with all five markers used. These data supported those reported previously (Tomlinson et al. 1999), in that limited LOH was observed. The reasons for the low frequency of LOH—for example, dominant germline mutation, haploinsufficiency, or “second hit” by promoter methylation—are currently unclear.
Discussion
We have shown that the HMPS gene is not located at 6q16–61, as previously reported, but at 15q13–q14. Furthermore, analysis of the 15q haplotypes of families SM96, SM1311, and SM2952, together with the significant LOD scores from two-point and multipoint analyses, shows that the region containing the HMPS gene overlaps that containing the CRAC1 gene. Comparison of data derived from the five families (SM96, SM1311, SM2952, SMU, and RF) revealed that all affected individuals who were tested shared the same haplotype in this region. The CRAC1 gene is therefore highly likely to be identical to the HMPS gene. The shared haplotype is therefore most probably derived from a common founder. Although the precise origin of families SM1311, SM2592, and RF are unknown, families SM96 and SMU originate from Lithuania, which is consistent with recent common ancestry.
The phenotypes of the five families are very similar, with members having developed multiple, classical colorectal adenomas and carcinomas (Tomlinson et al. 1999). Members of SM96, SM1311, and RF have also developed early-onset tumors which have features of both ‘classical’ adenomas and hyperplastic polyps, referred to variously as “dysplastic hyperplastic polyps,” “mixed hyperplastic/adenomatous polyps,” or “serrated adenomas.” No member of SM1311, SM2952, SMU, or RF has been reported as developing the atypical juvenile polyps reported to occur in a few members of SM96 (Whitelaw et al. 1997).
Together, these data provide compelling evidence for a high-penetrance colorectal tumor predisposition gene, HMPS/CRAC1, at 15q13–q14. This region contains few good candidate genes, a small number of genes with known function, and several predicted genes/proteins. HMPS/CRAC1 may explain some of the increased population prevalence of colorectal tumors in the Ashkenazi population. Although the multiple adenoma phenotype in this group may also result from mutations in APC, MYH, and other uncharacterized genes, all five multiple adenoma families which we have typed (and from which APC mutations have been excluded) probably result from HMPS/CRAC1 mutations. It remains to be seen whether HMPS/CRAC1 is also important in other ethnic groups.
Acknowledgments
We thank all the families involved in this study. We also thank the Cancer Research UK Equipment Park for their valuable help. This work was supported principally by the Bobby Moore Fund and Cancer Research UK and by the Fifth European Community Framework Program (grant QLG 2-CT-2001-01861). Thanks also go to Carrie Drovdlic and Heather Hampel for assistance with family studies. C.E. is partially funded by grant P30CA16058 from the National Cancer Institute (to The Ohio State University Comprehensive Cancer Center).
Electronic-Database Information
Accession numbers and URLs for data presented herein are as follows:
- Genetic Location Database, http://cedar.genetics.soton.ac.uk/public_html/ldb.html (for marker selection)
- Genome Database, http://gdbwww.gdb.org/ (for marker allele frequencies)
- Online Mendelian Inheritance in Man (OMIM),http://www.ncbi.nlm.nih.gov/Omim/ (for HMPS) [PubMed]
References
- Cottingham R, Idury R, Schaeffer A (1993) Faster sequential genetic linkage computations. Am J Hum Genet 53:252–263 [PMC free article] [PubMed] [Google Scholar]
- Gruber S, Entius M, Petersen G, Laken S, Longo P, Boyer R, Levin A, Mujumdar U, Trent J, Kinzler K, Vogelstein B, Hamilton S, Polymeropoulos M, Offerhaus G, Giardiello F (1998) Pathogenesis of adenocarcinoma in Peutz-Jeghers syndrome. Cancer Res 58:5267–5270 [PubMed] [Google Scholar]
- Lathrop G, Lalouel J, Julier C, Ott J (1984) Stategies for multilocus linkage analysis in humans. Proc Natl Acad Sci USA 81:3443–3446 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marsh D, Coulon V, Lunetta K, Rocca-Serra P, Dahia P, Zheng Z, Liaw D, et al (1998) Mutation spectrum and genotype-phenotype analyses in Cowden disease and Bannayan-Zonana syndrome, two hamartoma syndromes with germline PTEN mutation. Hum Mol Genet 7:507–515 [DOI] [PubMed] [Google Scholar]
- O’Connell J, Weeks D (1995) The VITESSE algorithm for rapid exact multilocus linkage analysis via genotype set-recoding and fuzzy inheritance. Nat Genet 11:402–408 [DOI] [PubMed] [Google Scholar]
- Thomas H, Whitelaw S, Cottrell S, Murday V, Tomlinson I, Markie D, Jones T, Bishop D, Hodgson S, Sheer D, Northover J, Talbot I, Solomon E, Bodmer W (1996) Genetic mapping of hereditary mixed polyposis syndrome to chromosome 6q. Am J Hum Genet 58:770–776 [PMC free article] [PubMed] [Google Scholar]
- Tomlinson I, Rahman N, Frayling I, Mangion J, Barfoot R, Hamoudi R, Seal S, Northover J, Thomas H, Neale K, Hodgson S, Talbot I, Houlston R, Stratton M (1999) Inherited susceptibility to colorectal adenomas and carcinomas: evidence for a new predisposition gene on15q14–q22. Gastroenterology 116:789–795 [DOI] [PubMed] [Google Scholar]
- Wang Z, Ellis I, Zauber P, Iwama T, Marchese C, Talbot I, Xue W, Yan Z, Tomlinson I (1999) Allelic imbalance at the LKB1 (STK11) locus in tumours from patients with Peutz-Jeghers’ syndrome provides evidence for a hamartoma-(adenoma)-carcinoma sequence. J Pathol 188:9–13 [DOI] [PubMed] [Google Scholar]
- Whitelaw SC, Murday VA, Tomlinson IP, Thomas HJ, Cottrell S, Ginsberg A, Bukofzer S, Hodgson SV, Skudowitz RB, Jass JR, Talbot IC, Northover JM, Bodmer WF, Solomon E (1997) Clinical and molecular features of the hereditary mixed polyposis syndrome. Gastroenterology 112:327–334 [DOI] [PubMed] [Google Scholar]
- Woodford-Richens K, Bevan S, Churchman M, Dowling B, Jones D, Norbury CG, Hodgson SV, et al (2000) Analysis of genetic and phenotypic heterogeneity in juvenile polyposis. Gut 46:656–660 [DOI] [PMC free article] [PubMed] [Google Scholar]