Abstract
Attention-deficit/hyperactivity disorder (ADHD [MIM 143465]) is a common, highly heritable neurobehavioral disorder of childhood onset, characterized by hyperactivity, impulsivity, and/or inattention. As part of an ongoing study of the genetic etiology of ADHD, we have performed a genomewide linkage scan in 204 nuclear families comprising 853 individuals and 270 affected sibling pairs (ASPs). Previously, we reported genomewide linkage analysis of a “first wave” of these families composed of 126 ASPs. A follow-up investigation of one region on 16p yielded significant linkage in an extended sample. The current study extends the original sample of 126 ASPs to 270 ASPs and provides linkage analyses of the entire sample, using polymorphic microsatellite markers that define an ∼10-cM map across the genome. Maximum LOD score (MLS) analysis identified suggestive linkage for 17p11 (MLS=2.98) and four nominal regions with MLS values >1.0, including 5p13, 6q14, 11q25, and 20q13. These data, taken together with the fine mapping on 16p13, suggest two regions as highly likely to harbor risk genes for ADHD: 16p13 and 17p11. Interestingly, both regions, as well as 5p13, have been highlighted in genomewide scans for autism.
Introduction
Attention-deficit/hyperactivity disorder (ADHD [MIM 143465]) is a pervasive neurobehavioral disorder affecting ∼5% of children and adolescents and ∼3% of adults (Wolraich et al. 1996; Goldman et al. 1998; Swanson et al. 1998; Scahill and Schwab-Stone. 2000). A growing body of epidemiological and clinical data indicate globally similar prevalence rates across diverse geographic and cultural settings, including studies in Brazil, Germany, New Zealand/Australia, Turkey, the United Kingdom, and the United States (Anderson et al. 1987; Baumgaertel et al. 1995; Wolraich et al. 1996; Gomez et al. 1999; Rohde et al. 1999; Tahir et al. 2000; Wilens et al. 2002). ADHD is more frequently diagnosed in boys, with male:female ratios between 3:1 and 4:1 (Cantwell 1996; Swanson et al. 1998). ADHD is defined by the presence of six or more symptoms of inattention and/or hyperactivity-impulsivity that lead to significant impairment in at least two settings and have their onset by age 7 years (American Psychiatric Association 1994). Under criteria of the Diagnostic and Statistical Manual of Mental Disorders, 4th edition (DSM-IV) (American Psychiatric Association 1994), three subtypes are recognized: inattentive (I), hyperactive-impulsive (HI), and combined (C), reflecting the presence of six or more symptoms on dimensions of inattention, hyperactivity-impulsivity, or both, respectively. Comorbidity is common in ADHD, with oppositional disorders, mood disorders, anxiety, and learning disabilities being the most prevalent conditions, likely reflecting the complexity of the biological etiology (August and Garfinkel 1989; Pelham et al. 1992; Biederman et al. 1996; Wolraich et al. 1998; Brown et al. 2001; Wilens et al. 2002).
ADHD has a significant genetic component. Data from clinical studies consistently support the familial nature of ADHD (Biederman et al. 1992; Faraone and Biederman 1994). Twin studies of categorically defined ADHD and/or continuous rating scales of hyperactivity, impulsivity, and inattention lead to estimates of heritability of ∼60%–90% (Levy et al. 1997; Smalley 1997; Faraone and Doyle 2001) and reported sibling relative-risk ratios (λs) of 4.0–8.0 (Smalley et al. 1997; Faraone et al. 2000). In addition, adoption studies demonstrate an increased frequency of ADHD diagnoses in biological relatives of probands (Morrison and Stewart 1973; Cantwell 1975; van den Oord et al. 1994), consistent with genetic underpinnings. Although segregation analysis (Deutsch et al. 1990; Faraone et al. 1992) suggested a possible major-gene effect, there are indications of multiple genes, with minor-to-moderate effect sizes, from molecular studies of candidate genes (Faraone et al. 2001) and genomewide exclusion mapping (Fisher et al. 2002).
Molecular genetic studies have primarily focused on candidate genes involved in catecholamine processes, synaptic transmission, and serotonin pathways. The selection of candidate genes has been driven by biological observations, such as the treatment efficacy of stimulants, and by the notion that the primary behavioral and cognitive deficits in ADHD are related to functions associated with the frontal-cortex and frontal-striatal networks (Barkley 1997; Yamasake et al. 2002). However, the results from >50 candidate gene studies are equivocal, and the effect sizes of purported susceptibility alleles are small (Comings et al. 1995; Cook et al. 1995; Lahoste et al. 1996; Gill et al. 1997; Smalley et al. 1998; Daly et al. 1999; Palmer et al. 1999; Comings et al. 2000; McCracken et al. 2000; Barr et al. 2001; Curran et al. 2001; Payton et al. 2001). Although polymorphisms in the dopamine D4 receptor gene (DRD4) and/or the dopamine transporter protein (DAT-1) have shown association in independent studies (Cook et al. 1995; Lahoste et al. 1996; Gill et al. 1997; Smalley et al. 1998; Daly et al. 1999; Faraone et al. 2001; Mill et al. 2001), the estimated effect sizes are small (genotype relative risks of ∼1.5 or less), and other studies have failed to replicate the observed associations, despite adequate power (Palmer et al. 1999; Holmes et al. 2000).
The alternative and complementary strategy of genomewide linkage analysis enables the detection of susceptibility loci without any a priori knowledge regarding the specific genes involved in the disease state. We have employed an affected-sibling-pair (ASP) strategy, with parental genotyping, and have reported elsewhere the results of a genomewide scan in a set of 126 ASPs (Fisher et al. 2002). Linkage analysis yielded multipoint maximum LOD scores (MLSs) ⩾1.0 in 12 genomic regions, including 16p13. In a follow-up investigation of 277 ASPs, using a dense set of microsatellite and SNP markers on 16p, fine-mapping yielded significant linkage (MLS 4.22; Smalley et al. 2002). Here, we present the results of a genomewide linkage analysis (∼10-cM density) in an extended cohort of 270 ASPs. In addition to the linkage we previously detected on 16p13, we find suggestive linkage for a region on 17p11 (near marker D17S1857; MLS 2.98) and nominal support (MLS ⩾1.0) for four genomic regions >1.0 on 5p13, 6q14, 11q25, and 20q13.
Subjects and Methods
Ascertainment and Diagnostic Procedures
Families that included at least two children with ADHD were recruited from multiple sources, including previous research studies of ADHD, clinics, hospitals, and schools in the greater Los Angeles area (Smalley et al. 2000). Families were evaluated at the University of California, Los Angeles (UCLA), and both consent and assent forms (approved by the UCLA institutional review board) were signed during the initial visit. Assessment of psychiatric disorders was performed using semistructured interviews, the Schedule for Affective Disorders and Schizophrenia for School-Age Children–Present and Lifetime version (KSADS-PL) (Kaufman et al. 1997) for subjects 5–17 years of age and the Schedule fo Affective Disorders and Schizophrenia–Lifetime Version, modified for the study of anxiety disorder (Fyer et al. 1995), with the Disruptive Disorders section of the KSADS-PL for subjects aged ⩾18 years. Interviews were conducted by clinical psychologists and highly trained interviewers and were administered to the mother as well as the child if the child was ⩾8 years of age. In addition, the Swanson, Nolan, and Pelham, version IV (SNAP-IV) (parent and teacher versions; Swanson 1995), the Child Behavior Checklist, and the Teacher’s Report Form (Achenbach 1993) were utilized to supplement information obtained in the direct interviews. Diagnosis was determined using a best-estimate procedure, with senior psychiatrists (J.J.M. and J.T.M.) reviewing all positive diagnoses. The mean weighted κ for diagnoses was 0.84 (SD=0.14), including values for ADHD (1.0), oppositional defiant disorder (0.93), and conduct disorder (1.0).
ADHD was diagnosed according to DSM-IV criteria and was defined as “definite” when all criteria were met or as “probable” when the subject met only five criteria, instead of the six required for definite diagnosis, but was accompanied by significant impairment. In the present study, we have qualitatively assigned all individuals with definite and probable diagnoses as affected, similar to the “broad” definition used in the first genomewide scan (Fisher et al. 2002). Families were excluded from analysis if an affected child met criteria for autism or schizophrenia. Full-scale IQ was measured using the Wechsler Intelligence Scale for Children, 3rd edition (Wechsler 1991), and academic achievement was determined using the Peabody Individual Achievement Test–Revised (Markwardt 1989). Affected children were excluded from analysis if their full-scale IQ was <70. A thorough description of the cohort and all associated procedures has been reported elsewhere (Smalley et al. 2000).
Demographic and Clinical Characteristics
The total sample includes 204 families, comprising 270 possible ASPs and 234 independent pairs (table 1). The initial set of 104 families, termed “wave 1,” included 126 ASPs (Fisher et al. 2002). We extended the sample with an additional set of 101 families, termed “wave 2” and comprising 140 ASPs. Together, the wave 1 and wave 2 families constitute the “combined” set. Family 580, a four-sibling family, contributed a single ASP to both wave 1 and wave 2, because of the logistics of data collection. The cohort is 72% male and 80% white, with the average age of affected children being 11.1 years (SD=3.6) and the average full-scale IQ being 105.9 (SD=14.2). The majority of the sample met “definite” diagnostic criteria (95%), with 24 affected siblings classified as “probable” (5%). All ASP families had at least one member with a definite diagnosis. As shown in table 2, the distribution of subtypes and comorbid conditions in the sample is similar to that expected on the basis of clinical and epidemiological studies of ADHD.
Table 1.
ASP Families Included in Genomewide Scan
| Wave 1a | Wave 2 | Combinedb | |
| No. of families with: | |||
| Two affected sibs | 96 | 86 | 180 |
| Three affected sibs | 6 | 12 | 18 |
| Four affected sibs | 2 |
3 |
6 |
| Total | 104 | 101 | 204 |
| Total independent ASPsc | 114 | 119 | 234 |
| Total ASPsd | 126 | 140e | 270 |
Subset of families reported elsewhere (Fisher et al. 2002).
The number of possible ASPs in the combined set reflects the sum of possible pairs in wave 1 and wave 2, plus an additional ASP created from the addition of 2 affected siblings from a wave 1 family that were collected after wave 1 was completed. The combined set includes this ASP, while wave 2 does not, so that the families in wave 1 and wave 2 are completely independent. Thus, in the combined data set there is 1 additional independent ASP and 4 additional total ASPs over and above the sum of wave 1 and wave 2.
A family with n sibs contributes n−1 independent pairs.
A family with n sibs contributes n(n−1)/2 possible pairs.
Of these 140 ASPs, 107 were included in a previous analysis of chromosome 16p (Smalley et al. 2002).
Table 2.
Demographics and Clinical Characteristics of 438 Affected Siblings
| Characteristic | No. of AffectedSiblings (%) |
| Sex: | |
| Male | 315 (72) |
| Female | 123 (28) |
| Ethnicity: | |
| White | 351 (80) |
| Latino | 20 (5) |
| Asian | 11 (3) |
| Othera | 56 (12) |
| Socioeconomic statusb: | |
| I | 97 (22) |
| II | 153 (35) |
| III | 135 (31) |
| IV | 46 (10) |
| V | 7 (2) |
| ADHD diagnosis: | |
| Definite | 414 (95) |
| Probable | 24 (5) |
| ADHD subtype: | |
| Combinedc | 211 (48) |
| Inattentived | 196 (45) |
| Hyperactive-impulsivee | 31 (7) |
| Comorbidity: | |
| Oppositional defiant disorder | 239 (45) |
| Conduct disorder | 64 (12) |
| Moodf | 97 (18) |
| Anxietyg | 45 (9) |
Includes subjects whose parents are of different ethnicities.
According to Hollingshead (1957).
Subjects exceed symptom thresholds in both inattentive and hyperactive-impulsive domains.
Subjects with inattentive ADHD exceed symptom thresholds in the inattentive domain but not in the hyperactive-impulsive domain.
Subjects with hyperactive-impulsive ADHD exceed symptom thresholds in the hyperactive-impulsive domain, but not in the inattentive domain.
Includes major depression, dysthymia, and/or bipolar disorder.
Includes two or more of the following: panic disorder, social or simple phobia, obsessive-compulsive disorder, and/or agoraphobia.
The extended sample of 277 ASPs reported elsewhere (Smalley et al. 2002) for fine-mapping on 16p overlaps with the current data set by 237 ASPs. The lack of overlap of 73 ASPs in the fine-mapping sample and the current sample was due to logistic issues stemming from the timing of the genotyping efforts at the two independent laboratories: the 10-cM markers (Wellcome Trust Centre for Human Genetics) and fine-mapping markers on 16p (UCLA). Specifically, 40 ASPs unique to the 16p fine-mapping sample were not genotyped for the 10-cM grid, and 33 ASPs included in the 10-cM genotyping were not genotyped for the fine-mapping analyses.
Genotyping and Laboratory Procedures
Affected siblings and their parents were genotyped with 423 polymorphic microsatellite markers, spanning the entire human genome. Both parents were genotyped in 188 (92%) of the 204 families, and a single parent was genotyped in the remaining 16 families. The majority of autosomal markers represent the ABI Prism Linkage Mapping Set (LMS) 2.5 panels, supplemented with additional markers to compensate for large intervals and/or poorly amplifying markers. X-chromosome markers were taken from the LMS 2.5 panels and from the Cooperative Human Linkage Center (CHLC)/Weber Human Screening Set version 6 (Research Genetics). Sex-averaged marker maps were determined from CHLC, Généthon (Dib et al. 1996), Marshfield, and maps generated from recombination fractions within the sample, using both Genehunter version 2.1 (Kruglyak et al. 1996) and ASPEX version 2.3. The average intermarker interval was 9.2 cM. Fine-mapping data for 16p13, from the previously published study (Smalley et al. 2002), were combined with the present data set to present the most complete analysis possible. The incorporated data set consists of 10 additional markers spanning 20 cM on 16p13, described in detail elsewhere (Smalley et al. 2002).
PCR was performed in 9-μl volumes in 96-well plates. Each reaction contained ∼24 ng of genomic DNA, 0.2 mM MgCl2, 0.2 mM dNTPs, and 0.25 U of AmpliTaq gold DNA polymerase (ABI). Standard thermocycling protocols were performed in 96-well MJ Research tetrad thermocyclers. For a minority of DNA samples with low yields, primer extension preamplification (PEP) was performed as described elsewhere (Zhang et al. 1992). PCR products were run on ABI 3700 capillary-action sequencers, and trace files were analyzed in Genotyper version 3.7. Random subsets of trace files were independently analyzed and reviewed by both M.N.O. and I.L.M., to ensure accurate and consistent allele designation. GAS version 2.0 (A. Young; Genotype Linkage Analysis–GAS Web site) was used to eliminate violations of Mendelian inheritance and to assign global allele designations. Haplotypes were generated in Genehunter version 2.1 and all double recombinants, as well as any chromosome with an excessive number of crossovers, were reviewed extensively. LINKAGE format files were generated with the use of MAKEPED and RECODE version 1.4.
ASP Linkage Analysis
MLS analysis (Risch 1990) was performed under a multiplicative model, utilizing the restrictions of the “possible triangle” (Holmans 1993). Such ASP likelihood-ratio methods are based on the comparison of the observed maximum-likelihood estimates of allele-sharing with allele-sharing proportions derived under the null hypothesis. Autosomal single-point MLSs were generated in Genehunter version 2.1, using Mapmaker/SIBS options (Kruglyak and Lander 1995). X-chromosome single-point MLS values were generated in Mapmaker/SIBS. To facilitate combined analysis of the entire data set, wave 2 alleles were globally binned to ensure that allele designations were consistent throughout both waves. All allele frequencies were derived from founders and random inidividuals in RECODE version 1.4 (Dan Weeks). Multipoint MLS analysis was performed using ASPEX 2.3 sib_ibd, with incremental estimates set to ∼1-cM intervals. Values for locus-specific sibling relative-risk ratios (λs) were calculated from the Z0 parameter, assuming a recombination fraction of 0 (λs=0.25/Z0; Risch, 1990). Simulations to determine the empirical significance of MLS values attained in the current study were performed using Simulate (Terwilliger et al. 1993). One thousand replicates of the entire autosomal genome were generated under the null hypothesis of no linkage, using actual marker parameters (e.g., allele number and frequency and recombination fraction) and genotype recovery frequencies. All replicates were analyzed in ASPEX 2.3, and the resultant MLS values were analyzed at various thresholds to determine empirical P values. Exclusion mapping was performed in ASPEX sib_ibd under defined λs values, assuming no dominance variance. Any region yielding a LOD score ⩽−2.00 was considered excluded. Exclusion mapping compares the likelihood of the genotype data under the assumption of specified values of λs with the likelihood under the null hypothesis of no linkage.
Results
Multipoint MLS Linkage Analysis
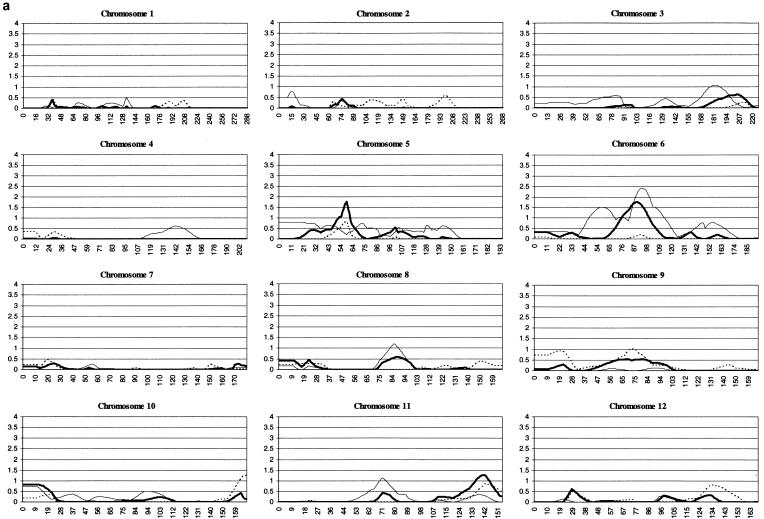
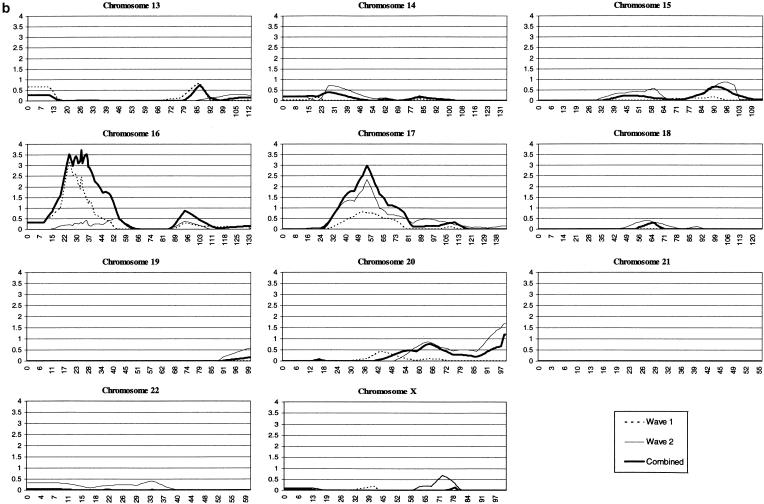
Multipoint MLS analysis of the combined data set yielded six genomic regions with values >1.0 (table 3). These constitute the six most promising regions for susceptibility loci within our sample: 5p13, 6q14, 11q25, 16p13, 17p11, and 20q13. Regions 16p13 (MLS = 3.73) and 17p11 (MLS = 2.98) yielded the strongest evidence for linkage in the present sample. To indicate relative contributions to the combined MLS values, separate analyses of wave 1, wave 2, and the combined data sets are presented (fig. 1). Region 17p11 is the only one that yields nominal evidence (MLS >0.74; Nyholt 2000) of linkage in the multipoint analysis of both wave 1 (MLS = 0.79) and wave 2 (MLS = 2.33) data sets. The sibling relative-risk ratios estimated from the Z0 parameter are as follows: 5p13, λs = 1.3; 6q14, λs = 1.4; 11q25, λs = 1.3; 16p13, λs = 1.5; 17p11, λs = 1.5; and 20q13, λs = 1.2.
Table 3.
Regions Yielding Multipoint LOD Scores >1 in MLS Analyses of ASPs with ADHD
|
Location |
MLS |
||||||||
| Chromosome | NearestMarker | Cytogenetica | Genetic(cM)b | Wave 1c | Pd | Wave 2 | Pd | Combined | Pd |
| 5 | D5S418 | 5p13 | 59 | .82 | .02599 | .61 | .04686 | 1.77 | .00215 |
| 6 | D6S460 | 6q14 | 79 | .19 | .17479 | 2.42 | .00042 | 1.75 | .00226 |
| 11 | D11S1320 | 11q25 | 133 | .85 | .02394 | .34 | .10541 | 1.27 | .00779 |
| 16 | D16S3060 | 16p13 | 18 | 3.13 | .00007 | .50 | .06458 | 3.73 | .00002 |
| 17 | D17S1857 | 17p11 | 46 | .79 | .02824 | 2.33 | .00053 | 2.98 | .00011 |
| 20 | D20S196 | 20q13 | 90 | .01 | .41504 | 1.68 | .00271 | 1.19 | .00962 |
The most likely cytogenetic location of marker, according to draft genome sequence data.
Distance from the most p-terminal genome-scan marker of the chromosome.
Subset of families previously reported. The current wave 1 MLS values differ slightly from those published elsewhere (Fisher et al. 2002), because of additional markers (e.g., 16p13) and the use of ASPEX rather than Genehunter as the analysis program.
LOD scores were converted into nominal P values by multiplying by 2loge10 and determining significance from χ2 tables, taking into account the one-sided nature of the linkage test, as described elsewhere (Lander and Kruglyak 1995).
Figure 1.
MLS values for individual chromosomes. The Y-axis indicates the MLS values and the X-axis indicates the positions in cM. The dotted line indicates the wave 1 set of families, the thin line represents the wave 2 set, and the thick line represents the combined set. All graphs begin at 0 cM, which was designated as 10 cM distal to the most p-terminal marker. The current wave 1 MLS values differ slightly from those reported elsewhere (Fisher et al. 2002), because of the presence of additional markers (e.g., 16p13) and the use of ASPEX, rather than Genehunter, for the analysis.
Single-Point MLS Linkage Analysis
Single-point MLS analysis in the combined set supports the multipoint MLS findings for four of the regions highlighted (table 4). Specifically, single-point MLS values are >1.0 for markers on 5p13 (D5S418), 6p12-6q14 (D6S257 and D6S460), 16p13 (D16S3047, rs153783, rs127293, and D16S3060) and 17p11 (D17S799 and D17S1857). In addition, 11q13 (D11S978) and 15q26 (D15S127) both yielded single-point MLS values >1.0, but the multipoint MLS values were <1.0. The most striking single-point MLS was on chromosome 5 at marker D5S418 (MLS 3.09). D5S418 is the only marker in the present study that yielded single-point MLS values >1.0 in both wave 1 (MLS 2.089) and wave 2 (MLS 1.158) data sets.
Table 4.
Markers Yielding Single-Point LOD Scores >1 in MLS Analyses of ASPs with ADHD [Note]
|
Location |
MLS |
||||||||
| Chromosomeand Marker | Genetic(cM)a | Cytogeneticb | Heterozygosityc(%) | Wave 1 | Pd | Wave 2 | Pd | Combined | Pd |
| 5: | |||||||||
| D5S418 | 61 | 5p13 | 84 | 2.089 | .00094 | 1.158 | .01046 | 3.091 | .00008 |
| 6: | |||||||||
| D6S257 | 69 | 6p12 | 73 | .617 | .04587 | 1.581 | .00349 | 1.837 | .00181 |
| D6S460 | 79 | 6q14 | 81 | .267 | .13357 | .638 | .04327 | 1.067 | .01333 |
| 11: | |||||||||
| D11S987 | 64 | 11q13 | 88 | .137 | .21336 | 1.057 | .01369 | 1.172 | .01009 |
| 15: | |||||||||
| D15S127 | 76 | 15q26 | 81 | .444 | .07634 | 1.176 | .00998 | 1.287 | .00745 |
| 16: | |||||||||
| D16S3114 | 13 | 16p13 | 79 | 1.942 | .00139 | .043 | .32778 | 1.738 | .00233 |
| D16S3047 | 13 | 16p13 | 73 | 1.741 | .00232 | .257 | .13831 | 1.501 | .00427 |
| rs153783 | 18e | 16p13 | 36 | .645 | .04231 | 1.409 | .00543 | 2.436 | .00040 |
| rs127293 | 18e | 16p13 | 47 | 1.014 | .01536 | .253 | .14016 | 1.555 | .00373 |
| D16S3060 | 18 | 16p13 | 81 | 1.268 | .00784 | .053 | .31297 | 1.421 | .00526 |
| 17: | |||||||||
| D17S799 | 33 | 17p12 | 76 | .625 | .04493 | .477 | .06917 | 1.061 | .01355 |
| D17S1857 | 45 | 17p11 | 71 | .441 | .07708 | 1.247 | .00771 | 1.326 | .00637 |
Note.— Table includes only markers yielding an MLS >1 in the combined data set.
Distance from the most p-terminal genome-scan marker of the chromosome.
The most likely cytogenetic location of marker, according to draft genome sequence data (Ensembl and UCSC).
Heterozygosity of each marker, as estimated from the entire study sample.
LODs were converted into nominal P values by multiplying by 2loge10 and then determining significance from χ2 tables, taking into account the one-sided nature of the linkage test, as described elsewhere (Lander and Kruglyak 1995).
SNP distances are based on physical distances (UCSC Genome Bioinformatics and Celera).
Exclusion Mapping
Exclusion mapping under a λs of 1.5 excluded 56.3% of the genome, indicating that the current study lacks sufficient power to effectively exclude loci with minor-effect size from approximately half of the genome. Exclusion mapping under a λs of 2.0 excluded 94.3% of the genome, indicating that it is unlikely that any loci with a moderate-effect size reside outside 10 discrete genomic regions: 5p13, 6p21-6q14, 8p23, 8q21, 11q25, 15q26, 16p13, 16q21, 17p11, and 20q13. Under a λs of 2.5, 99.8% of the genome was excluded, with only four intervals yielding LOD scores >−2.00: 5p13, 6p21-6q14, 16p13, and 17p11.
Discussion
A genomewide linkage scan in 270 ASPs represents the largest linkage study in ADHD to date. Previously, our group reported linkage analysis in an initial set of 126 ASPs (104 families) and identified 12 genomic regions yielding multipoint MLS values above a defined nominal threshold of 1.0 (Fisher et al. 2002). We extended the sample by 144 ASPs (101 families) and found multipoint MLS values >1.0 in six regions, of which three show support (MLS ⩾0.5) in both wave 1 and wave 2 families. We previously reported a significant linkage of the 16p13 region (MLS 4.22) (Smalley et al. 2002), and we observe a slight difference (MLS 3.73) in the present study, because of the incomplete overlap between the two sets of ASPs.
The current analysis shows that the second strongest region for linkage is on 17p11, where suggestive evidence (MLS 2.98) is seen in the combined sample and where support is present in both wave 1 and wave 2 families. An additional region on chromosome 5p13 shows support in wave 1 and wave 2, with a combined multipoint MLS of 1.77, falling short of suggestive linkage. The other three regions, 6q14, 11q25, and 20q13, show support in only one of the two waves of families. When a nominal LOD threshold of 1.0 is used, the present sample of 204 families has >90% power to detect susceptibility loci with λs of 1.6 (Risch 1990; Weeks and Lathrop 1995).
The three regions of strongest linkage in the present study, 5p13, 16p13, and 17p11, overlap those found in studies of autism (MIM 209850 and MIM 607373). Although ADHD and autism are clinically distinct disorders, the symptoms overlap (Smalley et al. 2000). As discussed elsewhere (Smalley et al. 2002), the region of 16p13 has been highlighted in three independent genomewide scans (Philippe et al. 1999; International Molecular Genetic Study of Autism Consortium [IMGSAC] 2001; Liu et al. 2001). Interestingly, the 1-LOD support interval of the new region on 17p11 overlaps the IMGSAC study (MLS 2.34) (2001), and a QTL analysis of language-related endophenotypes in the Liu et al. 2001 data set yields additional support for linkage to autism (Z >3.0; D. Geschwind and M. Alarcon, personal communication). Lastly, the 5p13 region highlighted in the current scan yielded the most significant evidence of linkage (MLS 2.55) in the Liu et al. (2001) genomewide scan and yielded nominal evidence (nonparametric linkage score of 1.65; P=.05) in a second independent linkage study in autism (Buxbaum et al. 2001). The convergence of these three linkage peaks (16p13, 17p11, and 5p13) in ADHD and autism, as well as the overlap for two other regions (7p and 15q) with autism in an independent ASP linkage study in ADHD (Bakker et al. 2003), suggests further research on the overlap is strongly warranted.
The candidate region on 17p is broad, with a 1-LOD support interval spanning 25 cM and covering 17p11-17q11. The serotonin transporter gene (5HTT [MIM 182138]) resides on 17q11 beneath the 1-LOD support described in the present analysis (fig. 1), ∼28 Mb from the p-telomere and 8 cM from D17S1857 (Marshfield Center for Medical Genetics; UCSC Genome Bioinformatics). The product of 5HTT is a sodium-calcium–dependent transporter that clears serotonin from the synaptic cleft into presynaptic neurons and is believed to be the site of action of antidepressants and amphetamines. Notably, three independent studies have demonstrated association of an insertion-deletion polymorphism in the promoter of 5HTT with ADHD (Manor et al. 2001; Seeger et al. 2001; Kent et al. 2002).
Region 5p13 yielded the third highest multipoint peak in the combined data set (MLS 1.77) and is the only genomic interval yielding nominal evidence of linkage in both the present study and the Bakker et al. (2003) linkage scan in ADHD (MLS 1.43). Marker D5S418 yielded a single-point MLS of 3.09 in the combined data set, the highest single-point MLS in the present study, and is the only marker yielding single-point MLS values >1.0 in both the wave 1 (MLS 2.089) and wave 2 (MLS 1.158) data sets. Interestingly, glial cell line–derived neurotrophic factor (GDNF), a gene necessary for the development of the sympathetic, parasympathetic, and enteric ganglia, is located in this region. GDNF is a protein that is crucial to the development of the peripheral autonomic nervous system (Pattyn et al. 1999), and it promotes survival, differentiation, and dopamine reuptake in dopaminergic neurons (Schaar et al. 1993; Beck et al. 1995).
The multipoint linkage peak on chromosome 6 (MLS 1.75), the fourth-highest peak in the combined analysis, contains a 30-cM 1-LOD support interval spanning 6p21-6q14. Although the combined linkage peak is centered at 6q14, wave 2 single-point MLS values indicate excess sharing across a 50-cM region spanning 6p22 (D6S422; MLS 1.14) to 6q14 (D6S460; MLS 0.64). Molecular genetic studies into reading disability (RD [MIM 600202]), or dyslexia, have converged on a putative susceptibility locus on 6p21, within the 1-LOD support interval presented here. A significant proportion of individuals affected with ADHD also have learning disabilities (Brown et al. 2001), and evidence indicates high rates of comorbidity with RD (Gilger et al. 1992; Semrud-Clikeman et al. 1992; Willcutt and Pennington. 2000). Cardon et al. (1994) first reported the QTL contributing to RD on 6p21, and genetic studies in three independent RD samples have produced evidence of linkage to the same region (Fisher et al. 1999; Gayan et al. 1999; Grigorenko et al. 2000). In addition, this region on 6p21 has recently been implicated as a susceptibility locus for ADHD (Willcutt et al. 2002) in a study of ADHD within sib pairs identified for RD. It has also been suggested, by another independent linkage study in RD (Petryshen et al. 2001), that a distinct susceptibility locus for RD may exist on 6q12.
The 6q region highlighted by the combined analysis contains two serotonin receptors. Serotonin receptor 1B is located 85 cM from the p-telomere, directly under the combined peak. Serotonin receptor 1E is located ∼95 cM from the p-telomere, at the distal edge of the 1-LOD support interval. The human leukocyte antigen (HLA) region resides on 6p21 and is just distal to the wave 2 1-LOD support interval presented here. Association of ADHD and two genes in the HLA has been reported; Warren et al. (1995) found association with the null allele of the C4B gene, and Odell et al. (1997) reported significant association with both the null allele of the C4B gene and the β1 allele of the dopamine receptor gene.
Bakker et al. (2003) present a genomewide scan in a large independent ADHD sib pair cohort. Notably, their analyses also revealed nominal evidence for linkage on 5p13 (MLS 1.42); however, the results of MLS analysis in the two studies showed little concordance for other regions. The discordance between linkage findings in the two studies may reflect variation between the clinical samples in the two studies, the polygenic nature of ADHD, and/or stochastic fluctuations. There are notable differences in the clinical samples. The sample reported by Bakker et al. (2003) consists of fewer individuals with the inattentive subtype of ADHD (12.6%) than our sample (45%) (P<.0001), and the Bakker et al. (2003) broad definition of ADHD includes individuals with clear-cut autism spectrum disorder (33%). In contrast, all autistic phenotypes are excluded in our sample. Whether these clinical differences account for the lack of repeatability of linkage findings across samples requires additional investigation. The polygenic nature of ADHD suggests that replication of true linkages will likely require substantially larger samples (Suarez 1994) than those of Bakker et al. (2003) or of the present study. A direct comparison of results from the two studies is complicated by the variability in fine-mapping data available in each (∼5-cM resolution on 7p13, 9q33, 10cen, and 15q15 in the sample reported by Bakker et al. [2003], and ∼1-cm resolution on 16p13 in our sample). Linkage signals will vary substantially on the basis of marker density, as is evident when our own analysis of the 16p13, using only the 10-cM marker set (MLS .92), is compared with the dense, ∼1-cM marker set (MLS 3.73); the variation is due, in part, to increased information content and inclusion of SNP data. We are in the process of pooling data across the two samples, to evaluate the 10-cM dense marker set within a single large sample and to fine-map a common set of regions on the basis of these analyses. Lastly, the discordance across studies may be due to stochastic variation. Some positive linkage signals within each study are likely to be false-positive results, and further evaluation of specific regions within a joint analysis of the two samples, as well as independent samples, will likely uncover true signals within the highlighted regions. Notably, the region on 5p, which is highlighted in both scans, may ultimately prove to be a true linkage signal, despite the finding that the MLS values found for this region in each study are <2.0. Further support for the possibility that some of the current peaks are true linkages stems from simulations (Wiltshire et al. 2003). Simulations performed within the unique parameters of our data set demonstrate that the observed number (n=6) of independent regions of linkage at the nominal pointwise P value ⩽.01 (MLS >1.18) is greater than the empirical expectation (n=2.8). The empirical P value of obtaining an MLS of 2.98 (17p) is P=.03, and that of obtaining an MLS of 3.73 (16p) is P=.01, indicating that both are significant. Overall, the variability of findings across the two available genomewide scans in ADHD reflect the need for larger ASP cohorts and population samples, providing both power to detect loci of minor effect and coverage of the genetic heterogeneity present in ADHD.
Exclusion mapping at λs of 1.5 indicates that the current study has sufficient power to exclude loci of minor-effect size from only 56.3% of the genome. Several proposed susceptibility genes in ADHD (e.g., DAT-1 and DRD4) have estimated effect sizes (genotype relative risks) that would correspond to λs, <1.5 (Waldman et al. 1998; Faraone et al. 2001) and would likely not be detected in the current sample, using an identity-by-descent sharing methodology. Furthermore, at a λs of 1.5, two of the nine regions highlighted by Bakker et al. (2003) fall within excluded regions, but under a λs of 1.4, we cannot exclude any of the nine regions identified by Bakker et al. (2003). Finally, exclusion mapping was performed under an assumption of no dominance variance and should not be overinterpreted beyond the constraints of the model.
Although the present study indicates that there is unlikely to be a major-gene effect in ADHD, we have identified four relatively strong candidate regions on the basis of linkage findings in the present data set, as well as reports from other investigations of ADHD (5p13), comorbid RD (6p21-6q14), and autism (5p13, 16p13, and 17p11). Replication of the present findings is needed in larger samples, and fine-mapping is required to identify allelic variants contributing to susceptibility to ADHD. The data also highlight possible pleiotropic effects of putative loci underlying ADHD, autism, and RD. Until the specific risk loci are identified, it is impossible to determine whether the overlap in linkage peaks reflects pleiotropy or distinct loci in close proximity to one another. However, the emerging pattern of substantial overlap of regions for these three disorders is certainly intriguing. Efforts focused on identifying genetic variants that contribute to common phenotypic presentations across distinct clinical conditions (e.g., RD and ADHD) may provide a powerful approach to gene detection. Furthermore, a neurobiological model that provides a unifying mechanism of ADHD, autism, and RD, such as variations in cerebral asymmetry (Geschwind and Miller 2001; Rinehart et al. 2002), may prove useful in ranking potential candidate genes within chromosome locations suggested by linkage studies.
Acknowledgments
This work was supported by National Institute of Mental Health grants MH58277 (to S.L.S.), MH01969 (to J.J.M.), and MH01805 (to J.T.M.); by University of California Life Sciences Informatics grant L9808 (to S.F.N.); by USPHS National Research Service Award GM07104 (to M.N.O.); and by the Wellcome Trust (support to A.P.M.). A.P.M. is a Wellcome Trust Principal Research Fellow. S.E.F. is a Royal Society Research Fellow. Thanks to all the families who participated in this research; to Elva Rios, Tae Kim, Laura Combs, Leah Pressman, Ph.D., and Deborah Lynn, M.D., for their help in data collection; to Lori Crawford, for technical assistance; and to Dennis Cantwell, M.D., who inspired our work on ADHD.
Electronic-Database Information
URLs for data presented herein are as follows:
- ASPEX Linkage Analysis Package, ftp://lahmed.stanford.edu/pub/aspex/index.html
- Celera, http://www.celera.com/
- Center for Medical Genetics, Marshfield Medical Research Foundation http://research.marshfieldclinic.org/genetics/
- Cooperative Human Linkage Centre, http://gai.nci.nih.gov/CHLC/
- Ensembl, http://www.ensembl.org/ (for cytogenetic markers)
- Généthon, http://ftp.genethon.fr/php/index.php
- Genotype Linkage Analysis–GAS, http://hpcio.cit.nih.gov/lserver/GAS.html
- Kruglyak Laboratory, http://www.fhcrc.org/labs/kruglyak/Downloads/ (for Genehunter)
- Mapmaker/SIBS, http://www-genome.wi.mit.edu/ftp/distribution/software/sibs/
- Online Mendelian Inheritance in Man (OMIM), http://www.ncbi.nlm.nih.gov/Omim/ (for ADHD, autistic disorder, and RD)
- Research Genetics, ftp://ftp.resgen.com/pub/mappairs/humanset/
- Statgen Software, http://watson.hgen.pitt.edu/register (for RECODE)
- UCSC Genome Bioinformatics, http://genome.cse.ucsc.edu/
References
- Achenbach, TM (1993) Empirically based taxonomy: how to use syndromes and profile types derived from the CBCL from 4 to 18, TRF, and WSR. University of Vermont Department of Psychiatry, Burlington, VT [Google Scholar]
- American Psychiatric Association (1994) DSM-IV: Diagnostic and statistical manual of mental disorders, 4th ed. American Psychiatric Association, Washington, DC [Google Scholar]
- Anderson JC, Williams S, McGee R, Silva PA (1987) DSM-III disorders in preadolescent children. Prevalence in a large sample from the general population. Arch Gen Psychiatry 44:69–76 [DOI] [PubMed] [Google Scholar]
- August GJ, Garfinkel BD (1989) Behavioral and cognitive subtypes of ADHD. J Am Acad Child Adolesc Psychiatry 28:739–748 [DOI] [PubMed] [Google Scholar]
- Bakker SC, van der Meulen DM, Buitelaar JK, Sandkuijl LA, Pauls DL, Monsuur AJ, van 't Slot R, Minderaa RB, Gunning WB, Pearson PL, Sinke RJ (2003) A whole-genome scan in 164 Dutch sib pairs with attention-deficit/hyperactivity disorder: suggestive evidence for linkage on chromosomes 7p and 15q. Am J Hum Genet 72:1251–1260 (in this issue) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barkley RA (1997) Attention-deficit/hyperactivity disorder, self-regulation, and time: toward a more comprehensive theory. J Dev Behav Pediatr 18:271–279 [PubMed] [Google Scholar]
- Barr, CL, Wigg K, Zai G, Roberts W, Malone M, Schachar R, Tannock R, Kennedy JL (2001) Attention-deficit hyperactivity disorder and the adrenergic receptors alpha 1C and alpha 2C. Mol Psychiatry 6:334–337 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baumgaertel A, Wolraich ML, Dietrich M (1995) Comparison of diagnostic criteria for attention deficit disorders in a German elementary school sample. J Am Acad Child Adolesc Psychiatry 34:629–638 [DOI] [PubMed] [Google Scholar]
- Beck KD, Valverde J, Alexi T, Poulsen K, Moffat B, Vandlen RA, Rosenthal A, Hefti F (1995) Mesencephalic dopaminergic neurons protected by GDNF from axotomy-induced degeneration in the adult brain. Nature 373:339–341 [DOI] [PubMed] [Google Scholar]
- Biederman, J, Faraone SV, Keenan K, Benjamin J, Krifcher B, Moore C, Sprich-Buckminster S, Ugaglia K, Jellinek MS, Steingard R (1992) Further evidence for family-genetic risk factors in attention deficit hyperactivity disorder: patterns of comorbidity in probands and relatives psychiatrically and pediatrically referred samples. Arch Gen Psychiatry 49:728–738 [DOI] [PubMed] [Google Scholar]
- Biederman J, Faraone S, Milberger S, Guite J, Mick E, Chen L, Mennin D, Marrs A, Ouellette C, Moore P, Spencer T, Norman D, Wilens T, Kraus I, Perrin J (1996) A prospective 4-year follow-up study of attention-deficit hyperactivity and related disorders. Arch Gen Psychiatry 53:437–446 [DOI] [PubMed] [Google Scholar]
- Brown RT, Freeman WS, Perrin JM, Stein MT, Amler RW, Feldman HM, Pierce K, Wolraich ML (2001) Prevalence and assessment of attention-deficit/hyperactivity disorder in primary care settings. Pediatrics 107:E43 [DOI] [PubMed] [Google Scholar]
- Buxbaum JD, Silverman JM, Smith CJ, Kilifarski M, Reichert J, Hollander E, Lawlor BA, Fitzgerald M, Greenberg DA, Davis KL (2001) Evidence for a susceptibility gene for autism on chromosome 2 and for genetic heterogeneity. Am J Hum Genet 68:1514–1520 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cantwell DP (1975) Genetics of hyperactivity. J Child Psychol Psychiatry 16:261–264 [DOI] [PubMed] [Google Scholar]
- ——— (1996) Attention deficit disorder: a review of the past 10 years. J Am Acad Child Adolesc Psychiatry 35:978–987 [DOI] [PubMed] [Google Scholar]
- Cardon LR, Smith SD, Fulker DW, Kimberling WJ, Pennington BF, DeFries JC (1994) Quantitative trait locus for reading disability on chromosome 6. Science 266:276–279 [DOI] [PubMed] [Google Scholar]
- Comings DE, Gade-Andavolu R, Gonzalez N, Wu S, Muhleman D, Blake H, Chiu F, Wang E, Farwell K, Darakjy S, Baker R, Dietz G, Saucier G, MacMurray JP (2000) Multivariate analysis of associations of 42 genes in ADHD, ODD and conduct disorder. Clin Genet 58:31–40 [DOI] [PubMed] [Google Scholar]
- Comings DE, Gade R, Muhleman D, Sverd J (1995) No association of a tyrosine hydroxylase gene tetranucleotide repeat polymorphism in autism, Tourette syndrome, or ADHD. Biol Psychiatry 37:484–486 [DOI] [PubMed] [Google Scholar]
- Cook EH Jr, Stein MA, Krasowski MD, Cox NJ, Olkon DM, Kieffer JE, Leventhal BL (1995) Association of attention-deficit disorder and the dopamine transporter gene. Am J Hum Genet 56:993–998 [PMC free article] [PubMed] [Google Scholar]
- Curran S, Mill J, Tahir E, Kent L, Richards S, Gould A, Huckett L, Sharp J, Batten C, Fernando S, Ozbay F, Yazgan Y, Simonoff E, Thompson M, Taylor E, Asherson P (2001) Association study of a dopamine transporter polymorphism and attention deficit hyperactivity disorder in UK and Turkish samples. Mol Psychiatry 6:425–428 [DOI] [PubMed] [Google Scholar]
- Daly G, Hawi Z, Fitzgerald M, Gill M (1999) Mapping susceptibility loci in attention deficit hyperactivity disorder: preferential transmission of parental alleles at DAT1, DBH and DRD5 to affected children. Mol Psychiatry 4:192–196 [DOI] [PubMed] [Google Scholar]
- Deutsch CK, Matthysse S, Swanson JM, Farkas LG (1990) Genetic latent structure analysis of dysmorphology in attention deficit disorder. J Am Acad Child Adolesc Psychiatry 29:189–194 [DOI] [PubMed] [Google Scholar]
- Dib C, Faure S, Fizames C, Samson D, Drouot N, Vignal A, Millasseau P, Marc S, Hazan J, Seboun E, Lathrop M, Gyapay G, Morissette J, Weissenbach J (1996) A comprehensive genetic map of the human genome based on 5,264 microsatellites. Nature 380:152–154 [DOI] [PubMed] [Google Scholar]
- Faraone SV, Biederman J (1994) Is attention deficit hyperactivity disorder familial? Harv Rev Psychiatry 1:271–287 [DOI] [PubMed] [Google Scholar]
- ——— (1998) Neurobiology of attention-deficit hyperactivity disorder. Biol Psychiatry 44:951–958 [DOI] [PubMed] [Google Scholar]
- Faraone SV, Biederman J, Chen WJ, Krifcher B, Keenan K, Moore C, Sprich S, Tsuang MT (1992) Segregation analysis of attention deficit hyperactivity disorder: evidence for single gene transmission. Psychiatr Genet 2:257–275 [Google Scholar]
- Faraone SV, Biederman J, Monuteaux MC (2000) Toward guidelines for pedigree selection in genetic studies of attention deficit hyperactivity disorder. Genet Epidemiol 18:1–16 [DOI] [PubMed] [Google Scholar]
- Faraone SV, Doyle AE (2001) The nature and heritability of attention-deficit/hyperactivity disorder. Child Adolesc Psychiatry Clin N Am 10:299–316 [PubMed] [Google Scholar]
- Faraone SV, Doyle AE, Mick E, Biederman J (2001) Meta-analysis of the association between the 7-repeat allele of the dopamine D(4) receptor gene and attention deficit hyperactivity disorder. Am J Psychiatry 158:1052–1057 [DOI] [PubMed] [Google Scholar]
- Fisher SE, Francks C, McCracken JT, McGough JJ, Marlow AJ, MacPhie IL, Newbury DF, Crawford LR, Palmer CG, Woodward JA, Del’Homme M, Cantwell DP, Nelson SF, Monaco AP, Smalley SL (2002) A genomewide scan for loci involved in attention-deficit/hyperactivity disorder. Am J Hum Genet 70:1183–1196 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fisher SE, Marlow AJ, Lamb J, Maestrini E, Williams DF, Richardson AJ, Weeks DE, Stein JF, Monaco AP (1999) A quantitative-trait locus on chromosome 6p influences different aspects of developmental dyslexia. Am J Hum Genet 64:146–156 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fyer AJ, Mannuzza S, Chapman TF, Martin LY, Klein DF (1995) Specificity in familial aggregation of phobic disorders. Arch Gen Psychiatry 52:564–573 [DOI] [PubMed] [Google Scholar]
- Gayan J, Smith SD, Cherny SS, Cardon LR, Fulker DW, Brower AM, Olson RK, Pennington BF, DeFries JC (1999) Quantitative-trait locus for specific language and reading deficits on chromosome 6p. Am J Hum Genet 64:157–164 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geschwind DH, Miller BL (2001) Molecular approaches to cerebral laterality: development and neurodegeneration. Am J Med Genet 101:370–381 [PubMed] [Google Scholar]
- Gilger JW, Pennington BF, DeFries JC (1992) A twin study of the etiology of comorbidity: attention-deficit hyperactivity disorder and dyslexia. J Am Acad Child Adolesc Psychiatry 31:343–348 [DOI] [PubMed] [Google Scholar]
- Gill M, Daly G, Heron S, Hawi Z, Fitzgerald M (1997) Confirmation of association between attention deficit hyperactivity disorder and a dopamine transporter polymorphism. Mol Psychiatry 2:311–313 [DOI] [PubMed] [Google Scholar]
- Goldman LS, Genel M, Bezman RJ, Slanetz PJ (1998) Diagnosis and treatment of attention-deficit/hyperactivity disorder in children and adolescents. Council on Scientific Affairs, American Medical Association. JAMA 279:1100–1107 [DOI] [PubMed] [Google Scholar]
- Gomez R, Harvey J, Quick C, Scharer I, Harris G (1999) DSM-IV AD/HD: confirmatory factor models, prevalence, and gender and age differences based on parent and teacher ratings of Australian primary school children. J Child Psychol Psychiatry 40:265–274 [PubMed] [Google Scholar]
- Grigorenko EL, Wood FB, Meyer MS, Pauls DL (2000) Chromosome 6p influences on different dyslexia-related cognitive processes: further confirmation. Am J Hum Genet 66:715–723 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holmans P (1993) Asymptotic properties of affected-sib-pair linkage analysis. Am J Hum Genet 52:362–374 [PMC free article] [PubMed] [Google Scholar]
- Holmes J, Payton A, Barrett JH, Hever T, Fitzpatrick H, Trumper AL, Harrington R, McGuffin P, Owen M, Ollier W, Worthington J, Thapar A (2000) A family-based and case-control association study of the dopamine D4 receptor gene and dopamine transporter gene in attention deficit hyperactivity disorder. Mol Psychiatry 5:523–530 [DOI] [PubMed] [Google Scholar]
- International Molecular Genetic Study of Autism Consortium (2001) A genomewide screen for autism: strong evidence for linkage to chromosomes 2q, 7q, and 16p. Am J Hum Genet 69:570–581 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaufman J, Birmaher B, Brent D, Rao U, Flynn C, Moreci P, Williamson D, Ryan N (1997) Schedule for Affective Disorders and Schizophrenia for School-Age Children-Present and Lifetime Version (K-SADS-PL): initial reliability and validity data. J Am Acad Child Adolesc Psychiatry 36:980–988 [DOI] [PubMed] [Google Scholar]
- Kent L, Doerry U, Hardy E, Parmar R, Gingell K, Hawi Z, Kirley A, Lowe N, Fitzgerald M, Gill M, Craddock N (2002) Evidence that variation at the serotonin transporter gene influences susceptibility to attention deficit hyperactivity disorder (ADHD): analysis and pooled analysis. Mol Psychiatry 7:908–912 [DOI] [PubMed] [Google Scholar]
- Kruglyak L, Daly MJ, Reeve-Daly MP, Lander ES (1996) Parametric and nonparametric linkage analysis: a unified multipoint approach. Am J Hum Genet 58:1347–1363 [PMC free article] [PubMed] [Google Scholar]
- Kruglyak L, Lander ES (1995) Complete multipoint sib-pair analysis of qualitative and quantitative traits. Am J Hum Genet 57:439–454 [PMC free article] [PubMed] [Google Scholar]
- LaHoste GJ, Swanson JM, Wigal SB, Glabe C, Wigal T, King N, Kennedy JL (1996) Dopamine D4 receptor gene polymorphism is associated with attention deficit hyperactivity disorder. Mol Psychiatry 1:121–124 [PubMed] [Google Scholar]
- Levy F, Hay DA, McStephen M, Wood C, Waldman I (1997) Attention-deficit hyperactivity disorder: a category or a continuum? Genetic analysis of a large-scale twin study. J Am Acad Child Adolesc Psychiatry 36:737–744 [DOI] [PubMed] [Google Scholar]
- Liu J, Nyholt DR, Magnussen P, Parano E, Pavone P, Geschwind D, Lord C, Iversen P, Hoh J, Ott J, Gilliam TC (2001) A genomewide screen for autism susceptibility loci. Am J Hum Genet 69:327–340 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manor I, Eisenberg J, Tyano S, Sever Y, Cohen H, Ebstein RP, Kotler M (2001) Family-based association study of the serotonin transporter promoter region polymorphism (5-HTTLPR) in attention deficit hyperactivity disorder. Am J Med Genet 105:91–95 [PubMed] [Google Scholar]
- Markwardt FC (1989) Examiner’s manual: Peabody Individual Acheivement Test—Revised. American Guidance Service, Circle Pines, MN [Google Scholar]
- McCracken JT, Smalley SL, McGough JJ, Crawford L, Del’Homme M, Cantor RM, Liu A, Nelson SF (2000) Evidence for linkage of a tandem duplication polymorphism upstream of the dopamine D4 receptor gene (DRD4) with attention deficit hyperactivity disorder (ADHD). Mol Psychiatry 5:531–536 [DOI] [PubMed] [Google Scholar]
- Mill J, Curran S, Kent L, Richards S, Gould A, Virdee V, Huckett L, Sharp J, Batten C, Fernando S, Simanoff E, Thompson M, Zhao J, Sham P, Taylor E, Asherson P (2001) Attention deficit hyperactivity disorder (ADHD) and the dopamine D4 receptor gene: evidence of association but no linkage in a UK sample. Mol Psychiatry 6:440–444 [DOI] [PubMed] [Google Scholar]
- Morrison JR, Stewart MA (1973) The psychiatric status of the legal families of adopted hyperactive children. Arch Gen Psychiatry 28:888–891 [DOI] [PubMed] [Google Scholar]
- Nyholt DR (2000) All LODs are not created equal. Am J Hum Genet 67:282–288 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Odell JD, Warren RP, Warren WL, Burger RA, Maciulis A (1997) Association of genes within the major histocompatibility complex with attention deficit hyperactivity disorder. Neuropsychobiology 35:181–186 [DOI] [PubMed] [Google Scholar]
- Palmer CG, Bailey JN, Ramsey C, Cantwell D, Sinsheimer JS, Del’Homme M, McGough J, Woodward JA, Asarnow R, Asarnow J, Nelson S, Smalley SL (1999) No evidence of linkage or linkage disequilibrium between DAT1 and attention deficit hyperactivity disorder in a large sample. Psychiatr Genet 9:157–160 [DOI] [PubMed] [Google Scholar]
- Pattyn A, Morin X, Cremer H, Goridis C, Brunet JF (1999) The homeobox gene Phox2b is essential for the development of autonomic neural crest derivatives. Nature 399:366–370 [DOI] [PubMed] [Google Scholar]
- Payton A, Holmes J, Barrett JH, Hever T, Fitzpatrick H, Trumper AL, Harrington R, McGuffin P, O’Donovan M, Owen M, Ollier W, Worthington J, Thapar A (2001) Examining for association between candidate gene polymorphisms in the dopamine pathway and attention-deficit hyperactivity disorder: a family-based study. Am J Med Genet 105:464–470 [DOI] [PubMed] [Google Scholar]
- Pelham WE Jr, Gnagy EM, Greenslade KE, Milich R (1992) Teacher ratings of DSM-III-R symptoms for the disruptive behavior disorders. J Am Acad Child Adolesc Psychiatry 31:210–218 [DOI] [PubMed] [Google Scholar]
- Petryshen TL, Kaplan BJ, Fu Liu M, de French NS, Tobias R, Hughes ML, Field LL (2001) Evidence for a susceptibility locus on chromosome 6q influencing phonological coding dyslexia. Am J Med Genet 105:507–517 [DOI] [PubMed] [Google Scholar]
- Philippe A, Martinez M, Guilloud-Bataille M, Gillberg C, Rastam M, Sponheim E, Coleman M, Zappella M, Aschauer H, Van Maldergem L, Penet C, Feingold J, Brice A, Leboyer M, van Malldergerme L (1999) Genome-wide scan for autism susceptibility genes. Paris Autism Research International Sibpair Study. Hum Mol Genet 8:805–812 [DOI] [PubMed] [Google Scholar]
- Rinehart NJ, Bradshaw JL, Brereton AV, Tonge BJ (2002) Lateralization in individuals with high-functioning autism and Asperger’s disorder: a frontostriatal model. J Autism Dev Disord 32:321–331 [DOI] [PubMed] [Google Scholar]
- Risch N (1990) Linkage strategies for genetically complex traits. II. The power of affected relative pairs. Am J Hum Genet 46:229–241 [PMC free article] [PubMed] [Google Scholar]
- Rohde LA, Biederman J, Busnello EA, Zimmermann H, Schmitz M, Martins S, Tramontina S (1999) ADHD in a school sample of Brazilian adolescents: a study of prevalence, comorbid conditions, and impairments. J Am Acad Child Adolesc Psychiatry 38:716–722 [DOI] [PubMed] [Google Scholar]
- Scahill L, Schwab-Stone M (2000) Epidemiology of ADHD in school-age children. Child Adolesc Psychiatr Clin N Am 9:541–555 [PubMed] [Google Scholar]
- Schaar DG, Sieber BA, Dreyfus CF, Black IB (1993) Regional and cell-specific expression of GDNF in rat brain. Exp Neurol 124:368–371 [DOI] [PubMed] [Google Scholar]
- Seeger G, Schloss P, Schmidt MH (2001) Marker gene polymorphisms in hyperkinetic disorder: predictors of clinical response to treatment with methylphenidate? Neurosci Lett 313:45–48 [DOI] [PubMed] [Google Scholar]
- Semrud-Clikeman M, Biederman J, Sprich-Buckminster S, Lehman BK, Faraone SV, Norman D (1992) Comorbidity between ADDH and learning disability: a review and report in a clinically referred sample. J Am Acad Child Adolesc Psychiatry 31:439–448 [DOI] [PubMed] [Google Scholar]
- Smalley SL (1997) Genetic influences in childhood-onset psychiatric disorders: autism and attention-deficit/hyperactivity disorder. Am J Hum Genet 60:1276–1282 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smalley SL, Bailey JN, Palmer CG, Cantwell DP, McGough JJ, Del’Homme MA, Asarnow JR, Woodward JA, Ramsey C, Nelson SF (1998) Evidence that the dopamine D4 receptor is a susceptibility gene in attention deficit hyperactivity disorder. Mol Psychiatry 3:427–430 [DOI] [PubMed] [Google Scholar]
- Smalley SL, Kustanovich V, Minassian SL, Stone JL, Ogdie MN, McGough JJ, McCracken JT, MacPhie IL, Francks C, Fisher SE, Cantor RM, Monaco AP, Nelson SF (2002) Genetic linkage of attention-deficit/hyperactivity disorder on chromosome 16p13, in a region implicated in autism. Am J Hum Genet 71:959–963 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smalley SL, McGough JJ, Del’Homme M, NewDelman J, Gordon E, Kim T, Liu A, McCracken JT (2000) Familial clustering of symptoms and disruptive behaviors in multiplex families with attention-deficit/hyperactivity disorder. J Am Acad Child Adolesc Psychiatry 39:1135–1143 [DOI] [PubMed] [Google Scholar]
- Suarez BK, Hampe CL, Van Eerdewegh P (1994) Problems of replicating linkage claims in psychiatry. American Psychiatric Association, Washington, DC [Google Scholar]
- Swanson JM (1995) SNAP-IV Scale. University of California Child Development Center, Irvine, CA [Google Scholar]
- Swanson JM, Sergeant JA, Taylor E, Sonuga-Barke EJ, Jensen PS, Cantwell DP (1998) Attention-deficit hyperactivity disorder and hyperkinetic disorder. Lancet 351:429–433 [PubMed] [Google Scholar]
- Tahir E, Yazgan Y, Cirakoglu B, Ozbay F, Waldman I, Asherson PJ (2000) Association and linkage of DRD4 and DRD5 with attention deficit hyperactivity disorder (ADHD) in a sample of Turkish children. Mol Psychiatry 5:396–404 [DOI] [PubMed] [Google Scholar]
- Terwilliger JD, Speer M, Ott J (1993) Chromosome-based method for rapid computer simulation in human genetic linkage analysis. Genet Epidemiol 10: 217–224 [DOI] [PubMed] [Google Scholar]
- van den Oord EJ, Boomsma DI, Verhulst FC (1994) A study of problem behaviors in 10- to 15-year-old biologically related and unrelated international adoptees. Behav Genet 24:193–205 [DOI] [PubMed] [Google Scholar]
- Warren RP, Odell JD, Warren WL, Burger RA, Maciulis A, Daniels WW, Torres AR (1995) Reading disability, attention-deficit hyperactivity disorder, and the immune system. Science 268:786–788 [DOI] [PubMed] [Google Scholar]
- Wechsler D (1991) Examiner’s manual: Wechsler Intelligence Child Development Center, Irvine, CA, Scale For Children, 3rd ed. Psychological Corporation, New York [Google Scholar]
- Weeks DE, Lathrop GM (1995) Polygenic disease: methods for mapping complex disease traits. Trends Genet 11:513–519 [DOI] [PubMed] [Google Scholar]
- Wilens TE, Biederman J, Spencer TJ (2002) Attention deficit/hyperactivity disorder across the lifespan. Annu Rev Med 53:113–131 [DOI] [PubMed] [Google Scholar]
- Willcutt EG, Pennington BF (2000) Psychiatric comorbidity in children and adolescents with reading disability. J Child Psychol Psychiatry 41:1039–1048 [PubMed] [Google Scholar]
- Willcutt EG, Pennington BF, Smith SD, Cardon LR, Gayan J, Knopik VS, Olson RK, DeFries JC (2002) Quantitative trait locus for reading disability on chromosome 6p is pleiotropic for attention-deficit/hyperactivity disorder. Am J Med Genet 114:260–268 [DOI] [PubMed] [Google Scholar]
- Wolraich ML, Hannah JN, Baumgaertel A, Feurer ID (1998) Examination of DSM-IV criteria for attention deficit/hyperactivity disorder in a county-wide sample. J Dev Behav Pediatr 19:162–168 [DOI] [PubMed] [Google Scholar]
- Wolraich ML, Hannah JN, Pinnock TY, Baumgaertel A, Brown J (1996) Comparison of diagnostic criteria for attention-deficit hyperactivity disorder in a county-wide sample. J Am Acad Child Adolesc Psychiatry 35:319–324 [DOI] [PubMed] [Google Scholar]
- Yamasaki H, LaBar KS, McCarthy G (2002) Dissociable prefrontal brain systems for attention and emotion. Proc Natl Acad Sci USA 99:11447–11451 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang L, Cui X, Schmitt K, Hubert R, Navidi W, Arnheim N (1992) Whole genome amplification from a single cell: implications for genetic analysis. Proc Natl Acad Sci USA 89:5847–5851 [DOI] [PMC free article] [PubMed] [Google Scholar]