Abstract
Cosegregation of profound, congenital deafness with markers on chromosome 6q13 in three Pakistani families defines a new recessive deafness locus, DFNB37. Haplotype analyses reveal a 6-cM linkage region, flanked by markers D6S1282 and D6S1031, that includes the gene encoding unconventional myosin VI. In families with recessively inherited deafness, DFNB37, our sequence analyses of MYO6 reveal a frameshift mutation (36-37insT), a nonsense mutation (R1166X), and a missense mutation (E216V). These mutations, along with a previously published missense allele linked to autosomal dominant progressive hearing loss (DFNA22), provide an allelic spectrum that probes the relationship between myosin VI dysfunction and the resulting phenotype.
Autosomal recessive deafness is a genetically heterogeneous neurosensory disorder for which 54 distinct loci have been published and 32 genes have been identified (Petit et al. 2001; Griffith and Friedman 2002). Most autosomal recessive deafness is clinically indistinguishable, so genetic loci are most often identified by linkage studies using large, usually consanguineous pedigrees (Friedman et al. 2000). We ascertained a large Pakistani family, PKDF10 (fig. 1), with six individuals who have bilateral, profound, sensorineural, congenital hearing loss segregating as an autosomal recessive disorder. In addition to deafness, vestibular dysfunction and mild facial dysmorphology also occur in this family. One hearing-impaired individual (IV:19; table 1) has retinitis pigmentosa (RP) along with a vestibular abnormality. The latter two signs, when co-occuring with deafness, constitute Usher syndrome, which is also genetically heterogeneous (Hereditary Hearing Loss Homepage). However, the other clinical phenotypes were mild and did not occur in all deaf individuals (table 1).
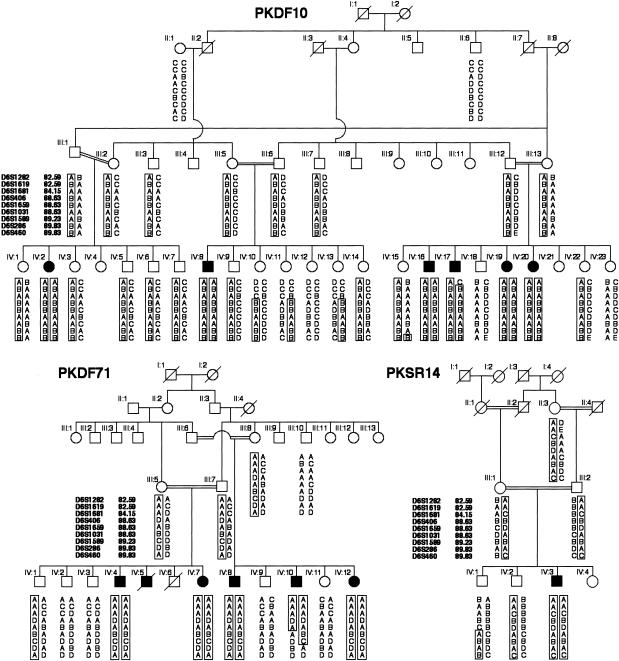
Figure 1.
Haplotypes of markers showing linkage to DFNB37 at 6q13 for three families segregating profound, sensorineural, recessive hearing loss. Affected individual IV:17 in family PKDF10 provided the proximal recombination breakpoint at marker D6S1282 (82.59 cM). The distal recombination is provided by affected individual IV:10 of family PKDF71 at marker D6S1031 (88.63 cM). National Institutes of Health (OH93-N-016) and National Centre of Excellence in Molecular Biology (CEMB) institutional review board approval and written informed consent were obtained from all participating subjects. DNA was extracted from either peripheral blood samples or buccal swabs and was amplified using fluorescent-labeled primers for STR markers linked to reported nonsyndromic recessive deafness (DFNB) loci. Amplimers were visualized by gel electrophoresis on ABI 377 DNA sequencers, and genotypes were determined using Genescan and Genotyper software (Applied Biosystems). The genetic linkage distances are from the Center for Medical Genetics Web server.
Table 1.
Clinical Description of PKDF10[Note]
|
Vestibular Phenotype |
Eye Phenotype |
||||||
| Subjecta | Age(years) | Hearing Phenotype | Age atAmbulation | ENGResults | FundoscopyResults | ERGResults | Other Findings |
| IV:16 | 20 | Profound hearing loss | 6 years | Abnormal | Congenital stationary night blindness | Normal | Retinal pigment epithelial changes; flat feet and prominence of the talus due to lack of arches in the feet; atrophy of the muscles around the Achilles tendon; minor facial dysmorphic features |
| IV:17 | 9 | Profound hearing loss | 14 mo | NA | No abnormality detected | Normal | History of hysteric fits from the age of 1 mo; flat feet and prominence of talus due to lack of arches in the feet; minor facial dysmorphic features; lordotic posture but no muscular weakness |
| IV:19 | 10 | Profound hearing loss | 15 mo | NA | RP | RP | Alternating squint; difficulties in tandem gait |
| IV:20 | 16 | Profound hearing loss | 18 mo | NA | No abnormality detected | Normal | History of hysteric fits from the age of 1 mo; alternating squint |
| IV:8 | 13 | Profound hearing loss | 18 mo | NA | No abnormality detected | Normal | Alternating squint; flat feet and prominence of talus due to lack of arches in the feet |
| IV:2 | 18 | Profound hearing loss | 30 mo | Abnormal | No abnormality detected | NA | |
| IV:23 | 17 | Normal | 3 years | NA | No abnormality detected | NA | Weakness in the right leg |
| IV:22 | 11 | Normal | 15 mo | NA | No abnormality detected | NA | |
| IV:18 | 24 | Normal | 18 mo | NA | No abnormality detected | NA | |
Note.— ENG = electronystagmography; ERG = electroretinography; NA = not available. All examinations were performed by local physicians in Pakistan, and the neurological examinations were videotaped. All results were reviewed by the clinicians at the National Institutes of Health who are also coauthors of the present article (A.J.G., R.C.C., and A.G.).
All subjects had normal reflexes and ambulated normally during the examination.
A new autosomal recessive deafness locus, DFNB37, was defined in family PKDF10, by virtue of exclusion of linkage of the deafness phenotype to markers linked to known DFNB loci. A genomewide scan using the Weber 9 marker panel revealed cosegregation with markers at 6q13 (LOD score [Z] 7.10 for D6S1589, at a recombination fraction [θ] of 0; see table 2). Haplotype analysis defined a proximal recombination at marker D6S1282 (82.59 cM) in affected individual IV:17 and a distal recombination in individual IV:15, who has normal hearing and is homozygous for marker D6S460 (89.63 cM) (fig. 1). Markers in the DFNB37 interval were used to screen 250 Pakistani and 100 Indian families segregating recessive deafness. Two additional families with DFNB37 linkage were identified, PKDF71 and PKSR14 (fig. 1; table 2). Affected individuals from family PKDF71 have profound sensorineural hearing loss, and the affected individual from family PKSR14 has severe-to-profound hearing loss. There were no obvious clinically relevant traits segregating in the families, other than deafness. Haplotype analysis of affected individual IV:10 from family PKDF71 defined a distal recombination breakpoint, reducing the linkage region for DFNB37 to ∼6 cM, bounded by markers D6S1282 (82.59 cM) and D6S1031 (88.63 cM) (fig. 1). This interval includes MYO6 (GenBank accession number AB002387), which encodes unconventional myosin VI.
Table 2.
Two-Point LOD Scores (at θ = 0)
|
LOD Score in Familya |
|||
| Marker | PKDF10 | PKDF71 | PKSR14 |
| D6S1282 | −∞ | 1.47 | 1.08 |
| D6S1619 | 4.84 | 4.03 | .98 |
| D6S1681 | 3.16 | 3.01 | .92 |
| D6S406 | 6.44 | 4.22 | .38 |
| D6S1659 | 6.53 | 3.90 | 1.23 |
| D6S1031 | 4.67 | 2.05 | 1.02 |
| D6S1589 | 6.99 | 2.27 | .18 |
| D6S286 | 6.44 | 1.63 | .34 |
| D6S460 | 5.35 | 1.35 | .41 |
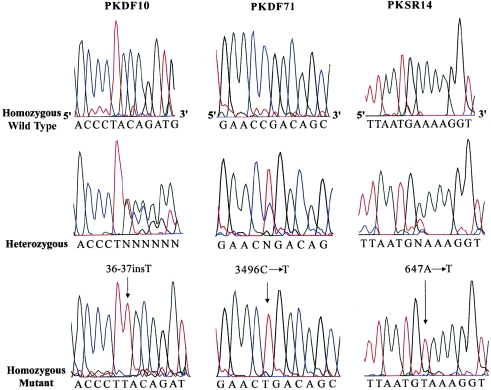
We screened for mutations in MYO6 by sequencing the 1 noncoding and 32 coding exons in the affected individuals from families PKDF10, PKDF71, and PKSR14. All exons were amplified by PCR from genomic DNA in a 20-μl reaction volume. Primers were designed to flank all of the exon-intron boundaries (see appendix A, table A1). PCR amplification, sequencing reactions, and mutational analyses were performed as described elsewhere (Ahmed et al. 2001). In all affected individuals of family PKDF10, we found a homozygous single-base-pair insertion (36-37insT) in the second exon of MYO6 (fig. 2). This insertion is predicted to cause a frameshift and premature translation termination after the first 12 amino acids of myosin VI. In family PKDF71, affected individuals are homozygous for a transition mutation, 3496C→T (fig. 2), resulting in a nonsense codon (R1166X) in exon 32, which encodes part of the globular domain of the tail region (fig. 3). These two mutations were not found in 100 ethnically matched control DNA samples from Pakistan.
Figure 2.
MYO6 mutations segregating in three Pakistani families. Left, Electropherograms of amplimers from genomic DNA templates, illustrating homozygosity for a single-base-pair insertion mutation in an affected individual, heterozygosity in an obligate carrier, and homozygosity for the wild-type allele in an unaffected individual from family PKDF10. An arrow indicates the site of the 36-37insT in the second exon. Center, Electropherograms illustrating genotypes of a homozygous wild-type allele, a 3496C→T heterozygote, and a person homozygous for 3496C→T of family PKDF71. Right, Electropherograms are shown for transversion mutation 647A→T, a carrier, and the wild-type allele of family PKSR14. All the mutations described here are numbered from start codon ATG (GenBank accession number AB002387).
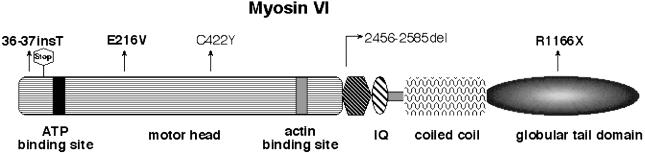
Figure 3.
A drawing of myosin VI, showing the locations of the mutations causing deafness in humans and mice. The three mutant alleles reported in this study are shown in bold letters, whereas C442Y and 2456-2585del are from the studies by Melchionda et al. (2001) and Avraham et al. (1995), respectively. Shown also is the predicted stop codon after twelve out-of-frame amino acids due to 36-37insT.
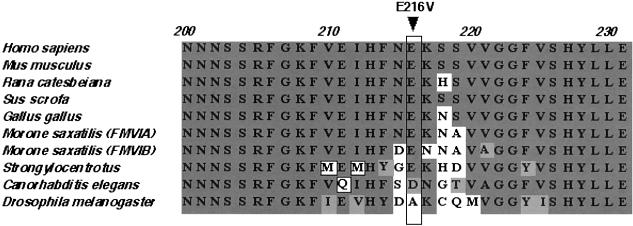
The single affected individual in family PKSR14 is homozygous for a missense mutation (E216V) in the motor domain of myosin VI caused by a transversion mutation, 647A→T (fig. 2). The E216V mutation substitutes a valine (nonpolar) for glutamate (polar, negatively charged). This glutamate residue is conserved in myosin VI proteins from human, mouse, chicken, pig, striped bass, and sea urchin (fig. 4). In Caenorhabditis elegans myosin VI, there is an aspartate residue (also polar, negatively charged) at this position, but Drosophila melanogaster myosin VI has a nonpolar amino acid (alanine) at this position. The 647A→T transversion segregating in family PKSR14 was not found among 270 normal representative DNA samples (540 chromosomes) from the same ethnic group or from the 81 DNA samples (162 chromosomes) of a Human Diversity panel (Coriell Cell Repositories).
Figure 4.
Alignment of a portion of myosin VI proteins from various species, showing conservation of the glutamate residue at position 216 in the motor domain among seven myosin VI proteins from Homo sapiens, Mus musculus, Rana catesbeiana, Sus scrofa, Gallus gallus, Morone saxatilis, and Strongylocentrotus. The conserved amino acids are shown with dark gray background, similar amino acids are shown with a light gray background, and the nonconserved amino acids are shown with a white background.
Myosins are motor proteins that hydrolyze ATP and translocate along actin filaments (Sellers 1999; Berg et al. 2001). Mutations of myosins IIA, IIIA, VIIA, and XVA are associated with hearing loss in humans (Gibson et al. 1995; Weil et al. 1995; Liu et al. 1997a, 1997b; Probst et al. 1998; Wang et al. 1998; Lalwani et al. 2000; Liburd et al. 2001; Walsh et al. 2002). Unlike these other actin-based motors, myosin VI moves toward the minus end of F-actin filaments (Wells et al. 1999; Nishikawa et al. 2002). Myosin VI is involved in many processes, including membrane trafficking, recycling, cell movement, and endocytosis (Mermall and Miller 1995; Bohrmann 1997; Mermall et al. 1998; Buss et al. 2001; Morris et al. 2002). In the inner and outer hair cells of the organ of Corti, myosin VI is highly expressed at the base of stereocilia rootlets and in the pericuticular necklace (Avraham et al. 1997; Hasson et al. 1997; Self et al. 1999).
Myosin VI is abundantly expressed in the retina (Breckler et al. 2000), and it has been speculated that mutations of MYO6 might cause RP (Ahituv et al. 2000). Interestingly, one hearing-impaired individual (IV:19, age 10 years; table 1) among the families with DFNB37 was found to have RP. No ocular abnormalities were detected in the older deaf individuals, and electroretinography results were normal among the other eight affected individuals (aged 9–21 years) from the three families with DFNB37. We cannot rule out the possibility of an atypical Usher syndrome in family PKDF10, since most of the affected individuals are too young to exhibit the ocular phenotype or there could be a modifier altering the effect of a null mutation in retina. In addition to deafness in family PKDF10, vestibular dysfunction and mild facial dysmorphology also occur, but not in all of the deaf individuals (table 1). Late ambulation, which may or may not be related to a vestibular dysfunction, was also found in individual IV:23 (table 1), who has normal hearing and is a noncarrier of the MYO6 mutant allele. The small sample size makes it difficult to determine whether there is reduced penetrance for these and other clinical findings that can be attributed to a null mutation of MYO6.
Two recessive putative null mutations of mouse Myo6 are responsible for deafness and vestibular dysfunction in Snell’s waltzer mice (Avraham et al. 1995), and a missense allele (C422Y) of MYO6 cosegregates with nonsyndromic, dominantly inherited, progressive hearing loss in a single family that defined the DFNA22 locus (MIM 606346) (Melchionda et al. 2001). The predicted structural motifs and the known, postulated mutant alleles of myosin VI that are associated with hearing loss are shown in figure 3. Since the DFNB37 alleles appear to be functional null alleles and since the heterozygous carriers of DFNB37 mutations of MYO6 have normal hearing, the putative DFNA22 mutation likely acts via a dominant-negative or gain-of-function mechanism. There is a possibility that two missense substitutions (C422Y and E216V) found in MYO6 may have nothing to do with hearing loss and may be in linkage disequilibrium with actual mutations. Biophysical measurements, such as a motility assay with these substitutions in the motor domain of MYO6, may help to address the pathogenic potential, if any exists. Nevertheless, two of the DFNB37 alleles (36-37insT and R1166X) reported herein constitute convincing genetic evidence that disablement of MYO6 causes recessively inherited deafness in humans.
Acknowledgments
We thank the families for their participation in this study. We appreciate the help of R. Siddique and the CEMB Deafness Research Group in Lahore, Pakistan, for their help in ascertaining these families. We also thank Barbara Ploplis, for her technical assistance, and Doris Wu and Alain Dabdoub, for critical reviews of the manuscript. This study was supported by the National Institute on Deafness and Other Communication Disorders intramural research project grants ZO1 DC00035-06 and ZO1 DC000039-06 (both to T.B.F.) and ZO1 DC000064-02 (to A.J.G.). The part of this study that was performed in Lahore was supported by Pakistan Atomic Energy Commission research grant 11(4)/91-iv.
Appendix A
Table A1.
MYO6 Intronic Primer Pairs Flanking Known Exons[Note]
|
Primer(5′→3′) |
|||
| Exon | Forward | Reverse | Product Length(bp) |
| 1 | aac aag aac tcc cgg ctt gt | cat cca tca cct gct tct cc | 589 |
| 2 | ggc aga tgt gtt tgt tag ttg g | cct agg gca cat acc tct gat t | 452 |
| 3 | tat gca acc aat taa gcc ctt c | ttc tga acc cgc aca gtg tat | 253 |
| 4 | agg atg agt caa agt gat tca ga | tct att caa gag gct cag atc aa | 291 |
| 5 | gga ctg att tgg gag tct tag tt | cca aga ggt ata cag ttt ctc caa | 391 |
| 6 | tga ttt ctt taa gag taa gtg gtc ct | aga atg agg tgg aac agt gg | 364 |
| 7 | tga tga tct agg ttt cag ttt tat atg | aag aga gtc ttt tgc atc tct ga | 258 |
| 8 | tgg aga tat acc atg cat att ttg | tcc tgc aac cat cta aag taa ca | 303 |
| 9 | aac ctc ttt gat aga caa atg gta tt | aag tat tag gct tga tgg caa tta t | 651 |
| 10 | tct tca tgg ttg gca cta ttt g | gtt aga act ctt act tgg gct cta aaa | 279 |
| 11 | agt gca tta att gac ctg gtg t | cat tct tca ttt ggg aga ttc a | 423 |
| 12 | aag cct tgc cta tta tat ggt ttt t | cca agc tca ggt ttt aaa cag aa | 352 |
| 13 | tgt gcc tat tct cac atg acc t | cat caa gta aaa tga gtt aca aag gag | 394 |
| 14 | gcc att att aca att aca ttt tat cct | ctt ccg tta tac acc atc cat c | 293 |
| 15 | gtt cag aaa cag tgc aaa att ca | gca cag aaa aag cac taa aca ca | 218 |
| 16 | tga tca gtc ctt gaa atc tgt ga | tgc acc ttt tta ata atg tct cgt | 286 |
| 17 | tga aaa gtg tga aaa ttt cct gt | aat aac cac gtg aaa taa tta ata acc | 320 |
| 18 | gga gaa aac cat ttc atg ttg a | ggg tct gct ctt gaa ctt cac t | 585 |
| 19 | cac tgt gta ctt tgg ctt ttt ga | tgc cat gtc tgg aga atg tta c | 442 |
| 20 | tgc ttt gaa agt tgc agg tat t | tgc act gta aaa taa tca aga taa tgg | 289 |
| 21 | ttt tgg act tcc gaa cag tga t | caa agc ttt aaa caa agc ctg aa | 295 |
| 22 | tat aaa tgc cac cca aat tga a | tgt tgt tag tga cca tat aat tca aga | 289 |
| 23 | ttg aca tgt gac cat ttt cag ac | caa cac tcc aca aac cat ctt g | 313 |
| 24 | ttg agc att act ttg tga aaa tga | gaa aac ctg agt atc caa act gc | 286 |
| 25 | gaa gtg aaa tac cct gtt tag ca | ttt gaa aaa cta agg acg ttt tct | 346 |
| 26 | ttt tgc tgt att tgc ata ttg ga | aaa atc gat tga acc cga gag | 575 |
| 27 | ctt ctg aag gat tct tta ttt tct gtt | cca tgt ccg gct aat ttg tt | 328 |
| 28 | aag ggg agt gat caa gta aac aa | tca ttc act tga gta tga aag tcg | 398 |
| 29 | aac aca aat ttg cac aat cca g | cca atg aga aaa cta ctc cca aa | 176 |
| 30 | tgt gtt acg gct aga ttt gtt ga | cat gta aca ggt tct ggt cca a | 308 |
| 31 | gca tgc ttt ctt gcc tct tta g | cat aac tat gtg gga tcc tct gg | 450 |
| 32 | aaa aat ctc ctt atg cga cag aat | tgg gac att tta ccc atg tct t | 314 |
| 33 | act ttt cag tca cca cct cga t | cca ctg aaa att gta gca aaa ca | 238 |
| 34 | tta cta tta ggg acc ttt ctt ctt ttt | ccc tca acc ctg aaa tgt aat a | 446 |
| 35a | aat agg tat ttc agg cat aca act g | tag tct tct ggc aaa gga tga g | 568 |
| 35b | cat cct ttg cca gaa gac tac c | gaa aca act tgg aca aga ttc tga | 619 |
| 35c | ggc ata gtg gct taa ctg gac t | caa aca cta gtc agc tgg gaa a | 554 |
Note.— Primers were designed through use of the Primer3 Web-Based Server.
Electronic-Database Information
The accession number and URLs for data presented herein are as follows:
- Center for Medical Genetics, Marshfield Medical Research Foundation, http://research.marshfieldclinic.org/genetics/
- Hereditary Hearing Loss Homepage, http://www.uia.ac.be/dnalab/hhh/
- GenBank, http://www.ncbi.nlm.nih.gov/Genbank/ (for MYO6 [accession number AB002387])
- Online Mendelian Inheritance in Man (OMIM), http://www.ncbi.nlm.nih.gov/Omim/ (for DFNA22) [PubMed]
- Primer3 Web-Based Server, http://www.genome.wi.mit.edu/cgi-bin/primer/primer3_www.cgi
References
- Ahituv N, Sobe T, Robertson NG, Morton CC, Taggart RT, Avraham KB (2000) Genomic structure of the human unconventional myosin VI gene. Gene 261:269–275 [DOI] [PubMed] [Google Scholar]
- Ahmed ZM, Riazuddin S, Bernstein SL, Ahmed Z, Khan S, Griffith AJ, Morell RJ, Friedman TB, Wilcox ER (2001) Mutations of the protocadherin gene PCDH15 cause Usher syndrome type 1F. Am J Hum Genet 69:25–34 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Avraham KB, Hasson T, Sobe T, Balsara B, Testa JR, Skvorak AB, Morton CC, Copeland NG, Jenkins NA (1997) Characterization of unconventional MYO6, the human homologue of the gene responsible for deafness in Snell’s waltzer mice. Hum Mol Genet 6:1225–1231 [DOI] [PubMed] [Google Scholar]
- Avraham KB, Hasson T, Steel KP, Kingsley DM, Russell LB, Mooseker MS, Copeland NG, Jenkins NA (1995) The mouse Snell’s waltzer deafness gene encodes an unconventional myosin required for structural integrity of inner ear hair cells. Nat Genet 11:369–375 [DOI] [PubMed] [Google Scholar]
- Berg JS, Powell BC, Cheney RE (2001) A millennial myosin census. Mol Biol Cell 12:780–794 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bohrmann J (1997) Drosophila unconventional myosin VI is involved in intra- and intercellular transport during oogenesis. Cell Mol Life Sci 53:652–662 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bork JM, Peters LM, Riazuddin S, Bernstein SL, Ahmed ZM, Ness SL, Polomeno R, et al (2001) Usher syndrome 1D and nonsyndromic autosomal recessive deafness DFNB12 are caused by allelic mutations of the novel cadherin-like gene CDH23. Am J Hum Genet 68:26–37 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Breckler J, Au K, Cheng J, Hasson T, Burnside B (2000) Novel myosin VI isoform is abundantly expressed in retina. Exp Eye Res 70:121–134 [DOI] [PubMed] [Google Scholar]
- Buss F, Arden SD, Lindsay M, Luzio JP, Kendrick-Jones J (2001) Myosin VI isoform localized to clathrin-coated vesicles with a role in clathrin-mediated endocytosis. EMBO J 20:3676–3684 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friedman T, Battey J, Kachar B, Riazuddin S, Noben-Trauth K, Griffith A, Wilcox E (2000) Modifier genes of hereditary hearing loss. Curr Opin Neurobiol 10:487–493 [DOI] [PubMed] [Google Scholar]
- Gibson F, Walsh J, Mburu P, Varela A, Brown KA, Antonio M, Beisel KW, Steel KP, Brown SD (1995) A type VII myosin encoded by the mouse deafness gene shaker-1. Nature 374:62–64 [DOI] [PubMed] [Google Scholar]
- Griffith AJ, Friedman TB (2002) Autosomal and X-linked auditory disorders. In: Keats BJB, Popper AN, Far RR (eds) Genetic and auditory disorders, vol 14. Springer, New York, pp 121–227 [Google Scholar]
- Hasson T, Gillespie PG, Garcia JA, MacDonald RB, Zhao Y, Yee AG, Mooseker MS, Corey DP (1997) Unconventional myosins in inner-ear sensory epithelia. J Cell Biol 137:1287–1307 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lalwani AK, Goldstein JA, Kelley MJ, Luxford W, Castelein CM, Mhatre AN (2000) Human nonsyndromic hereditary deafness DFNA17 is due to a mutation in nonmuscle myosin MYH9. Am J Hum Genet 67:1121–1128 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liburd N, Ghosh M, Riazuddin S, Naz S, Khan S, Ahmed Z, Liang Y, Menon PS, Smith T, Smith AC, Chen KS, Lupski JR, Wilcox ER, Potocki L, Friedman TB (2001) Novel mutations of MYO15A associated with profound deafness in consanguineous families and moderately severe hearing loss in a patient with Smith-Magenis syndrome. Hum Genet 109:535–541 [DOI] [PubMed] [Google Scholar]
- Liu XZ, Walsh J, Mburu P, Kendrick-Jones J, Cope MJ, Steel KP, Brown SD (1997a) Mutations in the myosin VIIA gene cause non-syndromic recessive deafness. Nat Genet 16:188–190 [DOI] [PubMed] [Google Scholar]
- Liu XZ, Walsh J, Tamagawa Y, Kitamura K, Nishizawa M, Steel KP, Brown SD (1997b) Autosomal dominant non-syndromic deafness caused by a mutation in the myosin VIIA gene. Nat Genet 17:268–269 [DOI] [PubMed] [Google Scholar]
- Melchionda S, Ahituv N, Bisceglia L, Sobe T, Glaser F, Rabionet R, Arbones ML, Notarangelo A, Di Iorio E, Carella M, Zelante L, Estivill X, Avraham KB, Gasparini P (2001) MYO6, the human homologue of the gene responsible for deafness in Snell’s waltzer mice, is mutated in autosomal dominant nonsyndromic hearing loss. Am J Hum Genet 69:635–640 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mermall V, Miller KG (1995) The 95F unconventional myosin is required for proper organization of the Drosophila syncytial blastoderm. J Cell Biol 129:1575–1588 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mermall V, Post PL, Mooseker MS (1998) Unconventional myosins in cell movement, membrane traffic, and signal transduction. Science 279:527–533 [DOI] [PubMed] [Google Scholar]
- Morris SM, Arden SD, Roberts RC, Kendrick-Jones J, Cooper JA, Luzio JP, Buss F (2002) Myosin VI binds to and localises with Dab2, potentially linking receptor-mediated endocytosis and the actin cytoskeleton. Traffic 3:331–341 [DOI] [PubMed] [Google Scholar]
- Nishikawa S, Homma K, Komori Y, Iwaki M, Wazawa T, Hikikoshi Iwane A, Saito J, Ikebe R, Katayama E, Yanagida T, Ikebe M (2002) Class VI myosin moves processively along actin filaments backward with large steps. Biochem Biophys Res Commun 290:311–317 [DOI] [PubMed] [Google Scholar]
- Petit C, Levilliers J, Hardelin JP (2001) Molecular genetics of hearing loss. Annu Rev Genet 35:589–646 [DOI] [PubMed] [Google Scholar]
- Probst FJ, Fridell RA, Raphael Y, Saunders TL, Wang A, Liang Y, Morell RJ, Touchman JW, Lyons RH, Noben-Trauth K, Friedman TB, Camper SA (1998) Correction of deafness in shaker-2 mice by an unconventional myosin in a BAC transgene. Science 280:1444–1447 [DOI] [PubMed] [Google Scholar]
- Self T, Sobe T, Copeland NG, Jenkins NA, Avraham KB, Steel KP (1999) Role of myosin VI in the differentiation of cochlear hair cells. Dev Biol 214:331–341 [DOI] [PubMed] [Google Scholar]
- Sellers JR (1999) Myosins. Oxford University Press, Oxford [Google Scholar]
- Walsh T, Walsh V, Vreugde S, Hertzano R, Shahin H, Haika S, Lee MK, Kanaan M, King MC, Avraham KB (2002) From flies’ eyes to our ears: mutations in a human class III myosin cause progressive nonsyndromic hearing loss DFNB30. Proc Natl Acad Sci USA 99:7518–7523 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang A, Liang Y, Fridell RA, Probst FJ, Wilcox ER, Touchman JW, Morton CC, Morell RJ, Noben-Trauth K, Camper SA, Friedman TB (1998) Association of unconventional myosin MYO15 mutations with human nonsyndromic deafness DFNB3. Science 280:1447–1451 [DOI] [PubMed] [Google Scholar]
- Weil D, Blanchard S, Kaplan J, Guilford P, Gibson F, Walsh J, Mburu P, Varela A, Levilliers J, Weston MD, Kelley PM, Kimberling WJ, Wagenaar M, Levi-Acobas F, Larget-Plet D, Munnich A, Steel KP, Brown SDM, Petit C (1995) Defective myosin VIIA gene responsible for Usher syndrome type 1B. Nature 374:60–61 [DOI] [PubMed] [Google Scholar]
- Wells AL, Lin AW, Chen LQ, Safer D, Cain SM, Hasson T, Carragher BO, Milligan RA, Sweeney HL (1999) Myosin VI is an actin-based motor that moves backwards. Nature 401:505–508 [DOI] [PubMed] [Google Scholar]