To the Editor:
In a recent report, Silva et al. (2002) provided partial (8.8 kb) information on the mtDNA coding region (within the region 7148–15946, in the numbering of the Cambridge reference sequence [CRS]; Anderson et al. [1981]) in 40 individuals from Brazil. On the basis of the similarity in nucleotide diversity and age estimates of the four founder haplogroups A, B, C, and D, they claimed to have added new evidence for a single early entry of the founder populations into America. However, a site-by-site audit of the data reveals that their sequences are not of high enough quality to justify such statements. The authors failed to realize that a large number of mutations associated with basal branches of the worldwide mtDNA phylogeny (Finnilä et al. 2001; Maca-Meyer et al. 2001; Torroni et al. 2001; Derbeneva et al. 2002; Herrnstadt et al. 2002; Kivisild et al. 2002) were not correctly scored in their data set.
In the case of the hypervariable segments of the mtDNA control region, Bandelt et al. (2001, 2002) have highlighted lab-specific idiosyncrasies through comparative phylogenetic analysis. For the coding region, the task of identifying anomalies and reconstructing their potential causes is somewhat easier because the vast majority of sites there do not appear to undergo frequent mutations. The coding region well supports a basal nesting of (monophyletic) haplogroups, many of which had already been identified through RFLP analysis and sequencing of the hypervariable segments (Richards and Macaulay 2001). For example, the basal division of Eurasian mtDNAs into macrohaplogroups M and N is amazingly clear cut. The Eurasian mtDNA phylogeny that emerges from the phylogenetic analysis of the complete mtDNA database is detailed (for east Asia) in figure 1 of Kivisild et al. (2002), which attempts a reconstruction of the mutational history. The African mtDNA phylogeny has also been well documented in recent papers (Maca-Meyer et al. 2001; Torroni et al. 2001; Herrnstadt et al. 2002).
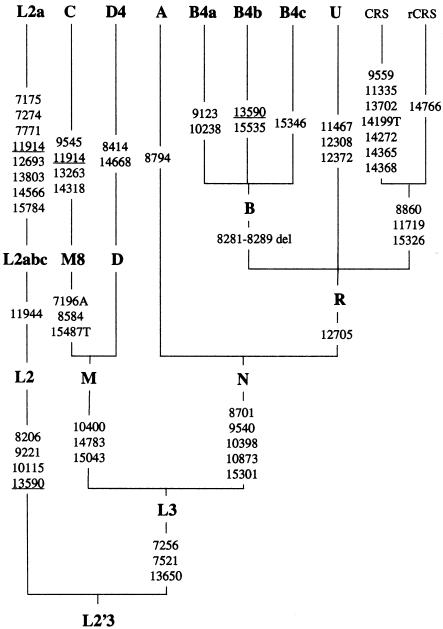
Silva et al. (2002) reported 40 mtDNAs, of which they assigned 31 to the Native American haplogroups A, B, C, and D (according to their fig. 1). The remaining nine mtDNAs can be assigned unambiguously to the Asian haplogroups B4 and D4, the Eurasian haplogroup U, and the African haplogroup L2a (table 1), as we will argue below. Figure 1 displays the truncation (relative to the 8.8-kb fragment under study) of the rooted phylogeny that is relevant for assigning these 40 mtDNAs to their respective haplogroups. This phylogeny is unanimously supported by the earlier publications. (However, note that mutations at 15301 and 11944 were not reconstructed most parsimoniously along the African mtDNA tree shown in fig. 1 of Herrnstadt et al. [2002]). The only instances of recurrent mutations (real or not) for the mutations and haplogroups highlighted in figure 1 are then as follows: the transversion 15487T is missing in the single haplogroup C lineage of Maca-Meyer et al. (2001); in the data of Herrnstadt et al. (2002), the B4b lineage 375 has experienced a transition at 14766, the L2a lineage 223 lacks the 7521 transition, and the 14566 transition is missing in the L2a lineage 165, which is closely related to another L2a lineage (bearing the 14566 mutation) from Torroni et al. (2001) in that they both share additional mutations at 3010 and 6663.
Table 1.
| Sample ID | Haplogroup | Sequence on Region 7148–15976a | Basal Mutations Missedb | Accession Number |
| GRC0149 | A | 7369 7522G 8027 8794 8860 11335 12007 12705 15326 15524 |
11719 | AF465949 |
| KTN0130 | A | 8794 8860 11129 11288 11719 12007 12406 12705 14178 14755 14861 | 15326 | AF465956 |
| KPO0013 | A | 8764 8794 9392 9966 11335 11719 12007 12292G 12314G 12705 13708 14566 |
8860 15326 | AF465957 |
| PTJ0003 | A |
8794 11719 11944 12007 12705 |
8860 15326 | AF465962 |
| WTE1182 | A | 8794 8860 11617G 11719 12007 12292G 12618 12705 15326 | … | AF465972 |
| WPI0167 | A |
8794 8860 10398 11719 12007 12705 14978 |
15326 | AF465974 |
| YAN0623 | A | 8794 8860 10694 11719 12007 12705 13928C 15317 15326 | … | AF465975 |
| YAN0665 | A | 8794 8860 11719 12007 12705 13928C | 15326 | AF465976 |
| KCR0029 | A |
8794 8860 9192 103981040011335 12007 12314G 12705 |
11719 15326 | AF465950 |
| GRC0169 | B4b | 7626 8860 9950 11335 11719 11821 13590 15326 15535 |
8281–8289del | AF465953 |
| KTN0209 | B4b | 8860 11150 11719 13590 14645 14647 15535 15914C | 8281–8289del 15326 | AF465955 |
| KPO0001 | B4b | 7369 7522G 8281–8289del 8860 9950 11335 11719 13590 15535 |
15326 | AF465958 |
| KPO0039 | B4b | 8736 8860 9950 10954 11335 11719 13590 15535 |
8281–8289del 15326 | AF465959 |
| KPO0023 | B4b | 8552 10604 11719 13590 13708 15535 | 8281–8289del 8860 15326 | AF465960 |
| QUE1876 | B4b | 8020 8860 11335 11719 12618 13590 |
8281–8289del 15326 15535 | AF465964 |
| QUE1881 | B4b | 8860 9950 11335 11719 13590 15043 15535 |
8281–8289del 15326 | AF465965 |
| YAN0637 | B4b | 8860 9950 11177 11719 12155C 13590 13708 15106 15535 | 8281–8289del 15326 | AF465980 |
| KRC0033 | B4b | 7227T 7251 8860 9950 103981040011335 11719 13590 |
8281–8289del 15535 15326 | AF465951 |
| QUE1880 | B4b | 7231C 8860 9950 103981040011335 11719 12192 13590 15326 |
8281–8289del 15535 | AF465968 |
| JAP1044 | B4c/B4a |
1011510238del 10398 11335 11719 15326 15346 |
8281–8289del 8860 | AF465948 |
| ARL0058 | C | 7196A 8078 8584 8701 9540 9545 10398 10400 10873 11719 11914 12705 13263 14783 15043 15301 15326 | 8860 14318 15487T | AF465945 |
| PTJ0068 | C | 8701 8860 9540 9545 10873 11719 11914 12705 13263 14318 14783 14788 15043 15914C | 7196A 8584 10398 10400 15301 15487T 15326 | AF465961 |
| QUE1875 | C |
7196A 8584 8701 8860 9540 9545 11335 11719 11914 12705 13263 13656 14783 15043 15301 |
10398 10400 10873 14318 15487T 15326 | AF465966 |
| QUE1878 | C |
8584 8701 9540 9545 10873 11335 11719 11914 12705 13263 13545 14783 15043 15191 |
7196A 8860 10398 10400 14318 15301 15487T 15326 | AF465967 |
| YAN0669 | C | 8701 8848 8860 9540 9545 10310 10398 10400 11719 11914 12705 13263 13326 14318 14783 15043 15326 | 7196A 8584 10873 15301 15487T | AF465977 |
| YAN0591 | C | 8584 8701 8848 8860 9540 9545 10873 11719 11914 12705 13263 13326 14783 15043 | 7196A 10398 10400 14318 15301 15487T 15326 | AF465978 |
| YAN0650 | C | 7196A 8701 8848 8860 9540 9545 10398 10873 11617G 11719 11914 12705 13263 13326 14318 14783 15043 15301 | 8584 10400 15487T 15326 | AF465979 |
| JAP1045 | D4 | 8701 8860 8964 9296 9540 9824A 10115 10398 10873 11719 12705 14783 15043 15301 15326 |
8414 10400 14668 | AF465947 |
| GRC0131 | D4 | 8701 8860 9540 10816T 10873 11335 11914 12705 13059 13067 14783 15043 |
8414 10398 10400 11719 14668 15301 15326 | AF465952 |
| JAP1043 | D4 | 8701 8860 9540 10398 10400 10873 11215 11719 12705 14783 15043 15301 15326 15874 | 8414 14668 | AF465946 |
| KTN0018 | D | 8701 8860 9540 10873 10874 12705 14687 14783 15043 | 10398 10400 11719 15301 15326 | AF465954 |
| PTJ0001 | D | 8701 8860 9540 10398 10400 10873 11150 11719 12705 14783 15043 15106 15301 | 15326 | AF465963 |
| TYR0004 | D | 8701 8860 9540 10398 10400 11719 12406 12705 12810 15301 | 10873 14783 15043 15326 | AF465969 |
| TYR0016 | D | 8701 8860 10398 10400 10819 10873 10874 11719 12406 12705 12810 | 9540 14783 15043 15301 15326 | AF465970 |
| NGR0524 | L2a |
7175 7256 7274 7521 8047del 8701 8860 9221 9540 10115 10398 10873 11719 11914 11944 12314G 12693 12705 13590 13650 |
7771 8206 13803 14566 15301 15326 15784 | AF465941 |
| NGR0522 | L2a | 7256 7274 7521 7771 8701 8860 9221 9540 10873 10994C 11029T 11335 11719 11914 11944 12292G 12693 12705 13590 13650 13803 15784 15802del 15848del |
7175 8206 10115 10398 14566 15301 15326 | AF465942 |
| NGR0475 | L2a |
7175 7256 7274 7521 7771 8701 8860 9221 9540 10373 10873 11719 11914 11944 12693 12705 13590 13650 13803 14668 15784 |
8206 10115 10398 14566 15301 15326 | AF465943 |
| NGR0510 | L2a | 7256 7274 7521 7771 8701 8860 9221 9540 10115 10398 10873 11617G 11719 11914 11944 12693 12705 13590 13650 13803 15784 | 7175 8206 14566 15301 15326 | AF465944 |
| WTE1150 | L2a |
7175 7256 7274 7521 7771 8701 8860 9221 10115 10398 10873 11335 11719 11914 11944 12693 12705 13194 13590 13650 13803 15301 15326 15784 |
8206 9540 14566 | AF465973 |
| WTE1145 | U | 7220A 7227T 7642 8860 9668 11467 11719 12308 12372 13590 15326 |
… | AF465971 |
Note .—Sites are numbered according to the revised reference sequence (Andrews et al. 1999); suffixes A, G, C, and T indicate transversions; “del” indicates a deletion. The mutations in boldface distinguish each sequence from the nearest mtDNA ancestor of haplogroups L2′3, M, N, and R. Potential reading errors or possible phantom mutations are italicized and underlined.
All bear 14766 in addition.
Basal polymorphisms that were undetected or omitted by Silva et al. (2002), including 11719 and the two rare mutations (8860 and 15326) in the CRS.
Figure 1.
Skeleton of the basal mtDNA phylogeny for the haplogroups identified in the data of Silva et al. (2002). “CRS” and “rCRS” refer to the reference sequence of Anderson et al. (1981) and the revised reference sequence of Andrews et al. (1999), respectively. The suffixes A, G, C, and T indicate transversions, and “del” indicates a deletion. Parallel mutations in different branches are underlined.
It is conspicuous that in all five haplogroup L2a mtDNAs of Silva et al. (2002), two of the basal transitions, 8206 and 14566, characteristic of L2 and L2a, respectively, are missed. Further L2a-diagnostic mutations, such as 7175, 7771, 13803, and 15784, are not always reported in the sequences (table 1). Moreover, the five L2a lineages have a total of only 11 other (private) mutations, comprising as many as five transversions, four deletions, and only two transitions. This pattern of private mutations differs from that in the three L2a lineages (nine transitions and no other mutations) of Ingman et al. (2000) and Torroni et al. (2001) in the same mtDNA region. It thus looks as though most of the real private mutations in the L2a mtDNAs were missed and that, instead, phantom mutations were scored.
The basal mutation 15487T of haplogroup M8 (which embraces haplogroups C and Z) is omitted in all seven C lineages of Silva et al.’s data (table 1). Other basal mutations for haplogroup C lineages are missing at sites 7196A, 8584, and 14318, in different combinations. It is remarkable that even deep mutations, such as 10400, 10873, and 15301 that distinguish macrohaplogroups M and N, were overlooked in six of the seven C lineages.
Among the seven D lineages in Silva et al. (2002), three sequences share mutations or motifs with D sequences reported elsewhere (Ingman et al. 2000; Derbeneva et al. 2002). The sequence JAP1045 (from an individual of Japanese origin) shares 8964, 9296, and 9824A with a Japanese mtDNA sequence from Ingman et al. (2000) and, therefore, definitely belongs to haplogroup D4, although the two characteristic D4 transitions (8414 and 14668) are not reported in the entire data set, except for one occurrence of 14668 in an L2a sequence! Similarly, the Japanese mtDNA sequence JAP1043 bears one of the mutations, 11215, found in Siberian mtDNAs of haplogroup D4 (Ingman et al. 2000; Derbeneva et al. 2002). The Guarani sequence GRC0131 of Silva et al. (2002) shares a rare transversion 10816T and a rare transition 13059 with the Guarani sequence of Ingman et al. (2000), but only the latter one has 8414 and 14668 and is thus confirmed as belonging to D4. These cases provide strong evidence for the systematic oversight of the basal mutations 8414 and 14668 in all haplogroup D lineages from Silva et al. (2002). Just as in the case of haplogroup C, several of the basal mutations that separate M and N are also missing in most of the D lineages.
Anomalies are also found in the nine sequences belonging to haplogroup A, although it was claimed by Silva et al. (2002) to be “the most homogeneous and best characterized” cluster in figure 1. Sample KCR0029 contains basal mutations 10398 and 10400 for haplogroup M. Sample KPO0013 has the 14566 mutation that is characteristic of haplogroup L2a. Sample PTJ0003 bears the L2abc-specific mutation 11944. Moreover, site 8027 is found mutated in only one A lineage, whereas this mutation was present in all the A sequences in Herrnstadt et al. (2002) and in one Chukchi sequence reported by Ingman et al. (2000).
In the 11 B lineages, only sample KPO0001 has the 9-bp deletion in the COII/tRNALys intergenic region, characteristic of haplogroup B. One or both of the basal mutations of B4b, 13590 and 15535, occur in all the samples (with the exception of JAP1044) and hint that they belong to B4b. It should be noted that in Herrnstadt et al. (2002), mutations 9950 and 11177 further defined a subhaplogroup of B4b that was baptized “B2.” We suggest that the 11177 mutation could have been omitted by Silva et al. (2002) as well. The Japanese B lineage JAP1044 could belong to haplogroup B4c or, alternatively, to B4a, as judged by the 15346 mutation or the 10238 transition, respectively (if the latter was simply misreported as a deletion). Two samples, KRC0033 and QUE1880, bear the 10400 mutation of haplogroup M, whereas sample QUE1881 harbors the 15043 mutation of M.
The U sequence in Silva et al. (2002) contains the full motif of haplogroup U, plus two transversions and three transitions not previously found in the published U sequences (Ingman et al. 2000; Finnilä et al. 2001; Maca-Meyer et al. 2001; Herrnstadt et al. 2002).
Rare deletions are found in two L2a and one B lineage of Silva et al. (2002). The 15802delA and 15848delA in the cytochrome b gene of sample NGR0522, 8047delT in the COII gene of sample NGR0524, and 10238delT in the ND3 gene of sample JAP1044 generate premature stop codons in these genes. These rare deletions all occur at a 2-bp repeat of the deleted base and might be generated by the Sequencer reading program. It is clear that the sequences of Silva et al. (2002) harbor more rare transversions and fewer private transitions than other reported sequences (Ingman et al. 2000; Finnilä et al. 2001; Maca-Mayer et al. 2001; Torroni et al. 2001; Herrnstadt et al. 2002). One cannot exclude the possibility that true transitions were erroneously scored as transversions or deletions by Silva et al. (2002). The two rare mutations 8860 and 15326 of the CRS are also missed in most of the sequences. The mutation 11335 in the CRS, which was found to be a sequencing error (Andrews et al. 1999), was present in 16 mtDNAs.
Processes that could account for these anomalies include the following:
-
1.
Only one strand of mtDNA was sequenced;
-
2.
Sequences were aligned with some variant of the CRS (a likely source of problems in the past; see Macaulay et al. [1999]);
-
3.
Sequences from different samples, especially those belonging to different haplogroups, were aligned together during the editing process (In this way, one might easily “borrow” a fragment of one sample into another when the sequences of the latter were not overlapping and, thus, introduce basal polymorphisms of one mtDNA lineage into another);
-
4.
Possible sample crossover or contamination during data collection;
-
5.
Relying just on the sequence scored by the Sequencer reading program without further manual checking of the chromatogram, especially relevant in the case of the rare deletions; and/or
-
6.
PCR errors during amplification.
In summary, we have every reason to mistrust the mtDNA sequences published by Silva et al. (2002). One cannot escape the conclusion that these data are seriously flawed or, at least, are not mtDNA as we know it.
References
- Anderson S, Bankier AT, Barrell BG, de Bruijn MH, Coulson AR, Drouin J, Eperon IC, Nierlich DP, Roe BA, Sanger F, Schreier PH, Smith AJ, Staden R, Young IG (1981) Sequence and organization of the human mitochondrial genome. Nature 290:457–465 [DOI] [PubMed] [Google Scholar]
- Andrews RM, Kubacka I, Chinnery PF, Lightowlers RN, Turnbull DM, Howell N (1999) Reanalysis and revision of the Cambridge reference sequence for human mitochondrial DNA. Nat Genet 23:147 [DOI] [PubMed] [Google Scholar]
- Bandelt H-J, Lahermo P, Richards M, Macaulay V (2001) Detecting errors in mtDNA data by phylogenetic analysis. Int J Legal Med 115:64–69 [DOI] [PubMed] [Google Scholar]
- Bandelt H-J, Quintana-Murci L, Salas A, Macaulay V (2002) The fingerprint of phantom mutations in mitochondrial DNA data. Am J Hum Genet 71:1150–1160 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Derbeneva OA, Sukernik RI, Volodko NV, Hosseini SH, Lott MT, Wallace DC (2002) Analysis of mitochondrial DNA diversity in the Aleuts of the Commander Islands and its implications for the genetic history of Beringia. Am J Hum Genet 71:415–421 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finnilä S, Lehtonen MS, Majamaa K (2001) Phylogenetic network for European mtDNA. Am J Hum Genet 68:1475–1484 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herrnstadt C, Elson JL, Fahy E, Preston G, Turnbull DM, Anderson C, Ghosh SS, Olefsky JM, Beal MF, Davis RE, Howell N (2002) Reduced-median-network analysis of complete mitochondrial DNA coding-region sequences for the major African, Asian, and European haplogroups. Am J Hum Genet 70:1152–1171; 71:448–449 (erratum) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ingman M, Kaessmann H, Pääbo S, Gyllensten U (2000) Mitochondrial genome variation and the origin of modern humans. Nature 408:708–713 [DOI] [PubMed] [Google Scholar]
- Kivisild T, Tolk H-V, Parik J, Wang Y, Papiha SS, Bandelt H-J, Villems R (2002) The emerging limbs and twigs of the East Asian mtDNA tree. Mol Biol Evol 19:1737–1751 [DOI] [PubMed] [Google Scholar]
- Maca-Meyer N, González AM, Larruga JM, Flores C, Cabrera VC (2001) Major genomic mitochondrial lineages delineate early human expansions. BMC Genetics 2:13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Macaulay V, Richards M, Sykes B (1999) Mitochondrial DNA recombination: no need to panic. Proc R Soc Lond B 266:2037–2039 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richards M, Macaulay V (2001) The mitochondrial gene tree comes of age. Am J Hum Genet 68:1315–1320 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silva WA Jr, Bonatto SL, Holanda AJ, Ribeiro-dos-Santos AK, Paixão BM, Goldman GH, Abe-Sandes K, Rodriguez-Delfin L, Barbosa M, Paçó-Larson ML, Petzl-Erler ML, Valente V, Santos SEB, Zago MA (2002) Mitochondrial genome diversity of Native Americans supports a single early entry of founder populations into America. Am J Hum Genet 71:187–192 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Torroni A, Rengo C, Guida V, Cruciani F, Sellitto D, Coppa A, Luna Calderon F, Simionati B, Valle G, Richards M, Macaulay V, Scozzari R (2001) Do the four clades of the mtDNA haplogroup L2 evolve at different rates? Am J Hum Genet 69:1348–1356 [DOI] [PMC free article] [PubMed] [Google Scholar]