Abstract
Perturbation of origin firing in chromosome replication is a possible cause of spontaneous chromosome instability in multireplicon organisms. Here, we show that chromosomal abnormalities, including aneuploidy and chromosome rearrangement, were significantly increased in yeast diploid cells with defects in the origin recognition complex. The cell cycle of orc1-4/orc1-4 temperature-sensitive mutant was arrested at the G2/M boundary, after several rounds of cell division at the restrictive temperature. However, prolonged incubation of the mutant cells at 37°C led to abrogation of G2 arrest, and simultaneously the cells started to lose viability. A sharp increase in chromosome instability followed the abrogation of G2 arrest. In orc1-4/orc1-4 rad9Δ/rad9Δ diploid cells grown at 37°C, G2 arrest and induction of cell death were suppressed, while chromosome instability was synergistically augmented. These findings indicated that DNA lesions caused by a defect in Orc1p function trigger the RAD9-dependent checkpoint control, which ensures genomic integrity either by stopping the cell cycle progress until lesion repair, or by inducing cell death when the lesion is not properly repaired. At semirestrictive temperatures, orc2-1/orc2-1 diploid cells demonstrated G2 arrest and loss of cell viability, both of which require RAD9-dependent checkpoint control. However, chromosome instability was not induced in orc2-1/orc2-1 cells, even in the absence of the checkpoint control. These data suggest that once cells lose the damage checkpoint control, perturbation of origin firing can be tolerated by the cells. Furthermore, although a reduction in origin-firing capacity does not necessarily initiate chromosome instability, the Orc1p possesses a unique function, the loss of which induces instability in the chromosome.
Spontaneous genetic alterations leading to loss of heterozygosity (LOH) in diploid yeast cells occur 2 to 3 orders of magnitude more frequently than spontaneous mutations occurring in the corresponding gene in haploid cells (18). This is because genetic changes in diploid cells are mostly due to chromosomal abnormalities, such as loss of chromosome, allelic recombination, truncation, amplification, and translocation. Our recent studies using Saccharomyces cerevisiae diploid cells indicate that processes of homologous recombination play a significant role in the chromosomal abnormalities (18, 39). This implies that specific events triggering homologous recombination frequently occur during normal cell growth (15). It is possible that during chromosomal DNA replication, double-strand DNA breaks (DSB) are produced so often that some escape the accurate process of recombination repair between sister chromatids and result in erroneous recombination or chromosome destruction.
Extensive studies in Escherichia coli demonstrated that stall or collapse of the replication fork triggers the recombination process, which in turn facilitates recovery of the interrupted replication (20, 22, 26, 27, 32). In eukaryotes, replication progression is monitored by checkpoint control mechanisms, which stop or delay the cell cycle when the replication fork is blocked (17, 48). It has been suggested that the DNA damage checkpoint control facilitates recovery of the replication fork by increasing the time for repair, inducing genes related to the repair, and activating the repair mechanisms (31). Cells lacking checkpoint control are hypersensitive to agents that block the replication fork (44, 45) and exhibit an increased level of chromosome instability even when cells are not treated with such agents (28, 43). Cells defective in homologous recombination also exhibit elevated chromosome instability (6, 16, 24, 25, 49). Therefore, it is possible that spontaneous chromosome abnormality results, at least in part, from a failure in the recovery of replication fork progression after blockage. However, factors that cause the block of replication fork progression in normally growing cells are largely unknown.
Spontaneous DNA damages, such as those caused by active oxygen radicals and their processed intermediates during repair, are plausible candidates for the obstruction of the replication fork in normal cells (34). To date, the query on whether the absence of mechanisms of base or nucleotide excision repair, which eliminate spontaneous damage, increases chromosome instability remains to be elucidated. Other possible candidates are intrinsic barriers of the replication fork within the chromosome. DNA regions containing such replication barriers within the E. coli genome are in fact hot spot sites for recombination (29). In S. cerevisiae, the repetitive unit of ribosomal DNA contains a replication barrier that promotes recombination between the units with an unusually high frequency (4, 21). Although another replication fork barrier was recently identified near the HML locus in the yeast genome (42), it is uncertain whether replication fork barriers are distributed at numerous loci throughout the genome. In addition to such particular replication barriers, it is generally speculated that progression of the replication fork is discontinued or delayed on collision with transcription apparatus, topoisomerases, or other proteins moving on or binding the same DNA molecule (3). In fact, replication fork pause sites transiently arresting replication fork movement were mapped to tRNA genes of S. cerevisiae in vivo. Replication fork pause sites are polar, stalling replication forks only when they oppose the direction of tRNA transcription (8). The state of the chromatin structure may also affect the progress of the replication fork. We further speculated that cellular mechanisms to avoid such unfavorable collisions might involve the efficiency and timing of initiation of DNA replication for each replicon (13, 35, 47), so that the advancement of the replication fork could be least interfered with. To examine this hypothesis, we analyzed genetic instability in cells defective in the origin-firing process of chromosomal replication.
When origin firing is delayed or perturbed, the replication fork occasionally moves into adjacent replicons that may not be ready, and one half segment of such a replicon is to be replicated by the invading replication fork that proceeds in the unusual direction. These overrunning replication forks possibly increase the chance of fork arrest and may induce chromosome instability. Earlier studies demonstrate that temperature-sensitive alleles of MCM1 and CDC6, which are involved in origin firing in S. cerevisiae showed increased levels of spontaneous chromosome instability, in agreement with this hypothesis (5, 11). We employed temperature-sensitive alleles of ORC1 and ORC2 to examine the effects on chromosome instability in diploid cells. The ORC1 and ORC2 genes encode subunits of the origin recognition complex, which plays a central role in origin firing in S. cerevisiae (10).
We found that both orc1-4/orc1-4 and orc2-1/orc2-1 diploid cells showed cell cycle arrest at the G2/M boundary at the restrictive temperature, although the G2 arrest of orc2-1/orc2-1 cells was only observed at temperatures between 26 and 30°C. The orc2-1/orc2-1 cells were arrested at the G1/S boundary at temperatures above 32°C. It appeared that the G2 arrest of both mutants is mediated by the RAD9-dependent checkpoint control. In the absence of the damage checkpoint, orc1-4/orc1-4 cells continued to divide at the restrictive temperature. Growth defects of orc2-1/orc2-1 cells were also suppressed when the RAD9-dependent checkpoint control was absent. These findings indicate that some kind of DNA lesion is produced and triggers the damage checkpoint in both orc mutant cells after the temperature shift. As expected, chromosome instability was elevated in orc1 mutant cells, following prolonged incubation at the restrictive temperature. Furthermore, kinetic studies of the induction of chromosome instability in or1-4/orc1-4 cells demonstrated that this process was strongly suppressed by the RAD9-dependent checkpoint control and induced only after abrogation of G2 arrest, which was initiated around 10 h after the temperature shift. Therefore, it is plausible that perturbation of the origin-firing process induces DNA lesions, which cause spontaneous chromosome instability. However, chromosome instability was never elevated in orc2-1/orc2-1 cells even in the absence of the RAD9-dependent checkpoint control. This difference between the orc alleles suggests that Orc1p has a novel function, the loss of which leads to chromosome instability.
In the present study, we also found that the RAD9-dependent checkpoint control is required for the loss of cell viability induced in both orc1 and orc2 mutants at restrictive temperatures. In orc1-4/orc1-4 cells, loss of cell viability and chromosome instability was simultaneously initiated upon the abrogation of G2 arrest. Furthermore, it appeared that the cell death is not a direct outcome of the chromosome instability elevated in the mutant diploid cells. These findings suggest that the loss of cell viability induced in orc1-4/orc1-4 and orc2-1/orc2-1 cells is a task of the damage checkpoint control, and this phenotype of orc1-4 mutant is designated CICD (for checkpoint-dependent induction of cell death). We propose two distinct roles for the damage checkpoint control in ensuring genome integrity in normally growing cells; one is to provide time for the recovery of stalled replication fork by delaying cell cycle progress, and the other is to eliminate cells with potential chromosome instability from the population by killing such cells.
MATERIALS AND METHODS
Strains of S. cerevisiae.
All yeast strains used in this study were derivatives of YKU1 and YKU23, parental strains of which are FY23 and FY838, respectively (18), and are listed in Table 1. Standard genetic manipulations of yeast were performed as previously described (18). The orc1-3 and orc1-4 alleles are base substitutions in the ORC1 gene, resulting in the amino acid changes F171V and L134P, respectively. These alleles were initially obtained by the method of mutagenic PCR and screened for temperature-sensitive growth at 37°C (K. Shirahige and H. Yoshikawa, unpublished data). The orc1-3 and orc1-4 strains were constructed by integration and excision of the pRS406 plasmid (7, 36) containing the mutant orc1 genes. The orc2-1 mutation (12) was introduced into yeast strains by the same method. The rad9Δ strains were constructed by transformation with a PCR-derived kanMX module flanked by short terminal sequences homologous to the end of the RAD9 open reading frame (41).
TABLE 1.
Genotypes of yeast strains used in this study
| Strain | Genotypea | Reference or origin |
|---|---|---|
| Haploid | ||
| YKU1 | MATa lys2Δ202 leu2Δ1 ura3-52 trp1Δ63 | 18 |
| YKU23 | MATα lys2Δ202 leu2Δ1 ura3-52 his3Δ200 ade2Δ::hisG | 18 |
| YKU34 | YKU1 except LEU2 ade2Δ::hisG III-205::URA3 III-314::ADE2 | 18 |
| YMJ23 | YKU34 except orc1-3 | This study |
| YMJ24 | YKU23 except orc1-3 | This study |
| YMJ25 | YKU34 except orc1-4 | This study |
| YMJ26 | YKU23 except orc1-4 | This study |
| YMJ29 | YKU34 except orc2-1 | This study |
| YMJ30 | YKU23 except orc2-1 | This study |
| YMJ31 | YKU34 except rad9Δ | This study |
| YMJ32 | YKU23 except rad9Δ | This study |
| YMJ33 | YMJ23 except rad9Δ | This study |
| YMJ34 | YMJ24 except rad9Δ | This study |
| YMJ35 | YMJ25 except rad9Δ | This study |
| YMJ36 | YMJ26 except rad9Δ | This study |
| YMJ39 | YMJ29 except rad9Δ | This study |
| YMJ40 | YMJ30 except rad9Δ | This study |
| Diploid | ||
| RD301 | MATa/MATα lys2Δ202/lys2Δ202 LEU2/leu2Δ1 ura3-52/ura3-52 trp1Δ63/TRP1 HIS3/his3Δ200 ade2Δ::hisG/ade2Δ::hisG III-205::URA3 III-314::ADE2 | YKU23 × YKU34 |
| RD602 | RD301 except orc1-3/orc1-3 | YMJ23 × YMJ24 |
| RD603 | RD301 except orc1-4/orc1-4 | YMJ25 × YMJ26 |
| RD605 | RD301 except orc2-1/orc2-1 | YMJ29 × YMJ30 |
| RD611 | RD301 except rad9Δ/rad9Δ | YMJ31 × YMJ32 |
| RD612 | RD301 except orc1-3/orc1-3 rad9Δ/rad9Δ | YMJ33 × YMJ34 |
| RD613 | RD301 except orc1-4/orc1-4 rad9Δ/rad9Δ | YMJ35 × YMJ36 |
| RD615 | RD301 except orc2-1/orc2-1 rad9Δ/rad9Δ | YMJ39 × YMJ40 |
III-205::URA3 signifies that the URA3 fragment is inserted at kb 205 of chromosome III. III-314::ADE2 signifies that the ADE2 fragment is inserted at kb 314 of chromosome III.
Growth of yeast cells.
Media for yeast strains, including complex glucose (YPD), synthetic complete (SC) and various dropout media, were prepared as described previously (18). For YPAD medium, adenine sulfate was added to YPD to a final concentration of 0.004%. Cells were precultured at the permissive temperature (23°C) in SC medium depleted of uracil, leucine, and adenine until midlog phase (1.0 × 106 to 1.0 × 107 cells/ml) and subsequently inoculated at a density of 2.0 × 105 cells/ml into YPAD medium supplemented with uracil (20 μg/ml). Cultures were incubated (zero time point) with agitation at nonpermissive (37°C) or seminonpermissive temperatures (26 to 32°C for orc2-1/orc2-1). For determination of cell viability, cells were collected at appropriate time points, diluted, sonicated, and incubated on YPD plates at 23°C for 5 to 7 days.
Analysis of LOH events in diploid yeast cells.
Analysis of LOH was performed as described previously for strain RD301 (18) with minor modifications. Cells were precultured, grown, and collected as described above. 5-Fluoro-orotic acid (5-FOA) plates were prepared as described (18). After dilution and sonication, samples were spread on plates of YPD, 5-FOA, 5-FOA depleted of leucine, 5-FOA depleted of leucine and adenine and incubated at 23°C for 5 to 7 days. At least four independent experiments were performed for each strain, and LOH frequency was presented as an average of these values.
Flow cytometry analysis and microscopic examination.
For DNA content measurement, cells were stained with propidium iodide and analyzed by fluorescence-activated cell sorting (19) with a Becton Dickinson FACScan and Cell Quest software. Cell nuclei were stained with DAPI (4′,6-diamidino-2-phenylindole) and examined using a Zeiss fluorescence microscope.
Analysis of yeast chromosomes by pulsed-field gel electrophoresis (PFGE).
PFGE analysis of yeast chromosomes was performed, using the standard conditions described previously (18). Electrophoresis was conducted on 1% PFGE-certified agarose (Bio-Rad) in 0.5× TBE buffer at 14°C, with a CHEF Mapper XA Pulsed Field Electrophoresis System (Bio-Rad). Following electrophoresis, the gel was stained with ethidium bromide (0.5 μg/ml) for 30 to 60 min, destained in deionized water for 20 to 60 min, and photographed using a charge-coupled device video camera (Atto) with DensitoGRAPH software (Atto).
RESULTS
Growth inhibition and loss of cell viability due to defects in the origin recognition complex in diploid yeast cells.
The ORC1 gene is essential for the growth of yeast cells (19). To obtain further insights into the cellular functions of this gene product, we constructed a series of temperature-sensitive orc1 mutants by PCR mutagenesis of the ORC1 gene (K. Shirahige and H. Yoshikawa, unpublished data). Among these, orc1-3 and orc1-4 were employed to examine defects in cell growth and effects of the mutations on chromosome instability. Since we intended to use diploid cells for examining chromosome instability in mutant strains, cells homozygous for each allele were constructed and their growth defects were studied at the nonpermissive temperature.
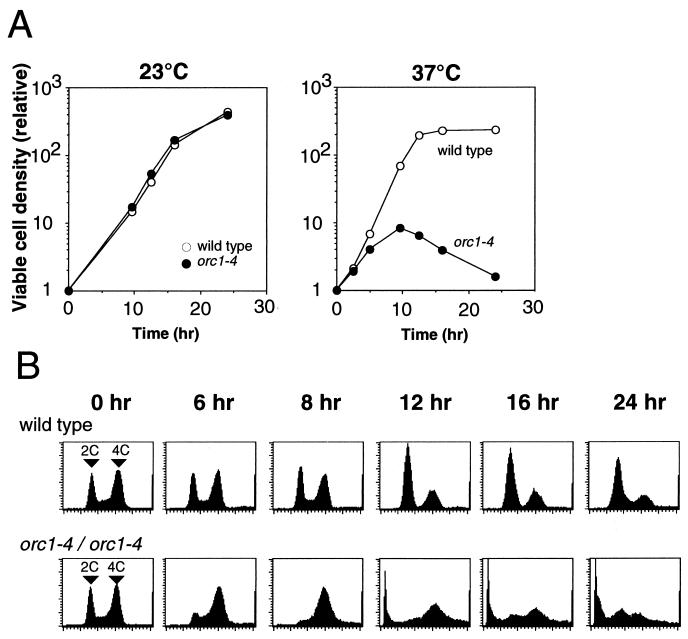
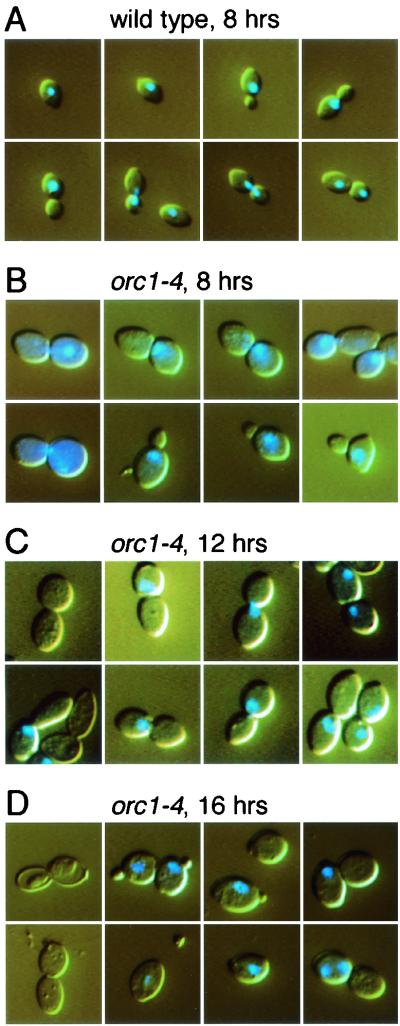
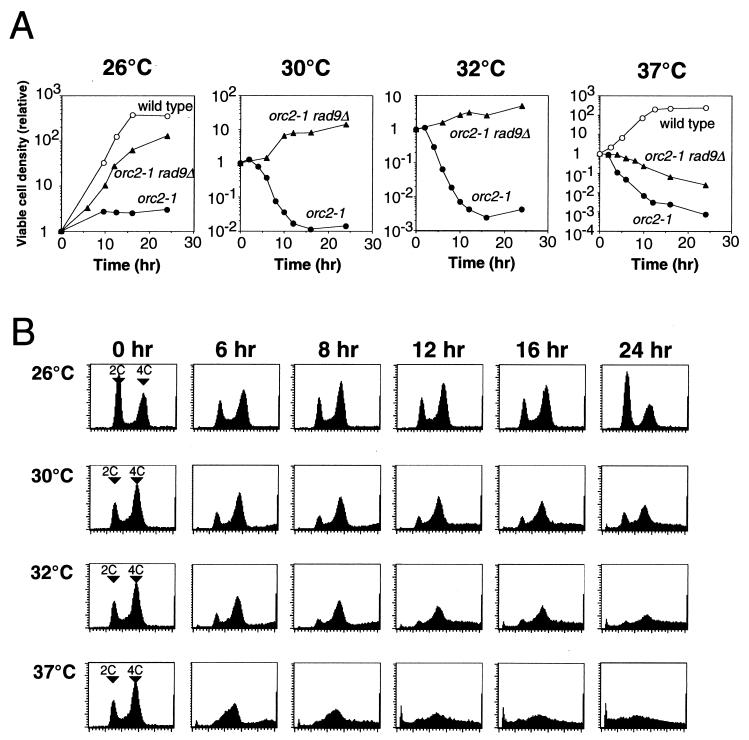
The orc1-4/orc1-4 homozygous cells were unable to form colonies at 37°C, indicative of a temperature-sensitive growth phenotype. This phenotype of orc1-4 allele was shown to be recessive to the wild-type allele. Notably, orc1-4/orc1-4 diploid cells did not immediately stop their division after shifting the growth temperature to 37°C but continued to grow for 10 h at the restrictive temperature, resulting in an eightfold increase in the number of viable cells at this time point (Fig. 1A). During this period, the growth rate was gradually decreased and the contents of G2 or M phase cells were increased (Fig. 1B). The typical morphology of cells at the 8-h time point was dumbbell shaped with an undivided nucleus (Fig. 2B), indicating that the cells were mostly in the G2 phase. After the 10-h time point, the diploid cells started to lose their viability (Fig. 1A). At the 24-h time point, the number of viable cells was decreased to about 10% of that observed at the 10-h time point. Interestingly, cell cycle arrest at the G2/M boundary was abrogated after the 10-h (catastrophic) time point, and cells or their remains with very low DNA content started to emerge (Fig. 1B). In contrast to the orc1-4/orc1-4 cells at the 8-h time point, dumbbell-shaped cells lacking nuclei emerged at the 12-h time point, and large single cells with small buds came out at the 16-h time point (Fig. 2C and D). Abnormal cells that lost the nuclei or possessed two nuclei were also observed at the 16-h time points. These observations by microscopic analyses were consistent with the data obtained from fluorescence-activated cell sorting analyses.
FIG. 1.
Induction of growth arrest and lethality in orc1-4/orc1-4 cells at nonpermissive temperature. Cells were precultured at 23°C, diluted with fresh medium as described in Materials and Methods, and grown with vigorous shaking at 23 or 37°C for 24 h. (A) Diploid strains, RD301 (wild-type) and RD603 (orc1-4/orc1-4), were grown at 23 and 37°C. At indicated time points, aliquots were withdrawn, appropriately diluted with YPD medium, and cultured on YPD plates at 23°C for 5 to 7 days. The relative density of viable cells was calculated by normalizing with the density of viable cells at the zero time point. (B) Cell cycle progression in diploid strains, RD301 (wild-type) and RD603 (orc1-4/orc1-4), grown at 37°C was analyzed by flow cytometry.
FIG. 2.
Cell morphology and nucleus of orc1-4/orc1-4 diploid strain grown at nonpermissive temperature. Diploid strains, RD301 (wild-type) and RD603 (orc1-4/orc1-4), were grown as described in Materials and Methods. Cells were withdrawn at indicated times after temperature shift to 37°C, stained with DAPI, and photographed with the same magnification of a microscope. (A) Wild-type cells grown for at 37°C 8 h. (B through D) orc1-4/orc1-4 cells grown at 37°C for indicated time.
The orc1-3/orc1-3 strain showed a temperature-sensitive growth phenotype (data not shown). Following temperature shift, division of the mutant cells ceased more rapidly than that of orc1-4/orc1-4 cells, and orc1-3/orc1-3 cells were arrested at the G2/M boundary. The viable cell number increased twofold within the first 10 h after the temperature shift. Similar to orc1-4/orc1-4 cells, viability of orc1-3/orc1-3 cells started to decline at the 10-h time point, and the cell death was accompanied with abrogation of G2 arrest of the cell cycle.
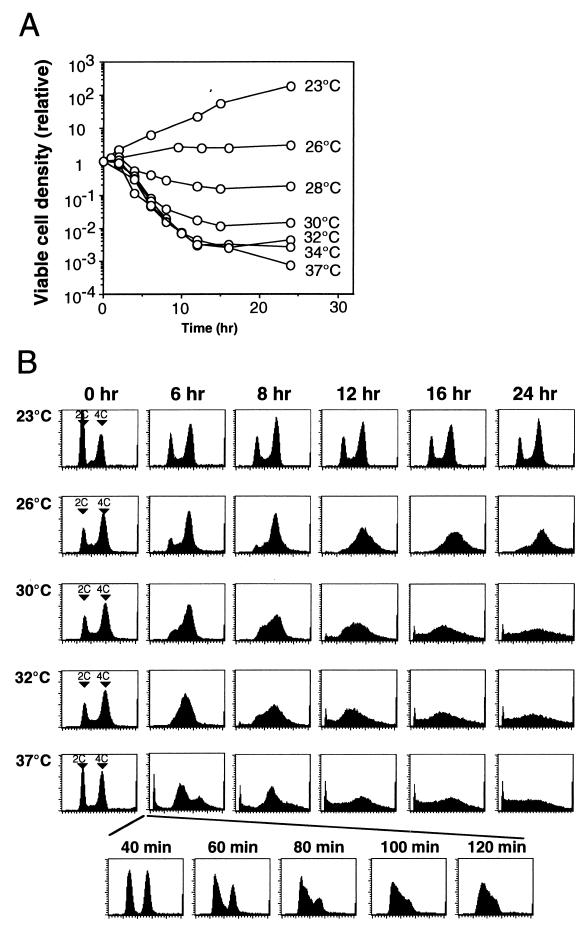
Temperature-sensitive mutants of other ORC genes have been previously reported (1, 9, 12). Haploid strains with orc2-1 or orc5-1 mutations showed loss of cell viability at the restrictive temperature, and cell death started at the 1-h time point subsequent to the temperature shift. We constructed an orc2-1/orc2-1 homozygous diploid strain coisogenic to orc1 mutant strains. As shown in Fig. 3A, orc2-1/orc2-1 cells displayed a more pronounced temperature-sensitive growth phenotype than orc1-4/orc1-4 cells at 37°C. The viability of orc2-1/orc2-1 cells decreased significantly upon temperature shift to 37°C with a time lag of 1 h, and most cells were arrested at the G1/S boundary at an early period of the heat treatment (Fig. 3B [37°C at the 80-min time point]). When the growth temperature was lowered, the loss of viability of orc2-1/orc2-1 cells was weakened, although this was still observed at early time points following the temperature shift (Fig. 3A). Unexpectedly, the phase of cell cycle arrest changed when lowering the growth temperature (Fig. 3B). At 30 and 26°C, cells were arrested at the G2/M boundary. However, abrogation of the cell cycle arrest was not apparent on prolonged heat treatment of the cells. Distribution of DNA content in dying orc2-1/orc2-1 cells was also different from that observed with the orc1-4/orc1-4 cells. Therefore, the process of cell death in orc2-1/orc2-1 cells at the nonpermissive temperatures were different from that in orc1-4/orc1-4 cells, suggesting distinct effects of these orc mutations on progress of the cell cycle.
FIG. 3.
Induction of cell death and patterns of cell cycle arrest in orc2-1/orc2-1 diploid cells at different temperatures. RD605 (orc2-1/orc2-1) cells were precultured at 23°C, diluted with fresh medium as described in Materials and Methods, and grown with vigorous shaking at indicated temperatures (zero time) for 24 h. (A) At indicated time points, aliquots were withdrawn, appropriately diluted with YPD medium, and cultured on YPD plates at 23°C for 5 to 7 days. The relative density of viable cells was calculated by normalizing with the density of viable cells at the zero time point. (B) Cell cycle progression in RD605 (orc2-1/orc2-1) grown at indicated temperatures was analyzed by flow cytometry.
RAD9-dependent checkpoint control is required for both G2/M cell cycle arrest and loss of cell viability due to defects in ORC1 and ORC2 genes.
Since orc1-4/orc1-4 cells were able to proceed to at least three rounds of cell cycle after the temperature shift, the replication of chromosome DNA was not severely affected in the mutant cells. We suspected that the arrest of cell cycle in orc1 mutant cells at the nonpermissive temperature might not be a direct effect of the mutation. Instead, the orc1 mutations might cause a situation that activates the G2/M checkpoint control of cell cycle. To examine this possibility, we constructed orc1-4/orc1-4 rad9Δ/rad9Δ and orc1-3/orc1-3 rad9Δ/rad9Δ double mutant strains and studied their growth and cell cycle progress at 37°C.
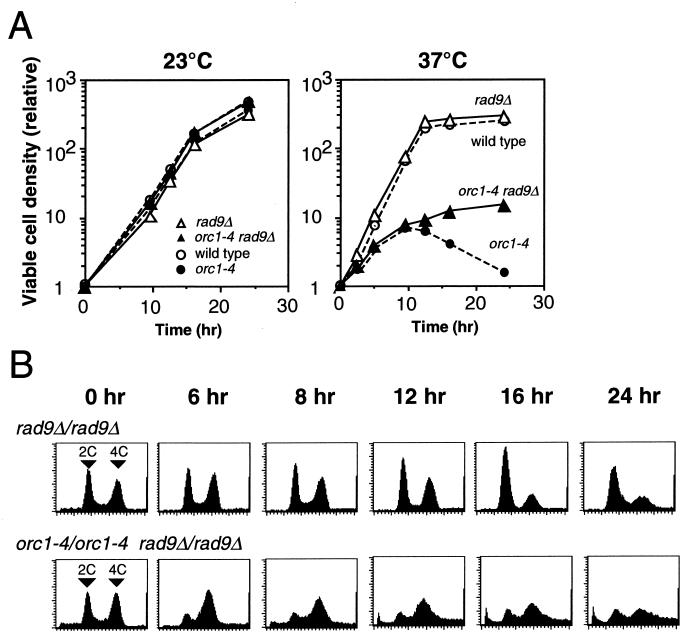
At the nonpermissive temperature, the rad9Δ mutation did not affect the growth of orc1-4/orc1-4 diploid cells until the 10-h time point, but suppressed the loss of cell viability after the catastrophic time point (Fig. 4A). The number of viable cells continued to increase until the 24-h time point, although the growth rate was gradually reduced. Colony-forming ability of the orc1-4/orc1-4 diploid strain was partially recovered by the rad9Δ mutation (Fig. 5). Similar to orc1-4/orc1-4 cells, the contents of G2 or M phase cells were increased in the double mutant cells following the temperature shift (Fig. 4B). However, in contrast to the orc1-4/orc1-4 strain, tight arrest of cell cycle at the G2/M boundary at the 8-h time point was not observed in the orc1-4/orc1-4 rad9Δ/rad9Δ strain. Accumulation of cells with very low DNA content was also suppressed by the rad9Δ mutation. Similar results were obtained with the orc1-3/orc1-3 rad9Δ/rad9Δ diploid strain (data not shown). From these observations, we concluded that the RAD9 gene is involved in the G2 arrest induced in orc1 diploid cells at the restrictive temperature. Furthermore, the RAD9-dependent G2/M checkpoint control is required for the loss of cell viability initiated at the catastrophic time point.
FIG. 4.
Effects of rad9Δ mutation on viability and cell cycle arrest of orc1-4/orc1-4 diploid cells at nonpermissive temperature. Experimental procedures were as described in the legend of Fig. 1. (A) Diploid strains, RD611 (rad9Δ/rad9Δ) and RD613 (orc1-4/orc1-4 rad9Δ/rad9Δ) were grown at 23 and 37°C. As controls, growth curves for RD301 (wild-type) and RD603 (orc1-4/orc1-4) are indicated by broken lines. (B) Cell cycle progression in diploid strains, RD611 (rad9Δ/rad9Δ) and RD613 (orc1-4/orc1-4 rad9Δ/rad9Δ), grown at 37°C was analyzed by flow cytometry.
FIG. 5.
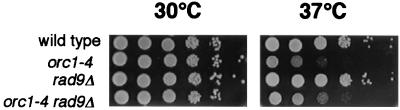
Effects of rad9Δ mutation on temperature-sensitive growth phenotype of orc1-4/orc1-4 mutant strain. Diploid strains, RD611 (rad9Δ/rad9Δ) and RD613 (orc1-4/orc1-4 rad9Δ/rad9Δ), were logarithmically grown in YPD medium at 23°C. Cells were stepwise 10-fold diluted with YPD medium, spotted onto YPD plates, and incubated at 30 or 37°C for 2 days. As controls, growth capabilities of wild-type diploid (RD301) and orc1-4/orc1-4 diploid (RD603) strains are also shown.
G2 arrest of orc2-1/orc2-1 cells at 26°C was also dependent on the RAD9 function. In diploid orc2-1/orc2-1 rad9Δ/rad9Δ cells, growth inhibition and G2 arrest at 26°C were almost completely suppressed (Fig. 6). This implied that the RAD9-dependent G2/M checkpoint control is required for the G2 arrest induced in orc2-1/orc2-1 cells at 26°C. In addition, it appeared that the loss of viability of orc2-1/orc2-1 cells under the restrictive condition is dependent on the RAD9 function. Cell death induced at temperatures above 30°C was significantly but only partially suppressed (Fig. 6). Therefore, at the higher temperatures, the induction of cell death in orc2-1/orc2-1 cells possibly involves both RAD9-dependent and RAD9-independent mechanisms.
FIG. 6.
Effects of rad9Δ mutation on viability and cell cycle arrest of orc2-1/orc2-1 diploid cells at different temperatures. Cells were precultured at 23°C, diluted with fresh medium as described in Materials and Methods, and grown with vigorous shaking at indicated temperatures (zero time) for 24 h. At indicated time points, aliquots were withdrawn, appropriately diluted with YPD medium, and plated onto YPD plates at 23°C for 5 to 7 days. The relative density of viable cells was calculated by normalizing with the density of viable cells at the zero time point. (A) RD301 (wild-type [open circles]), RD605 (orc2-1/orc2-1 [closed circles]), and RD615 (orc2-1/orc2-1 rad9Δ/rad9Δ [closed triangles]) were grown at 26, 30, 32, and 37°C. (B) Cell cycle progression in RD615 (orc2-1/orc2-1 rad9Δ/rad9Δ) grown at the indicated temperatures was analyzed by flow cytometry.
From the effects of rad9Δ mutation on growth defects caused by orc mutations, it was suggested that when functions of ORC are partially hampered, cells produce some kind of DNA lesion or an unusual DNA structure which activates the G2/M checkpoint control governed by the Rad9p. It is evident, however, that genome-wide DNA replication can be completed at the degree of malfunction of ORC in the temperature-sensitive orc mutants.
High incidence of chromosome loss and chromosome rearrangement in temperature-sensitive orc1 but not orc2 mutant cells after their treatment for 24 h under the restrictive conditions
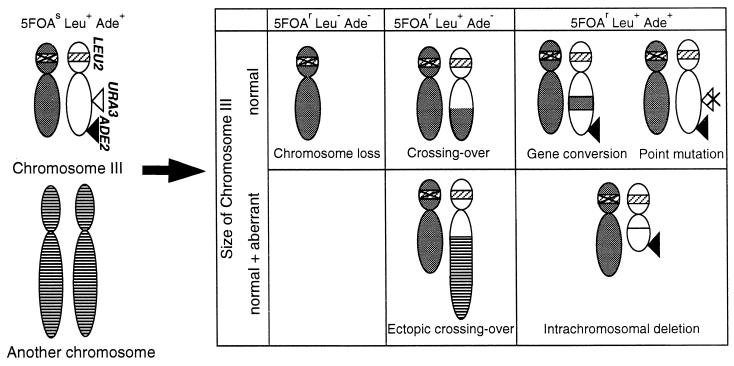
We expected that improper firing of replication origins during chromosome replication might induce chromosome instability in orc mutant cells. The observation that the G2/M checkpoint control was activated in orc mutant cells prompted us to examine survivors of orc mutant cells treated at restrictive temperatures for 24 h in their chromosomal integrity. To do this, we utilized URA3 hemizygote diploid strains in which the URA3 marker was placed on the right arm of chromosome III (18). LOH events leading to functional inactivation of the hemizygous URA3 marker are easily detected as 5-FOA-resistant clones. Our previous analyses indicated that the LOH events occurring in wild-type yeast diploid cells result from loss of chromosome, allelic recombination, intrachromosomal deletion, and interchromosomal ectopic recombination including translocation (18). Gene conversion and intragenic mutation were very rare events causing the LOH. These classes of LOH events can be distinguished by analyzing the status of other selectable heterozygous markers located on chromosome III and by directly examining the copy number and size of the chromosome III by PFGE (Fig. 7).
FIG. 7.
Assay system to screen and classify the genetic events leading to LOH. A pair of chromosomes III in parent strain (left of the arrow) and their possible alteration in 5-FOA-resistant (5-FOAr) convertants (right of the arrow) are illustrated with relative positions of the three markers used for the analysis. The 5-FOAr convertants are classified according to their indicated phenotypes and the altered patterns of chromosome III detected by PFGE, Southern hybridization, and PCR. The segments of chromosome III originally harboring the markers are shown by white, those of the homologous chromosome III are shown by gray, and those of another chromosome are shown by stripes. The URA3 insert at III-205 is indicated by an open triangle, the ADE2 insert at III-314 is shown by solid triangles, and the positions of intrinsic LEU2 loci are indicated by shaded bars, which are marked with a cross for the leu2 allele. A point mutation inactivating the URA3 insert is shown by a cross on the open triangle.
As shown in Table 2, LOH frequencies in orc mutant diploid strains grown at the permissive temperature were equal to that determined with the wild-type strain. When the orc1 mutant strains were treated at 37°C for 24 h, 20- to 30-fold increment of the LOH frequencies was observed. On the other hand, the orc2-1/orc2-1 diploid strain did not show any significant enhancement of LOH frequency at 26, 30, and 32°C. It was technically difficult to determine LOH frequency in the orc2-1/orc2-1 strain treated at 37°C for 24 h because of severe cell death.
TABLE 2.
Chromosome instability induced in orc mutant diploid cells treated at nonpermissive temperatures for 24 h
| Strain | Frequency of LOH (10−5)a at temp (°C)
|
||||
|---|---|---|---|---|---|
| 23 | 26 | 30 | 32 | 37 | |
| Wild type | 12 (1.0) | 19 (1.0) | 18 (1.0) | ND | 22 (1.0) |
| orc1-3/orc1-3 | 12 (1.0) | ND | ND | ND | 780 (35) |
| orc1-4/orc1-4 | 11 (0.92) | ND | ND | ND | 450 (20) |
| orc2-1/orc2-1 | 14 (1.2) | 22 (1.2) | 22 (1.2) | 37 | ND |
The frequency of 5-FOAr cells was determined for each strain after cultivation for 24 h at the indicated temperatures as described in Materials and Methods. Numbers in parentheses indicate increases in frequency of LOH relative to that of the wild-type strain grown at each temperature. ND, not determined.
Among the LOH clones screened from orc1-4/orc1-4 cells surviving after the heat treatment, those suffering from loss of chromosome III were predominant (Table 3). Hence, the frequency of loss of the chromosome in surviving orc1-4/orc1-4 cells was 60 times as high as that for the wild-type cells. This propensity of orc1-4/orc1-4 cells to spontaneously lose chromosome was not confined to chromosome III. In a direct examination of chromosome patterns of individual surviving clones by PFGE, various chromosomes were found to be lost frequently from the clones (data not shown). Seventeen out of 60 clones that survived after the heat treatment for 24 h appeared to have lost some chromosome, whereas no such aneuploid clone was detected among the 60 clones of untreated orc1-4/orc1-4 cells that were examined.
TABLE 3.
Class distribution of LOH events occurring in wild-type and mutant diploid cells during cultivation at 37°C for 24 h
| Classes | Frequency of LOH (10−5)a
|
|||
|---|---|---|---|---|
| Wild type | rad9Δ/rad9Δ | orc1-4/orc1-4 | orc1-4/orc1-4 rad9Δ/rad9Δ | |
| Chromosome loss | 6.6 (1.0) | 56 (8.5) | 400 (61) | 1,300 (200) |
| Allelic recombination | ||||
| Crossing over | 11 (1.0) | 34 (3.1) | 19 (1.7) | 150 (14) |
| Gene conversionb | 1.2 (1.0) | 1.5 (1.3) | 8.4 (2.8) | 1.7 (1.4) |
| Chromosome rearrangement | ||||
| Ectopic crossover | 2.4 (1.0) | 7.9 (3.3) | 19 (7.9) | 100 (42) |
| Intrachromosomal deletionc | 0.64 (1.0) | 2.5 (3.9) | 5.9 (9.2) | 8.0 (13) |
| Total | 22 (1.0) | 100 (4.5) | 450 (20) | 1,600 (73) |
The frequency of each class was estimated by multiplying the frequency of 5-FOA-resistant Leu−, 5-FOA-resistant Leu+ Ade−, and 5-FOA-resistant Leu+ Ade+ cells by the corresponding ratio of the examined clones of each phenotype, respectively (18).
This class is defined as LOH events leading to 5-FOA-resistant Leu+ Ade+ cells without showing HML-MAT deletion and, thus, includes intragenic point mutation. Since frequencies of such point mutations in wild-type and orc1-4 haploid strains were 0.097× 10−5 and 0.25 × 10−5, respectively, contribution of the intragenic point mutation to this class was expected to be less than 10%.
This class represents HML-MAT deletion (18).
LOH clones resulting from events other than chromosome loss were also increased significantly in the surviving orc1-4/orc1-4 cells. The frequency of LOH clones caused by intrachromosomal deletion, occurring exclusively between HMR and MAT loci, was about 10-fold higher than that for wild-type clones. Interchromosomal ectopic recombination, resulting in aberrant chromosomes, occurred about eight times more frequently in the surviving orc1-4/orc1-4 cells than wild-type cells. However, frequencies of LOH clones resulting from allelic recombination, both crossing over and gene conversion types, were only slightly elevated in the surviving orc1-4/orc1-4 cells. Similar patterns of class distribution of LOH clones were observed with orc1-3/orc1-3 strains when they were treated at 37°C for 24 h (data not shown).
Time course of chromosome instability in orc1 mutant cells at the restrictive temperature.
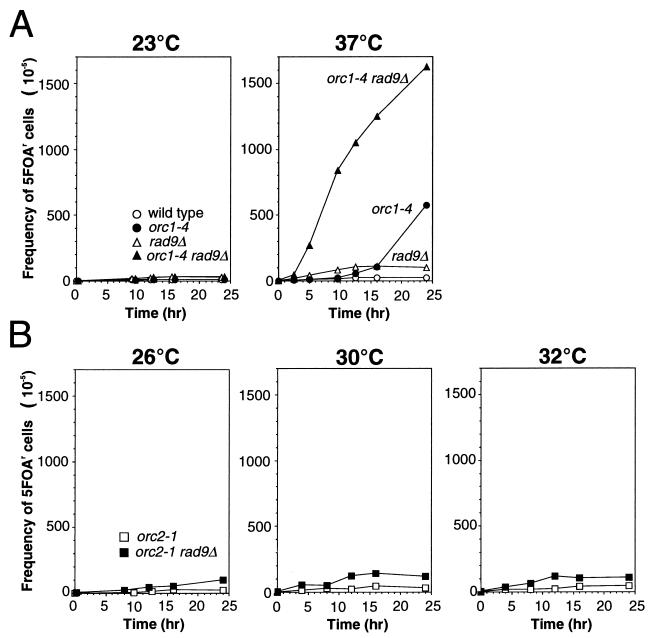
To obtain a clue to a relationship between growth defects and chromosome instability observed in the orc1 mutant diploid strains, the frequency of LOH was measured for the orc1-4/orc1-4 mutant cells at several time points after the temperature shift (Fig. 8A).
FIG. 8.
Induction of chromosome instability in orc1-4/orc1-4 and orc2-1/orc2-1 diploid cells in the presence and absence of Rad9p function after the temperature shift. Cells were grown at the indicated temperatures, and aliquots were withdrawn at the indicated time points and examined for frequencies of 5-FOA-resistant (5-FOAr) cells, as described in Materials and Methods. (A) RD301 (wild-type), RD603 (orc1-4/orc1-4), RD611 (rad9Δ/rad9Δ), and RD613 (orc1-4/orc1-4 rad9Δ/rad9Δ) were grown at 23 and 37°C. (B) RD605 (orc2-1/orc2-1) and RD615 (orc2-1/orc2-1 rad9Δ/rad9Δ) were grown at 26, 30, and 32°C.
In orc1-4/orc1-4 cells, LOH frequencies were not significantly increased for the first 10 h after the temperature shift. Then, beyond the catastrophic time point, LOH frequencies were sharply increased. Similar results were obtained with the orc1-3/orc1-3 diploid strain (data not shown). Chromosome instability caused by the defect in Orc1p thus appeared to be accompanied with the abrogation of G2 arrest in the mutant cells.
Roles of RAD9-dependent cell cycle checkpoint control in the maintenance of chromosomes in orc1 temperature-sensitive mutant cells.
Since no significant induction of chromosome instability was observed until the incubation of orc1-4/orc1-4 cells at 37°C was past the catastrophic time point, the defects in Orc1p seemed not to result directly in chromosome instability. However, it seemed likely that the chromosome instability under nonpermissive conditions was suppressed by some cellular function in the orc1-3/orc1-3 and orc1-4/orc1-4 cells. If this were the case, the suppression of chromosome instability would be released when the cell cycle arrest was abrogated. From this reason, we expected that the RAD9-dependent checkpoint control might be involved in the suppression of chromosome instability in the mutant cells. Similarly, we speculated that no apparent induction of the chromosome instability in the orc2-1/orc2-1 cells under nonpermissive conditions might be due to strong suppression of chromosome instability by the RAD9-dependent checkpoint control. To examine these possibilities, we measured the LOH frequencies of orc1-4/orc1-4 rad9Δ/rad9Δ and orc2-1/orc2-1 rad9Δ/rad9Δ diploid cells after the temperature shift.
The ORC-proficient rad9Δ/rad9Δ strain showed a slightly increased level of LOH frequency (Table 3). The rad9Δ mutations affected all major classes of the LOH events that occurred in wild-type cells. LOH frequency was further increased when the orc1-4/orc1-4 rad9Δ/rad9Δ double mutant strain was grown at 37°C (Fig. 8A). In this strain, chromosome instability was induced shortly after the temperature shift, demonstrating LOH frequencies 8- and 30-fold higher than those of the rad9Δ/rad9Δ and orc1-4/orc1-4 strains, respectively, at the 10-h time point. As shown in Table 3, the high LOH frequency of the double mutant strain was due to synergistic effects of rad9Δ and orc1-4 mutations. About 80% of the LOH events were chromosome loss in the orc1-4/orc1-4 rad9Δ/rad9Δ cells. Interestingly, LOH caused by allelic recombination, especially crossover type, was significantly enhanced in the double mutant cells. These data clearly indicated that the RAD9-dependent checkpoint control suppresses chromosome instability induced by the defect in Orc1p function and that the suppression was almost complete until the catastrophic 10-h time point. Furthermore, it was evident that DNA lesions caused by the defect in Orc1p function that triggers the RAD9-dependent checkpoint control directly leads to the chromosome instability.
In contrast to the synergistic effects of rad9Δ and orc1-4 mutations on LOH frequency, the orc2-1/orc2-1 rad9Δ/rad9Δ double mutant strain grown at 26, 30, or 32°C showed the same level of LOH frequency as that determined with the orc2-1/orc2-1 strain (Fig. 8B). Under these conditions, the orc2-1/orc2-1 rad9Δ/rad9Δ strain grew normally but at slightly lower rates (Fig. 6A). However, as described earlier, the defect in Orc2p function triggers the RAD9-dependent checkpoint control at temperatures above 26°C. Therefore, it was suggested that DNA lesions caused by the orc2-1 mutation are recognized by the checkpoint control system but do not lead to the chromosome instability that can be detected by the experimental system used in this study. The different effects of orc1-4 and orc2-1 on chromosome instability implied that the nature of DNA lesions caused by orc1-4 was distinct from those caused by the orc2-1 mutation.
DISCUSSION
RAD9-dependent checkpoint control is required for G2/M cell cycle arrest and induction of cell death caused by temperature-sensitive orc1 mutations.
Arrest of cell cycle induced in orc2-1 and orc5-1 temperature-sensitive mutants has been presumed to be a direct effect of the defect in each ORC subunit protein, and the cell death in these orc mutants has been considered to result from a chromosome catastrophe by incomplete replication or from permanent arrest of cells in M phase (1, 10, 12). In the present study, we demonstrate that growth defects in orc1 temperature-sensitive mutant cells are distinct from those in orc2-1 and orc5-1 strains and involve the action of RAD9-dependent damage checkpoint control, which executes the cell cycle arrest and is required for the induction of cell death. These findings indicated that some kind of DNA lesion, readily recognized by the damage checkpoint control, is produced in the orc1-4/orc1-4 mutant cells at the restrictive temperature.
Using synchronized orc1-4 haploid cells, it was shown that efficiency of origin firing for each replicon is reduced in general and the progression of S phase slows down in the cells under restrictive conditions (Y. Kato, K. Shirahige, and H. Yoshikawa, unpublished results). Consequently, in such a situation, the average span of a replicon is enlarged, and a replication fork traverses a longer distance and proceeds with unusual directionality in many regions of chromosomes, possibly increasing the chance of the replication fork to stall. Most recently, van Brabant and colleagues reported that yeast cells carrying an artificial chromosome with an ∼170-kb origin-free region induced the RAD9-dependent checkpoint control (40). The artificial chromosome was unstable in a rad9Δ strain, undergoing deletions within the origin-free region. The DNA lesion spontaneously arising within the unusually extended replicon triggers the checkpoint control and induces the chromosome instability, resembling that induced in orc1-4/orc1-4 cells at the restrictive temperature.
Abrogation of G2 arrest followed by induction of cell death during a prolonged heat-treatment of orc1-4/orc1-4 cells.
We have shown that orc1-4/orc1-4 cells arrested at the G2/M boundary resumed the cell cycle around the 10-h time point after the temperature shift. The abrogation of G2 arrest observed with the orc1-4/orc1-4 diploid cells is probably relevant to a phenomenon called adaptation, in which cells escape from the RAD9-dependent G2 arrest caused by an irreparable DSB after 8 to 12 h of the G2 arrest (14, 23, 31, 33). The duration of G2 arrest until the start of adaptation is almost the same as that until the abrogation of G2 arrest was initiated in the heat-treated orc1-4/orc1-4 cells. The increased level of chromosome instability in the orc1-4/orc1-4 cells that survived after a prolonged heat treatment indicates that DNA lesions remain unrepaired in the mutant cells. However, the adaptation to the damage checkpoint has been clearly documented in the response of yeast cells to X-ray irradiation, endonucleolytic break, and damage generated by the cdc13 mutation, all of which generate DSB. Since we provided no direct evidence for induction of DSB in the orc1-4/orc1-4 cells, we reserve use of the term “adaptation” for the abrogation of G2 arrest in the heat-treated orc1-4/orc1-4 cells. We have to await future studies to determine the relevance between the phenomena. The use of an adaptation-deficient cdc5 allele would be helpful in such studies.
In the orc1 mutant cells, loss of cell viability was induced only after the G2 arrest was abrogated. Upon the abrogation of G2 arrest, cells or their remains with a trace amount of DNA started to emerge. It is therefore likely that the chromosome DNA is degraded in the dying cells. Since the orc1-4/orc1-4 rad9Δ/rad9Δ cells continued mitotic growth for at least 24 h under the restrictive condition, it is apparent that loss of cell viability in the orc1-4/orc1-4 cells is not simply due to unrepaired DNA lesions or the enhanced chromosome instability. The adaptation of RAD9-dependent G2 arrest is an actively promoted cellular process (37). Similarly, the induction of cell death as well as the abrogation of G2 arrest in orc1-4/orc1-4 cells might be an active process in which the RAD9-dependent checkpoint control is involved. From this point of view, we propose that the induction of cell death is an intrinsic task of the RAD9-dependent damage checkpoint control and term this phenotype of orc1 mutants CICD (for checkpoint-dependent induction of cell death). Bennett et al. reported their findings relevant to the CICD, in which a single irreparable DSB in a dispensable plasmid leads to RAD9-dependent lethality in yeast cells (2). In mammalian cells, checkpoint control mechanisms are involved in the cellular apoptosis as well as the arrest of cell cycle (48). It is also widely known that the abrogation of cell cycle arrest is prerequisite to initiation of the apoptotic process caused by DNA damages (30). Although the molecular mechanisms of CICD is unknown, the dual roles of the damage checkpoint control are presumably common in both monocellular and multicellular eukaryotes.
Roles of RAD9-dependent checkpoint control in growth defects of orc2-1/orc2-1 cells.
The RAD9-dependent checkpoint control may be triggered in cells carrying other temperature-sensitive orc alleles at the restrictive temperature. This is true at least in the case of orc2-1. Upon shifting to 26°C, orc2-1/orc2-1 cells were not arrested at the G1/S boundary but at the G2/M boundary. Loss of cell viability was also noted under this condition. Both cell cycle arrest and cell death at the semirestrictive temperature were almost fully suppressed by rad9Δ mutation, implying that DNA lesions arising in the orc2-1/orc2-1 cells are capable of triggering the RAD9-dependent checkpoint control. It is also clear that loss of viability in orc2-1/orc2-1 cells requires the RAD9-dependent checkpoint control at least under the semirestrictive condition. Therefore, we conclude that the induction of cell death in orc2-1/orc2-1 cells is a CICD phenotype of this allele.
At temperatures higher than 26°C, a more pronounced cell death was observed and its suppression in the absence of Rad9p was only partial, indicating involvement of RAD9-independent mechanisms in the loss of viability in the orc2-1/orc2-1 cells at higher temperatures. It should be noted that in orc2-1/orc2-1 cells the phase of cell cycle arrest was changed when increasing the growth temperature; intra-S arrest was predominant at 30°C, while G1/S arrest was predominant at 37°C. Since the orc2-1/orc2-1 cells were not permanently arrested at these phases of the cell cycle under the restrictive conditions, the mutant cells may undergo loss of viability upon abrogation of the intra-S or G1/S arrest. Given that the cell cycle arrest at G1 or S phase is abrogated, mitotic growth of orc2-1/orc2-1 cells would probably be restrained by G1/M checkpoint when the initiation of DNA replication is severely inhibited (38, 46). The G1/M checkpoint may be related to the RAD9-independent lethality observed in orc2-1/orc2-1 cells at higher temperatures.
Chromosome instability caused by orc1 temperature-sensitive alleles.
DNA lesions induced in orc1-4/orc1-4 cells at the restrictive temperature lead to chromosome instability. In Rad9p-proficient orc1-4/orc1-4 cells, chromosome instability was suppressed until the 10-h time point, after which cell cycle arrest was abrogated. When Rad9p was absent, chromosome instability in the diploid cells was induced shortly after the temperature shift. Therefore, the DNA lesion that triggers RAD9-dependent checkpoint control is possibly a direct cause of chromosome instability. Loss of chromosome III is the most prominent LOH event in orc1-4/orc1-4 cells at the restrictive temperature. This may be due to a mitotic problem of incompletely replicated sister chromatids, following the abrogation of G2 arrest. Another possible cause of chromosome loss in orc1-4/orc1-4 cells is the failure of homologous recombination between sister chromatids or homologous chromosomes in the S and/or the G2 phase (18). It was clearly demonstrated that the DNA lesions produced in orc1-4/orc1-4 cells lead to chromosome aberrations involving intrachromosomal recombination and allelic or ectopic recombination between chromosomes. Assuming that the spontaneously stalled replication fork is the DNA lesion itself, orc1-4/orc1-4 cells require processes of recombination repair to solve the problem.
In contrast to orc1-4/orc1-4, chromosome instability was never induced in orc2-1/orc2-1 cells, even in the absence of RAD9-dependent checkpoint control. Therefore, DNA lesions induced in orc2-1/orc2-1 cells may be efficiently repaired whenever the RAD9-dependent checkpoint control works or not. In orc2-1/orc2-1 cells, however, the DNA lesions are constantly produced at a level sufficient to activate continuously the RAD9-dependent checkpoint control. Furthermore, growth defect in orc2-1/orc2-1 cells is severer than that observed with orc1-4/orc1-4 cells. One possible explanation for these puzzling observations is that DNA lesions produced in both alleles differ in structure or in distribution within the genome. Although the reason for these differences between orc1-4 and orc2-1 alleles is unclear, it is suggested that Orc1p possesses a unique function, the absence of which leads to severe chromosome instability.
In multireplicon organisms, the order of origin firing is regulated by unknown mechanisms (35, 47). However, the regulation seems not to be completely strict, and utilization of origins in a single round of the cell cycle differs between individual cells. Therefore, DNA lesions may be caused by a spontaneous perturbation of origin firing even in wild-type yeast cells, although its frequency is lower than in the orc1 alleles. Since rad9Δ/rad9Δ cells showed a moderate level of chromosome instability, the RAD9-dependent checkpoint control seems to play the same role in both wild-type and orc1 alleles. Hence, it is probable that the perturbation of origin firing is monitored by the checkpoint control and, responding to the level of perturbation, cells are arrested at the G2/M boundary or eliminated from the population by the checkpoint mechanisms. When cells lose the checkpoint control, the perturbation of origin firing can be tolerated by the cells and results in chromosome instability in the cells.
Acknowledgments
We are indebted to H. Araki, S. Maki, K. Sugimoto, and H. Yoshikawa for valuable discussions.
This work was financially supported by the Grant-in-Aid for Scientific Research on Priority, areas B (11239208 and 13141204 to K.U.) and C (12213082 to H.M.), from the Ministry of Education, Culture, Sports, Science, and Technology.
REFERENCES
- 1.Bell, S. P., R. Kobayashi, and B. Stillman. 1993. Yeast origin recognition complex functions in transcription silencing and DNA replication. Science 262:1844-1849. [DOI] [PubMed] [Google Scholar]
- 2.Bennett, C. B., A. L. Lewis, K. K. Baldwin, and M. A. Resnick. 1993. Lethality induced by a single site-specific double-strand break in a dispensable yeast plasmid. Proc. Natl. Acad. Sci. USA 90:5613-5617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Brewer, B. J. 1988. When polymerases collide: replication and the transcriptional organization of the E. coli chromosome. Cell 53:679-686. [DOI] [PubMed] [Google Scholar]
- 4.Brewer, B. J., and W. L. Fangman. 1988. A replication fork barrier at the 3′ end of yeast ribosomal RNA genes. Cell 55:637-643. [DOI] [PubMed] [Google Scholar]
- 5.Bruschi, C. V., J. N. McMillan, M. Coglievina, and M. S. Esposito. 1995. The genomic instability of yeast cdc6-1/cdc6-1 mutants involves chromosome structure and recombination. Mol. Gen. Genet. 249:8-18. [DOI] [PubMed] [Google Scholar]
- 6.Chen, C., K. Umezu, and R. D. Kolodner. 1998. Chromosomal rearrangements occur in S. cerevisiae rfa1 mutator mutants due to mutagenic lesions processed by double-strand-break repair. Mol. Cell 2:9-22. [DOI] [PubMed] [Google Scholar]
- 7.Christianson, T. W., R. S. Sikorski, M. Dante, J. H. Shero, and P. Hieter. 1992. Multifunctional yeast high-copy-number shuttle vectors. Gene 110:119-122. [DOI] [PubMed] [Google Scholar]
- 8.Deshpande, A. M., and C. S. Newlon. 1996. DNA replication fork pause sites dependent on transcription. Science 272:1030-1033. [DOI] [PubMed] [Google Scholar]
- 9.Dillin, A., and J. Rine. 1998. Roles for ORC in M phase and S phase. Science 279:1733-1737. [DOI] [PubMed] [Google Scholar]
- 10.Dutta, A., and S. P. Bell. 1997. Initiation of DNA replication in eukaryotic cells. Annu. Rev. Cell. Dev. Biol. 13:293-332. [DOI] [PubMed] [Google Scholar]
- 11.Elble, R., and B. K. Tye. 1992. Chromosome loss, hyperrecombination, and cell cycle arrest in a yeast mcm1 mutant. Mol. Biol. Cell 3:971-980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Foss, M., F. J. McNally, P. Laurenson, and J. Rine. 1993. Origin recognition complex (ORC) in transcriptional silencing and DNA replication in S. cerevisiae. Science 262:1838-1844. [DOI] [PubMed] [Google Scholar]
- 13.Friedman, K. L., B. J. Brewer, and W. L. Fangman. 1997. Replication profile of Saccharomyces cerevisiae chromosome VI. Genes Cells 2:667-678. [DOI] [PubMed] [Google Scholar]
- 14.Galgoczy, D. J., and D. P. Toczyski. 2001. Checkpoint adaptation precedes spontaneous and damage-induced genomic instability in yeast. Mol. Cell. Biol. 21:1710-1718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Haber, J. E. 1999. DNA recombination: the replication connection. Trends Biochem. Sci. 24:271-275. [DOI] [PubMed] [Google Scholar]
- 16.Haber, J. E., and M. Hearn. 1985. Rad52-independent mitotic gene conversion in Saccharomyces cerevisiae frequently results in chromosomal loss. Genetics 111:7-22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hartwell, L. H., and T. A. Weinert. 1989. Checkpoints: controls that ensure the order of cell cycle events. Science 246:629-634. [DOI] [PubMed] [Google Scholar]
- 18.Hiraoka, M., K. Watanabe, K. Umezu, and H. Maki. 2000. Spontaneous loss of heterozygosity in diploid Saccharomyces cerevisiae cells. Genetics 156:1531-1548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hori, Y., K. Shirahige, C. Obuse, T. Tsurimoto, and H. Yoshikawa. 1996. Characterization of a novel CDC gene (ORC1) partly homologous to CDC6 of Saccharomyces cerevisiae. Mol. Biol. Cell 7:409-418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Horiuchi, T., and Y. Fujimura. 1995. Recombinational rescue of the stalled DNA replication fork: a model based on analysis of an Escherichia coli strain with a chromosome region difficult to replicate. J. Bacteriol. 177:783-791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kobayashi, T., M. Hidaka, M. Nishizawa, and T. Horiuchi. 1992. Identification of a site required for DNA replication fork blocking activity in the rRNA gene cluster in Saccharomyces cerevisiae. Mol. Gen. Genet. 233:355-362. [DOI] [PubMed] [Google Scholar]
- 22.Kogoma, T. 1997. Stable DNA replication: interplay between DNA replication, homologous recombination, and transcription. Microbiol. Mol. Biol. Rev. 61:212-238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kramer, K. M., and J. E. Haber. 1993. New telomeres in yeast are initiated with a highly selected subset of TG1-3 repeats. Genes Dev. 7:2345-2356. [DOI] [PubMed] [Google Scholar]
- 24.Malone, R. E., B. A. Montelone, C. Edwards, K. Carney, and M. F. Hoekstra. 1988. A reexamination of the role of the RAD52 gene in spontaneous mitotic recombination. Curr. Genet. 14:211-223. [DOI] [PubMed] [Google Scholar]
- 25.McDonald, J. P., and R. Rothstein. 1994. Unrepaired heteroduplex DNA in Saccharomyces cerevisiae is decreased in RAD1 RAD52-independent recombination. Genetics 137:393-405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Michel, B. 2000. Replication fork arrest and DNA recombination. Trends Biochem. Sci. 25:173-178. [DOI] [PubMed] [Google Scholar]
- 27.Michel, B., M. J. Flores, E. Viguera, G. Grompone, M. Seigneur, and V. Bidnenko. 2001. Rescue of arrested replication forks by homologous recombination. Proc. Natl. Acad. Sci. USA 98:8181-8188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Myung, K., A. Datta, and R. D. Kolodner. 2001. Suppression of spontaneous chromosomal rearrangements by S phase checkpoint functions in Saccharomyces cerevisiae. Cell 104:397-408. [DOI] [PubMed] [Google Scholar]
- 29.Nishitani, H., M. Hidaka, and T. Horiuchi. 1993. Specific chromosomal sites enhancing homologous recombination in Escherichia coli mutants defective in RNase H. Mol. Gen. Genet. 240:307-314. [DOI] [PubMed] [Google Scholar]
- 30.Orr-Weaver, T. L., and R. A. Weinberg. 1998. A checkpoint on the road to cancer. Nature 392:223-224. [DOI] [PubMed] [Google Scholar]
- 31.Paulovich, A. G., D. P. Toczyski, and L. H. Hartwell. 1997. When checkpoints fail. Cell 88:315-321. [DOI] [PubMed] [Google Scholar]
- 32.Rothstein, R., B. Michel, and S. Gangloff. 2000. Replication fork pausing and recombination or “gimme a break.” Genes Dev. 14:1-10. [PubMed] [Google Scholar]
- 33.Sandell, L. L., and V. A. Zakian. 1993. Loss of a yeast telomere: arrest, recovery, and chromosome loss. Cell 75:729-739. [DOI] [PubMed] [Google Scholar]
- 34.Schar, P. 2001. Spontaneous DNA damage, genome instability, and cancer—when DNA replication escapes control. Cell 104:329-332. [DOI] [PubMed] [Google Scholar]
- 35.Shirahige, K., Y. Hori, K. Shiraishi, M. Yamashita, K. Takahashi, C. Obuse, T. Tsurimoto, and H. Yoshikawa. 1998. Regulation of DNA-replication origins during cell-cycle progression. Nature 395:618-621. [DOI] [PubMed] [Google Scholar]
- 36.Sikorski, R. S., and P. Hieter. 1989. A system of shuttle vectors and yeast host strains designed for efficient manipulation of DNA in Saccharomyces cerevisiae. Genetics 122:19-27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Toczyski, D. P., D. J. Galgoczy, and L. H. Hartwell. 1997. CDC5 and CKII control adaptation to the yeast DNA damage checkpoint. Cell 90:1097-1106. [DOI] [PubMed] [Google Scholar]
- 38.Toyn, J. H., A. L. Johnson, and L. H. Johnston. 1995. Segregation of unreplicated chromosomes in Saccharomyces cerevisiae reveals a novel G1/M-phase checkpoint. Mol. Cell. Biol. 15:5312-5321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Umezu, K., M. Hiraoka, M. Mori, and H. Maki. Structural analysis of aberrant chromosomes that occur spontaneously in diploid Saccharomyces cerevisiae: retrotransposon Ty1 plays a crucial role in chromosomal rearrangements. Genetics, in press. [DOI] [PMC free article] [PubMed]
- 40.van Brabant, A. J., C. D. Buchanan, E. Charboneau, W. L. Fangman, and B. J. Brewer. 2001. An origin-deficient yeast artificial chromosome triggers a cell cycle checkpoint. Mol. Cell 7:705-713. [DOI] [PubMed] [Google Scholar]
- 41.Wach, A., A. Brachat, R. Pohlmann, and P. Philippsen. 1994. New heterologous modules for classical or PCR-based gene disruptions in Saccharomyces cerevisiae. Yeast 10:1793-1808. [DOI] [PubMed] [Google Scholar]
- 42.Wang, Y., M. Vujcic, and D. Kowalski. 2001. DNA replication forks pause at silent origins near the HML locus in budding yeast. Mol. Cell. Biol. 21:4938-4948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Weinert, T. A., and L. H. Hartwell. 1990. Characterization of RAD9 of Saccharomyces cerevisiae and evidence that its function acts posttranslationally in cell cycle arrest after DNA damage. Mol. Cell. Biol. 10:6554-6564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Weinert, T. A., and L. H. Hartwell. 1988. The RAD9 gene controls the cell cycle response to DNA damage in Saccharomyces cerevisiae. Science 241:317-322. [DOI] [PubMed] [Google Scholar]
- 45.Weinert, T. A., G. L. Kiser, and L. H. Hartwell. 1994. Mitotic checkpoint genes in budding yeast and the dependence of mitosis on DNA replication and repair. Genes Dev. 8:652-665. [DOI] [PubMed] [Google Scholar]
- 46.Weinreich, M., C. Liang, H. H. Chen, and B. Stillman. 2001. Binding of cyclin-dependent kinases to ORC and Cdc6p regulates the chromosome replication cycle. Proc. Natl. Acad. Sci. USA 98:11211-11217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Yamashita, M., Y. Hori, T. Shinomiya, C. Obuse, T. Tsurimoto, H. Yoshikawa, and K. Shirahige. 1997. The efficiency and timing of initiation of replication of multiple replicons of Saccharomyces cerevisiae chromosome VI. Genes Cells 2:655-665. [DOI] [PubMed] [Google Scholar]
- 48.Zhou, B. B., and S. J. Elledge. 2000. The DNA damage response: putting checkpoints in perspective. Nature 408:433-439. [DOI] [PubMed] [Google Scholar]
- 49.Zou, H., and R. Rothstein. 1997. Holliday junctions accumulate in replication mutants via a RecA homolog-independent mechanism. Cell 90:87-96. [DOI] [PubMed] [Google Scholar]