Abstract
Age-related maculopathy (ARM) is a leading cause of visual impairment among the elderly in Western populations. To identify ARM-susceptibility loci, we genotyped a subset of subjects from the Beaver Dam (WI) Eye Study and performed a model-free genomewide linkage analysis for markers linked to a quantitative measure of ARM. We initially genotyped 345 autosomal markers in 325 individuals (N=263 sib pairs) from 102 pedigrees. Ten regions suggestive of linkage with ARM were observed on chromosomes 3, 5, 6, 12, 15, and 16. Prior to fine mapping, the most significant regions were an 18-cM region on chromosome 12, near D12S1300 (P=.0159); a region on chromosome 3, near D3S1763, with a P value of .0062; and a 6-cM region on chromosome 16, near D16S769, with a P value of .0086. After expanding our analysis to include 25 additional fine-mapping markers, we found that a 14-cM region on chromosome 12, near D12S346 (located at 106.89 cM), showed the strongest indication of linkage, with a P value of .004. Three other regions, on chromosomes 5, 6, and 15, that were nominally significant at P⩽.01 are also appropriate for fine mapping.
Introduction
Age-related maculopathy (ARM) is an important cause of severe visual impairment (R. Klein et al. 1995; Tielsch 1995). Although the natural history of ARM is becoming better understood, its pathogenesis remains unknown (Schick et al. 2001). Data from most family studies of ARM have suggested a strong genetic component (Meyers and Zachary 1988; Dosso and Bovet 1992; Small et al. 1992, 1999; Stone et al. 1992; Meyers 1994; Meyers et al. 1995; Hoyng et al. 1996; M. L. Klein et al. 1998; Weeks et al. 2000).
ARM is typically characterized by the presence of large, soft, yellow-white drusen. Such drusen may be distributed either individually or in close proximity to adjacent drusen. These drusen usually first appear in the 5th decade of life and become more pronounced with age (R. Klein et al. 1997). Progression of the disease is marked by disruption of cells in the outer layer of the retina, characterized by the appearance of grayish-blackish deposits in the deep retina, accompanied by depigmentation of the retinal pigment epithelium (RPE), and may take on several different forms. One form of late ARM, known as “geographic atrophy” or “dry” ARM, is usually characterized by a sharply defined, round area of drop out of the outer segments of the retinal rod and cone photoreceptors and the RPE. Another form, labeled as “neovascular” or “wet” ARM, involves the new growth of small choroidal blood vessels into the subretinal space.
Data from several population-based studies have provided information on the prevalence, incidence, and rates of progression of ARM (Kahn et al. 1977a, 1977b; B. E. Klein and R. Klein 1982; Gibson et al. 1985; Bressler et al. 1989, 1995; B. E. Klein et al. 1990; R. Klein et al. 1992, 1997, 1999a, 1999b; Mitchell et al. 1995; Schachat et al. 1995; Vingerling et al. 1995; Cruickshanks et al. 1997; Sparrow et al. 1997). One such study was conducted in a large (N = 4,926), mostly (99%) white population of persons 43–86 years of age living in Beaver Dam, WI (R. Klein et al. 1992, 1997). In Beaver Dam, persons ⩾75 years of age had a prevalence of late ARM of 7.1% and a 5-year incidence of 5.4% (R. Klein et al. 1992, 1997). Heiba et al. (1994) performed a complex segregation analysis of the sibships identified in this population, using Bonney’s (1984) class D regressive model. They found a quantitative measure for ARM based on the Wisconsin Age-Related Maculopathy Grading Scheme protocol (R. Klein and B. E. K. Klein 1991; R. Klein et al. 1991a, 1991b, 1991c, 1992) and adjusted for age- and age2-suggested evidence of major gene segregation. The results were consistent with a major Mendelian gene effect contributing ∼55% and ∼57% of the total variability in the right and left eyes, respectively. Following up this evidence, we performed a genomewide screen in a sample of 105 sibships from this population, to identify loci segregating for ARM. In the present article, we describe a model-free linkage analysis of these sibship data.
Families and Methods
Families
The Beaver Dam Eye Study (BDES) comprises a community sample of 4,926 subjects between 43 and 86 years of age who are of mostly western European extraction (R. Klein et al. 1992, 1997). Beaver Dam was selected for a study of maculopathy because it had a high participation rate in a previous population-based study of diabetic retinopathy and because it had a relatively stable population (N=17,179 at the time of the 1980 census) (R. Klein et al. 1991c). Eligibility requirements for entry into the study included living in the city or township of Beaver Dam and being in the target age range at the time of a private census performed in 1987–88. Of 6,612 households identified, 3,715 contained at least one person who satisfied the age criterion. Of 5,924 persons who were eligible for participation, we examined 4,926 persons over a 30-mo period, 337 permitted an interview only, 226 died before the examination, 39 moved out of the area, 18 could not be located, and 378 refused to participate. Family relationships were ascertained during the study evaluation, and pedigrees were constructed. Of the 4,926 observations at baseline examination, 2,783 were distributed among 602 pedigrees. Severity scores for ARM that were based on the Wisconsin Age-Related Maculopathy Grading Scheme (R. Klein and B. E. K. Klein 1991; R. Klein et al. 1991a, 1991b, 1991c, 1992; Heiba et al. 1994) were assigned for the right and left eye of each participant. There were 105 sibships selected for genotyping and linkage analysis. These sibships had to contain one member who had a mean severity score, for the two eyes, of ⩾4.5. This criterion was chosen because, on the basis of the prior segregation analysis (Heiba et al. 1994), such individuals would have only a 2.5% probability of not having two copies of a putative recessive severity-increasing allele. In addition, the sample was augmented by sibships that had been selected, on the basis of severity, for nuclear sclerosis and cortical cataracts. The genotyped data set contains 325 individuals. The sibships range in size from two to six, with an average of 2.46 persons per sibship. Similar to the original data set, the subset contains slightly more females (N=176) than males (N=149). Those genotyped comprise a total of 263 sib pairs from 105 sibships representing 102 pedigrees.
Phenotypic Evaluation
Although earlier clinical studies of ARM relied on direct and indirect ophthalmoscopy for the detection of this condition, later studies found that stereoscopic color fundus photographs of the macula provide a more objective, sensitive, and reproducible method for the documentation of ARM (R. Klein and B. E. K. Klein 1991; R. Klein et al. 1991a, 1991b, 1991c; Bird et al. 1995). The photographs are graded using standardized protocols to detect and classify the presence and severity of ARM lesions. Details of the grading procedure have been described elsewhere (R. Klein et al. 1991a, 1991b). In brief, a circular grid was placed on the photographic slide, dividing the macular area into nine subfields, consisting of central (a single subfield), inner (four inner subfields), and outer (four outer subfields) circles. Some lesions were graded in each subfield, other lesions were graded only in the subfield defining the macular area, and still others were graded in additional fields outside the macular area. For the purpose of the present article, only measurements made within the nine subfields defined by the grid are presented. Circles of defined size (63, 125, 175, 250, 322, 350, and 644 μm in diameter) that were printed on clear plastic were used to estimate the size of drusen and areas involved by drusen, increased retinal pigment, and RPE depigmentation.
Two gradings were performed for each eye (R. Klein and B. E. K. Klein 1991, 1995; R. Klein et al. 1991a, 1991b; 1992, 1997, 2001, 2002). First, a preliminary masked grading was done by one of two senior graders. Second, detailed gradings were performed by one of three other experienced graders. For detailed grading, each eye was graded independently of the fellow eye. The assessment consisted of a subfield-by-subfield, lesion-by-lesion evaluation of each photograph set, using the Wisconsin Age-Related Maculopathy Grading Scheme (R. Klein et al. 1991a, 1991b). After both gradings were performed, a series of edits and reviews were performed. The presence and severity of specific lesions at the third examination (e.g., maximum drusen size, type, area, and pigmentary abnormalities), as determined by detailed grading, were compared to those of the preliminary grading. Standardized edit rules were used to adjudicate disagreements (R. Klein et al. 1991a, 1997). Finally, the detailed graders were asked to make side-by-side comparisons between baseline, 5-year follow-up, and 10-year follow-up photographs for eyes that showed change in ARM lesions between baseline and follow-up. These edits were masked as to whether the photographs were taken at baseline or follow-up.
A 15-level severity scale based on pigmentary abnormalities, geographic atrophy, signs of exudative macular degeneration, and drusen size, type, and area was constructed (table 1). The ordering of the scale was based on associations of the presence and absence of early signs of ARM with the incidence of progression to geographic atrophy or exudative macular degeneration (R. Klein and B. E. K. Klein 1995).
Table 1.
Revised Wisconsin Age-Related Maculopathy Coding Protocol[Note]
| Level | Drusen Diameter(μm) | Drusen Typea | Drusen Area(μm2) | Increased Retinal Pigment(μm) | RPE Depigmentation | Late ARM |
| 1 | None/questionable | None/HI | None | None | None | None |
| 2 | <63 | HD | <9,160 | None | None | None |
| 3 | <63 | HD | ⩾9,160 | None | None | None |
| 4 | ⩾63 | HD/SD | <196,350 | None | None | None |
| 5 | <63 | HD | Any size | Any size | None | None |
| <63 | HD | Any size | None | Any size | None | |
| <63 | HD | Any size | Any size | Any size | None | |
| 6 | ⩾63 | HD/SD | ⩾196,350 | None | None | None |
| ⩾63 | HD/SD | <196,350 | <125 | None | None | |
| 7 | ⩾63 | HD/SD | ⩾196,350 | <125 | None | None |
| ⩾63 | HD/SD | <196,350 | ⩾125 in ⩾1 subfields or <125 in ⩾2 subfields | None | None | |
| ⩾63 | SI/Ret | <196,350 | None or <125 | None | None | |
| 8 | ⩾63 | HD/SD | ⩾196,350 | ⩾125 in ⩾1 subfields or <125 in ⩾2 subfields | None | None |
| ⩾63 | HD/SD | <196,350 | None or any size | <6.25% | None | |
| ⩾63 | SI/Ret | ⩾196,350 | None or <125 | None | None | |
| ⩾63 | SI/Ret | <196,350 | ⩾125 in ⩾1 subfields or <125 in ⩾2 subfields | None | None | |
| 9 | ⩾63 | HD/SD | ⩾196,350 | None or any size | <6.25% | None |
| ⩾63 | HD/SD | <196,350 | None or any size | ⩾6.25% of ⩾1 subfields or <6.25% of ⩾2 subfields | None | |
| ⩾63 | SI/Ret | ⩾196,350 | ⩾125 in ⩾1 subfields or <125 in ⩾2 subfields | None | None | |
| ⩾63 | SI/Ret | <196,350 | None or any size | <6.25% of subfield | None | |
| 10 | ⩾63 | HD/SD | ⩾196,350 | None or any size | ⩾6.25% of ⩾1 subfields or <6.25% of ⩾2 subfields | None |
| ⩾63 | SI/Ret | ⩾196,350 | None or any size | <6.25% of subfield | None | |
| ⩾63 | SI/Ret | <196,350 | None or any size | ⩾6.25% of ⩾1 subfields or <6.25% of ⩾2 subfields | None | |
| 11 | ⩾63 | SI/Ret | ⩾196,350 | None or any size | ⩾6.25% of ⩾1 subfields or <6.25% of ⩾2 subfields | None |
| 12 | NA | NA | NA | NA | NA | Geographic atrophyb |
| 13 | NA | NA | NA | NA | NA | Exudative, detachmentc |
| 14 | NA | NA | NA | NA | NA | Exudative, advancedc |
| 15 | NA | NA | NA | NA | NA | Geographic atrophy, exudative |
Data given represent maximum allowable in any gradable subfield. NA = not applicable.
HI = hard indistinct; HD = hard distinct; SD = soft distinct; SI = soft indistinct; Ret = reticular.
In the absence of exudative ARM.
In the absence of geographic atrophy.
ARM severity was determined on each eye at three time points spaced at 5-year intervals, and, at each time point, the scores for the right and left eyes were averaged. However, if a score was missing for either eye, then the score for the available eye was substituted for the missing score (this occurred for 7.9% of the persons in the study). Then, because the scores for at least two time points were available for all of the persons in the linkage-analysis data set, the scores at two time points were averaged to produce the final score. When scores at all three time points were available, which occurred for 74.46% of the participants, the first and last scores were averaged; otherwise, the scores at the two available time points were averaged. In this way, all of the scores in the analysis were based on the same or a similar amount of information. We then used multiple regression analysis to investigate the effects of age, age2, and sex—together with their interactions—as well as to investigate the effects that alcohol consumption, smoking and multiple-vitamin usage (as represented by categorical variables), and sunlight exposure had on this ARM score. As has previously been observed by Heiba et al. (1994), who used the Wisconsin Age-Related Maculopathy Grading Scheme (R. Klein and B. E. K. Klein 1991; R. Klein et al. 1991a, 1991b, 1991c), we found age and age2 to be nominally significant in predicting maculopathy, assuming independence among persons, and the residuals were used as our trait value—that is, ARMresidual = [(ARMfirst examination + ARMlast examination)/2] − 8.6124 + (0.2269 × age) − (0.0023 × age2).
Molecular Methods
Genome scan.—We genotyped 345 markers from 22 autosomal chromosomes by using the Weber panel 8 marker set, which has an average marker spacing of 11 cM. We also genotyped two additional markers, D1S406 and D1S236, contained in a 14-cM candidate-gene region spanning the ABCR (retina-specific ABC transporter) gene, which is suggested to be the gene responsible for Stargardt disease, a recessive macular dystrophy that has been considered to be related to ARM (Allikmets et al. 1997). Another four markers—D1S466, D1S202, D1S2625, and D1S413—in a 26-cM candidate-gene region, on 1q21-q35, that is postulated to contain an autosomal dominant locus related to a dry type of ARM (M. L. Klein et al. 1998) were also genotyped. Thus, 351 markers were initially genotyped.
After extracting DNA from the blood samples, we used a fluorescence-based genotyping method for the genome scan. PCR primers conjugated with fluorescent dyes were purchased from IDT Technology and Applied Biosystems. Genomic DNA (at 10 ng/μl, 3 μls) was PCR amplified using 0.225 U of Platinum Taq DNA Polymerase (Gibco), Tris-HCl (200 mM, pH 8.4), dNTPs (200 μM each), MgCl2 (1.0–3.0 mM), and forward and reverse PCR primers (0.2 μM). The final reaction volume was 12 μl, and the reactions were performed in 96-well plates on an MJ Research Tetrad DNA Engine. The amplification reactions were optimized for the fluorescent-dye–labeled primers by using the published conditions as the initial value. After PCR, 8–10 markers were pooled to create multiplexed panels. Each panel was size separated on an ABI 377 (Applied Biosystems) DNA Sequencer running the GeneScan 3.1 software for Macintosh. Tamra 500 size standards were run in each lane, and the data were collected and analyzed using the Genotyper 2.5 software. Besides the in-lane standards, each 96-well plate contained four CEPH controls (two samples each from individuals 1331–01 and 1331–02). The CEPH controls were used to standardize allele calling across plates, with the replicates being the safeguard for allele-calling reliability.
Fine mapping.—After reviewing the first-stage genome-scan linkage results, we extended our coverage of chromosome 12 to include an additional 25 markers over an area of suggestive significance that overlapped a region previously reported to show linkage with ARM (Weeks et al. 2000). Our fine-mapped region of interest on chromosome 12 has an average spacing density of 2.2 cM. We also decided to fine map areas of suggestive significance indicated by our genome scan, on chromosome 3, 5, and 16. We extended our coverage to include an additional 8 markers on chromosome 3, in two candidate-gene areas (one in a 16.48-cM region between D3S1304 and D3S3038 and the other in a 29.45-cM region between D3S1764 and D3S3053); 10 markers on chromosome 5, in an 88.10-cM region between D5S1457 and D5S1480; and four markers on chromosome 16, in a 46.33-cM region between D16S764 (18.51 cM) and D16S753.
Genotyping protocols for fine mapping were similar to those for the genome scan. Multiplexed panels of markers were size separated on an ABI 3700 (Applied Biosystems) capillary gel by running the GeneScan 3.5 software for Windows NT. Rox 500 size standards were run in each lane, and the data were collected and analyzed using the Genotyper 3.6 software for Windows NT. Initial marker placements were based on locations provided at the Marshfield Center for Medical Genetics Web site, although other placements were also investigated.
Error Checking
After each round of genotyping, the allelic data were checked for Mendelian inconsistencies, using the program Markerinfo (S.A.G.E., version 4.2). Family members whose data were inconsistent with Mendelian transmission were referred back to the laboratory for verification. We also compared the observed number of homozygotes to the expected number of homozygotes based on the estimated allele frequencies and Hardy-Weinberg equilibrium proportions. A significant inequality between the observed and expected numbers of homozygotes could indicate that some heterozygotes were miscalled as homozygotes (Gomes et al. 1999). However, we found no significant departure from Hardy-Weinberg proportions in our data. After regenotyping and final verification, allelic inconsistencies that could not be reconciled (0.68%) were changed to missing values for the purpose of analysis. Consistent estimates of allele frequencies for each genetic marker were then established by gene counting.
Prior to performing the linkage analysis, we reclassified the sib pairs in each pedigree according to their likely true relationship, using the program Reltest (S.A.G.E., version 4.2). Reltest makes use of a Markov-process model of allele sharing along the chromosomes, to classify relationships (Olson 1999). The probability of misclassification depends on the total length of the genome scan and overall marker informativeness. It is possible for individual pairs to be misclassified if one or both members have a high proportion of missing genotypes. We reclassified 6 individuals in 3 full sibships as unrelated and 21 individuals in 12 full sibships as half-sibs. There was only one reclassification in which both members of a sib pair had as much as 10% missing data.
Linkage Analysis
It is known that the power of a model-free quantitative-trait linkage analysis depends on the scale of measurement (Wilson et al. 1991). To perform a Box-Cox power transformation of the data (Box and Cox 1964), which requires that all the measures be positive, we adjusted the quantitative scores to 80 years old, as done by Heiba et al. (1994). We repeated the segregation analysis performed by Heiba et al. (1994) on these new adjusted scores, specifically in the subsample that was genotyped, and we obtained results similar to those originally reported (data not shown), finding that the most parsimonious model was the two mean, recessive model, although the estimated allele frequency for high values was larger, also expected because of the ascertainment bias. In particular, a Box-Cox power transformation parameter λ1=0.759 gave the best indication of genetic involvement under the recessive model. Thus, the age-adjusted scores were raised to this power prior to linkage analysis.
We used Sibpal (S.A.G.E., version 4.2), a model-free linkage program, to perform a multipoint linkage analysis. The reported P values were obtained assuming the usual asymptotic distribution of the likelihood-ratio test statistics, but all critical results were verified by a permutation test: the estimates of allele sharing were permuted among sibships of the same size, as well as within sibships in the case of sibships with more than two sibs.
A weighted average, of the squared trait difference and the squared mean-corrected trait sum, that is further adjusted for the nonindependence of the sib pairs (Elston and Shete 2000; Shete et al., in press) was used as the dependent variable to perform the linear regressions (option W4 in Sibpal). In this method, the optimal weights are inversely proportional to the residual variances that are obtained from two different regressions, based respectively on the squared sib-pair trait differences and the mean-corrected trait sums. Control of the type I error probabilities is achieved by using a generalized-estimating-equation approach with a robust sandwich estimate of the variance (Huber 1967; Gourieroux et al. 1984). Although this method is very efficient (Putter et al. 2002), it can become numerically unstable and require caution when the residual correlations among the squared sib-pair trait differences and the squared mean-corrected sib-pair trait sums become large. In these linkage analyses, we did not observe any numerical instability.
Results
Genome Scan
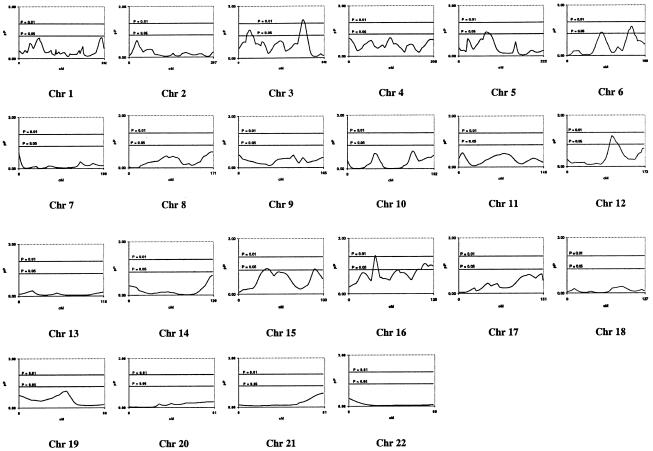
The results of the whole-genome scan are presented in figure 1. We observed 14 markers on six chromosomes (3, 5, 6, 12, 15, and 16), with a significance level of P⩽.05 (table 2). Two possible regions of linkage, containing four markers altogether, are located on chromosome 3, with D3S1763 having the greatest significance (P=.0081). Our analysis suggests that D5S2500 (P=.0382)—on chromosome 5, in a region initially suggested by Weeks et al. (2000) but later lost when more sib pairs were added (Weeks et al. 2001)—may be marginally significant for linkage whereas two markers (D6S1031 and GATA165G02) on chromosome 6, each located in a different region of the chromosome, show slight significance. We found suggestions of linkage to ARM at a P value ⩽.05 over an 18-cM region, on chromosome 12, that includes D12S1300 (P=.0159) and PAH (P=.0354), which are located within 20 cM of a previous linkage report (Weeks et al. 2000). Chromosome 15 exhibits marginal evidence of linkage to ARM in two regions located 62 cM apart, at D15S659 and D15S816, whereas chromosome 16 suggests evidence of linkage at D16S769 (P=.0086), located at 39 cM, and at a second region, located between 117 and 124 cM, that contains D16S539 and D16S621.
Figure 1.
Multipoint results of the genomewide linkage scan for ARM in the BDES linkage analysis, using the Weber panel 8 map spacing. For each chromosome (Chr), genetic distance (cM) and −log10(P) value (pP) are plotted on the X- and Y-axes, respectively. Horizontal lines are drawn at P=.05 and P=.01.
Table 2.
Multipoint Estimates and P Values on Chromosomes 3, 5, 6, 12, 15, and 16, Showing Possible Linkage (P⩽.05) with ARM in the BDES Genome Scan
| Location(cM) | Marker | P |
| Chromosome 3: | ||
| 24.0 | .0368 | |
| 26.0 | GATA164B08 | .0323 |
| 28.0 | .0269 | |
| 30.0 | .0237 | |
| 32.0 | .0234 | |
| 34.0 | .0267 | |
| 36.0 | .0338 | |
| 38.0 | D3S1259 | .0444 |
| 40.0 | .0460 | |
| 172.0 | .0486 | |
| 174.0 | .0318 | |
| 175.0 | D3S1744 | .0272 |
| 176.0 | .0209 | |
| 178.0 | .0121 | |
| 180.0 | .0077 | |
| 182.0 | .0062 | |
| 184.0 | .0066 | |
| 186.0 | D3S1763 | .0081 |
| 188.0 | .0109 | |
| 190.0 | .0169 | |
| 192.0 | .0289 | |
| Chromosome 5: | ||
| 68.0 | .0474 | |
| 70.0 | .0392 | |
| 72.0 | .0371 | |
| 74.0 | D5S2500 | .0382 |
| 76.0 | .0398 | |
| 78.0 | .0419 | |
| 80.0 | .0450 | |
| 82.0 | .0495 | |
| Chromosome 6: | ||
| 84.0 | .0439 | |
| 86.0 | D6S1031 | .0415 |
| 88.0 | .0444 | |
| 152.0 | .0390 | |
| 154.0 | .0306 | |
| 156.0 | GATA165G02 | .0286 |
| 158.0 | .0218 | |
| 160.0 | .0192 | |
| 162.0 | .0213 | |
| 164.0 | .0289 | |
| 166.0 | .0429 | |
| Chromosome 12: | ||
| 94.0 | .0497 | |
| 96.0 | .0296 | |
| 98.0 | .0192 | |
| 99.0 | D12S1300 | .0159 |
| 100.0 | .0165 | |
| 102.0 | .0185 | |
| 104.0 | .0222 | |
| 106.0 | .0278 | |
| 108.0 | PAH | .0354 |
| Chromosome 15: | ||
| 30.0 | D15S659 | .0477 |
| 32.0 | .0463 | |
| 34.0 | .0413 | |
| 36.0 | .0422 | |
| 92.0 | D15S816 | .0444 |
| Chromosome 16: | ||
| 36.0 | .0413 | |
| 38.0 | .0130 | |
| 39.0 | D16S769 | .0086 |
| 40.0 | .0113 | |
| 42.0 | .0242 | |
| 108.0 | .0360 | |
| 110.0 | .0273 | |
| 112.0 | .0241 | |
| 114.0 | .0257 | |
| 116.0 | .0308 | |
| 117.0 | D16S539 | .0344 |
| 118.0 | .0319 | |
| 120.0 | .0290 | |
| 122.0 | .0291 | |
| 124.0 | D16S621 | .0317 |
Fine Mapping on Chromosome 12
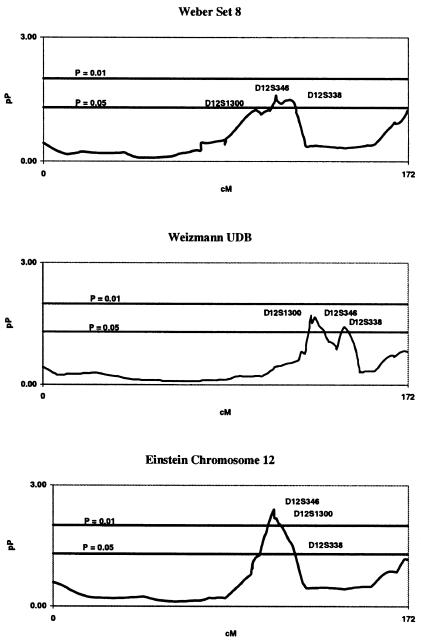
We fine mapped the area of significance on chromosome 12 indicated by our genome scan, because there was significant overlap between this region and a previous report of ARM linkage (Weeks et al. 2000). We extended our coverage to include an additional 25 markers across a 54-cM region on 12q21-q24, according to the marker locations provided at the Marshfield Center for Medical Genetics Web site. A subsequent review of the public databases led us to conclude that a consensus fine map did not exist for chromosome 12, and the structure and size of our sibship sample was not sufficient to independently derive map order. Thus, we conducted linkage analyses using two additional map orders based on physical maps provided at the Weizmann Institute Unified Database for Human Genome Mapping Web site and the Harvard Partners Genome Center Web site, to improve map order. Figure 2 shows a comparison of the fine-mapping linkage results obtained by using each of the three maps. Using the map positions provided by the Harvard Partners map, we narrowed our region of significance to a 14-cM area, on 12q22-q23, that contained five markers indicated to be significantly linked with ARM at P=.01 (table 3). The peak marker in this region is D12S346 (P=.004).
Figure 2.
Fine-mapping multipoint results on chromosome 12 for the BDES linkage analysis, compared using the Weber panel 8, Weizmann Institute Unified Database (UDB), and Harvard Partners fine-mapping marker positions. Genetic distance (cM) and −log10(P) value (pP) are plotted on the X- and Y-axes, respectively.
Table 3.
Multipoint Estimates and P Values on Chromosome 12, Showing Possible Linkage (P⩽.05) with ARM in the BDES Genome Scan[Note]
| Location(cM) | Marker | P |
| 102.00 | .0206 | |
| 103.20 | D12S309 | .0140 |
| 104.00 | .0098 | |
| 106.00 | .0048 | |
| 106.89 | D12S346 | .0040 |
| 107.00 | D12S1671 | .0064 |
| 107.00 | D12S1300 | .0063 |
| 107.32 | D12S1588 | .0067 |
| 108.00 | .0066 | |
| 109.06 | D12S1727 | .0088 |
| 110.00 | .0095 | |
| 112.00 | .0139 | |
| 113.19 | PAH | .0190 |
| 114.00 | .0223 | |
| 114.06 | D12S360 | .0229 |
| 114.71 | D12S338 | .0251 |
| 116.00 | .0324 |
Note.— Marker positions were obtained using the Harvard Partners fine map of chromosome 12. P values and distances between markers were interpolated by S.A.G.E., version 4.2.
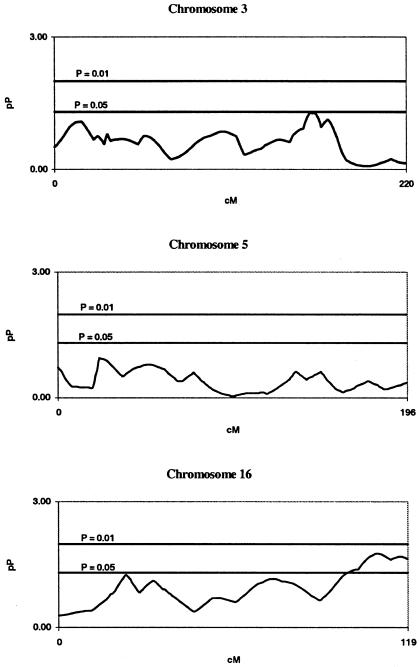
Fine Mapping on Chromosomes 3, 5, and 16
After reviewing the results of our follow-up mapping of chromosomes 3, 5, and 16 (fig. 3), we found that, in contrast to our fine mapping of chromosome 12, the linkage signals in our first candidate region, on chromosome 3, had weakened significantly whereas our signal in our second candidate region had become marginal at best. Our signal on chromosome 5 virtually disappeared, and, on chromosome 16, the signal in our fine-mapped region around D16S769 had diminished to the point of marginal significance. We attempted to improve results for these chromosomes by using alternative marker placements, but few differences were observed in the results.
Figure 3.
Fine-mapping multipoint results on chromosomes 3, 5, and 16 for the BDES linkage analysis, using the Weber panel 8 marker placements. Genetic distance (cM) and −log10(P) value (pP) are plotted on the X- and Y-axes, respectively.
Candidate-Gene Regions on Chromosome 1
We were also interested in the relationship between ARM in the Beaver Dam study and the dry form of ARM expressed by mutations in the recently discovered ABCR gene (Allikmets et al. 1997). This gene was refined to a 2-cM region located between the polymorphic markers D1S406 and D1S236, on 1p21-p13, in a set of families with Stargardt disease. Although this group has reported additional data in support of their original findings (Allikmets et al. 2000), other studies have given negative results (De La Paz et al. 1999; Souied et al. 2000; Yates and Moore 2000). Furthermore, a candidate gene for a dry form of ARM has been reported to be located within a 9-cM region on 1q25-q31, between D1S466 and D1S413, and to segregate as an autosomal dominant trait (M. L. Klein et al. 1998). Accordingly, we also fine mapped a 12-cM region that included D1S466, D1S518, D1S202, D1S2625, D1S413, and D1S1660. Using the Weber panel 8 marker locations, we found weak evidence with GATA129H04 (P=.07) in our genomewide multipoint analysis to support a hypothesis of linkage to ARM, but this marker was 56.9 cM away from the ABCR gene and was neither in the ABCR region (Allikmets et al. 1997) nor in the region specified by M. L. Klein et al. (1998) (table 4). To completely exclude these two regions, we performed tests that allowed for both additive and dominant components (Elston et al. 2000). Since the P values for all these tests were <.5 in these regions, we cannot exclude linkage. However, the evidence on 1p is weak at best (P>.25).
Table 4.
Chromosome 1 Candidate-Gene Regions, Multipoint Estimates, and P Values Associated with Each Marker
| Marker | Estimate ± SE | P |
| Allikmets et al. (1997) region: | ||
| D1S406 | .1485 ± .2770 | .2962 |
| D1S236 | −.0368 ± .2794 | .5523 |
| M. L. Klein et al. (1998) region: | ||
| D1S466 | .1052 ± .2544 | .3399 |
| D1S518 | −.0112 ± .2576 | .5174 |
| D1S202 | .0739 ± .2555 | .3864 |
| D1S2625 | .1468 ± .2354 | .2667 |
| D1S413 | .2095 ± .2489 | .2003 |
| D1S1660 | −.0072 ± .2600 | .5111 |
Discussion
We performed a whole-genome scan for ARM by using a quantitative trait that is representative of the continuum of severity for macular degeneration—ranging from drusen, which are an early characteristic of ARM and other macular diseases (Stone et al. 1999; Tarttelin et al. 2001), to late ARM (table 1). To map susceptibility genes, we selected sibships for genotyping that would have higher mean and variance of age-adjusted ARM scores than the general BDES population, although we did not select specifically for any particular form of ARM, wet or dry. Prior to linkage analysis, we investigated the effects of covariates and found only age and age2 to be nominally significant in the prediction of maculopathy. We used a power-transformed ARM score after adjusting for age and age2 as the chosen ARM trait for linkage analysis. Other correlates (e.g., smoking, sunlight exposure, and vitamin use) have been reported to be associated with ARM (Taylor et al. 1992). However, these covariates are generally associated with the most severe forms of the disease. Using the BDES population data (N=4,926), we did not observe any evidence that these variables play a role in the full disease spectrum that included the less severe forms. Nevertheless, we performed an additional linkage analysis that included covariates for drinking, smoking, sunlight exposure, and vitamin use and found no found no significant differences in the results.
We genotyped an additional 25 markers in the region of interest on chromosome 12. After the addition of these markers, the linkage signal increased from a multipoint P value of .0159, for D12S1300, when the marker order in the Marshfield map for Weber panel 8 was used, to a multipoint P value of .004, for D12S346, when the markers were reordered using the chromosome 12 physical map provided by the Harvard Partners (fig. 2). After adding more samples and markers, Weeks et al. (2001) obtained weaker evidence for linkage with ARM on chromosome 12, with an Sall score of 1.5. Although their results are not inconsistent with ours, we also observed that correct specification of the marker order during the fine-mapping stage was very important. For example, if either the Marshfield map or the Weizmann map were utilized during the fine-mapping stage, we lost some of the linkage signal (fig. 2). Although Weeks et al. (2001) used Simwalk2 to identify excessive numbers of double recombinants, they apparently did not investigate alternative map orders in the fine-mapping step, and this may be one cause for a weaker linkage signal in the denser scan. An alternative explanation is locus and phenotypic heterogeneity between their sample and ours.
The phenotype that we describe in the present article is not the same as that described in previous genome scans for ARM (Weeks et al. 2000, 2001). Weeks et al. (2000) collected a set of 364 families (2,129 individuals) from vitreoretinal practices and utilized a dichotomous trait, with a large percentage of affected individuals having evidence of exudative disease, for their model-based and model-free linkage analyses. With the most liberal criteria that they used for affection status, Weeks et al. (2000, 2001) would have classified only those individuals with severity scores of ⩾12 on our scale as affected. By using a quantitative trait and a power transformation, rather than a binary trait, we might expect to increase power to detect linkage, because we no longer have to classify each individual into one of two categories (affected or unaffected) with the attendant probability of misclassifying individuals into the wrong category (Elston 1979).
Weeks et al. (2000) performed an affected-only analysis. They computed LOD scores while assuming, a priori, a single simple dominant model (disease-allele frequency 0.0001; f0 = 0.01, f1 = 0.90, and f2 = 0.90, where fi is the penetrance for an individual with i susceptibility alleles), allowing for heterogeneity under an admixture model. They used three different models to define affection status. Their first model, A, classified as affected only the population subset that included individuals clearly affected with ARM as determined on the basis of drusen, pigmentary changes, and the presence of end-stage disease; all others were classified as unknown. In their second model, B, those individuals classified as affected under model A were augmented by individuals who were probably affected with ARM, with all others again being classified as unknown. The third model, C, adds to those affected individuals defined in model B by also classifying as affected those individuals who lacked sufficient evidence to rule out ARM, with all others being classified as unknown. When the Gene-Hunter Plus software package was used, model C demonstrated possible linkage near D12S2070, in a region on 12q21-q24, bounded by D12S1052 and D12S1045, with a nonparametric LOD score of 1.43. This signal, based on a larger number of affected persons in the analysis, can be interpreted as detecting a locus that may have any effect, moderate or severe. This area is the same general area, on chromosome 12, that we found to be linked to general ARM susceptibility in the present study.
The approach of Weeks et al. (2000, 2001) was geared toward identifying the most severe phenotype and is therefore likely to lead to susceptibility loci for the most severe forms of ARM. In contrast, our approach focuses on the identification of susceptibility genes for the whole range of disease, from mild to severe. Although not all drusen progress to late ARM, soft, large drusen are thought to represent the first stage in a multistep ARM progression (R. Klein et al. 2002). Identification of loci for inception or early stages of the disease is just as important as identification of genes for severe forms of the disease. Using the chosen quantitative trait, we found several chromosomal areas suggestive of linkage, including regions on chromosomes 3, 5, 12, 15, and 16 (fig. 1). Our conclusion would be that any concordance in linkage results for the most severe disease (model A; Weeks et al. 2000, 2001), versus the full spectrum (present analysis), would mean that we have replicated evidence for loci that predispose to greater severity.
Weeks et al. (2000) also reported a region of interest near D5S1480, on 5q32, where glutathione peroxidase 3 has been localized (Yoshimura et al. 1994); however, this region is located at the opposite end of chromosome 5 from where our study suggests linkage, near D5S2500, on 5p15. Our other linkage signals resided on regions other than 1q31, 9p13, 10q26, and 17q25, where Weeks et al. (2001) obtained maximal evidence for linkage with ARM. The result on 1q31, found in the interval between D1S1660 and D1S1647 (Weeks et al. 2001), is consistent with the ARM linkage originally reported by M. L. Klein et al. (1998), with a LOD score of 2.46 in a large family. In contrast, in the Beaver Dam sample, we found only weak evidence at best to support a hypothesis of linkage between ARM and either 1q25-q31, as postulated by M. L. Klein et al. (1998), or 1p21-p13, as suggested by Allikmets et al. (1997), and we conclude that a gene in either region would have to have a very small effect on ARM susceptibility, when compared to the regions on chromosomes 3, 5, 12, and 16. As pointed out by De La Paz et al. (1999), the difference may be due either to the way in which the disease was defined in our study, as opposed to the studies conducted by Allikmets et al. (1997) or M. L. Klein et al. (1998), or to both the difference in the population samples being studied and locus heterogeneity.
A candidate gene for apoptotic protease activating factor 1 has been mapped to a location slightly upstream from D12S346 (P=.0040) (see the Ensembl Genome Browser Web site). A precondition for ARM may be an accumulation of the age pigment lipofuscin in lysosomes of the RPE cells (Suter et al. 2000). These cells appear to die by apoptosis, with subsequent death of photoreceptor cells. The loss of these cells may result in the more severe forms of ARM that are expressed by geographic atrophy and/or exudative detachment of the retina. It has been suggested that the mitochondria function as cellular sensors of stress, into which different apoptosis-induction pathways converge (Kroemer and Reed 2000). As the mitochondria become destabilized, they release apoptosis-inducing proteins such as cytochrome c, which activates apoptosis-specific proteases called “caspases,” in addition to apoptosis-inducing factors that induce nuclear apoptosis independently of caspase activation (Leist et al. 1999; Susin et al. 1999; Daugas et al. 2000). Apoptotic protease activating factor 1 may be important in the pathophysiology of cell death related to age-related macular degeneration and, in the future, for its pharmacological control. Other potential candidate genes related to our chromosome 12 region of interest are given in table 5.
Table 5.
Candidate Genes Located Proximal and Distal to D12S346 (P=.004) after Fine Mapping[Note]
| Locus | Gene Name |
| DSPG3 (MIM 601657) | Dermatan sulphate proteoglycan 3 |
| KERA (MIM 603288) | Keratocan |
| LUM (MIM 600616) | Lumican |
| DCN (MIM 125255) | Decorin |
| BTG1 (MIM 109580) | B-cell translocation gene |
| EEA1 (MIM 605070) | Early endosome antigen 1 |
| SOC2_Human | Unknown |
| MRPL42 | Unknown |
| UBE2N (MIM 603679) | Ubiquitin-conjugating enzyme |
| PGAM1 (MIM 172250) | Phosphoglyceric acid mutase |
| CRADD (MIM 603454) | Casp2 and Ripk1 domain-containing adaptor with death domain |
| PLXNC1 (MIM 604259) | Plexin C1 |
| NR2C1 (MIM 601529) | Nuclear receptor subfamily 2, group C, member 1 |
| N7BM_Human | Unknown |
| VEZA_Human | Unknown |
| NTN4 | Netrin 4 |
| SNRPF (MIM 603541) | Small nuclear ribonucleoprotein polypeptide F |
| HAL (MIM 235800) | Histidinemia |
| LTA4H (MIM 151570) | Leukotriene A4 hydroxylase |
| ELK3 (MIM 600247) | ETS-domain protein |
| APAF1 (MIM 602233) | Apoptotic protease activating factor 1 |
| D12S346 | |
| NR1H4 (MIM 603826) | Nuclear receptor subfamily 1, group H, member 4 |
| ARL1 | ADP-ribosylation factor-like 1 |
| MYBPC1 (MIM 160794) | Myosin-binding protein C, slow type |
| CHPT1 | Unknown |
| AD16_Human | Unknown |
| IGF1 (MIM 147440) | Insulin-like growth factor 1 |
| PAH (MIM 261600) | Phenylalanine hydroxylase |
| ASCL1 (MIM 100790) | Achaete-scute complex, Drosophila, homolog-like 1 |
| TRA1 (MIM 191175) | Tumor rejection antigen-1 |
| TDG (MIM 601423) | Thymine-DNA glycosylase |
| PGAM1 (MIM 172250) | Phosphoglycerate mutase 1 |
| NFYB (MIM 189904) | Nuclear transcription factor Y, β |
| TXNRD1 (MIM 601112) | Thioredoxin reductase 1 |
Note.— The given loci are located between 90.4 and 103.9 Mb on the chromosome 12 Ensembl physical map.
In summary, our linkage analysis of ARM in the BDES population appears to provide evidence that markers located on chromosome 12 may be linked to ARM. Our chromosome 12 results are supported by a previous study of possible linkage between ARM and markers located in a similar region (Weeks et al. 2000). Furthermore, because we observed a weak signal on chromosome 1, we were unable to exclude either candidate-gene region on chromosome 1, but we conclude that these regions do not have a major effect on ARM pathogenesis in the Beaver Dam population.
Acknowledgments
This study was supported, in part, by U.S. Public Health Service research grants GM28356, from the National Institute of General Medical Sciences, and U10-EY06594 and EY10605, from the National Eye Institute; training grant HL07567, from the National Heart, Lung, and Blood Institute; and resource grant RR03655, from the National Center for Research Resources.
Electronic-Database Information
URLs for data presented herein are as follows:
- Center for Medical Genetics, http://research.marshfieldclinic.org/genetics/
- Ensembl Genome Browser, http://www.ensembl.org/
- Harvard Partners Genome Center, http://www.hpcgg.org/Sequence/human.html
- Online Mendelian Inheritance in Man (OMIM), http://www.ncbi.nlm.nih.gov/Omim/(for candidate genes related to the chromosome 12 region of interest)
- Unified Database for Human Genome Mapping, The, http://genecards.weizmann.ac.il/udb/
References
- Allikmets R, International ABCR Screening Consortium (2000) Further evidence for an association of ABCR alleles with age-related macular degeneration. Am J Hum Genet 67:487–491 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allikmets R, Sing N, Sun H, Shroyer NF, Hutchinson A, Chidambaram A, Gerrard B, Baird L, Stauffer D, Peiffer A, Rattner A, Smallwood P, Li Y, Anderson KL, Lewis RA, Nathans J, Leppert M, Dean M, Lupski JR (1997) A photoreceptor cell-specific ATP-binding transporter gene (ABCR) is mutated in recessive Stargardt macular dystrophy. Nat Genet 15:236–246 [DOI] [PubMed] [Google Scholar]
- Bird AC, Bressler NM, Bressler SB, Chisholm IH, Coscas G, Davis MD, de Jong PT, Klaver CC, Klein BEK, Klein R, Mitchell P, Sarks JP, Sarks SH, Soubrane G, Taylor HR, Vingerling JR (1995) An international classification and grading system for age-related maculopathy and age-related macular degeneration: the International ARM Epidemiological Study Group. Surv Ophthalmol 39:367–374 [DOI] [PubMed] [Google Scholar]
- Bonney GE (1984) On the statistical determination of major gene mechanisms in continuous human traits: regressive models. Am J Med Genet 18:731–739 [DOI] [PubMed] [Google Scholar]
- Box GEP, Cox DR (1964) An analysis of transformations (with discussion). J R Stat Soc B 26:211–252 [Google Scholar]
- Bressler NM, Bressler SB, West SK, Fine SL, Taylor HR (1989) The grading and prevalence of macular degeneration in Chesapeake Bay watermen. Arch Ophthalmol 107:847–852 [DOI] [PubMed] [Google Scholar]
- Bressler NM, Munoz B, Maguire MG, Vitale SE, Schein OD, Taylor HR, West SK (1995) Five-year incidence and disappearance of drusen and retinal pigment epithelial abnormalities: Waterman Study. Arch Ophthalmol 113:301–308 [DOI] [PubMed] [Google Scholar]
- Cruickshanks KJ, Hamman RF, Klein R, Nondahl DM, Shetterly SM (1997) The prevalence of age-related maculopathy by geographic region and ethnicity: the Colorado-Wisconsin Study of Age-Related Maculopathy. Arch Ophthalmol 115:242–250 [DOI] [PubMed] [Google Scholar]
- Daugas E, Susin SA, Zamzami N, Ferri KF, Irinopoulou T, Larochette N, Prevost MC, Leber B, Andrews D, Penninger J, Kroemer G (2000) Mitochondrio-nuclear translocation of AIF in apoptosis and necrosis. FASEB J 14:729–739 [PubMed] [Google Scholar]
- De La Paz MA, Guy VK, Abou-Donia S, Heinis R, Bracken B, Vance JM, Gilbert JR, Gass JDM, Haines JL, Pericak-Vance MA (1999) Analysis of the Stargardt disease gene (ABCR) in age-related macular degeneration. Ophthalmology 106:1531–1536 [DOI] [PubMed] [Google Scholar]
- Dosso AA, Bovet J (1992) Monozygotic twin brothers with age-related macular degeneration. Ophthalmologica 205:24–28 [DOI] [PubMed] [Google Scholar]
- Elston RC (1979) Major locus analysis for quantitative traits. Am J Hum Genet 31:655–661 [PMC free article] [PubMed] [Google Scholar]
- Elston RC, Buxbaum S, Jacobs KB, Olson JM (2000) Haseman and Elston revisited. Genet Epidemiol 19:1–17 [DOI] [PubMed] [Google Scholar]
- Elston RC, Shete S (2000) Adding power to Haseman and Elston’s (1972) method. GeneScreen 1:63–65 [Google Scholar]
- Gibson JM, Rosenthal AR, Lavery J (1985) A study of the prevalence of eye disease in the elderly in an English community. Trans Ophthalmol Soc UK 104:196–203 [PubMed] [Google Scholar]
- Gomes I, Collins A, Lonjou C, Thomas NS, Wilkinson J, Watson M, Morton N (1999) Hardy-Weinberg quality control. Ann Hum Genet 63:535–538 [DOI] [PubMed] [Google Scholar]
- Gourieroux C, Monfort A, Trognon A (1984) Pseudo maximum likelihood methods: theory. Econometrica 52:682–700 [Google Scholar]
- Heiba IM, Elston RC, Klein BEK, Klein R (1994) Sibling correlations and segregation analysis of age-related maculopathy: the Beaver Dam Eye Study. Genet Epidemiol 11:51–67 [DOI] [PubMed] [Google Scholar]
- Hoyng CB, Poppelaars F, van de Pol TJR, Kremer H, Pickers AJLG, Deutman AF, Cremers FPM (1996) Genetic fine mapping of the gene for recessive Stargardt disease. Hum Genet 98:500–504 [DOI] [PubMed] [Google Scholar]
- Huber PJ (1967) The behaviour of maximum likelihood estimates under nonstandard conditions. In: Proceedings of the Fifth Berkeley Symposium on Mathematical Statistics and Probability, University of California Press, Berkeley, pp 221–223 [Google Scholar]
- Kahn HA, Leibowitz HM, Ganley JP, Kini MM, Colton T, Nickerson RS, Dawber TR (1977a) The Framingham Eye Study. I. Outline and major prevalence findings. Am J Epidemiol 106:17–32 [DOI] [PubMed] [Google Scholar]
- ——— (1977b) The Framingham Eye Study. II. Association of ophthalmic pathology with single variables previously measured in the Framingham Heart Study. Am J Epidemiol 106:33–41 [DOI] [PubMed] [Google Scholar]
- Klein BE, Klein R (1982) Cataracts and macular degeneration in older Americans. Arch Ophthalmol 100:571–573 [DOI] [PubMed] [Google Scholar]
- Klein BE, Klein R, Linton KLP, Magli YL, Neider MW (1990) Assessment of cataracts from photographs in the Beaver Dam Eye Study. Ophthalmology 97:1428–1433 [DOI] [PubMed] [Google Scholar]
- Klein ML, Schultz DW, Edwards A, Matise TC, Rust K, Berselli CB, Trzupek K, Weleber RG, Ott J, Wirtz MK, Acott TS (1998) Age-related macular degeneration: clinical features in a large family and linkage to chromosome 1q. Arch Ophthalmol 116:1082–1088 [DOI] [PubMed] [Google Scholar]
- Klein R, Clegg L, Cooper LS, Hubbard LD, Klein BEK, King WN, Folsom AR (1999a) Prevalence of age-related maculopathy in the Atherosclerosis Risk in Communities Study. Arch Ophthalmol 117:1203–1210 [DOI] [PubMed] [Google Scholar]
- Klein R, Davis MD, Magli YL, Klein BEK (1991a) Wisconsin Age-Related Maculopathy Grading System. Department of Ophthalmology, University of Wisconsin School of Medicine, Madison, WI (National Technical Information Service accession number PB91-184267/AS) [Google Scholar]
- Klein R, Davis MD, Magli YL, Segal P, Hubbard L, Klein BEK (1991b) The Wisconsin Age-Related Maculopathy Grading System. Ophthalmology 98:1128–1134 [DOI] [PubMed] [Google Scholar]
- Klein R, Klein BEK (1991) Beaver Dam Eye Study: manual of operations. U.S. Department of Commerce, Springfield, VA (National Technical Information Service accession number PB91149823) [Google Scholar]
- ——— (1995) Beaver Dam Eye Study II: manual of operations. U.S. Department of Commerce, Springfield, VA (National Technical Information Service accession number PB95273827) [Google Scholar]
- Klein R, Klein BEK, Jensen SC, Mares-Perlman JA, Cruickshanks KJ, Palta M (1999b) Age-related maculopathy in a multiracial United States population: the National Health and Nutrition Examination Survey III. Ophthalmology 106:1056–1065 [DOI] [PubMed] [Google Scholar]
- Klein R, Klein BE, Jensen SC, Meuer SM (1997) The five-year incidence and progression of age-related maculopathy: the Beaver Dam Eye Study. Ophthalmology 104:7–21 [DOI] [PubMed] [Google Scholar]
- Klein R, Klein BEK, Lee KE, Cruickshanks KJ, Chappell RJ (2001) Changes in visual acuity in a population over a 10-year period: the Beaver Dam Eye Study. Ophthalmology 108:1757–1766 [DOI] [PubMed] [Google Scholar]
- Klein R, Klein BEK, Linton KLP (1992) Prevalence of age-related maculopathy: the Beaver Dam Eye Study. Ophthalmology 99:933–943 [DOI] [PubMed] [Google Scholar]
- Klein R, Klein BEK, Linton KLP, De Mets DL (1991c) The Beaver Dam Eye Study: visual acuity. Ophthalmology 98:1310–1315 [DOI] [PubMed] [Google Scholar]
- Klein R, Klein BEK, Tomany SC, Meuer SM, Huang G-H (2002) Ten-year incidence and progression of age-related maculopathy: the Beaver Dam Eye Study. Ophthalmology 109:1767–1779 [DOI] [PubMed] [Google Scholar]
- Klein R, Wang Q, Klein BE, Moss SE, Meuer SM (1995b) The relationship of age-related maculopathy, cataract, and glaucoma to visual acuity. Invest Ophthalmol Vis Sci 36:182–191 [PubMed] [Google Scholar]
- Kroemer G, Reed JC (2000) Mitochondrial control of cell death. Nat Med 6:513–519 [DOI] [PubMed] [Google Scholar]
- Leist M, Single B, Naumann H, Fava E, Simon B, Kuhnle S, Nicotera P (1999) Inhibition of mitochondrial ATP generation by nitric oxide switches apoptosis to necrosis. Exp Cell Res 249:396–403 [DOI] [PubMed] [Google Scholar]
- Meyers SM (1994) A twin study on age-related macular degeneration. Trans Am Ophthalmol Soc 92:775–843 [PMC free article] [PubMed] [Google Scholar]
- Meyers SM, Greene T, Gutman FA (1995) A twin study of age-related macular degeneration. Am J Ophthalmol 120:757–766 [DOI] [PubMed] [Google Scholar]
- Meyers SM, Zachary AA (1988) Monozygotic twins with age-related macular degeneration. Arch Ophthalmol 106:651–653 [DOI] [PubMed] [Google Scholar]
- Mitchell P, Smith W, Attebo K, Wang JJ (1995) Prevalence of age-related maculopathy in Australia: the Blue Mountains Eye Study. Ophthalmology 102:1450–1460 [DOI] [PubMed] [Google Scholar]
- Olson JM (1999) A general conditional-logistic model for affected-relative-pair linkage studies. Am J Hum Genet 65:1760–1769 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Putter H, Sandkuijl LA, van Houwelingen JC (2002) Score test for detecting linkage to quantitative traits. Genet Epidemiol 22:345–355 [DOI] [PubMed] [Google Scholar]
- Schachat AP, Hyman L, Leske MC, Connell AM, Wu SY (1995) Features of age-related macular degeneration in a black population: the Barbados Eye Study Group. Arch Ophthalmol 113:728–735 [DOI] [PubMed] [Google Scholar]
- Schick JH, Iyengar SK, Elston RC, Fijal BA, Klein BEK, Klein R (2001) The genetic epidemiology of age-related maculopathy. Int J Hum Genet 1:11–24 [Google Scholar]
- Shete S, Jacobs KB, Elston RC. Adding further power to the Haseman and Elston method for detecting linkage in larger sibships: weighting sums and differences. Hum Hered (in press) [DOI] [PubMed] [Google Scholar]
- Small KW, Udar N, Yelchits S, Klein R, Garcia C, Gallardo G, Puech B, Puech V, Saperstein D, Lim J, Haller J, Flaxel C, Kelsell R, Hunt D, Evans K, Lennon F, Pericak-Vance M (1999) North Carolina macular dystrophy (MCDR1) locus: a fine resolution genetic map and haplotype analysis. Mol Vis 5:38 [PubMed] [Google Scholar]
- Small KW, Weber JL, Roses A, Lennon F, Vance JM, Pericak-Vance MA (1992) North Carolina macular dystrophy is assigned to chromosome 6. Genomics 13:681–685 [DOI] [PubMed] [Google Scholar]
- Souied EH, Ducroq D, Rozet JM, Gerber S, Perrault I, Munnich A, Coscas G, Soubrane G, Kaplan J (2000) ABCR gene analysis in familial exudative age-related macular degeneration. Invest Ophthalmol Vis Sci 41:244–247 [PubMed] [Google Scholar]
- Sparrow JM, Dickinson AJ, Duke AM, Thompson JR, Gibson JM, Rosenthal AR (1997) Seven year follow-up of age-related maculopathy in an elderly British population. Eye 11:315–324 [DOI] [PubMed] [Google Scholar]
- Stone EM, Lotery AJ, Munier FL, Héon E, Piguet B, Guymer RH, Vandenburgh K, Cousin P, Nishimura D, Swiderski RE, Silvestri G, Mackey DA, Hageman GS, Bird AC, Sheffield VC, Schorderet DF (1999) A single EFEMP1 mutation associated with both Malattia Leventinese and Doyne honeycomb retinal dystrophy. Nat Genet 22:199–202 [DOI] [PubMed] [Google Scholar]
- Stone EM, Nichols BE, Streb LM, Kimura AE, Sheffield VC (1992) Genetic linkage of vitelliform macular degeneration (Best’s disease) to chromosome 11q13. Nat Genet 1:246–250 [DOI] [PubMed] [Google Scholar]
- Susin SA, Lorenzo HK, Zamzami N, Marzo I, Snow BE, Brothers GM, Mangion J, Jacotot E, Costantini P, Loeffler M, Larochette N, Goodlett DR, Aebersold R, Siderovski DP, Penninger JM, Kroemer G (1999) Molecular characterization of mitochondrial apoptosis-inducing factor. Nature 397:441–446 [DOI] [PubMed] [Google Scholar]
- Suter M, Remé C, Grimm C, Wenzel A, Jäättela M, Esser P, Kociok N, Leist M, Richter C (2000) Age-related macular degeneration: the lipofuscin component N-retinyl-N-retinylidene ethanolamine detaches proapoptotic proteins from mitochondria and induces apoptosis in mammalian retinal pigment epithelial cells. J Biol Chem 275:39625–39630 [DOI] [PubMed] [Google Scholar]
- Tarttelin EE, Gregory-Evans CY, Bird AC, Weleber RG, Klein ML, Blackburn J, Gregory-Evans K (2001) Molecular genetic heterogeneity in autosomal dominant drusen. J Med Genet 38:381–384 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor HR, West S, Munoz B, Rosenthal FS, Bressler SB, Bressler NM (1992) The long-term effects of visible light on the eye. Arch Ophthalmol 110:99–104 [DOI] [PubMed] [Google Scholar]
- Tielsch JM (1995) Vision problems in the U.S.: a report on blindness and vision impairment in adults age 40 and older. Society to Prevent Blindness, Schaumburg, IL, pp 1–20 [Google Scholar]
- Vingerling JR, Dielemans I, Bots ML, Hofman A, Grobbee DE, de Jong PT (1995) Age-related macular degeneration is associated with atherosclerosis: the Rotterdam Study. Am J Epidemiol 142:404–409 [DOI] [PubMed] [Google Scholar]
- Weeks DE, Conley YP, Tsai HJ, Mah TS, Rosenfeld PJ, Paul TO, Eller AW, Morse LS, Dailey JP, Ferrell RE, Gorin MB (2001) Age-related maculopathy: an expanded genome-wide scan with evidence of susceptibility loci within the 1q31 and 17q25 regions. Am J Ophthalmol 132:682–692 [DOI] [PubMed] [Google Scholar]
- Weeks DE, Conley YP, Mah TS, Paul TO, Morse L, Ngo-Chang J, Dailey JP, Ferrell RE, Gorin MB (2000) A full genome scan for age-related maculopathy. Hum Mol Genet 9:1329–1349 [DOI] [PubMed] [Google Scholar]
- Wilson AF, Elston RC, Tran LD, Siervogel RM (1991) Use of the robust sib-pair method to screen for single-locus, multiple-locus, and pleiotropic effects: application to traits related to hypertension. Am J Hum Genet 48:862–872 [PMC free article] [PubMed] [Google Scholar]
- Yates JRW, Moore AT (2000) Genetic susceptibility to age related macular degeneration. J Med Genet 37:83–87 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshimura S, Suemizu H, Taniguchi Y, Arimori K, Kawabe N, Moriuchi T (1994) The human plasma glutathione peroxidase-encoding gene: organization, sequence and localization to chromosome 5q32. Gene 145:293–297 [DOI] [PubMed] [Google Scholar]