Abstract
Minisatellite MS1 (locus D1S7) is one of the most unstable minisatellites identified in humans. It is unusual in having a short repeat unit of 9 bp and in showing somatic instability in colorectal carcinomas, suggesting that mitotic replication or repair errors may contribute to repeat-DNA mutation. We have therefore used single-molecule polymerase chain reaction to characterize mutation events in sperm and somatic DNA. As with other minisatellites, high levels of instability are seen only in the germline and generate two distinct classes of structural change. The first involves large and frequently complex rearrangements that most likely arise by recombinational processes, as is seen at other minisatellites. The second pathway generates primarily, if not exclusively, single-repeat changes restricted to sequence-homogeneous regions of alleles. Their frequency is dependent on the length of uninterrupted repeats, with evidence of a hyperinstability threshold similar in length to that observed at triplet-repeat loci showing expansions driven by dynamic mutation. In contrast to triplet loci, however, the single-repeat changes at MS1 exclusively involve repeat deletion, and can be so frequent—as many as 0.7–1.3 mutation events per sperm cell for the longest homogeneous arrays—that alleles harboring these long arrays must be extremely ephemeral in human populations. The apparently impossible existence of alleles with deletion-prone uninterrupted repeats therefore presents a paradox with no obvious explanation.
Introduction:
Human GC-rich minisatellites are highly variable tandem-repetitive DNA sequences located predominantly in subtelomeric regions of the genome (Jeffreys et al. 1985; Royle et al. 1988). Alleles can be as long as 30 kb, with repeat-unit lengths of 6–100 bp (Jeffreys et al. 1995). New alleles at these highly unstable loci generally arise by germline-specific complex recombination events, including both interallelic gene conversions and intra-allelic rearrangements (Armour et al. 1993; Buard and Vergnaud 1994; Jeffreys et al. 1994; Tamaki et al. 1999, reviewed by Bois and Jeffreys [1999]), with most loci showing a bias toward mutation in the male germline (Henke and Henke 1995; Dubrova et al. 1997). Mutants are also generated via a crossover pathway, but at a lower frequency (Jeffreys et al. 1998b; Buard et al. 2000b). Some loci show polarity, with mutation events targeted to one end of the repeat array (Jeffreys et al. 1994). Analysis of one minisatellite has shown that this polarity correlates with a highly localized meiotic recombination hotspot, adjacent to the repeat DNA, which appears to drive repeat-DNA instability (Jeffreys et al. 1998a). New alleles are also generated in somatic cells but are far less frequent than in the germline and arise by simple intra-allelic duplications and deletions (May et al. 1996; Jeffreys and Neumann 1997; Buard et al. 2000a). Germline mutation frequencies vary not only between minisatellites but also between alleles at a given locus (Monckton et al. 1994; May et al. 1996; Buard et al. 1998; Tamaki et al. 1999), and, in one case, this variation has been linked to a flanking polymorphism that appears to influence hotspot activity (Jeffreys et al. 1998a). The most dramatic variation is observed at CEB1, where sperm mutation rates vary from <0.05% to 25% per allele, the latter being the highest rate yet observed at a minisatellite (Buard et al. 1998).
Minisatellite MS1 (D1S7) is one of the most unstable human minisatellites yet identified and is unusual in having a short repeat unit of 9 bp. It is located in the subtelomeric region of chromosome 1 (1p33-35) and shows alleles ranging from 60 to >1,000 repeats, with an allele-length heterozygosity >99% (Wong et al. 1987; Royle et al. 1988). The germline mutation rate has been estimated in families at 5.2% per gamete (Jeffreys et al. 1988), although this will be an underestimate, given the short repeat unit and long arrays, which make the detection of small mutational changes extremely difficult. In contrast to most minisatellites, mutation occurs with similar frequencies in both the male and female germline (Jeffreys et al. 1988; Henke and Henke 1995; Dubrova et al. 1997). MS1 has been extensively used in forensic analyses (Wong et al. 1987) and is the only minisatellite known to be unstable in colon cancer cells that show microsatellite instability (Hoff-Olsen et al. 1995; see Mitchell et al. 2002), suggesting that replication or repair errors might contribute to MS1 repeat instability.
MS1 instability processes have been studied only in Saccharomyces cerevisiae, after integration of the human minisatellite near a recombination hotspot upstream of the yeast LEU2 locus (Cederberg et al. 1993; Maleki et al. 1997; Berg et al. 2000). In yeast, MS1 shows both mitotic and meiotic instability at high frequencies. This is in contrast to other minisatellites studied in the same system, such as MS32 (Appelgren et al. 1997) and MS205 (He et al.1999), which mutate at high frequency only in meiosis. In mitosis, MS1 instability is related not only to allele length, with evidence for an instability threshold of 0.75 kb (Maleki et al. 1997), but also to the internal structure of the alleles. Tetrad analysis showed that mutants arising at meiosis are generated by both intra- and interallelic recombination events and that most mutants arise by gene conversions that include the minisatellite plus neighboring DNA (Berg et al. 2000).
The evidence, both from yeast and from tumors, therefore suggests that minisatellite MS1 may be unusual in showing both meiotic and mitotic repeat instability. However, there is no direct information available on MS1 mutation processes in humans. We have therefore used both small-pool (SP) PCR (Jeffreys et al. 1994) and single-molecule (SM) PCR (Yauk et al. 2002) to analyze the incidence of de novo mutant molecules in sperm and somatic DNA. We then used minisatellite variant repeat (MVR) mapping by PCR (Jeffreys et al 1991; Berg et al. 2000) to characterize the structural basis of mutation within the repeat array. Although this work has shown that MS1 mutation does share some features in common with other GC-rich minisatellites, it has also revealed an entirely novel germline-specific instability process that rapidly eliminates certain classes of alleles, rendering their existence in human populations an enigma.
Material and Methods
Mutant Detection and Purification
All DNAs were prepared, as described elsewhere, under conditions designed to minimize the risk of DNA contamination (Jeffreys et al. 1994). Genomic DNAs were digested with MboI prior to use. DNA concentrations were estimated by use of UV spectrometry, and the number of amplifiable molecules of each MS1 allele was confirmed by Poisson analysis of limiting single-molecule dilutions of digested DNA (Jeffreys et al. 2000). Extreme dilutions of genomic DNA were made in 5 mM Tris-HCl (pH 7.5) and 5 μg/ml carrier herring sperm DNA prior to PCR amplification.
SP-PCR analyses were performed on multiple aliquots of digested genomic DNA, each containing 30–80 amplifiable molecules of each MS1 allele (0.22–0.58 ng DNA per PCR). DNA was amplified by long PCR using Taq plus Pfu polymerases, as described elsewhere (Jeffreys et al. 1998b), with 0.2 μM primers MS1−420 (5′-AGGTCTCTAGCATAGTGCTTGGCACAG-3′) and MS1+280 (5′-CATTTTACAGATAGGGAAACTGACAGC-3′). These primers generate an MS1 amplicon containing 410 bp of DNA 5′ to the repeat array plus 310 bp of DNA 3′ to the array. Cycling was for 1 min at 96°C, followed by 25 cycles at 96°C for 20 s, 60°C for 30 s, and 70°C for 2.5 min. PCR products were electrophoresed through a 40-cm 0.8% SeaKem LE (FMC Bioproducts) agarose gel in 44 mM Tris-borate (pH 8.3) and 1 mM EDTA and was then transferred to a nylon membrane (Hybond N, Amersham) and hybridized with a 32P-labeled MS1 probe (Wong et al. 1987). SM-PCR analyses were similarly performed on multiple aliquots of genomic DNA, each containing 4.0 pg DNA (0.55 amplifiable molecule), with DNA amplified for 25 cycles. Positive SM-PCRs were identified by Southern blot hybridization with the MS1 probe; they were then reamplified for 11–14 cycles, using the nested primers MS1−350 (5′-CCTTTGCCATTTCCATAAACACGTATC-3′) and MS1-B (5′-AAGAAGCATATGCAACCCATGAGG-3′), and PCR products were electrophoresed side by side on 40-cm gels, as for SP-PCR analyses.
Mutants detected in SP-PCRs were purified away from the progenitor allele by two or three rounds of agarose gel electrophoresis, collection of size fractions around the position of the mutant, limited reamplification of each fraction for 6–15 cycles with primers MS1−420 and MS1+280, and analysis of PCR products by gel electrophoresis and hybridization. Final mutant products were at least 95% pure and were reamplified for 11–24 cycles with the nested primers prior to MVR analysis. Full details of the purification procedure were reported elsewhere (Jeffreys and Neumann 1997).
MVR-PCR Analysis
MVR-PCR analysis of progenitor alleles, purified mutants, and PCR products from individual SM-PCR was performed as described elsewhere (Berg et al. 2000), with some modifications. All primers were purchased from Scandinavian Gene Synthesis and were >99% full length. Forward mapping used 1 μM flanking primer 1-HF (5′-CACGCTCATTTGCCATTGATTTTAAGT-3′), located 263 bp into the 5′ flanking region of MS1, together with 2–5 fg PCR product, 0.05 U/μl Taq polymerase, 10 nM 1-TAG-A primer, 1 nM 1-TAG-B primer, 1 nM 1-TAG-K primer or 10–20 nM 1-TAG-C primer depending on the number of C-type repeats, plus 1 μM TAG (5′-TCATGCGTCCATGGTCCGGA-3′). The MVR primers 1-TAG-A, -B, -K, and -C are described elsewhere (Berg et al. 2000). DNA was amplified at 96°C for 15 s, 50°C for 35 s, and 70°C for 3.3 min for 4 cycles, and then underwent 13 cycles of 96°C for 15 s, 61°C for 35 s, and 70°C for 3.3 min, followed by a chase at 70°C for 10 min. Reverse mapping was performed as for forward mapping, with the same primer concentrations, using the following primers: 3′ flanking primer 1-HR (5′-AGGACCACCCAATCTGGGCTCCCA-3′), located 25 bp downstream of MS1, plus TAG and one of the following A, B, C, or K repeat–specific primers (the synthetic TAG 5′ extension is indicated in lowercase): 1-R-TAG-A (5′-tcatgcgtccatggtccggaCCCT[A/G]TCCACCCT[A/G]TCCACCCTA-3′; 1-R-TAG-B (5′-tcatgcgtccatggtccggaCCCT[A/G]TCCACCCT[A/G]TCCACCCTC-3′); 1-R-TAG-C (5′-tcatgcgtccatggtccggaCCCT[A/G]TCCACCCT[A/G]TCCACCCTG-3′); or 1-R-TAG-K (5′-tcatgcgtccatggtccggaCCT[A/G]TCCACCCT[A/G]TCCACCCTAA-3′). MVR-PCR products from the four different reactions were analyzed side by side by agarose gel electrophoresis and Southern blot hybridization with the 32P-labeled MS1 probe. Forward and reverse MVR codes read from autoradiographs were merged into a complete allele structure. Repeat units that failed to amplify with any of the MVR primers (because of the presence of additional variant[s] that block primer binding) were classified as O-type repeats. MVR maps are oriented as in the article by Gray and Jeffreys (1991).
Results
Detecting MS1 Mutants in Human DNA
Given the small size of the MS1 repeat unit, we focused our attention on short alleles to maximize the resolution of mutants with altered numbers of repeats. We screened a panel of 98 semen donors of northern European origin and identified 11 men who were heterozygous for a large allele plus a short allele of <160 repeats suitable for mutation analysis. Similarly, analysis of 68 Zimbabwean semen donors identified 11 short alleles, with one man heterozygous for two different short alleles.
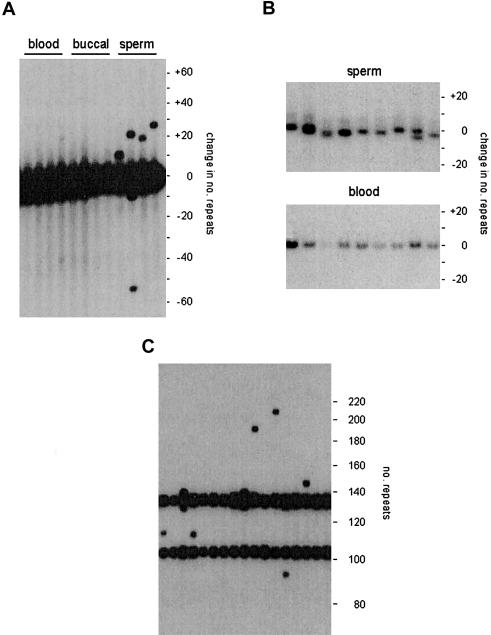
We used SP-PCR (Jeffreys et al. 1994) to analyze sperm DNA from one of the northern European donors (man 1) for whom blood and buccal DNA was also available. Man 1 was heterozygous for an allele 140 repeats long plus a second allele of ∼1,000 repeats, too large for mutation analysis (fig. 1A). Mutants of the short allele involving the gain or loss of at least five repeats (the resolution limit for detecting changes in man 1 by SP-PCR) could be detected at a frequency of 1.5% per sperm cell (54 mutants in 3,500 progenitor molecules screened). Mutational changes of the small allele ranged from a loss of 54 repeats to a gain of 110 repeats. Mutation was biased toward expansion, with 85% of mutants showing gains of repeats. Analysis of blood DNA and buccal DNA (5,100 and 2,900 molecules screened respectively) revealed no detectable mutants, indicating a low frequency of changes in somatic DNA (<0.06% and <0.1% for blood and buccal DNA, respectively; P>.95). Thus, as with other minisatellites, instability at MS1 resulting in large length changes is mainly restricted to the germline.
Figure 1.
Detecting MS1 mutants in genomic DNA. A, Detection of large mutational changes by SP-PCR. Aliquots of blood, buccal, and sperm DNA from the same individual, each containing 50 amplifiable molecules of each allele (140 and ∼1,000 repeats), were amplified by PCR, using primers flanking the minisatellite, and the products were analyzed by agarose gel electrophoresis and Southern blot hybridization, with a 32P-labeled MS1 probe. The numbers of repeats gained or lost, relative to the size of the smaller allele, are shown at right. B, Detection of small length changes by SM-PCR. Multiple aliquots of sperm and blood DNA, each containing 0.55 amplifiable molecules of each allele, were amplified and tested for positive reactions, which were then analyzed side by side, as in A. C, SP-PCR analysis of sperm DNA from man 2, who was heterozygous for 104 and 134 repeat MS1 alleles. Each PCR contained 16 amplifiable molecules of each allele.
We used SM-PCR (Yauk et al. 2002) to analyze smaller length changes at MS1. This method uses amplification of minisatellite molecules from extreme dilutions of genomic DNA such that 98% of reactions will contain, at most, only one or two amplifiable molecules. Analysis of 120 molecules from sperm DNA revealed considerable allele length heterogeneity (fig. 1B), with molecules varying in length by as many as four repeats and with some PCRs showing two different length molecules. Although this heterogeneity indicates considerable instability, the small length changes made it impossible to identify progenitor alleles with confidence, preventing the estimation of mutation rate from allele length changes. In contrast, analysis of blood DNA (120 molecules screened; fig. 1B) and buccal DNA (54 molecules analyzed; data not shown) showed no evidence of such small length changes. Thus, as with large mutational changes, these small rearrangements appear to be largely, if not completely, restricted to sperm DNA.
The Structural Basis of Large Mutational Changes in Sperm DNA
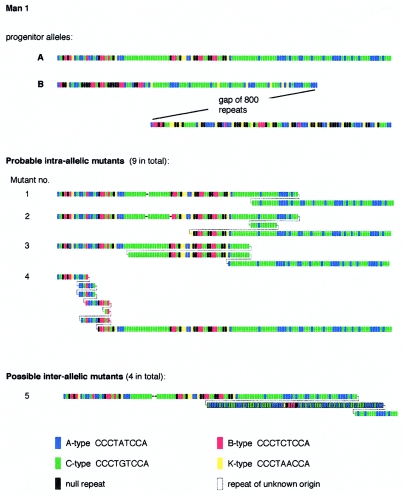
We used an MVR-PCR system for analyzing the interspersion patterns of five variant repeat types (A, B, C, K, and O) along MS1 alleles (Berg et al. 2000) to compare the structures of 13 different sperm mutants identified by SP-PCR analysis with the structures of the progenitor alleles (fig. 2). The shorter progenitor allele A of 140 repeats was mapped in its entirety, whereas only the terminal 100 repeats from each end of the larger ∼1,000-repeat allele B could be mapped. Comparison of mutant and progenitor alleles showed that most mutants appeared to be derived by intra-allelic rearrangements within the smaller allele. In some cases (e.g., mutant 1; fig. 2), the rearrangement involved a simple and perfect duplication of a block of repeats. In most cases, however, these apparently intra-allelic rearrangements were complex, with multiple imperfect reduplications of blocks of repeats that appeared to be derived solely from the smaller allele (e.g., mutant 4). Four mutants showed possible evidence for interallelic transfer of information, as indicated by rearrangements within the 3′ region of the smaller allele, which consists exclusively of A and C repeats, involving insertion of a block of repeats of unclear origin, including non-A/C repeat types (e.g. mutant 5). The incomplete structure of the larger progenitor allele, however, made it impossible to confirm that these mutation events involved interallelic transfer of repeats.
Figure 2.
Structures of large size-change MS1 mutations isolated from sperm DNA of man 1. Representative examples of the 13 mutants analyzed are shown, with different repeat types color-coded as indicated. The very large progenitor allele B was only partially mapped. Deletions are indicated by dashes, and duplications by splitting the mutant structure to show likely or definite alignments between duplicated regions identified by eye. Repeats of uncertain origin are outlined in black.
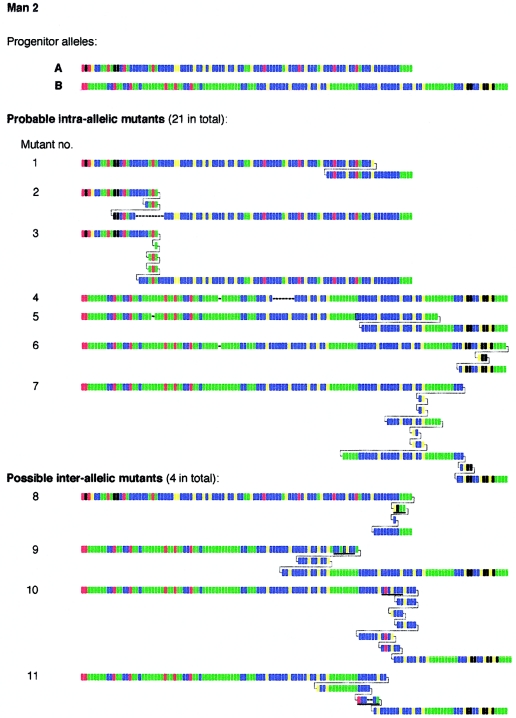
To investigate large rearrangements in more detail, we analyzed sperm DNA from the Zimbabwean man (man 2) who was heterozygous for two short MS1 alleles, of 104 and 134 repeats. Both alleles in man 2 were MVR mapped in their entirety, revealing very different repeat structures (fig. 3). SP-PCR analysis of 5,000 amplifiable molecules of each progenitor allele yielded 98 sperm mutants with gains or losses of at least four repeats, giving a mean sperm mutation rate of 1.0% per allele, similar to that seen in man 1. MVR analysis of 25 mutants from man 2 showed that 21 appeared to have arisen by sometimes complex intra-allelic rearrangements within one or another allele (fig. 3). The remaining four mutants, however, did show some evidence of interallelic transfer, with short anomalous blocks of repeats within regions of complex intra-allelic duplication. When the 3′ ends of the two progenitor alleles were aligned, these anomalous blocks matched corresponding blocks in the other allele. Similar, though more definitive, in-register transfers of repeats between end-aligned alleles have been seen during sperm mutation at other minisatellites (Jeffreys et al. 1994).
Figure 3.
Representative examples of large size change MS1 sperm mutations recovered from man 2. Underlined blocks of repeats in putative interallelic mutants are those that match the other allele when both alleles are aligned at their 3′ ends.
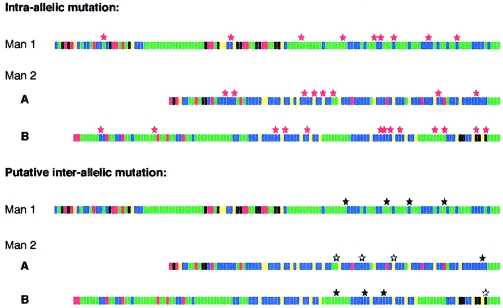
Intra-allelic rearrangements in both men appeared to be fairly randomly scattered along the alleles, but putative interallelic transfers were concentrated toward the 3′ end of the repeat array (fig. 4). However, these intra- and interallelic distributions are not significantly different (one-sample runs test and Kolmogorov-Smirnov test P>.05), and the apparent 3′ clustering of interallelic events may be due, in part, to biased ascertainment, particularly in man 1, in whom the restricted repertoire of A and C repeat types near the 3′ end of the allele facilitates the detection of interallelic transfers.
Figure 4.
Distribution of rearrangement breakpoints along alleles. Breakpoints are shown separately for apparently intra-allelic rearrangements (red stars) and for putative interallelic transfers (black stars). The locations of matching regions in the donor allele in interallelic mutants are also indicated (white stars). Locations are defined as the center point of the repeat block involved in the rearrangement. If a mutant showed multiple events, then the central repeat of the entire region involved was taken as the breakpoint location.
Structural Analysis of Small Mutational Changes in Sperm DNA
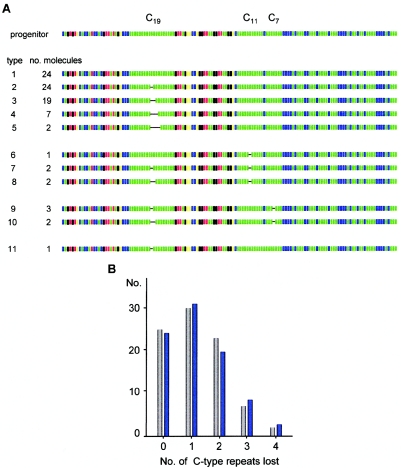
The small mutational changes detected in sperm DNA from man 1 (fig. 1B) were further characterized by MVR-PCR analysis of SM-PCR products, after eliminating reactions that contained more than one length class of molecule (fig. 1B). In total, 87 sperm molecules selected at random were analyzed. Comparison of minisatellite structures with the progenitor allele (defined as the allele structure determined from blood DNA) showed that only 22 of the 87 molecules were nonmutant, giving a mutation frequency of 75% for this allele. The 65 mutant molecules carried a total of 81 distinct mutations that altered repeat copy number. All of these mutations were deletions and were highly nonrandomly distributed, with 72 located in homogeneous stretches of C-type repeats (fig. 5A) and a further 5 affecting repeat units immediately adjacent to these stretches (results not shown). This clustering in C arrays, which make up only 26% of the allele, is highly significant (χ2=63[1df]; P<.001).
Figure 5.
Hyperdeletion within an MS1 allele. A, Structures of randomly chosen MS1 allele A molecules characterized, by use of SM-PCR, in sperm DNA from man 1. Note that 2 of the 24 nonmutants have lost a repeat unit outside the C arrays (not shown). B, Incidence of deletions in the C19 repeat array in this allele. The numbers of single molecules that carry each size of deletion are shown (gray bars), together with the least-squares best-fit Poisson distribution of mutants predicted if each mutation event involves the loss of only a single repeat (blue bars). Under this model, each sperm cell will have accumulated an average of 1.28 mutation events in the C19 repeat array.
The three different C arrays (C19, C11, and C7) in this allele showed very different levels of instability. The C11 and C7 arrays each contained five deletion events, which, in all cases, involved a single repeat unit, giving a mutation rate of 6% (5 mutants in 87 molecules) in each of these homogeneous arrays. Instability in the C19 array was far higher, with 62 of the 87 molecules carrying deletions of one to four repeats, giving a mutation rate of 71%. Nine of the molecules showed deletions both in the C19 array and in either the C11 or C7 array; these double-mutation frequencies are those expected if each class of mutant arises independently in the germline. Similarly, 10 of the 13 large rearrangements characterized in this man (fig. 2) were accompanied by C-array deletions distal to the site of large rearrangement, with 9 of these mapping to the C19 array. The proportion of large mutation molecules carrying C19 deletions (9/13) is not significantly different from the proportion of randomly selected molecules carrying such deletions (62/87) (Fisher's exact test, two-sided, P=1). This suggests that C-array instability can also occur independently of the larger, more complex rearrangements. It therefore follows that multiple sequential mutations could also occur in the C19 array, perhaps by a process involving the strict loss of a single repeat per mutation event, as is seen in the other C arrays. The observed distribution of length changes in the C19 array fits well with this model (fig. 5B) and predicts that an average of 1.3 single-repeat deletion events per sperm cell accumulate in this region. Together with other modes of mutation, the overall level of instability in this allele is extreme, with a combined mutation rate of 80% per sperm cell and perhaps as high as 140%, depending on whether multiple mutations occur within the C19 array.
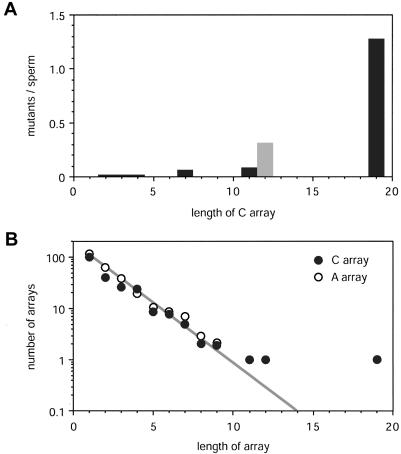
C-array instability in this allele is not atypical. Of the 25 mutants characterized in man 2 (fig. 3), 6 contained single-repeat deletions distal to the site of complex rearrangement. All of these microdeletions were in the 16 mutants derived from allele B, which contains five C arrays ranging from C6 to C12; none were in mutants from allele A, which is devoid of C arrays. All deletions were within the C arrays, with five of the six in the longest C12 array. The deletion rate in C12 is therefore ∼31% (5/16), compared with a mean rate of ∼1.6% in the other four C arrays. The relationship between C-array length and the frequency of deletion in sperm DNA is shown in figure 6, together with the abundance of C arrays in 13 short MS1 alleles analyzed in human populations.
Figure 6.
Instability of C arrays and their prevalence in MS1 alleles. A, Sperm mutation rates in C arrays deduced from SM-PCR analysis of allele A in man 1, under the assumption that mutation steps always involve the loss of a single C-type repeat. No deletions were seen in the two C2, three C3, and two C4 arrays in this allele, giving a mean mutation rate of <0.005 per array (P>.95). The estimate for C12 arrays was taken from the incidence of deletions in this array in large mutants characterized in man 2 (see fig. 3). B, Abundance of C arrays in MS1 alleles. The distribution was determined from alleles in men 1 and 2 and from 10 additional short alleles (64–155 repeats) from northern European men who were fully typed by MVR-PCR (1,720 repeats typed in total, data not shown). For comparison, the abundance of A-repeat clusters is also shown. The log-linear relationship (gray line) between abundance and short array length (<10 repeats) is the same for C- and A-type repeats.
Discussion
Like other GC-rich minisatellites (Jeffreys et al. 1994; May et al. 1996; Buard et al. 1998, 2000a; Tamaki et al. 1999), MS1 shows a mode of repeat unit turnover that is restricted to the germline and frequently involves complex rearrangements biased toward gains of repeat units and resulting in relatively large changes in allele size. All mutants analyzed have different structures, as predicted for products of meiotic recombination, with no evidence of germinal mosaicism that might signal premeiotic mutation. Most of these large mutations appear to be intra-allelic, involving sometimes complex duplications and deletions of blocks of repeat units. There is, however, evidence in man 2 that some mutation events may involve the transfer of short blocks of repeats between alleles aligned at their 3′ ends, accompanied by complex rearrangements in the recipient allele at the site of insertion. Similar, though more definitive, examples of in-phase interallelic gene conversion have been seen at several other human minisatellites, and, for some but not all loci, represent the major mode of germline mutation (Jeffreys et al. 1994; Buard et al. 1998). These putative interallelic events at MS1 appear to be targeted toward the 3′ end of the repeat array, reminiscent of a polarized mutation seen at some minisatellites that in one case has been linked to the presence of a flanking recombination hotspot (Jeffreys et al. 1998a). The more frequent intra-allelic rearrangements may also arise by aberrant processing of recombination-initiating events (Buard et al. 1998).
The structure of these putative interallelic events in sperm DNA contrasts with the repeat turnover processes seen when MS1 is integrated near a hotspot for double-strand breaks upstream of LEU2 in S. cerevisiae (Berg et al. 2000). Meiotic instability in yeast frequently results in recombinant arrays consisting of the 5′ end of one allele fused to the 3′ end of the other allele; similar recombinants are also seen at other human minisatellites integrated into yeast at the same position as MS1 (Appelgren et al. 1997; He et al. 1999). However, there are no examples of such recombinants in the 38 sperm mutants characterized in the present study. Similarly, analysis of other human minisatellites has shown that most sperm recombinants involve DNA transfers solely within the repeat array and that relatively few show fused 5′-3′ recombinant structures (Buard and Vergnaud 1994; Jeffreys et al. 1994, 1998a, 1998b; Buard et al. 1998, 2000b; Tamaki et al. 1999). This difference in the meiotic behavior of minisatellites in humans and yeast could reflect either species-specific differences in the way that recombination-initiating events are subsequently processed in tandem repeat DNA or an effect of genome position (e.g., location of the minisatellite with respect to a recombination hotspot).
Unlike all other minisatellites characterized to date, MS1 shows a second novel mode of instability involving repeat unit deletion within sequence-homogeneous regions of alleles. These deletions are targeted to the longest arrays of C-type repeats. The alleles tested for mutation did not contain long homogeneous arrays of other types of repeats—the longest is an A8 array in allele A of man 2—and, although none of the nine mutants characterized from this allele showed mutation within the A8 array, this could still be compatible with a mutation rate as high as 28% (P=.05) (fig. 3). It is therefore unclear whether instability is targeted specifically to C repeats. Indeed, MS1 alleles contain a similar abundance of A and C arrays, declining in frequency with increasing array length (fig. 6B), suggesting that they are subjected to similar population turnover processes and thus that A arrays might also be highly unstable.
These deletion events in C arrays appear to be restricted to the germline and seem to arise independently of large-scale rearrangements. Only 1 of the 13 large rearrangements characterized in man 1 shows a breakpoint located within the C19 array, compared with 9 of these mutants carrying a C19 deletion remote from the site of complex rearrangement (Fisher's exact test, P=.004). Thus, C arrays are targets for microdeletion but do not promote large rearrangements. Deletions always involve the loss of a single repeat unit in shorter C arrays and may well do so in the longest C19 array. C-array instability increases markedly with array length (fig. 6A) and becomes intense at the longest array, with a nominal mutation rate of 71% (62/87 sperm molecules carry a deletion in C19) and possibly as high as 130% per sperm cell if mutation proceeds by single-repeat deletions. This is by far the highest mutation rate ever recorded for a minisatellite. The effect of array length on instability also implies that microdeletion rates will vary enormously between alleles, depending on their content of homogeneous blocks of repeats. We have confirmed this prediction by SM-PCR analysis of sperm DNA from a third man with a 60-repeat MS1 allele containing only short homogeneous arrays (one C3, three C4, and two C6 arrays). The screening of 140 molecules showed no evidence of instability (data not shown), indicating that the microdeletion rate at this allele is at least 30-fold lower than for allele A in man 1.
The relationship between C-array length and microdeletion rate shows evidence of a hyperinstability threshold at ∼12 repeat units (108 bp), with a significant difference in rates between C11 and C12 arrays (Fisher's exact test, P=.015) (fig. 6A). This is very similar to the instability threshold of 34–38 uninterrupted CGG repeats (102–114 bp) at the fragile-X microsatellite (Eichler et al. 1994). Another parallel with triplet repeats emerges when comparing the MS1 C19 array with the C11 and C7 arrays in the same allele; the C11 and C7 arrays together constitute a C19 array interrupted by a single A-type repeat (fig. 2). The perfect C array shows a microdeletion rate of 71%–130%, compared with just 11% in the imperfect array. This stabilization of an array when homogeneity is interrupted is similar to that seen at microsatellites associated with fragile-X syndrome (Eichler et al. 1994; Kunst et al. 1994), spinocerebellar ataxia type 1 and 2 (Chung et al. 1993; Imbert et al. 1996; Pulst et al. 1996; Sanpei et al. 1996), and Friedreich ataxia (Montermini et al. 1997). However, triplet repeat instability is heavily biased toward expansion, in stark contrast to hyperdeletion in minisatellite MS1. Homogeneity-dependent instability has also been detected at human minisatellite CEB1 (although it leads, in general, to gains of repeats [Buard et al. 1998]) and at mouse expanded simple tandem repeat loci, resulting in both gains and losses (Bois et al. 2001).
The mechanism of MS1 hyperdeletion remains unknown. The apparent absence of such instability in blood and buccal DNA suggests that replication slippage (Pearson et al. 1998; Sinden et al. 2002) is unlikely to be the process. Instead, the apparent germline specificity of these sperm deletions is consistent with their being generated by slippage during meiotic recombination. Another possibility is postmeiotic repair, as seen in mice transgenic for Huntington disease CAG repeats, in which expansions arise in postmeiotic haploid cells via repair of DNA breaks generated during sperm maturation (Kovtun and McMurray 2001).
The existence of the MS1 hyperdeletion mechanism creates a paradox concerning the existence of alleles containing long homogeneous arrays. Allele A in man 1 is so unstable that the C19 array cannot survive for more than three generations (P>.95) without undergoing deletion, and even a four-repeat deletion (the largest seen in our survey of mutants) would still leave the array prone to further rapid deletion. The length distribution of C arrays in alleles also shows that the C19 array is remarkably long (fig. 6B). This leads to the question of how such long arrays can come into existence in the face of this intense pressure to delete, which is substantial even in shorter subthreshold arrays, with C7 and C11 arrays showing a mutation rate of 6% per sperm cell and a strong (>3.2:1, P>.95) bias in favor of deletion (fig. 6A). One formal, though highly unlikely, possibility is that these microdeletions render sperm cells inviable or infertile and that transmitted alleles will be purged of most of the microdeletions detected in sperm DNA. The second possibility is that some individuals may carry modifiers that promote compensating expansions or that such expansions are generated in the female germline premeiotically or by meiotic recombination or, possibly, by repair in mature oocytes (Kaytor et al. 1997). However, this compensation would have to be exquisitely balanced to prevent either rapid deletion or runaway expansion. The third possibility is that other mutation events in males create long arrays de novo. Two examples of such a creation have been observed in the MS1 mutants characterized. One is a complex duplication with breakpoints in the C19 and C11 arrays in man 1 that creates a perfect C25 array at the duplication junction (mutant 3; fig. 2). The second is a single A→C switch that converts the C11+C7 array into a C19 array (mutant 11; fig. 5). However, both of these events that create long homogeneous arrays require the preexistence of C arrays of significant length (>6 repeats) that would still show substantial instability and a strong bias to deletion. We therefore have no explanation of how such hyperdeleting arrays—or, indeed, their putative shorter progenitor arrays, which exist at a significant population frequency (fig. 6B)—can come into existence and persist in human populations.
Acknowledgments
We thank J. Blower, S. B. Kanoyangwa, and volunteers, for providing semen, blood, and buccal samples; S. Mistry, for oligonucleotide synthesis; and our colleagues, for helpful discussions. This work was supported by Swedish Radiation Protection Institute and National Board for Laboratory Animals grants (to U.R.) and by Medical Research Council and Royal Society grants (to A.J.J.).
References
- Appelgren H, Cederberg H, Rannug U (1997) Mutations at the human minisatellite MS32 integrated in yeast occur with high frequency in meiosis and involve complex recombination events. Mol Gen Genet 256:7–17 [DOI] [PubMed] [Google Scholar]
- Armour JAL, Harris PC, Jeffreys AJ (1993) Allelic diversity at minisatellite MS205 (D16S309): evidence for polarized variability. Hum Mol Genet 2:1137–1145 [DOI] [PubMed] [Google Scholar]
- Berg I, Cederberg H, Rannug U (2000) Tetrad analysis shows that gene conversion is the major mechanism involved in mutation at the human minisatellite MS1 integrated in Saccharomyces cerevisiae. Genet Res 75:1–12 [DOI] [PubMed] [Google Scholar]
- Bois P, Jeffreys AJ (1999) Minisatellite instability and germline mutation. Cell Mol Life Sci 55:1636–1648 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bois PRJ, Southgate L, Jeffreys AJ (2001) Length of uninterrupted repeats determines instability at the unstable mouse expanded simple tandem repeat family MMS10 derived from independent SINE B1 elements. Mamm Genome 12:104–111 [DOI] [PubMed] [Google Scholar]
- Buard J, Bourdet A, Yardley J, Dubrova Y, Jeffreys AJ (1998) Influences of array size and homogeneity on minisatellite mutation. EMBO J 17:3495–3502 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buard J, Collick A, Brown J, Jeffreys AJ (2000a) Somatic versus germline mutation processes at minisatellite CEB1 (D2S90) in humans and transgenic mice. Genomics 65:95–103 [DOI] [PubMed] [Google Scholar]
- Buard J, Shone AC, Jeffreys AJ (2000b) Meiotic recombination and flanking marker exchange at the highly unstable human minisatellite CEB1 (D2S90). Am J Hum Genet 67:333–344 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buard J, Vergnaud G (1994) Complex recombination events at the hypermutable minisatellite CEB1 (D2S90). EMBO J 13:3203–3210 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cederberg H, Agurell E, Hedenskog M, Rannug U (1993) Amplification and loss of repeat units of the human minisatellite MS1 integrated in chromosome III of a haploid yeast strain. Mol Gen Genet 238:38–42 [DOI] [PubMed] [Google Scholar]
- Chung M-Y, Ranum LPW, Duvick LA, Servadio A, Zoghbi HY, Orr HT (1993) Evidence for a mechanism predisposing to intergenerational CAG repeat instability in spinocerebellar ataxia type I. Nat Genet 5:254–258 [DOI] [PubMed] [Google Scholar]
- Dubrova YE, Netserov VN, Krouchinsky NG, Ostapenko VA, Vergnaud G, Giraudeau F, Buard J, Jeffreys AJ (1997) Further evidence for elevated human minisatellite mutation rate in Belarus eight years after the Chernobyl accident. Mutat Res 381:267–278 [DOI] [PubMed] [Google Scholar]
- Eichler EE, Holden JJA, Popovich BW, Reiss AL, Snow K, Thibodeau SN, Richards CS, Ward PA, Nelson DL (1994) Length of uninterrupted CGG repeats determines instability in the FRM1 gene. Nat Genet 8:88–94 [DOI] [PubMed] [Google Scholar]
- Gray IC, Jeffreys AJ (1991) Evolutionary transience of hypervariable minisatellites in man and the primates. Proc R Soc Lond B 243:241–253 [DOI] [PubMed] [Google Scholar]
- He Q, Cederberg H, Armour JAL, May CA, Rannug U (1999) Cis-regulation of inter-allelic exchanges in mutation at the human minisatellite MS205 in yeast. Gene 232:143–153 [DOI] [PubMed] [Google Scholar]
- Henke J, Henke L (1995) Recent observations in human DNA-minisatellite mutations. Int J Legal Med 107:204–208 [DOI] [PubMed] [Google Scholar]
- Hoff-Olsen P, Meling GI, Olaisen B (1995) Somatic mutations in VNTR-locus D1S7 in human colorectal carcinomas are associated with microsatellite instability. Hum Mutat 5:329–332 [DOI] [PubMed] [Google Scholar]
- Imbert G, Saudou F, Yvert G, Devys D, Trottier Y, Garnier J-M, Weber C, Mandel J-L, Cancel G, Abbas N, Dürr A, Didierjean O, Stevanin G, Agid Y, Brice A (1996) Cloning of the gene for spinocerebellar ataxia 2 reveals a locus with high sensitivity to expanded CAG/glutamine repeats. Nat Genet 14:285–291 [DOI] [PubMed] [Google Scholar]
- Jeffreys AJ, Allen MJ, Armour JAL, Collick A, Dubrova Y, Fretwell N, Guram T, Jobling M, May CA, Neil DL, Neumann R (1995) Mutation processes at human minisatellites. Electrophoresis 16:1577–1585 [DOI] [PubMed] [Google Scholar]
- Jeffreys AJ, MacLeod A, Tamaki K, Neil DL, Monckton DG (1991) Minisatellite repeat coding as a digital approach to DNA typing. Nature 354:204–209 [DOI] [PubMed] [Google Scholar]
- Jeffreys AJ, Murray J, Neumann R (1998a) High-resolution mapping of crossovers in human sperm defines a minisatellite-associated recombination hotspot. Mol Cell 2:267–273 [DOI] [PubMed] [Google Scholar]
- Jeffreys AJ, Neil DL, Neumann R (1998b) Repeat instability at human minisatellites arising from meiotic recombination. EMBO J 17:4147–4157 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeffreys AJ, Neumann R (1997) Somatic mutation processes at a human minisatellite. Hum Mol Genet 6:129–136 [DOI] [PubMed] [Google Scholar]
- Jeffreys AJ, Ritchie A, Neumann R (2000) High-resolution analysis of haplotype diversity and meiotic crossover in the human TAP2 recombination hotspot. Hum Mol Genet 9:725–733 [DOI] [PubMed] [Google Scholar]
- Jeffreys AJ, Royle NJ, Wilson V, Wong Z (1988) Spontaneous mutation rates to new length alleles at tandem-repetitive hypervariable loci in human DNA. Nature 332:278–281 [DOI] [PubMed] [Google Scholar]
- Jeffreys AJ, Tamaki K, MacLeod A, Monckton DG, Neil DL, Armour JAL (1994) Complex gene conversion events in germline mutation at human minisatellites. Nat Genet 6:136–145 [DOI] [PubMed] [Google Scholar]
- Jeffreys AJ, Wilson V, Thein SL (1985) Hypervariable “minisatellite” regions in human DNA. Nature 314:67–73 [DOI] [PubMed] [Google Scholar]
- Kaytor MD, Burright EN, Duvick LA, Zoghbi HY, Orr HT (1997) Increased trinucleotide repeat instability with advanced maternal age. Hum Mol Genet 6:2135–2139 [DOI] [PubMed] [Google Scholar]
- Kovtun IV, McMurray CT (2001) Trinucleotide expansion in haploid germ cells by gap repair. Nat Genet 27:407–411 [DOI] [PubMed] [Google Scholar]
- Kunst CB, Warren ST (1994) Cryptic and polar variation of the fragile X repeat could result in predisposing normal alleles. Cell 77:853–861 [DOI] [PubMed] [Google Scholar]
- Maleki S, Cederberg H, Rannug U (1997) Mutations occurring at the human minisatellite MS1 integrated in haploid yeast are similar to MS1 mutations in humans. Mol Gen Genet 254:37–42 [DOI] [PubMed] [Google Scholar]
- May CA, Jeffreys AJ, Armour JAL (1996) Mutation rate heterogeneity and the generation of allele diversity at the human minisatellite MS205 (D16S309). Hum Mol Genet 5:1823–1833 [DOI] [PubMed] [Google Scholar]
- Mitchell RJ, Farrington SM, Dunlop MG, Campbell H (2002) Mismatch repair genes hMLH1 and hMSH2 and colorectal cancer: a HuGE review. Am J Epidemiol 156:885–902 [DOI] [PubMed] [Google Scholar]
- Monckton DG, Neumann R, Guram T, Fretwell N, Tamaki K, MacLeod A, Jeffreys AJ (1994) Minisatellite mutation rate variation associated with a flanking DNA sequence polymorphism. Nat Genet 8:162–170 [DOI] [PubMed] [Google Scholar]
- Montermini L, Andermann E, Labuda M, Richter A, Pandolfo M, Cavalcanti F, Pianese L, Iodice L, Farina G, Monticelli A, Turano M, Filla A, De Michele G, Cocozza S (1997) The Friedreich ataxia GAA triplet repeat: premutation and normal alleles. Hum Mol Genet 6:1261–1266 [DOI] [PubMed] [Google Scholar]
- Pearson CE, Eichler EE, Lorenzetti D, Kramer SF, Zoghbi HY, Nelson DL, Sinden RR (1998) Interruptions in the triplet repeats of SCA1 and FRAXA reduce the propensity and complexity of slipped strand DNA (S-DNA) formation. Biochemistry 37:2701–2708 [DOI] [PubMed] [Google Scholar]
- Pulst S-M, Nechiporuk A, Nechiporuk T, Gispert S, Chen X-N, Lopes-Cendes I, Pearlman S, Starkman S, Orozco-Diaz G, Lunkes A, DeJong P, Rouleau GA, Auburger G, Korenberg JR, Figueroa C, Sahba S (1996) Moderate expansion of normally biallelic trinucleotide repeat in spinocerebellar ataxia type 2. Nat Genet 14:269–276 [DOI] [PubMed] [Google Scholar]
- Royle NJ, Clarkson RE, Wong Z, Jeffreys AJ (1988) Clustering of hypervariable minisatellites in the proterminal regions of human autosomes. Genomics 3:352–360 [DOI] [PubMed] [Google Scholar]
- Sanpei K, Takano H, Igarashi S, Sato T, Oyake M, Sasaki H, Wakisaka A, Tashiro K, Ishida Y, Ikeuchi T, Koide R, Saito A, Tanaka T, Hanyu S, Takiyama Y, Nishizawa M, Shimizu N, Nomura Y, Segawa M, Iwabuchi K, Eguchi I, Tanaka H, Takahashi H, Tsuji S (1996) Identification of the spinocerebellar ataxia type 2 gene using a direct identification of repeat expansion and cloning technique, DIRECT. Nat Genet 14:277–284 [DOI] [PubMed] [Google Scholar]
- Sinden RR, Potaman VN, Oussatcheva EA, Pearson CE, Lyubchenko YL, Shlyakhtenko LS (2002) Triplet repeat structures and human genetic disease: dynamic mutations from dynamic DNA. J Biosci 27:53–65 [DOI] [PubMed] [Google Scholar]
- Tamaki K, May CA, Dubrova YE, Jeffreys AJ (1999) Extremely complex repeat shuffling during germline mutation at human minisatellite B6.7. Hum Mol Genet 8:879–888 [DOI] [PubMed] [Google Scholar]
- Wong Z, Wilson V, Patel I, Povey S, Jeffreys AJ (1987) Characterization of a panel of highly variable minisatellites cloned from human DNA. Ann Hum Genet 51:269–288 [DOI] [PubMed] [Google Scholar]
- Yauk CL, Dubrova YE, Grant GR, Jeffreys AJ (2002) A novel single molecular analysis of spontaneous and radiation-induced mutation at a mouse tandem repeat locus. Mutat Res 500:147–156 [DOI] [PubMed] [Google Scholar]