Abstract
Uric acid nephrolithiasis (UAN) is a common disease with an established genetic component that presents a complex mode of inheritance. While studying an ancient founder population in Talana, a village in Sardinia, we recently identified a susceptibility locus of ∼2.5 cM for UAN on 10q21-q22 in a relatively small sample that was carefully selected through genealogical information. To refine the critical region and to identify the susceptibility gene, we extended our analysis to severely affected subjects from the same village. We confirm the involvement of this region in UAN through identical-by-descent sharing and autozygosity mapping, and we refine the critical region to an interval of ∼67 kb associated with UAN by linkage-disequilibrium mapping. After inspecting the genomic sequences available in public databases, we determined that a novel gene overlaps this interval. This gene is divided into 15 exons, spanning a region of ∼300 kb and generating at least four different proteins (407, 333, 462, and 216 amino acids). Interestingly, the last isoform was completely included in the 67-kb associated interval. Computer-assisted analysis of this isoform revealed at least one membrane-spanning domain and several N- and O-glycosylation consensus sites at N-termini, suggesting that it could be an integral membrane protein. Mutational analysis shows that a coding nucleotide variant (Ala62Thr), causing a missense in exon 12, is in strong association with UAN (P=.0051). Moreover, Ala62Thr modifies predicted protein secondary structure, suggesting that it may have a role in UAN etiology. The present study underscores the value of our small, genealogically well-characterized, isolated population as a model for the identification of susceptibility genes underlying complex diseases. Indeed, using a relatively small sample of affected and unaffected subjects, we identified a candidate gene for multifactorial UAN.
Introduction
Nephrolithiasis is a common multifactorial disorder of unknown etiology, which is characterized by the presence of stones in the urinary tract and known genetic involvement (Jaeger 1996; Curhan et al. 1997; Baggio 1999; Scheinman 1999). Kidney stones are estimated to affect 10% of the population (Serio and Fraioli 1999; Rivers et al. 2000). The major classes of stones are calcium oxalate, calcium phosphate, and uric acid. Uric acid nephrolithiasis (UAN [MIM 605990]), which occurs when the urine becomes overly concentrated with uric acid that may form small crystals (and, subsequently, stones), accounts for 20% of all stones. Every year a considerable number of people are hospitalized for the disease. An improved understanding of the genetic factors that contribute to the development of this disorder will lead to better diagnosis, treatment, and prevention and will also help to stem, or even to reverse, the rise in prevalence of this disease.
In humans and some primates, a renal-hematic urate homeostasis system was developed as a consequence of evolutionary loss of hepatic uricase present in other mammalian species (Wu et al. 1989, 1992; Abramson and Lipkowitz 1990). Uric acid is the end product of purine metabolism: two-thirds of daily production is excreted by the kidneys and one-third by the gastrointestinal tract (Roch-Ramel and Guisan 1999). Hyperuricemia (hyperuricosuria) can occur when there is excessive dietary consumption of purines from meat and fish or when there is altered urate elimination. Insight into the nature of urate homeostasis was derived from a recently identified renal urate–anion exchanger regulating blood urate levels (Enomoto et al. 2002). The molecular basis of urate handling by the human kidney is not yet clear, but new knowledge may come from the study of associated diseases, such as UAN.
A powerful approach to mapping complex disease genes is to study isolated, founder populations, in which genetic heterogeneity and environmental noise are likely to be reduced and where a substantial proportion of affected individuals are likely to share susceptibility variants identical by descent (IBD) from common ancestors (Wright et al. 1999; Peltonen et al. 2000). For our studies, we have selected residents of Talana, a particularly secluded village in Sardinia. This small, isolated population is characterized by higher prevalence of UAN than is seen in the rest of the island. The disease shows familial clustering in Talana, although the transmission of UAN does not follow a simple Mendelian inheritance pattern. Clinical studies have shown that other factors (such as low urinary pH and high levels of uricosuria) are present in these patients with UAN, thereby confirming the multifactorial etiology of the disease.
In a previous study, we performed a multistep genomewide search in several families belonging to a single nine-generation genealogical tree comprising 37 individuals with UAN. We identified a susceptibility locus on the chromosomal region 10q21-q22, with the highest evidence at marker D10S1652 (Ombra et al. 2001). We now describe the fine mapping of the identified critical region, using more-severely affected individuals from the same village. The fine mapping led to the detection of a 67-kb interval, around marker D10S1652, that is statistically associated with UAN. We identified a novel gene, called “ZNF365,” that encodes at least four different protein isoforms spanning this region. Interestingly, one of the isoforms resides entirely within the 67-kb interval. In addition, in this isoform, we identify a missense amino acid variant that alters the protein secondary structure and is associated with the disease, making it a strong candidate predisposing factor.
Material and Methods
Genealogical Data and Sample Collection
All individuals participating in the study were among the 1,200 inhabitants of Talana, a village in east-central Sardinia that is characterized by slow population growth and high levels of endogamy and inbreeding (Wright et al. 1999; Angius et al. 2001). Historical and archival data allowed the reconstruction of the genealogy of each individual as far back as the 17th century. Haplotype and phylogenetic analyses of the Y chromosome and characterization of mtDNA haplogroups were used to determine the number of ancestral founders. Approximately 80% of the present-day population derives from 8 paternal and 11 maternal ancestral lineages. All genealogical data, as well as phenotypic and genetic data, are stored in relational databases. The bioinformatic framework comprises several algorithms that allow interrogation and extraction of data for analyses.
Clinical features of the sample and details of recruitment and diagnostic procedures have been described elsewhere (Ombra et al. 2001). In brief, we collected information on age at diagnosis, medications, hospital admissions, and family history of kidney stones. UAN diagnoses were clinically confirmed by accurate physical examination and by renal ultrasonography. The original patient list included 134 affected subjects. For the present study, we selected 62 patients (mean age 58.4 years) with severe UAN. Disease severity was established on the basis of the following criteria: age at onset, number of episodes of abdominal colic, and number and diameter of expelled stones. Affected subjects with mild-to-moderate disease symptoms were excluded from the study. We identified 94 control subjects, who were examined by use of ultrasonography to exclude individuals with asymptomatic disease. The mean age at observation of unaffected controls was ∼50 years, which meant that those with a predisposition to disease were very likely to have developed stones, given that the mean age at onset for affected subjects is ∼38 years. All subjects gave written informed consent, and all samples were taken in accordance with the Helsinki declaration.
SNP Genotyping
To find common SNPs in the UAN critical region, we used Sequencher software to perform an extensive alignment of genomic sequences deposited in public databases. The screening panel included 10 unrelated Talana subjects. PCR amplicons of 800–1,000 nt containing the selected putative polymorphisms were designed using Oligo, version 4.0, software. PCR were prepared using 50 ng of genomic DNA template, 0.5 μM of each PCR primer, 1.5 mM MgCl2, 0.2 mM of each dNTP, 0.5 U Taq polymerase, and the buffer recommended by the supplier (Amersham), in a final volume of 25 μl. Thermocycling started with a single 2-min denaturation step at 94°C, followed by 35 cycles of 30 s denaturation at 94°C, 30 s annealing at melting temperature (MT), and a 45-s extension at 72°C. One final 7-min extension step at 72°C was added.
Samples were then sequenced using the Big Dye Terminator Ready Reaction kit (Applied Biosystems). Sequencing reactions were performed on a 9700 Thermal Cycler (Applied Biosystems) for 25 cycles of 95°C for 10 s, MT for 5 s, and 60°C for 2 min. After the sequencing, each reaction was precipitated with isopropanol to remove excess dye terminators. Sequencing of the products was performed on the ABI Prism 3100 Genetic Analyzer (Applied Biosystems). Polymorphisms were detected by multiple alignments of sequences, using the program Autoassembler (Applied Biosystems).
Mutational analysis of all 15 exons of the identified candidate gene was performed. Each PCR amplicon was designed for scanning of ZNF365 exons 1–15, including intron/exon boundaries. All PCRs and sequence reactions were performed as described above.
Genotyping of SNPs was performed by dot-blot hybridization of PCR products, comprising the polymorphisms, with short allele-specific oligonucleotide probes, using an established protocol (Ristaldi et al. 1989). To estimate the frequency of the identified variant Ala62Thr in an outbred population, we genotyped 200 individuals from other parts of Sardinia. The primers used to detect the Ala62Thr variant were 5′-CTCCACTCCACCTTTTTAAG-3′ and 5′-GCTGACATTGGTACTTACTG-3′.
Statistical Analysis
From the available genealogical information, we identified all pairwise connections between all pairs of affected and unaffected subjects and estimated the kinship coefficients, the number of meiotic steps separating each pair, and the nine condensed identity coefficients using an algorithm developed by Abney et al. (2000). To perform formal kinship analysis of our cases and controls, we first checked that there was not a significant difference in the distributions of number of meiotic steps between the affected and unaffected subjects, thus ensuring an unbiased sample selection. Differences between affected and unaffected subjects in kinship and identity coefficients were tested through 10,000 resampling cycles.
The genealogical information allows the identification of all common ancestors of the 62 affected individuals, linked in an extended complex pedigree comprising 710 subjects. Simwalk2 (Sobel and Lange 1996) was used in the linkage analysis to compute IBD-sharing statistics on the basis of the degree of marker-allele clustering among affected relatives in the extended pedigree. Homozygosity mapping was performed with the program Mapmaker/Homoz (Kruglyak et al. 1995), providing real inbreeding coefficients, estimated from the whole genealogy, for the parents of affected subjects or affected sibs.
Association analyses for a single locus were performed with the DISEQ program (Terwilliger 1995), that, applying a likelihood-ratio test for linkage disequilibrium (LD) with only 1 df, irrespective of the number of alleles at a given marker, does not require correction for multiple alleles. We also performed 10,000 Monte Carlo simulations to assess empirical significance of the departure of observed values from the expected values in the contingency table. Each simulation generated a table having the same marginal totals as the one under consideration, and empirical significance was derived by counting the number of times that a χ2 value associated with the real table was achieved by the randomly simulated data. Although multiple tests performed for each marker increase the false-positive rate, a Bonferroni correction of nominal values would likely be overly conservative, since the tests are highly correlated. It is unclear what multiple-test correction would be more appropriate.
We also performed multilocus haplotype analysis for association, to minimize the chance of type I error, thereby providing stronger statistical support for the region. We used Simwalk2 to generate the haplotypes with the highest likelihood, using all genealogical information about the subjects and all typed relatives. Differences in frequencies of haplotypes between cases and controls were tested for all possible haplotypes of adjacent markers. Only common haplotypes (frequency >5% in the whole sample) were compared between cases and controls. We performed 100,000 Monte Carlo simulations to evaluate empirical significance.
cDNA Clones
KIAA0844 was kindly provided by Kazusa DNA Research Institute. ZNF365A, ZNF365C, and ZNF365D cDNA sequences were obtained by RT-PCR/TOPO-cloning vector (Invitrogen) strategy, using pairs of primers designed in different exons of the gene in human kidney RNA (Clontech) and 5′– and 3′–rapid amplification cDNA end (RACE) strategy performed with human kidney Marathon-Ready cDNA kit. ZNF365B cDNA sequence was obtained by sequencing the cDNA clones 3069791 and 4821260 from the IMAGE Consortium.
Bioinformatic Predictions
Different modeling programs for predicting potential transmembrane domains, all with default parameters, were used: DAS (Cserzo et al. 1997), TopPred 2 (von Heijne 1992), TMPred (Hofmann and Stoffel 1993), PRED-TMR2 (Pasquier and Hamodrakas 1999), TMAP (Persson and Argos 1996). The α-helical predictions were obtained using the SecStr package. This package makes use of an algorithm that combines the results of the secondary structure prediction of six different methods (Burgess et al. 1974; Lim 1974; Dufton and Hider 1977; Nagano 1977; Chou and Fasman 1978; Garnier et al. 1978). The coiled-coil segments were predicted using two different programs: COILS (Lupas et al. 1991) and PAIRCOILS (Berger et al. 1995).
RNA Expression Studies
A human multiple-tissue northern blot (Clontech) was hybridized with ZNF365A, -B, -C, and -D transcripts. The northern blot was prehybridized, hybridized, and washed according to the manufacturer’s instructions (Clontech).
RT-PCR
For the expression pattern of the ZNF365 gene, RT-PCR was performed on a normalized cDNA panel (Clontech). Primers used to amplify the four transcripts were as follows: ZNF365A (5′-TCCTCAAATAGGAAGCCC-3′ and 5′-AAATTAGCTGAGGGGAAGGT-3′ in exons 4 and 5, respectively); ZNF365B (5′-AACAACTGAGTACCTGCCAT-3′ and 5′-TCGCCTGGTGTGCCAAAATG-3′ in exons 6 and 7, respectively); ZNF365C (5′-AACAACTGAGTACCTGCCAT-3′ and 5′-GGAGTCCTAAGACGTCCTTC-3′ in exons 6 and 13, respectively); and ZNF365D (5′-CTATGCAGGAGTTTCAATTC-3′ and 5′-GGAGTCCTAAGACGTCCTTC-3′ in exons 12 and 13, respectively). Primers for the G3PDH gene were obtained from Clontech. All primers were located in exons, and the included introns were sufficiently large that amplification from genomic DNA yielded either no PCR product or an easily distinguishable product. PCRs were performed using 1 μl of cDNA in a 10-μl PCR volume containing 1× PCR buffer (Amersham), 0.2 mM dNTPs, 0.5 U AmpliTaq polymerase (Amersham), and 0.5 μM each of the described primers. Using a DNA Thermal Cycler 9700 (Applied Biosystems), we performed 40 cycles of amplification for ZNF365A, -B, and -D transcripts, 45 cycles for ZNF365C, and 25 cycles for G3PDH. Cycles consisted of 30 s at 94°C, 30 s at MT, and 30 s at 72°C.
For gene characterization, RT-PCR was performed using 0.5-μg samples of human kidney (Clontech), adrenal gland (Clontech), and blood mRNA that were reverse-transcribed in a 50-μl reaction mixture containing 1× RT buffer (20 mM Tris-HCl [pH 8.4], 50 mM KCl, and 2.5 mM MgCl2), 10 mM DTT, 0.5 mM dNTP, 0.2 μg random hexamers (Roche), and 200 U SuperScript RT (Invitrogen). After a 60-min incubation at 37°C, 1 μg DNase-free RNase was added and incubated 10 min at 37°C. The cDNA formed was extracted twice with phenol/chloroform/isoamylalcohol (25:24:1). The cDNA was then precipitated overnight with 1:10 vol 3 M sodium acetate (pH 5.2) and 2.5 vol ethanol; 10-ng samples of each cDNA were used as templates in 25-μl PCRs containing 1× PCR buffer (Amersham), 0.2 mM dNTPs, 0.5 U AmpliTaq polymerase (Amersham), and 0.5 μM each of the primer sequences derived from cDNA. Using a DNA Thermal Cycler 9700 (Applied Biosystems), we performed amplification in 40 cycles of 30 s at 94°C, 30 s at MT, and 45 s at 72°C.
5′- and 3′-RACE strategy was performed using 5 μl Marathon human kidney cDNA (Clontech), 1× PCR buffer (Amersham), 0.2 mM dNTPs, 1.0 U AmpliTaq polymerase (Amersham), and 0.5 μM each of the gene-specific primer and AP1 primer (Clontech) in a final volume of 50 μl. Twenty-five amplification cycles were performed using 30 s at 94°C, 20 s at MT, and 4 min at 68°C. Nested PCRs were performed with the same protocol described for the first PCRs, using 1/250 of the first PCRs and gene-specific primers 2 and AP2 (Clontech), in a final volume of 50 μl. Thirty amplification cycles were performed at 30 s at 94°C, 20 s at MT, and 4 min at 68°C. Aliquots of the products of the first and second PCRs performed with different gene-specific primers were run on agarose gel and were blotted on nylon membrane, according to the manufacturer’s instructions (Amersham). Membranes were hybridized with a PCR product including exons 2–5 of the ZNF365D transcript. Positive PCR products were cloned in TOPO-vector (Invitrogen) and were sequenced with M13 forward and reverse primers, as described above.
Results
Linkage Analysis
We reported elsewhere the identification, in a 2.5-cM region flanked by markers D10S1719 and D10S1640, of a susceptibility locus for UAN on 10q21-q22 (Ombra et al. 2001). For the present study, we reconstructed the genealogical relationships of 62 severe cases and 94 normal subjects and tested the identified region through nonparametric linkage and autozygosity-mapping analyses.
Nonparametric linkage analysis, based on IBD sharing, was performed on a large pedigree connecting all affected subjects. Cases displayed greater IBD sharing than would be expected by chance in the region, where allele-sharing statistics A, C, and D of Simwalk2 resulted in P values <.05, again with a peak value at marker D10S1652. From formal kinship analysis, the UAN cases were found to have higher mean kinship coefficients than those of unaffected subjects (0.0150 vs. 0.0139), and the difference between the two groups was highly significant (empirical P<.0001, estimated from resampling procedure). UAN cases were also found to have a greater probability of sharing all four alleles IBD (condensed identity coefficient Δ1) and both alleles IBD (condensed identity coefficient Δ7) than did controls (empirical P<.0001, estimated from resampling procedure). Thus, we performed autozygosity mapping that yielded multipoint LOD scores >3.0 in the region flanked by D10S1652 and D10S1719. These results confirm the involvement of the region in UAN in this enlarged sample.
Transcriptional Map of UAN Locus
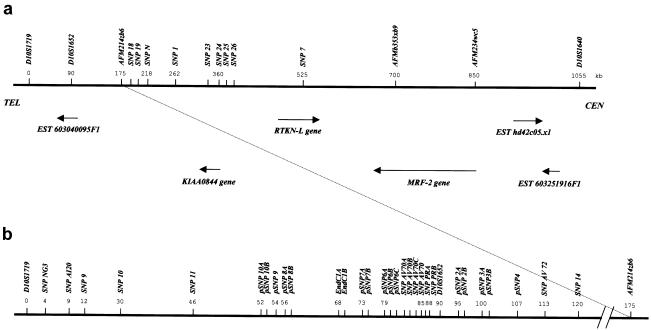
We obtained a consensus genomic sequence of the 1.1-Mb region, corresponding to the identified 2.5-cM critical interval, by alignment of partial sequences available in different databases. To identify the disease-susceptibility gene by use of the candidate gene approach, we developed a transcriptional map of this region. Using several sequence-analysis tools and database-mining procedures, we determined that the 1.1-Mb interval contained at least six novel genes, including two uncharacterized genes (significant similarity with proteins of known or inferred function) and four orphan genes (fig. 1A). The modulator-recognition factor 2 (MRF-2) gene is the ortholog of the mouse Desrt gene. Mice homozygous for mutations in this gene were retarded in their mental and sexual development, with transient immune abnormalities (Lahoud et al. 2001). The rhotekin-like (RTKN-L) gene is a homolog of RTKN gene, an inhibitor of ρ-GTPase activity localized on chromosome 2 (Fu et al. 2000). The ESTs 603251916F1 (UniGene cluster Hs.252954), hd42c05.x1, and 603040095F1, as well as KIAA0844 cDNA (Nagase et al. 1998), exhibit no significant similarity to any other protein deposited in public databases. In the absence of functional assignment, it is not possible to know which one of these six genes may have a role in UAN, although the EST 603040095F1 is the closest to marker D10S1652, which is associated with UAN.
Figure 1.
Physical and transcriptional maps. Markers of the physical map are indicated above the solid bar representing genomic DNA; orientations of genes are indicated (arrows). a, Physical and transcriptional map of the 1.1-Mb region corresponding to the UAN critical region. The known microsatellites markers are D10S1719, D10S1652, and D10S1640. All genes and ESTs located in this region are shown and their mapping position is in scale. b, SNP map of the region around marker D10S1652. Allele frequencies in the Talana population for all SNPs are available from the authors.
Fine Mapping
To fine map the identified locus and to refine the critical region, we performed LD-mapping analysis, generating an SNP map around the D10S1652 marker previously found to be significantly associated with UAN (Ombra et al. 2001) in the village of Talana. Comparison among genomic sequences deposited in various databases allowed us to identify 39 new SNPs in this region (fig. 1B). The sequence analysis of PCR amplicons (∼800–1,000 bp) containing the selected putative polymorphisms showed an average nucleotide diversity, of 0.2% (one variable site every 500 bp), across the region. We performed association analysis using a subset of 11 SNPs evenly spaced throughout the region, 2 newly localized microsatellite markers (AFM214zb6 and AFMA234wc5), and 3 markers that were included in our previous study (D10S1719, D10S1652, and D10S1640). This set of markers was genotyped in 62 affected and 94 unaffected subjects and in their close relatives, to infer the most likely phase and, hence, haplotypes. For affected subjects, 112 complete haplotypes were reconstructed (90% of the theoretical maximum [124]), and, for unaffected subjects, 142 complete haplotypes were reconstructed (76% of the theoretical maximum [188]).
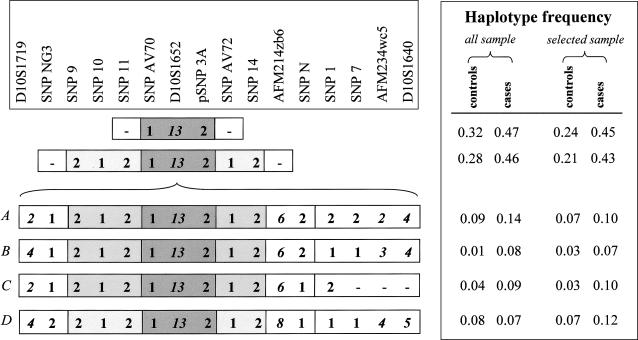
At the nominal significance level of <.05, two markers (namely, D10S1652 and pSNP3A) were associated with UAN in the likelihood-ratio test and were also significantly associated in the empirical test performed with the use of 10,000 Monte Carlo simulations (table 1). Since the case-control sample included related individuals (thereby increasing the type I error rate of the association test), we repeated the tests while controlling for close relationships among the subjects. From the complete set, a smaller sample of individuals was created by selecting cases and controls separated by more than three meiotic steps, without regard to genotypes. The resulting selected sample consisted of 34 cases and 53 controls separated by an average of eight meiotic steps. Also, in this cohort, the same two markers provided significant evidence of association, with the highest significance at marker D10S1652 (P=.0009; empirical P=.0031) (table 1). Haplotype frequencies in the whole sample and in the selected independent sample are shown in figure 2. The haplotype flanked by SNPs NG3 and AFM214zb6 (fig. 2) was the most common core haplotype among patients with UAN and was more frequent in cases than in controls. The four most common extended haplotypes (haplotypes A, B, C, and D) containing this core haplotype were observed among cases. In the selected sample, we performed haplotype-based LD-mapping analysis, in which we considered only the 11 biallelic polymorphisms, because STRPs present higher mutation rates (which may impede detection of ancient LD segments). The analysis was performed by testing all possible haplotypes of adjacent markers, using windows of variable sizes. Haplotype-association analysis identified a significant core region of 67 kb, flanked by SNPAV72 and SNP11 (fig. 2) and associated with disease susceptibility (empirical P<.05).
Table 1.
Results of the Association Tests[Note]
| Marker | Position(kb) | Frequencyin Controls | Frequencyin Cases | Pa | Empirical Pb,c | Pd | Empirical Pc,e |
| D10S1719 | 0 | … | … | ns | ns | ns | ns |
| SNP NG3 | 4 | .66 | .54 | ns | ns | ns | ns |
| SNP 9 | 12 | .74 | .79 | ns | ns | ns | ns |
| SNP 10 | 30 | .76 | .82 | ns | ns | ns | ns |
| SNP 11 | 46 | .77 | .82 | ns | ns | ns | ns |
| SNP AV70 | 85 | .68 | .76 | ns | ns | ns | ns |
| D10S1652 | 90 | … | … | .0006 | .0004 | .0009 | .0031 |
| pSNP 3A | 100 | .68 | .53 | .0123 | .0073 | .0146 | .0075 |
| SNP AV72 | 113 | .87 | .91 | ns | ns | ns | ns |
| SNP 14 | 120 | .87 | .90 | ns | ns | ns | ns |
| AFM214zb6 | 175 | … | … | .0213 | ns | ns | ns |
| SNP N | 216 | .52 | .49 | ns | ns | ns | ns |
| SNP 1 | 260 | .72 | .64 | ns | ns | ns | ns |
| SNP 7 | 527 | .69 | .57 | ns | ns | ns | ns |
| AFM234wc5 | 850 | … | … | ns | ns | ns | ns |
| D10S1640 | 1055 | … | … | ns | ns | ns | ns |
Note.— All markers tested were in the 1.1-Mb region, and the frequency of the most common allele for each SNP is shown. Two-point–association analysis was performed, with underrepresented alleles (<2.5% in the combined sample) grouped together; ns = not significant.
Two-point association test comparing 62 affected and 94 unaffected subjects, including related individuals. The tests were performed with the DISEQ program.
Empirical P values obtained by performing 10,000 Monte Carlo simulations between the 62 affected and 94 unaffected subjects.
Empirical P values obtained by performing repeated simulations to generate tables having the same marginal totals as the one under consideration, and counting the number of times that a χ2 value associated with the real table was achieved by the randomly simulated data.
Two-point association test performed between 34 cases and 53 controls separated by more than three meiotic steps (independent sample).
Empirical P values obtained by performing 10,000 Monte Carlo simulations in the independent case-control sample.
Figure 2.
Haplotype frequencies in the whole sample (all sample) and in the subsample obtained controlling for close relatedness among subjects (selected sample). Haplotype-association tests performed in the selected sample showed that the haplotype between SNP AV70 and pSNP 3A (dark gray) was significantly associated with the disease. The haplotype flanked by SNP NG3 and AFM214zb6 (light gray) was more frequent in cases than in controls. Microsatellite markers were not considered in the haplotype-association tests. Microsatellite alleles are shown in italics. The same allele at marker D10S1652 was always present in the associated haplotype (Ombra et al. 2001).
When we consider only individuals with complete haplotypes (56 cases and 71 controls), the most common 67-kb risk haplotype in patients with UAN was observed in 40 cases (71.4%), compared with 35 controls (49.3%). Homozygotes accounted for 32.5% (13/40) of these cases. The 67-kb haplotype was associated with a significantly increased risk of UAN (odds ratio [OR] 2.57; 95% CI 1.15–5.80).
Molecular Characterization of the UAN-Susceptibility Gene
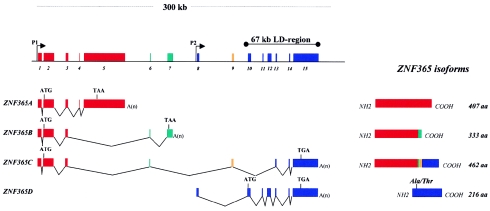
All previously described genes except the EST 603040095F1 were located outside the 67-kb associated interval; therefore, none of them were further analyzed. We focused our interest on the novel gene corresponding to this EST. To characterize the 5′ and 3′ regions, we performed RT-PCR and used RACE strategies, starting from the coding region. An extensive gene, spanning a 300-kb region and overlapping the 67-kb associated interval, was disclosed. Genomic and cDNA sequence comparisons showed that the gene consists of 15 exons and includes the EST 603040095F1 within the core interval and the KIAA0844 sequence outside it (figs. 1 and 3).
Figure 3.
Genomic structure of ZNF365, showing alternative splicing and alternative start sites that generate four different transcripts. Initiation codons (ATG) and stop codons (TAA or TGA) are shown for all isoforms. Two different promoters, CpG island (P1) and TATA box (P2) are indicated (arrows). Exons used for each transcript (colored boxes), together with the associated 67-kb interval, are indicated. The Ala62Thr missense variant found in the ZNF365D isoform is shown. Exons 1–5 and 10–13 identified the KIAA0844 transcript and the EST 603040095F1, respectively.
In the human kidney, we identified a complex pattern of alternative splicing and transcriptional start sites that generated different proteins of 407, 333, 462, and 216 amino acids, respectively (fig. 3).
ZNF365A transcript
This 1,798-bp transcript, containing the entire 1,224-bp ORF, was obtained by RT-PCR based on the information of the Kazusa DNA Research Institute for KIAA0844 cDNA and was deposited in the EMBL Nucleotide Sequence Database (Nagase et al. 1998). It was split in 5 exons encoding a putative protein of 407 amino acids with a molecular weight of 46,558 Da, and its putative promoter region contains a CpG island. The N-terminal region of this protein (amino acids 26–51) harbors the classical zinc finger domain of C2H2 family (fig. 4). This domain is found in numerous nucleic acid–binding proteins, but some investigators have suggested that it could be used in protein-protein interaction and in membrane association (Laity et al. 2001). This gene was renamed “zinc finger protein 365” (ZNF365A) by the HUGO Gene Nomenclature Committee. Secondary structure predictions for this protein, which were created using various programs, strongly suggest the presence of four α-helical coiled-coil domains that are found in various protein families and frequently used as oligomerization motifs (Jorde et al. 2000). Northern analysis showed that the transcript was ubiquitously expressed, with a 5-kb transcript present at low levels in all adult human tissues examined and with slightly higher levels in the brain (data not shown).
Figure 4.
Alignment of the amino acid sequences of ZNF365 protein isoforms. Amino acids are indicated by single-letter codes. Identical amino acids are indicated (gray boxes). The C2H2 zinc finger domain (solid line) and the coiled-coil segments (dotted line) are indicated. The predicted transmembrane domains are framed. ZNF365D has eight consensus sites for N/O-glycosylation (asterisks). The Ala62Thr missense variant found in the ZNF365D isoform is shown (arrow).
ZNF365B transcript
This 1,671-bp transcript, was identified by EST analysis and cDNA clone sequencing. The 333–amino acid protein was identical to the ZNF365A protein in the N-terminal region (exons 1–3) containing the zinc finger domain and coiled-coil segments, but it diverged at C-terminal region (exons 6–7) (figs. 3 and 4).
ZNF365C transcript
The ZNF365C transcript was isolated by RT-PCR using different combinations of primers anchored in different exons of the KIAA0844 transcript and EST 603040095F1. This 3,376-bp transcript overlapped the critical region, showing that the KIAA0844 transcript and the EST 603040095F1 belong to the same gene. ZNF365C has the first four exons and the same CpG island promoter as the ZNF365B transcript outside of the critical region and four different terminal exons (9, 13, 14, and 15). Exons 13, 14, and 15 are located in the associated region (fig. 3). Moreover, the encoded 462–amino acid protein was similar to ZNF365B protein but had a different C-terminal region. Computer-assisted analysis of this isoform, based on transmembrane domain prediction, revealed at least two membrane-spanning domains in this novel C-terminal region from amino acid 344 to 395, suggesting that it could be an integral membrane protein (fig. 4).
ZNF365D transcript
The ZNF365D transcript spans a region of ∼150 kb and uses its own specific TATA-box promoter, differing from ZNF365A, -B, and -C, which use the same CpG island promoter. Interestingly, all the transcript coding exons are included in the 67-kb associated interval (fig. 3). The full-length cDNA was obtained by 5′- and 3′-RACE strategies. The composite 2,695-bp cDNA sequence encoded a 216–amino acid protein completely different from the proteins encoded by ZNF365A and -B but identical to ZNF365C in the C-terminal region (figs. 3 and 4). Prediction analysis revealed a strong transmembrane domain in this isoform at positions 126–149, suggesting that, like ZNF365C, it could be an integral membrane protein with N-termini outside the cell. In the noncytosolic portion, several potential modification sites were detected, including possible N- and O-linked glycosylation sites at Asn-20, Asn-82, Thr-88, Thr-89, Ser-90, Ser-91, Ser-93, and Thr-97, suggesting that this outer membrane protein portion may be glycosylated.
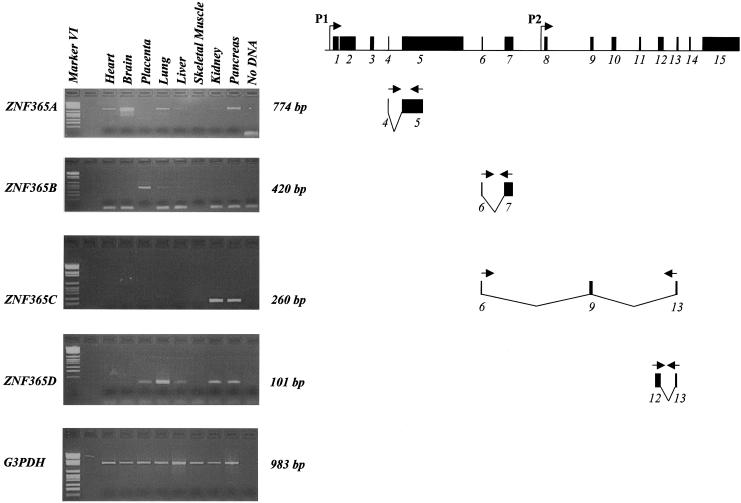
When all isoforms of 407, 333, 462, and 216 amino acids were used as a query, BLAST algorithms revealed no significant homology to other proteins deposited in public databases, suggesting that they could be a new class of proteins. ZNF365B, -C, and -D revealed no signal by northern analysis. RT-PCR was performed to study the expression profile of the ZNF365 gene in different human tissues, using a normalized cDNA panel. Significant expression was found in brain for ZNF365A transcript, as was observed in northern blot analysis (data not shown), and low-level expression occurred in other tissues. ZNF365B transcript was detected in placenta and at low levels in lung and liver, ZNF365C was detected only in kidney and pancreas, and ZNF365D was detected in placenta, lung, liver, kidney, and pancreas (fig. 5).
Figure 5.
RT-PCR analysis of the expression of ZNF365A, -B, -C, and- D transcripts in human heart, brain, placenta, lung, liver, skeletal muscle, kidney, and pancreas. G3PDH (bottom panel) was used as a control gene. The amplified fragments of the ZNF365 transcripts and the size of the amplifications are shown (right).
Identification of a Specific ZNF365D Susceptibility Variant
To test our hypothesis that ZNF365 may have a role in UAN, we focused on detecting possible sequence variations that alter the level or activity of the coded proteins, ultimately contributing to the development of the clinical phenotype. We resequenced all exons of ZNF365 and part of the intronic regions encompassing the critical region. Resequencing was performed in eight patients who carried the putative nephrolithiasis-susceptibility haplotype. Of special interest was the identification of two variants that cause missense in exon 5 (ZNF365A transcript), resulting in serine/alanine amino acid substitution (variant Ser337Ala), and in exon 12 (ZNF365D transcript), resulting in alanine/threonine amino acid substitution (variant Ala62Thr) (fig. 3). Moreover, we identified a number of polymorphisms in UTR and intronic regions of the gene.
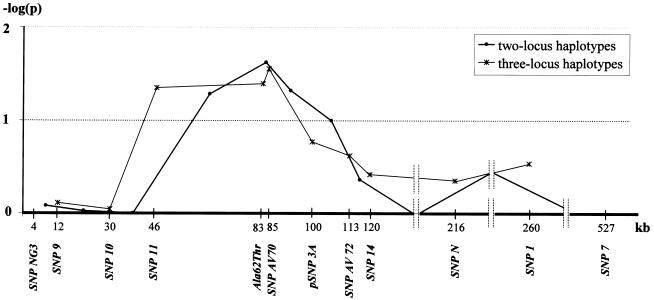
Association analysis was performed in the selected case-control sample to examine whether the presence of these alleles conferred increased risk of UAN. Comparison of allelic frequencies in cases and controls showed strong association between UAN and variant Ala62Thr (P=.0096; empirical P=.0051) with a significant increase in risk (OR 2.73; 95% CI 1.01–7.44). However, the Ser337Ala variant was not significantly associated. When we compared haplotype frequencies in cases and controls who carried the identified variant, we observed significant evidence of association with UAN for different haplotype windows that included the variant, whereas a sharp decrease of association was observed when moving away from the associated site (fig. 6). The variant Ala62Thr was invariably associated with the 67-kb core haplotype that confers increased risk. The estimated penetrances of Ala62Thr showed an increasing trend across genotypes (penetrance of Ala/Ala homozygotes 17.6%; penetrance of Ala/Thr heterozygotes 32.3%; penetrance of Thr/Thr homozygotes 44.8%), thus suggesting that the mutation may be causative.
Figure 6.
Results of the haplotype-association analysis performed in the selected sample comprising 64 case haplotypes and 82 control haplotypes. The curves represent the extent of association, expressed as −log(p), between UAN and multilocus haplotypes. For two-locus haplotypes, results are shown for the pair of adjacent markers, and points are drawn at the midpoint between the two. For three-locus haplotypes, results are shown for combinations of adjacent markers, and points are drawn at the position of the middle marker. Empirical significance was assessed through Monte Carlo simulations. Only common haplotypes (frequency >5% in the combined sample) were compared.
After the identification of the Ala62Thr mutation, we investigated the influence of this variant on the linkage findings. Employing a procedure like the one used by Hampe et al. (2002) to determine the contributions of a specific polymorphism to a linkage curve, we performed two complementary analyses by stratifying the family sample on affected subjects carrying and not carrying the variant. Affected subjects who did not have the selected genotype were coded as “unknown.” For these analyses, we used a large pedigree comprising 335 subjects and connecting 60 affected individuals (43 carrying the variant Ala62Thr and 17 not carrying it). When we considered only those subjects who carried the variant to be affected, we found that the evidence of linkage in the whole region was increased (peak P value of .0005 for Simwalk2 statistic D), whereas the evidence for linkage dropped steeply (P>.05 for all Simwalk2 statistics on the whole region) when only affected subjects not carrying the variant were considered to be affected, indicating a major influence of Ala62Thr on the linkage evidence in this region.
Given that some polymorphisms (e.g., a nonsynonymous change [missense mutation]) are more likely to alter the function of a protein, we investigated the possibility that the high-risk allele ZNF365D (Ala62Thr) may play a functional role in the UAN phenotype. Interestingly, computer-assisted analysis strongly predicted the secondary structural alteration of the ZNF365D protein induced by the Ala62Thr variation. Essentially, the third α-helical loop of the putative UAN-susceptibility allele (threonine) is not present compared to the not associated allele (alanine). These results suggest that threonine causes a significant conformational change that may have important implications for the biological function of this protein or its interaction with other proteins.
Discussion
UAN is a multifactorial disorder influenced by genetics and environmental factors. The clustering of the disease in families suggests a predisposing set of genes not yet characterized. On the other hand, a purine-rich diet and climatic factors have an equally important role in the etiology of the disease. Because UAN is a very common disorder, it must result from many different combinations of causative risk factors. Identification of groups of patients sharing the same predisposing genes and environment in open populations is therefore very difficult. The study of an isolated population characterized by genetic and environmental homogeneity presents potential advantages for mapping genes for common multifactorial diseases, such as UAN (Wright et al. 1999; Peltonen et al. 2000). The ideal isolated population should be a large pedigree with many generations (Jorde et al. 2000), in which allelic and locus homogeneity is likely to be maximized. Indeed, a powerful approach to LD mapping is the ascertainment of a set of distantly related affected individuals, which would enrich for genetic forms of multifactorial diseases and in which a large proportion of affected individuals are likely to share the same disease-predisposing allele inherited from common ancestors.
We performed our study in a particularly isolated village of a secluded area of east-central Sardinia, in which 80% of the present-day population appears to derive from 8 paternal and 11 maternal ancestral lineages (Angius et al. 2001). All genealogical data, as well as phenotypic and genetic data, are stored in relational databases that connect 5,219 individuals in a 16-generation pedigree. The bioinformatic framework comprises several algorithms that select from the database the subjects most suitable for the analysis.
We found an increased prevalence of UAN cases in Talana. Many cases cluster in families of varying sizes. Elsewhere, we reported results of a multistep genomewide search in an extended family comprising 37 individuals with UAN, and we identified a susceptibility locus of ∼2.5 cM on 10q21-q22, with the highest evidence at marker D10S1652 (Ombra et al. 2001).
In the present study, we selected, on the basis of phenotype severity, an additional 62 UAN cases; all were connected in an extended genealogical tree. Linkage analysis confirms the involvement of the identified susceptibility locus. Given the significant evidence, derived from formal kinship analysis, that UAN cases are more likely than controls to share all four alleles IBD (i.e., to be autozygous), we also performed homozygosity mapping in our sample of cases. Although homozygosity mapping is usually performed in the study of rare Mendelian recessive disorders, it has also been suggested as a powerful tool for mapping complex disease-predisposing genes (even those with minor effects) in inbred, isolated populations (Broman and Weber 1999).
To detect the causative gene, we constructed a transcriptional map of the 1.1-Mb consensus genomic sequence, which corresponds to the identified 2.5-cM 10q21-q22 UAN locus. No obvious candidate genes were found. We then applied fine-mapping approaches, to refine the critical region associated with UAN. Since an increased rate of false-positive results is expected in an association analysis in which cases are more closely related than controls, we carefully selected a subsample by use of the information from the genealogy data. In the resulting group, cases and controls present the same degree of relatedness and therefore do not inflate the significance of the test (Genin and Clerget-Darpoux 1996). We were able to identify a common core haplotype found in excess in cases compared with controls and to narrow, through multilocus LD analysis, the critical interval associated with UAN to ∼67 kb, encompassing marker D10S1652. This small region was included in a 300-kb interval in which we found the novel gene ZNF365, which encodes at least four different proteins. Although, at present, there is no indication as to the function of these proteins, one of these isoforms, which we named “Talanin,” resides entirely within the 67-kb region, representing a strong candidate risk factor for the disease. Through sequence analysis of the coding region and most of the introns, we detected several polymorphisms and only one coding variant, Ala62Thr, that provided the strongest evidence of association with UAN (P=.0051) and that segregates the original evidence for linkage to the region. We also obtained significant evidence of increased homozygosity in the associated region among affected individuals. This may suggest that the presence of two copies of the susceptibility allele would lead to a greater increase in risk of UAN than would the presence of a single copy, under the assumption that the susceptibility gene has a dosage effect.
The underlying mechanism by which the Ala62Thr variant confers susceptibility to UAN remains unclear; however, bioinformatic simulation predicts a change in the secondary structure of the protein, since one of the α-helixes disappears when alanine is changed to threonine. This suggests that the Ala62Thr variant may play a causative role. For the other polymorphism showing association with UAN (pSNP3A, which is contained in intron 9 of the gene), we did not identify a functional role. Therefore, its association with UAN could be due to its being in strong LD with Ala62Thr.
It is always difficult to prove the phenotypic effect of a missense mutation in a protein, such as Talanin, for which the function is not yet known. Until functional studies can explain the role of the Ala62Thr variant in UAN, it will be important to replicate these results in other populations. Preliminary results in other isolated villages in the same area of Sardinia indicate that the same Ala62Thr susceptibility variant is present in 12 of 14 unrelated patients with UAN, compared with 47 of 92 controls (P=.02). On the other hand, this variant is rather common in the general Sardinian population (frequency 32%).
Together, these results provide convergent evidence for the interpretation that Ala62Thr may contribute to risk of UAN. Ala62Thr provided the strongest evidence of association with UAN, although we cannot exclude the presence, in the associated interval, of another causative variant that is in LD with the identified Talanin polymorphism. Future genetic and functional studies are necessary to further validate these conclusions.
Our results suggest that Talana villagers and similar populations can be considered ideal for an efficient and parsimonious approach to the identification of genetic risk factors for complex traits. We believe that a study in which a small number of samples are phenotypically well characterized and strategically placed on extended genealogies can challenge studies based on much greater numbers in outbred populations.
Acknowledgments
We are grateful to the population of Talana and to all the individuals who participated in this study. We thank Anna Rosa Tegas for support in Talana.
Electronic-Database Information
Accession numbers and URLs for data presented herein are as follows:
- EMBL Nucleotide Sequence Database, http://www.ebi.ac.uk/embl/ (for mRNA sequences ZNF365A [accession number AJ505147], ZNF365B [accession number AJ505148], ZNF365C [accession number AJ505149], ZNF365D [accession number AJ505150], MRF-2 [accession number XM084482], RTKN-L [accession number BC025765], and KIAA0844 [accession number AB020651]; and ESTs 603251916F1 [accession number BI603606], hd42c05.x1 [accession number AW511012], and 603040095F1 [accession number BI822044])
- Online Mendelian Inheritance in Man (OMIM), http://www.ncbi.nlm.nih.gov/Omim/ (for UAN) [PubMed]
References
- Abney M, McPeek MS, Ober C (2000) Estimation of variance components of quantitative traits in inbred populations. Am J Hum Genet 66:629–650 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Abramson RG, Lipkowitz MS (1990) Basic principles in transport. In: Kinne RKH (ed) Comparative physiology, vol 3. Karger, Basel, pp 115–153 [Google Scholar]
- Angius A, Melis PM, Morelli L, Petretto E, Casu G, Maestrale GB, Fraumene C, Bebbere D, Forabosco P, Pirastu M (2001) Archival, demographic and genetic studies define a Sardinian sub-isolate as a suitable model for mapping complex traits. Hum Genet 109:198–209 [DOI] [PubMed] [Google Scholar]
- Baggio B (1999) Genetic and dietary factors in idiopathic calcium nephrolithiasis: what do we have, what do we need? J Nephrol 12:371–374 [PubMed] [Google Scholar]
- Berger B, Wilson DB, Wolf E, Tonchev T, Milla M, Kim PS (1995) Predicting coiled coils by use of pairwise residue correlations. Proc Natl Acad Sci USA 92:8259–8263 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Broman KW, Weber JL (1999) Long homozygous chromosomal segments in reference families from the Centre d’Etude du Polymorphisme Humain. Am J Hum Genet 65:1493–1500 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burgess AW, Ponnuswamy PK, Scheraga HA (1974) Analysis of conformations of amino acid residues and prediction of backbone topography in proteins. Isr J Chem 12: 239–286 [Google Scholar]
- Chou P, Fasman GD (1978) Prediction of the secondary structure of proteins from their aminoacid sequence. Adv Enzymol 47:45–148 [DOI] [PubMed] [Google Scholar]
- Cserzo M, Wallin E, Simon I, von Heijne G, Elofsson A (1997) Prediction of transmembrane alpha-helices in prokaryotic membrane proteins: the dense alignment surface method. Protein Eng 10:673–676 [DOI] [PubMed] [Google Scholar]
- Curhan GC, Willett WC, Rimm EB, Stampfer MJ (1997) Family history and risk of kidney stones. J Am Soc Nephrol 8:1568–1573 [DOI] [PubMed] [Google Scholar]
- Dufton MJ, Hider RC (1977) Snake toxin secondary structure predictions: structure activity relationships. J Mol Biol 115:117–193 [DOI] [PubMed] [Google Scholar]
- Enomoto A, Kimura H, Chairoungdua A, Shigeta Y, Jutabha P, Cha SH, Hosoyamada M, Takeda M, Sechine T, Igarashi T, Matsuo H, Kikuchi Y, Oda T, Ichida K, Hosoya T, Shimokata K, Niwa T, Kanai Y, Endou H (2002) Molecular identification of a renal urate-anion exchanger that regulates blood urate levels. Nature 417:447–452 [DOI] [PubMed] [Google Scholar]
- Fu Q, Yu L, Liu Q, Zhang J, Zhang H, Zhao S (2000) Molecular cloning, expression characterization, and mapping of a novel putative inhibitor of rho GTPase activity, RTKN, to D2S145–D2S286. Genomics 66:328–332 [DOI] [PubMed] [Google Scholar]
- Hampe J, Frenzel H, Mirza MM, Croucher PJ, Cuthbert A, Mascheretti S, Huse K, Platzer M, Bridger S, Meyer B, Nurnberg P, Stokkers P, Krawczak M, Mathew CG, Curran M, Schreiber S (2002) Evidence for a NOD2-independent susceptibility locus for inflammatory bowel disease on chromosome 16p. Proc Natl Acad Sci USA 99:321–326 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garnier J, Osguthorpe DJ, Robson B (1978) Analysis of the accuracy and implications of simple methods for predicting the secondary structure of globular proteins. J Mol Biol 120:97–120 [DOI] [PubMed] [Google Scholar]
- Genin E, Clerget-Darpoux F (1996) Association studies in consanguineous populations. Am J Hum Genet 58:861–866 [PMC free article] [PubMed] [Google Scholar]
- Hofmann K, Stoffel W (1993) TMbase: a database of membrane spanning proteins segments. Biol Chem 374:166 [Google Scholar]
- Jaeger P (1996) Genetic versus environmental factors in renal stone disease. Curr Opin Nephrol Hypertens 5:342–346 [DOI] [PubMed] [Google Scholar]
- Jorde LB, Watkins WS, Kere J, Nyman D, Eriksson AW (2000) Gene mapping in isolated populations: new roles for old friends? Hum Hered 50:57–65 [DOI] [PubMed] [Google Scholar]
- Kruglyak L, Daly MJ, Lander ES (1995) Rapid multipoint linkage analysis of recessive traits in nuclear families, including homozygosity mapping. Am J Hum Genet 56:519–527 [PMC free article] [PubMed] [Google Scholar]
- Lahoud MH, Ristevski S, Venter DJ, Jermiin LS, Bertoncello I, Zavarsek S, Hasthorpe S, Drago J, de Kretser D, Hertzog PJ, Kola I (2001) Gene targeting of Desrt, a novel ARID class DNA-binding protein, causes growth retardation and abnormal development of reproductive organs. Genome Res 11:1327–1334 [DOI] [PubMed] [Google Scholar]
- Laity JH, Lee BM, Wright PE (2001) Zinc finger proteins: new insights into structural and functional diversity. Curr Opin Struct Biol 11:39–46 [DOI] [PubMed] [Google Scholar]
- Lim V I (1974) Algorithms for prediction of alpha-helical and beta-structural regions in globular proteins. J Mol Biol 88:873–894 [DOI] [PubMed] [Google Scholar]
- Lupas A, Van Dyke M, Stock J (1991) Predicting coiled coils from protein sequences. Science 252:1162–1164 [DOI] [PubMed] [Google Scholar]
- Nagano K (1977) Logical analysis of the mechanism of protein folding. IV. Supersecondary structures. J Mol Biol 109:235–250 [DOI] [PubMed] [Google Scholar]
- Nagase T, Ishikawa K, Suyama M, Kikuno R, Hirosawa M, Miyajima N, Tanaka A, Kotani H, Nomura N, Ohara O (1998) Prediction of the coding sequences of unidentified human genes. XII. The complete sequences of 100 new cDNA clones from brain which code for large proteins in vitro. DNA Res 5:355–364 [DOI] [PubMed] [Google Scholar]
- Ombra MN, Forabosco P, Casula S, Angius A, Maestrale G, Petretto E, Casu G, Colussi G, Usai E, Melis P, Pirastu M (2001) Identification of a new candidate locus for uric acid nephrolithiasis. Am J Hum Genet 68:1119–1129 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pasquier C, Hamodrakas SJ (1999) An hierarchical artificial neural network system for the classification of transmembrane proteins. Protein Eng 12:631–634 [DOI] [PubMed] [Google Scholar]
- Peltonen L, Palotie A, Lange K (2000) Use of population isolates for mapping complex traits. Nat Rev Genet 1:182–190 [DOI] [PubMed] [Google Scholar]
- Persson B, Argos P (1996) Topology prediction of membrane proteins. Protein Sci 5:363–371 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ristaldi MS, Pirastu M, Rosatelli C, Monni G, Erlich H, Saiki R, Cao A (1989) Prenatal diagnosis of beta-thalassaemia in Mediterranean populations by dot blot analysis with DNA amplification and allele specific oligonucleotide probes. Prenat Diagn 9:629–638 [DOI] [PubMed] [Google Scholar]
- Rivers K, Shetty S, Menon M (2000) When and how to evaluate a patient with nephrolithiasis. Urol Clin North Am 27:203–213 [DOI] [PubMed] [Google Scholar]
- Roch-Ramel F, Guisan B (1999) Renal transport of urate in humans. News Physiol Sci 14:80–84 [DOI] [PubMed] [Google Scholar]
- Scheinman SJ (1999) Nephrolithiasis. Semin Nephrol 19:381–388 [PubMed] [Google Scholar]
- Serio A, Fraioli A (1999) Epidemiology of nephrolithiasis. Nephron Suppl 81:26–30 [DOI] [PubMed] [Google Scholar]
- Sobel E, Lange K (1996) Descent graphs in pedigree analysis: applications to haplotyping, location scores, and marker-sharing statistics. Am J Hum Genet 58:1323–1337 [PMC free article] [PubMed] [Google Scholar]
- Terwilliger JD (1995) A powerful likelihood method for the analysis of linkage disequilibrium between trait loci and one or more polymorphic marker loci. Am J Hum Genet 56:777–787 [PMC free article] [PubMed] [Google Scholar]
- von Heijne G (1992) Membrane protein structure prediction, hydrophobicity analysis and the positive-inside rule. J Mol Biol 225:487–494 [DOI] [PubMed] [Google Scholar]
- Wright AF, Carothers AD, Pirastu M (1999) Population choice in mapping genes for complex diseases. Nat Genet 23:397–404 [DOI] [PubMed] [Google Scholar]
- Wu X, Lee CC, Muzny DM, Caskey CT (1989) Urate oxidase: primary structure and evolutionary implications. Proc Natl Acad Sci USA 86:9412–9416 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu X, Muzny DM, Lee CC, Caskey CT (1992) Two independent mutational events in the loss of urate oxidase. J Mol Evol 34:78–84 [DOI] [PubMed] [Google Scholar]