Abstract
In the search for genes involved in type 1 diabetes (T1D), other than the well-established risk alleles at the human leukocyte antigen loci, we have investigated the association and interaction of polymorphisms in genes involved in the IL4/IL13 pathway in a sample of 90 Filipino patients with T1D and 94 controls. Ten single-nucleotide polymorphisms (SNPs), including two promoter SNPs in the IL4R locus on chromosome 16p11, one promoter SNP in the IL4 locus on chromosome 5q31, and four SNPs—including two promoter SNPs—in the IL13 locus on chromosome 5q31 were examined for association, linkage disequilibrium, and interaction. We found that both individual SNPs (IL4R L389L; odds ratio [OR] 0.34; 95% confidence interval [CI] 0.17–0.67; P=.001) and specific haplotypes both in IL4R (OR 0.10; 95% CI 0–0.5; P=.001) and for the five linked IL4 and IL13 SNPs (OR 3.47; P=.004) were strongly associated with susceptibility to T1D. Since IL4 and IL13 both serve as ligands for a receptor composed, in part, of the IL4R α chain, we looked for potential epistasis between polymorphisms in the IL4R locus on chromosome 16p11 and the five SNPs in the IL4 and IL13 loci on chromosome 5q31 and found, through use of a logistic-regression model, significant gene-gene interactions (P=.045, corrected for multiple comparisons by permutation analysis). Our data suggest that the risk for T1D is determined, in part, by polymorphisms within the IL4R locus, including promoter and coding-sequence variants, and by specific combinations of genotypes at the IL4R and the IL4 and IL13 loci.
Introduction
Type 1 diabetes (T1D [MIM 222100]) is a multifactorial autoimmune disease involving dysregulation of glucose homeostasis that results from destruction of the insulin-producing β cells within the pancreatic islets by autoreactive T cells (Tisch and McDevitt 1996; Azar et al. 1999). T1D, also known as “insulin-dependent diabetes mellitus” or “juvenile-onset diabetes,” has an environmental as well as a strong genetic component; the concordance among MZ twins is 30%–70%, depending on the human leukocyte antigen (HLA) type, and the sibling risk ratio (λs) is 15 (Risch 1987; Pociot and McDermott 2002). A variety of genomewide scans in multiplex families have identified a number of linkage regions for T1D (e.g., Cox et al. 2001), including the highly polymorphic HLA region on chromosome 6p21, which has been estimated to contribute >50% of the total genetic risk (Noble et al. 1996). Association studies in a number of populations have demonstrated that, although HLA-DRB1, DQA1, and DQB1 are the major genetic determinants of T1D susceptibility, multiple loci within the HLA region appear to contribute to disease risk (Erlich et al. 1996; Tisch and McDevitt 1996; Noble et al. 2000, 2002; Bugawan et al. 2002). Some HLA alleles and haplotypes appear to confer susceptibility (positive association) and some protection (negative association), whereas others are neutral. The identification of potentially disease-related alleles and haplotypes has been aided by the study of different human populations, with differing patterns of linkage disequilibrium (LD). Linkage and association studies outside the HLA region have, thus far, with the exception of the insulin locus (Bennett et al. 1996) and the CTLA-4 locus (Nistico et al. 1996; Larsen et al. 1999; Lee et al. 2001; Klitz et al. 2002), failed to identify individual genes and specific alleles that are reproducibly associated with T1D.
Association studies with candidate genes involved in immune responses, such as those encoding elements of the T cell activation pathway, represent one approach to finding T1D disease genes. IL4 and IL13 have been shown to protect against diabetes development in rodent models of T1D (Zaccone et al. 1999), suggesting the possibility that cytokines involved in the Th1 and/or Th2 pathways may play a role in T1D pathogenesis (Glimcher and Murphy 2000). IL4 (MIM 147780) and IL13 (MIM 147683) are key components in the induction of the Th2 lymphocyte phenotype and the downregulation of the Th1 lymphocyte phenotype. In humans, IL4 transcript levels are greatly reduced in new-onset T1D (Berman et al. 1996), and some authors have speculated that IL13, as an anti-inflammatory cytokine and a mediator of the Th2 pathway, represents a potential therapeutic approach in the prevention of T1D (Kretowski et al. 2000). The effects of IL4 and IL13 on Th1/Th2 balance and on other immune phenotypes are mediated via binding to a receptor containing the IL4R α chain. The heterodimeric receptor consists of a high-affinity α subunit, encoded by the IL4R locus (MIM 147781) on chromosome 16p11.2-12.1, and either the common γ subunit (IL2RG mapping to Xq13) or the IL13R α subunit (which maps to chromosome X). The IL13 receptor, also a heterodimer, is composed of one IL13R α subunit and either IL4R or an IL13R α2 subunit. (Kawakami et al. 2001).
Some IL4R SNPs have been reported to affect signal transduction via the IL4 receptor (Kruse et al. 1999) and the level of gene expression of IL4R (Hackstein et al. 2001). In addition, polymorphisms in the IL4R, IL4, and the IL13 loci have been reported to be associated with atopic asthma and serum immunoglobulin E (IgE) levels (Noguchi et al. 1998; van der Pow Kraan et al. 1999; Graves et al. 2000; Sandford et al. 2000; Howard et al. 2002). Thus, SNPs within the IL4R locus, as well as those within the genes encoding the ligands IL4 and IL13, represent a set of candidate gene polymorphisms for T1D susceptibility, and we have examined association and interaction of these SNPs in a Filipino population.
Subjects and Methods
Subjects
All patients and controls were from the southern region of Luzon Island, Philippines. Patients included in the study (n=90) were affected by T1D as defined by the 1997 American Diabetes Association classification (Expert Committee on the Classification of Diabetes Mellitus 1997). Patients were born in the Philippines, and all have two Filipino parents. In a previous study, these patients were characterized as having C-peptide levels <0.3 mmol/liter and were tested for the presence of antibodies to islet cell autoantigens, confirming that they were indeed patients with classical T1D (Medici et al. 1999). A total of 94 unaffected Filipino subjects without a family history of diabetes were collected as the control group.
Genotyping
Eight IL4R SNPs, two IL13 SNPs, and one IL4 SNP were genotyped using a linear-array (immobilized probe) method essentially as described by Mirel et al. (2002). In brief, ∼100 ng of genomic DNA was PCR amplified using nine biotinylated primer pairs in a single PCR. The labeled amplicons were hybridized to an immobilized sequence-specific oligonucleotide probe array on a nylon membrane. The presence of bound amplicon to a specific probe is detected using streptavidin–horseradish peroxidase and a soluble colorless substrate, 3,3′,5,5′-tetramethylbenzidine, which can be converted, in the presence of H2O2, to a blue precipitate. The two IL13 and two IL4R promoter SNPs (table 1) were genotyped using allele-specific PCR on a PE9700 thermal cycler (ABI), measuring SyBr Green (Molecular Probes) fluorescence (Higuchi et al. 1993).
Table 1.
SNPs Used in this Study, and Allele Frequencies in Patients with T1D and Controls
|
Allele |
Frequency in(%) |
|||||
| Geneand SNP | Reference | Variant | Controls | Patients | Pa | ORb (95% CI) |
| IL4R: | ||||||
| 5′ (−3223) | C | T | 41.9 | 51.1 | .10 | 1.45 (.96–2.19) |
| 5′ (−1914) | C | T | 44.1 | 42.7 | .86 | 1.06 (.70–1.60) |
| I50V | A | G | 44.1 | 53.9 | .06 | 1.48 (.98–2.23) |
| N142N | C | G | 1.1 | 0 | .17 | .52 (.00–2.13) |
| E375A | A | C | 20.7 | 11.7 | .02 | .50 (.28–.90) |
| L389L | G | T | 17.6 | 6.7 | .001 | .34 (.17–.67) |
| C406R | T | C | 19.1 | 11.7 | .05 | .56 (.31–.99) |
| S478P | T | C | 18.6 | 11.7 | .06 | .58 (.32–1.04) |
| Q551R | A | G | 27.7 | 23.3 | .34 | .80 (.50–1.27) |
| S761P | T | C | 100.0 | 100.0 | … | … |
| IL4: | ||||||
| 5′ (−524) | T | C | 30.9 | 34.4 | .46 | 1.18 (.76–1.82) |
| IL13: | ||||||
| 5′ (−1512) | A | C | 30.6 | 41.0 | .05 | 1.58 (1.03–2.42) |
| 5′ (−1112) | C | T | 23.1 | 30.9 | .12 | 1.49 (.94–2.37) |
| Intron 3 | C | T | 40.4 | 45.6 | .32 | 1.23 (.82–1.86) |
| R110Q | G | A | 39.9 | 46.1 | .23 | 1.29 (.85–1.95) |
Differences in reference allele frequencies between cases and controls were tested using a χ2 test. Values in boldface italic type are nominally significant P values (P<.05).
ORs refer to the variant allele. Where the frequency in patients was 0, the OR has been computed under the assumption that a single patient sample carried the variant allele.
Molecular Haplotyping
The seven-SNP haplotypes are labeled and identified by the allele of IL4R SNPs 4–10 present on that haplotype. Unambiguous haplotypes comprising seven IL4R SNPs were obtained by inference when fewer than two heterozygous SNPs were present in an individual. In other cases, a molecular haplotyping method was used, with details as follows: Two allele-specific PCRs were performed in parallel, such that the amplification product spanned two or more SNP loci. The choice of allele-specific and common 5′-biotinylated primers depended on which SNP loci were heterozygous but was limited to the SNPs within IL4R exon 12. The two biotin-labeled PCR products were subsequently hybridized separately to the immobilized probe linear array described in the “Genotyping” subsection, and the in-phase, linked SNP alleles were identified.
Statistical Haplotype Estimation
Maximum-likelihood haplotype frequencies were computed using an expectation-maximization (EM) algorithm (see Excoffier and Slatkin 1995), as implemented by the Arlequin software program (L. Excoffier).
Gene-Gene Interaction Modeling and Permutation Analysis
Gene-gene interaction modeling
Gene-by-gene interactions at SNPs in different genes were evaluated by assessing whether the genotype frequencies at unlinked loci were independent (i.e., the IL13 and IL4 SNPs on chromosome 5 and the IL4R SNPs on chromosome 16) among patients. These analyses were performed for each pair of unlinked SNPs, by means of a χ2 test in contingency tables with marginals defined by genotype counts either in patients or in controls. The χ2 values and the corresponding degrees of freedom were added over all IL4R SNP comparisons, and the P value of the sum of χ2 values was computed for each of the chromosome 5 SNPs.
For each IL4R SNP, the homozygote genotype with the highest odds ratio (OR) was given a value of 2, the heterozygote was given a value of 1, the other homozygote was 0. A logistic regression was performed on all nine IL4R polymorphisms in this way, and a new numerical variable, “il4r,” was derived as follows:
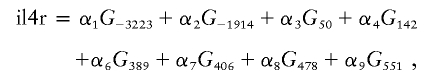 |
where Gi denotes the genotype (0, 1, or 2) at the ith SNP position and αj denotes the coefficient fitted by logistic regression (the coefficients fitted by the regression were α1=0.368, α2=0.053, α3=0.37, α4=0.061, α5=0.66, α6=1.08, α7=0.57, α8=0.54, and α9=0.22). Epistasis was then tested independently for each of the five chromosome 5 SNPs by fitting the following logistic regression model: P(T1D)=exp(X)/[1+exp(X)], where X=C+β1il4r+β2Gchr5i+β3(il4r×Gchr5i), C is the intercept, β represents the parameter estimates in the logistic regression model, and Gchr5i is the genotype of one of the chromosome 5 SNPs (values 0, 1, or 2).
Permutation analysis
Since we compared five different SNPs, it was important to correct for multiple tests. However, a Bonferroni or a Dunn-Sidak correction is not appropriate, since the IL4 and IL13 SNPs are not independent (see table A1 in appendix A). Therefore, permutation analysis was performed, keeping the patient and control genotype frequencies constant but permuting the chromosome 5 and chromosome 16 genotypes within the patient and control groups separately. In this way, only the epistatic interaction between the two genetic regions was tested, and not the individual chromosome 5 and chromosome 16 genetic associations. Two hundred permutations were performed to test for epistasis at all five chromosome 5 SNPs each time. Analyses were performed using S-Plus, version 6.0 Professional (Insightful Corporation).
Results
We have performed an association and interaction analysis of three genes in the IL4/IL13 pathway in a case-control study among Filipinos, including 10 SNPs in the IL4R locus on chromosome 16p11, 2 of which are in the promoter region. We also analyzed one SNP in the IL4 promoter and four SNPs in the IL13 locus, two of which are in the promoter region; these loci are closely linked on chromosome 5q31.
Individual SNPs
The distributions of alleles at the individual SNPs in the IL4R locus (n=10), the IL4 locus (n=1), and the IL13 locus (n=4) among patients and controls are shown in table 1. LD patterns can be estimated using maximum-likelihood approaches with individual genotype data from unrelated individuals (Slatkin and Excoffier 1996.) The patterns of pairwise LD for these SNPs inferred among the control population are shown in tables A1 and A2 in appendix A. Among the individual IL4R SNPs, three (E375A, L389L, and C406R) showed a nominally significant association (variant allele decreased) with T1D. The variant allele at I50V was slightly increased, whereas, at S478P, the variant allele was decreased in patients; both associations had a P value of .06. The two promoter SNPs were not significantly associated with T1D, although the variant allele of the −3223 SNP was slightly increased among patients (OR 1.45, P=.10). With the exception of this promoter SNP and the I50V SNP, with which it is in strong LD, the variant allele at each SNP is underrepresented among patients. Some of the polymorphic amino acid residues in this chain appear to be biologically important and affect IL-4 receptor signaling (Kruse et al. 1999). Of the 10 IL4R SNPs typed in this study, the L389L SNP, however, showed the strongest association with T1D in this population, with significantly lower frequencies among patients than controls (OR 0.34; P=.001). Since this is a silent (synonymous) polymorphism, it is unlikely that this SNP is responsible for the observed protective effect for T1D. We note that this SNP is in very strong LD (table A1 in appendix A) with the nonsynonymous flanking SNPs (E375A, C406R, S478P, and Q551R) and that these SNPs all show a trend toward protection (negative association). The L389L SNP is also in strong negative LD with the −3223 promoter SNP. We also note that the LD pattern between the variant allele at I50V and the other coding-sequence SNPs differ between Filipinos (table A1 in appendix A) and whites (Mirel et al. 2002).
In the comparison of genotypes at the individual IL4R SNPs (table 2), the protective effect appears to be dominant, in that the heterozygote for IL4R L389L has an OR of 0.29. Among the individual SNPs on chromosome 5q31, only the variant alleles at the two IL13 promoter SNPs were increased among patients (OR 1.58, P=.05 for −1512; and OR 1.49, P=.12 for −1112) (table 1). When genotype frequencies are compared, however, the IL13 R110Q SNP showed a nominally significant association in this population (P=.03; table 2) These data suggest that the variant homozygote, but not the heterozygote, may be at increased risk for T1D. Given the multiple comparisons and the relatively small sample size in this study, however, this association with the IL13 R110Q SNP should be seen as a hypothesis-generating observation that requires further testing in other studies.
Table 2.
Genotype Frequencies in Patients with T1D and Controls
|
Frequency in(%) |
||||
| SNP andGenotype | Controls | Patients | Fisher’s Exact Test Pa | OR (95% CI) |
| IL4R −3223: | ||||
| CC | 29.0 | 24.1 | .78 (.40–1.51) | |
| CT | 58.1 | 51.7 | .77 (.43–1.38) | |
| TT | 12.9 | 24.1 | .16 | 2.15 (.99–4.68) |
| IL4R −1914: | ||||
| CC | 31.2 | 36.0 | 1.24 (.67–2.29) | |
| CT | 49.5 | 42.7 | .76 (.43–1.36) | |
| TT | 19.4 | 21.3 | .66 | 1.13 (.55–2.33) |
| IL4R 50: | ||||
| AA | 34.7 | 23.6 | .58 (.30–1.11) | |
| AG | 43.2 | 44.9 | 1.08 (.60–1.93) | |
| GG | 22.1 | 31.5 | .18 | 1.62 (.84–3.13) |
| IL4R 142: | ||||
| CC | 97.9 | 100.0 | ||
| CG | 2.1 | 0 | ||
| GG | … | … | .5 | |
| IL4R 375: | ||||
| AA | 62.1 | 78.7 | 2.25 (1.17–4.33) | |
| AC | 34.7 | 19.1 | .44 (.23–.87) | |
| CC | 3.2 | 2.2 | .04 | .70 (.12–4.32) |
| IL4R 389: | ||||
| GG | 67.4 | 87.6 | 3.43 (1.60–7.37) | |
| GT | 30.5 | 11.2 | .29 (.13–.63) | |
| TT | 2.1 | 1.1 | .002 | .53 (.05–5.93) |
| IL4R 406: | ||||
| CC | 2.1 | 2.2 | 1.07 (.15–7.76) | |
| CT | 33.7 | 19.1 | .46 (.24–.92) | |
| TT | 64.2 | 78.7 | .06 | 2.05 (1.06–3.97) |
| IL4R 478: | ||||
| CC | 2.1 | 2.2 | 1.07 (.15–7.76) | |
| CT | 32.6 | 19.1 | .49 (.25–.96) | |
| TT | 65.3 | 78.7 | .09 | 1.96 (1.01–3.79) |
| IL4R 551: | ||||
| AA | 52.6 | 57.3 | 1.21 (.68–2.16) | |
| AG | 38.9 | 38.2 | .97 (.53–1.76) | |
| GG | 8.4 | 4.5 | .56 | .51 (.15–1.76) |
| IL4 −590: | ||||
| CC | 10.5 | 12.4 | 1.20 (.48–2.98) | |
| CT | 40.0 | 42.7 | 1.12 (.62–2.01) | |
| TT | 49.5 | 44.9 | .81 | .83 (.47–1.49) |
| IL13 intron 3: | ||||
| CC | 33.7 | 31.5 | .90 (.49–1.68) | |
| CT | 50.5 | 46.1 | .84 (.47–1.49) | |
| TT | 15.8 | 22.5 | .54 | 1.55 (.74–3.25) |
| IL13 110: | ||||
| AA | 12.6 | 27.0 | 2.55 (1.19–5.49) | |
| AG | 54.7 | 38.2 | .51 (.28–.92) | |
| GG | 32.6 | 34.8 | .02 | 1.10 (.60–2.03) |
| IL13 −1512: | ||||
| AA | 48.4 | 34.8 | .57 (.31–1.03) | |
| AC | 41.9 | 48.3 | 1.29 (.72–2.32) | |
| CC | 9.7 | 16.9 | .14 | 1.89 (.78–4.57) |
| IL13 −1112: | ||||
| CC | 60.2 | 51.7 | .71 (.39–1.27) | |
| CT | 33.3 | 34.8 | 1.07 (.58–1.97) | |
| TT | 6.5 | 13.5 | .24 | 2.26 (.81–6.31) |
Values in boldface italic type are nominally significant P values (P<.05).
Haplotypes
IL4R
IL4R haplotypes (complex alleles) were estimated using an EM method (Excoffier and Slatkin 1995) and were directly determined by molecular haplotyping methods. The molecular haplotyping method allowed the unambiguous assignment of phase for the six SNPs in exon 12. With the addition of inferred phase for N142N, seven different seven-SNP IL4R haplotypes were determined in this population, and their frequencies among patients and controls were compared (table 3). We focused initially on the seven-SNP haplotypes in our analysis, to allow comparison with the results of Mirel et al. (2002), our earlier study of a sample of white patients with T1D in which the two promoter SNPs of IL4R were not typed. One specific seven-SNP haplotype (CCTCCGT) was significantly underrepresented among Filipino patients (OR 0.4; P=.005). This same haplotype was found to be protective (significant negative association) by the TDT method in the Human Biological Data Interchange (HBDI), a repository of multiplex white families with T1D (Mirel et al. 2002). In the HBDI families, stratification based on the highest-risk HLA genotype (HLA-DRB1*0301-DQB1*0201/HLA-DRB1*04-DQB1*0302) was necessary to demonstrate the protective effect of the IL4R haplotype (Mirel et al. 2002). A significant negative association was found only among those families in which neither affected sib was DR3/4, presumably because the effect of the IL4R polymorphisms was relatively modest compared with the risk conferred by this high-risk HLA genotype (conferring a disease risk higher than in DR3/3 or DR4/4 homozygotes). Among Filipinos, a significant protective effect of a specific IL4R haplotype was observed without such HLA stratification (table 3). This may reflect the absence, among Filipinos, of a higher risk associated with DR3/4 than with DR3/3 or DR4/4 genotypes (Bugawan et al. 2002). We have attributed the lack of the “DR3/4 effect” among Filipinos, well established by many studies of T1D in whites, to the differing patterns of LD of DQB1 alleles with DRB1*04 alleles between Asians and whites (Bugawan et al. 2002).
Table 3.
Molecular IL4R Haplotypes (Complex Alleles) in Filipino Patients with T1D and Controls
|
Frequency in |
||||
| Seven-SNPHaplotype | Controls(N=188)c | Patients(N=178)c | χ2 Pa | ORb (95% CI) |
| CAGTTAT | 70.7 | 77.0 | .49 | 1.38 (.9–2.2) |
| CCTCCGT | 15.4 | 6.7 | .005 | .4 (.2–.8) |
| GAGTTAT | 1.1 | 0 | .50 | .51 (.0–5.8) |
| CAGTTGT | 7.4 | 11.2 | .23 | 1.57 (.8–3.2) |
| CCGCCGT | 3.2 | 5.1 | .38 | 1.62 (.6–4.6) |
| CCTTTGT | 1.6 | 0 | .27 | .35 (.0–3.5) |
| CCTGTAT | .5 | 0 | .98 | 1.12 (.0–15.8) |
| Total (6 df) | .08 | |||
The value in boldface italic type is a nominally significant P value (P<.05).
Where the frequency in patients was 0, the ORs have been computed under the assumption that a single patient sample carried the haplotype.
Sample size (N) refers to the number of chromosomes.
The molecular haplotyping approach used here did not allow the assignment of phase for the two promoter SNPs and the I50V SNP in the IL4R locus. Consequently, we used the EM approach to estimate frequencies for 10-SNP haplotypes, using these three individual SNPs and the seven-SNP haplotype previously determined by molecular methods (table 4). Of the 17 10-SNP haplotypes with an estimated frequency >1% in either group, only one 10-SNP haplotype containing the protective seven-SNP haplotype—CCA-CCTCCGT, labeled “CC(H-5)” in table 4—appeared strongly negatively associated (OR 0.10; 95% CI 0–0.5; P=.001) with disease. Interestingly, the other haplotype (CTA-CCTCCGT, labeled “CT[H-5]” in table 4), which contained the same seven-SNP haplotype and which differs only at the −1914 SNP, was not significantly associated with disease (OR 0.66; P=.33). Our data suggest that a specific combination of IL4R promoter SNPs with a particular coding-sequence allelic variant contributes to the risk for T1D.
Table 4.
Estimated Haplotype Frequencies in Patients with T1D and Controls, for 10-SNP IL4R Haplotypes, Based on EM Algorithm Analysis of the Seven-SNP Molecular Haplotype (Table 3) and the Three Additional 5′ SNPs[Note]
|
Frequency in % (SD)c in |
|||||
| HaplotypeLabela | IL4R 10-SNP Haplotypeb | Controls (N=188)f | Patients (N=180)f | ORd (95% CI) | Pe |
| CC(H-5) | CC-ACCTCCGT | 5.8 (1.7) | 0 | .09 (.0–.5) | .001 |
| CT(H-5) | CT-ACCTCCGT | 8.5 (2.3) | 5.8 (2.2) | .66 (.3–1.6) | .33 |
| CT(H-3) | CT-GCCTCCGT | .9 (.6) | 0 | .62 (.0–6.0) | .21 |
| TC(H-3) | TC-GCCTCCGT | 0 | .9 (1.1) | 1.70 | .19 |
Note.— Total χ2 (23 df) for all 10-SNP haplotypes = 36.03; P=.04.
The haplotype label names the 10-SNP haplotypes by the identity of the −3223 and −1914 alleles, followed, in parentheses, by the eight-SNP haplotype ID as presented by Mirel et al. (2002).
The hyphen in each haplotype separates the two promoter SNPs from the eight coding-sequence SNPs.
The SD was computed using 100 bootstrap replicates. Of the possible 512 haplotypes, 24 were observed.
Where the frequency in patients was 0, the ORs have been computed under the assumption that a single patient sample carried the haplotype. Where the frequency in controls was 0, the ORs have been computed under the assumption that a single control sample carried the haplotype.
Differences in allele frequencies between cases and controls were tested using a χ2 test.
Sample size (N) refers to the number of chromosomes.
IL4 and IL13
The IL4 and the four IL13 SNPs are in strong LD (appendix A). We estimated five-SNP haplotype frequencies and compared them between patients and controls (table 5); the overall distributions were different (P=.005), and one haplotype, TCTTA, was strongly associated with T1D (OR 3.47; P=.004). One surprising observation is that the four-SNP IL13 haplotype CTTA appears to be associated with disease only in combination with the T allele at the IL4 −524 promoter SNP, since the CCTTA haplotype shows no disease association. These data could reflect LD between the associated five-SNP haplotype and some nearby causal gene or that a particular combination of a promoter variant at IL4 and promoter and coding variants at IL13 are responsible for an elevated T1D risk (gene-gene interaction).
Table 5.
Estimated IL4 and IL13 Haplotype Frequencies in Patients with T1D and Controls, for Five SNPs on Chromosome 5q31
|
Frequency in %(SD)b in |
||||
| Five-SNPHaplotypea | Controls(N=188) | Patients(N=176)d | ORc | P |
| CACCA | 4.1 (1.3) | 3.5 (1.7) | .84 | |
| CACCG | 8.9 (2.2) | 10.6 (2.9) | 1.21 | |
| CACTA | 1.3 (1.1) | 3.0 (1.8) | 2.26 | |
| CCCCG | 1.5 (1.0) | .9 (.9) | .61 | |
| CCTCG | 1.1 (.9) | 5.5 (2.3) | 5.19 | .02 |
| CCTTA | 12.6 (2.4) | 9.3 (2.0) | .71 | |
| TACCG | 33.6 (3.8) | 21.9 (3.9) | .55 | .03 |
| TACTA | 16.4 (3.3) | 16.3 (3.0) | .99 | |
| TACTG | 5.0 (1.7) | 3.2 (1.4) | .62 | |
| TCCCG | 4.6 (1.8) | 9.8 (2.9) | 2.23 | .06 |
| TCCTA | 1.3 (1.1) | 0 | .42 | |
| TCTCG | 5.3 (2.4) | 1.7 (1.2) | .31 | .07 |
| TCTTA | 4.1 (2.1) | 12.8 (2.2) | 3.47 | .004 |
| Others | 0 | 1.7 | ||
| Total (12 df) | .001 | |||
In the table and text, the haplotypes are designated with the SNPs in the following order: IL4 −524, IL13 −1512, IL13 −1112, IL13 intron 3, and IL13 110. The physical order of the SNPs on 5q31 is IL13 −1512, IL13 −1112, IL13 intron 3, IL13 110, IL4 −524. Of the 17 inferred five-SNP haplotypes, only those with an estimated frequency of >1% are shown.
The SD was computed using 100 bootstrap replicates.
Where the frequency in patients was 0, the ORs have been computed under the assumption that a single patient sample carried the haplotype.
N is reduced because of missing genotypes.
Gene-Gene Interaction
Since IL4 and IL13 both serve as ligands for a receptor composed, in part, of the IL4R α chain, we looked for potential gene-gene interactions between polymorphisms in the IL4R locus on chromosome 16p11 and the five SNPs in the IL4 and IL13 loci on chromosome 5q31. In one approach, we examined the statistical independence for genotypes at the 10 IL4R SNPs and the genotypes at each of the IL4 and IL13 SNPs (table 6). No deviation from independence was found for these SNPs among controls, but a significant deviation was found for the IL4 −524 promoter SNP (P=.001) and the IL13 intron 3 SNP (P=.019) among patients.
Table 6.
Test for Independence Between Genotype Frequencies at IL4R SNPs and Genotype Frequencies at Five Chromosome 5 SNPs
|
P for |
||
| SNP | Controls | Patientsa |
| IL4 −524 | .15 | .001 |
| IL13 −1512 | .77 | .78 |
| IL13 −1112 | .93 | .73 |
| IL13 110 | .99 | .91 |
| IL13 intron3 | .99 | .02 |
Values in boldface italic type are nominally significant P values (P<.05).
To assess whether the IL4 or the IL13 SNPs modified the effect on T1D susceptibility due to IL4R SNPs, we modeled epistasis through use of a logistic-regression model. For each of the five chromosome 5q31 SNPs, we tested whether the effect of the combined IL4R SNP genotypes on T1D susceptibility differed depending on the chromosome 5q31 SNP genotype. The results (table 7) indicate that there is an epistatic interaction between the IL4R genotypes and IL4 and IL13 genotypes. To address the issue of multiple comparisons, we performed permutation analysis. In 22/200 permutations, one or more of the five SNP tests showed a P<.035; in 13/200, one or more of the SNP tests had a P<.035, and another one had a P<.075; and, in 9/200, one had a P<.035, another a P<.075, and a third a P<.135. Thus, the pattern observed in table 7 has a probability of P<.045. We conclude that the epistatic interaction observed between these chromosome 5 SNPs and the IL4R genotypes is statistically significant and indicates that, in this data set, the genotypes in the IL4-IL13 region affect the genetic susceptibility to T1D conferred by IL4R.
Table 7.
Epistasis between IL4R and Five Chromosome 5 SNPs[Note]
| SNP | β3 | SE | OR | Wald’sχ2 | NominalP Valuea |
| IL4 −524:il4r | −.22 | .10 | .81 | 4.55 | .03 |
| IL13 −1512:il4r | .04 | .15 | 1.04 | .06 | .81 |
| IL13 −1112:il4r | .10 | .16 | 1.10 | .37 | .55 |
| IL13 intron3:il4r | .17 | .09 | 1.18 | 3.19 | .07 |
| IL13 110:il4r | .14 | .09 | 1.15 | 2.24 | .14 |
Note.— The overall P value for all five tests by permutation analysis was P<.045 (see text).
The value in boldface italic type is a nominally significant P value (P<.05).
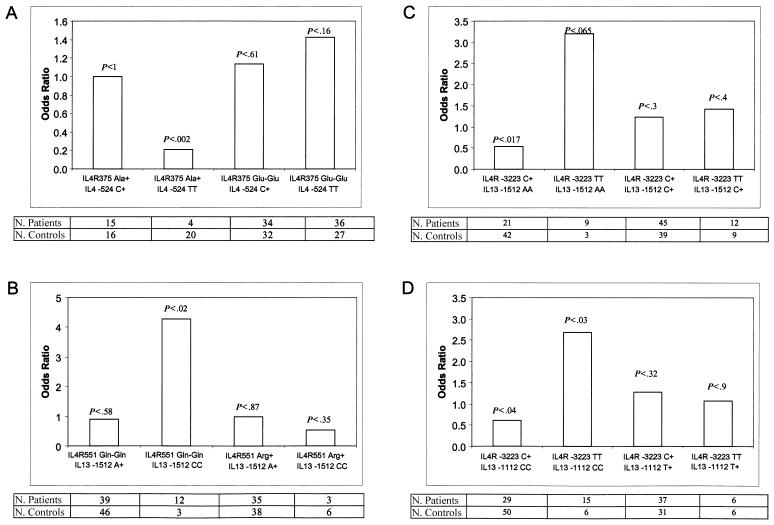
To illustrate this interaction, we also calculated the ORs for individual IL4R SNPs as a function of the IL4 and IL13 SNP genotype and plotted them in the same format used by Howard et al. (2002) to exemplify IL4R-IL13 epistasis with regard to susceptibility to asthma. The ORs and P values from the stratified contingency-table analyses are shown in figure 1. The differences among the ORs were greatest for the IL4R −3223 SNP and the four IL13 SNPs. For example, the IL13 −1512 SNP AA genotype has an OR of 3.21 (95% CI 0.84–12.23) with the TT IL4R −3223 genotype and an OR of 0.53 (95% CI 0.29–0.98) with the CC genotype (fig. 1C), whereas the IL13 −1112 SNP AA genotype has an OR of 2.67 (95% CI 1.01–7.05) with the TT IL4R −3223 genotype and an OR of 0.62 (95% CI 0.36–1.06) with the CC genotype (fig. 1D).
Figure 1.
Gene-gene interaction. ORs and patient and control counts of specific IL4R and IL4-IL13 genotypes are shown. A, IL4R E375A (Glu encoded by the A allele, Ala by the C allele) with IL4 T-524C. B, IL4R Q551R (Gln encoded by the A allele, Arg by the G allele) with IL13 A-1512C. C, IL4R C-3223T and IL13 A-1512C. D, IL4R C-3223T and IL13 C-1112T. The P value for the association of each genotype combination is shown above each OR bar.
Discussion
We have investigated the association and interaction of polymorphisms in three candidate genes (IL4R, IL4, and IL13) in a sample of unrelated Filipino patients with T1D and control individuals. Of the 10 individual IL4R SNPs examined in this study, the strongest association was with L389L (OR 0.34; P=.001), although the flanking SNPs, which show LD to it and to each other, are also associated. Analysis of IL4R SNP haplotypes or complex alleles suggested that one haplotype appeared to confer dominant protection. Initially, we focused on seven-SNP haplotypes, determined by a molecular method, to allow comparison with the results of Mirel et al. (2002). The same seven-SNP haplotype was associated negatively with T1D among Filipinos as in white multiplex families (Mirel et al. 2002). In general, the interpretation of disease association studies with multiple candidate SNPs is complicated by the issue of multiple testing and the consequent reduction in statistical power. In the case of this seven-SNP IL4R haplotype, we have observed a similar protective effect in two different populations and in two different study designs—namely, case-control and TDT. In addition to the biological plausibility (e.g., the expected functional consequences) of these SNPs, these observations strongly suggest that variants of the IL4R molecule influence susceptibility to T1D. In this study, we also examined two promoter SNPs, allowing estimation of 10-SNP haplotypes for IL4R. The analysis of 10-SNP IL4R haplotypes among Filipinos suggests that a specific promoter variant in combination with specific coding sequence variants may be responsible for the observed protection (table 4).
The observation of an interaction between polymorphisms in the IL13 and IL4 genes and polymorphism in the gene (IL4R) encoding the receptor for the products of these two represents an interesting and biologically plausible hypothesis that, given the multiple comparisons, requires further testing. Another example of epistasis was reported in a recently published study of patients with asthma that showed a gene-gene interaction between IL4R and IL13 in the determination of serum IgE levels (Howard et al. 2002.).
Several recent studies have shown that the reference (wild-type) allele at several of the IL4R SNPs examined here is associated with atopic asthma and increased IgE levels (Sandford et al. 2000; Howard et al. 2002). Thus, it appears that the same alleles at IL4R SNPs confer an increased risk for a canonical Th1 (T1D) and Th2 (atopic asthma) disease. If true, these associations argue against an effect on Th1/Th2 balance mediated by polymorphism in the IL4R gene and suggest instead that these variations may influence some aspect of immune regulation and homeostasis in both Th1 and Th2 pathways and possibly B cell activation, as well. Conceivably, the observed patterns of disease association reflect the effect of IL4R polymorphisms on the balance between the activation of Th1 and Th2 cells and that of T regulatory cells. Finally, the extent of risk for T1D may be determined by specific combinations of variants at the IL4R locus and at the genes encoding its two ligands, IL4 and IL13.
Acknowledgments
We thank Maria Alejandrino for expert technical assistance. This work is funded in part by National Institutes of Health grant R01 AI29042-09A1 (to H.A.E.) and by a grant from the Centro Nazionale Studio Diabetes (to P.P.).
Appendix A
Table A1.
Pairwise LD between SNPs in this Cohort (IL4R SNPs)[Note]
| LD |
|||||||||
| −3223 (T) | −1914 (T) | 50 (G) | 142 (G) | 375 (C) | 389 (T) | 406 (C) | 478 (C) | 551 (G) | |
| −3223 (T) | … | −.183*** | .176*** | .004 | −.063*** | −.068*** | −.061*** | −.059*** | −.068*** |
| −1914 (T) | −1.0 | … | −.068*** | −.005 | .105 | .029* | .014 | .011 | .027 |
| 50 (G) | .75 | −.81 | … | .006 | −.049** | −.067*** | −.043** | −.039** | −.079*** |
| 142 (G) | 1.0 | −1.0 | 1.0 | … | −.002 | −.002 | −.002 | −.002 | −.003 |
| 375 (C) | −.70 | .70 | −.53 | −1.0 | … | .139*** | .152*** | .147*** | .145*** |
| 389 (T) | −.93 | .29 | −.86 | −1.0 | 1.0 | … | .126*** | .122*** | .122*** |
| 406 (C) | −.74 | .13 | −.49 | −1.0 | 1.0 | .89 | … | .150*** | .133*** |
| 478 (C) | −.74 | .62 | −.48 | −1.0 | 1.0 | .85 | 1.0 | … | .135*** |
| 551 (G) | −.56 | .17 | −.65 | −1.0 | .96 | .96 | .96 | 1.0 | … |
Note.— Upper right triangle: D. Lower left triangle: D′ (normalized LD, D′=D/Dmax). All values refer to the variant allele indicated in the table. Statistically significant LD values are indicated as follows: *P<.05; **P<.01; ***P<.0001.
Table A2.
Pairwise LD between SNPs in this Cohort (IL4 and IL13 SNPs)[Note]
| LD |
|||||
| IL4 −524 (C) | IL13 −1512 (C) | IL13 −1112 (T) | IL13 intron3 (T) | IL13 110 (A) | |
| IL4 −524 (C) | … | .062*** | .069*** | .024 | .063*** |
| IL13 –1512 (C) | .29 | … | .163*** | .057*** | .058** |
| IL13 –1112 (T) | .41 | 1.0 | … | .077*** | .078*** |
| IL13 intron 3 (T) | .13 | .31 | .54 | … | .201*** |
| IL13 110 (A ) | .34 | .31 | .55 | .84 | … |
Note.— Upper right triangle: D. Lower left triangle: D′ (normalized LD, D′=D/Dmax). All values refer to the variant allele indicated in the table. Statistically significant LD values are indicated as follows: *P<.05; **P<.01; ***P<.0001.
Electronic-Database Information
The URL for data presented herein is as follows:
- Online Mendelian Inheritance in Man (OMIM), http://www.ncbi.nlm.nih.gov/Omim/ (for T1D, interleukin receptor 4, interleukin 4, and interleukin 13)
References
- Azar ST , Tamim H , Beyhum HN, Habbal MZ, Almawi WY (1999) Type I (insulin-dependent) diabetes is a Th1- and Th2-mediated autoimmune disease. Clin Diagn Lab Immunol 6:306–310 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bennett ST, Wilson AJ, Cucca F, Nerup J, Pociot F, McKinney PA, Barnett AH, Bain SC, Todd JA (1996) IDDM2-VNTR-encoded susceptibility to type 1 diabetes: dominant protection and parental transmission of alleles of the insulin gene-linked minisatellite locus. J Autoimmun 9:415–421 [DOI] [PubMed] [Google Scholar]
- Berman MA, Sandborg CI, Wang Z, Imfeld KL, Zaldivar F Jr, Dadufalza V, Buckingham BA (1996) Decreased IL-4 production in new onset type I insulin-dependent diabetes mellitus. J Immunol 157:4690–4696 [PubMed] [Google Scholar]
- Bugawan TL, Klitz W, Alejandrino M, Ching J, Panelo A, Solfelix CM, Petrone A, Buzzetti R, Pozzilli P, Erlich HA (2002) The association of specific HLA class I and II alleles with type 1 diabetes among Filipinos. Tissue Antigens 59:452–469 [DOI] [PubMed] [Google Scholar]
- Cox NJ, Wapelhorst B, Morrison VA, Johnson L Pinchuk L, Spielman RS, Todd JA, Concannon P (2001) Seven regions of the genome show evidence of linkage to type 1 diabetes in a consensus analysis of 767 multiplex families. Am J Hum Genet 69:820–830 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Erlich HA, Rotter JI, Chang JD, Shaw SJ, Raffel LJ, Klitz W, Bugawan TL, Zeidler A (1996) Related association of HLA-DPB1*0301 with IDDM in Mexican-Americans. Diabetes 45:610–614 [DOI] [PubMed] [Google Scholar]
- Excoffier L, Slatkin MW (1995) Maximum likelihood estimation of molecular haplotype frequencies in a diploid population. Mol Biol Evol 12:921–927 [DOI] [PubMed] [Google Scholar]
- Expert Committee on the Diagnosis and Classification of Diabetes Mellitus (1997) Report of the expert committee on the diagnosis and classification of diabetes mellitus. Diabetes Care 20:1183 [DOI] [PubMed] [Google Scholar]
- Glimcher LH, Murphy KM (2000) Lineage commitment in the immune system: the T helper lymphocyte grows up. Genes Dev 14:1693–1711 [PubMed] [Google Scholar]
- Graves PE, Kabesch M, Halonen M, Holberg CJ, Baldini M, Fritzsch C, Weiland SK, Erickson RP, von Mutius E, Martinez FD (2000) A cluster of seven tightly linked polymorphisms in the IL-13 gene is associated with total serum IgE levels in three populations of white children. J Allergy Clin Immunol 105:506–513 [DOI] [PubMed] [Google Scholar]
- Hackstein H, Hecker M, Kruse S, Bohnert A, Ober C, Deichmann KA, Bein G (2001) A novel polymorphism in the 5′ promoter region of the human interleukin-4 receptor alpha-chain gene is associated with decreased soluble interleukin-4 receptor protein levels. Immunogenetics 53:264–269 [DOI] [PubMed] [Google Scholar]
- Higuchi R, Fockler C, Dollinger G, Watson R (1993) Kinetic PCR analysis: real-time monitoring of DNA amplification reactions. Biotechnology (NY) 11:1026–1030 [DOI] [PubMed] [Google Scholar]
- Howard TD, Koppelman GH, Xu J, Zheng SL, Postma DS, Meyers DA, Bleecker ER (2002) Gene-gene interaction in asthma: IL4RA and IL13 in a Dutch population with asthma. Am J Hum Genet 70:230–236 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawakami, K, Taguchi J, Murata T, Puri RK (2001) The interleukin-13 receptor α2 chain: an essential component for binding and internalization but not for interleukin-13-induced signal transduction through the STAT6 pathway. Blood 97:2673–2679 [DOI] [PubMed] [Google Scholar]
- Klitz W, Bugawan TL, Panelo A, Solfelix CM, Buzzetti R, Pozzilli P, Steiner L, Alejandrino M, Erlich HA (2002) Association of CTLA-4 variation with type I diabetes in Filipinos. Immunogenetics 54:310–313 [DOI] [PubMed] [Google Scholar]
- Kretowski A, Mysliwiec J, Kinalska I (2000) In vitro interleukin-13 production by peripheral blood in patients with newly diagnosed insulin-dependent diabetes mellitus and their first degree relatives. Scand J Immunol 51:321–325 [DOI] [PubMed] [Google Scholar]
- Kruse S, Japha T, Tedner M, Sparholt SH, Forster J, Kuehr J, Deichmann KA (1999) The polymorphisms S503P and Q576R in the interleukin-4 receptor α gene are associated with atopy and influence the signal transduction. Immunology 96:365–371 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larsen ZM, Kristiansen OP, Mato E, Johannesen J, Puig-Domingo M, de Leiva A, Nerup J, Pociot F (1999) IDDM12 (CTLA4) on 2q33 and IDDM13 on 2q34 in genetic susceptibility to type 1 diabetes (insulin-dependent). Autoimmunity 31:35–42 [DOI] [PubMed] [Google Scholar]
- Lee YJ, Lo FS, Shu SG, Wang CH, Huang CY, Liu HF, Wu CC, Yang TY, Chang JG (2001) The promoter region of the CTLA4 gene is associated with type 1 diabetes mellitus. J Pediatr Endocrinol Metab 14:383–388 [DOI] [PubMed] [Google Scholar]
- Medici F, Hawa MI, Giorgini A, Panelo A, Solfelix CA, Leslie RD, Pozzilli P (1999) Antibodies to GAD65 and a tyrosine phosphatase–like molecule IA–2ic in Filipino type I diabetic patients. Diabetes Care 22:1458–1462 [DOI] [PubMed] [Google Scholar]
- Mirel D, Valdes AM, Reynolds RL, Lazzeroni L, Erlich HA, Noble JA (2002) Association of IL4R haplotypes with type 1 diabetes. Diabetes 51:3336 [DOI] [PubMed] [Google Scholar]
- Nistico L, Buzzetti R, Pritchard LE, Van der Auwera B, Giovannini C, Bosi E, Larrad MT, Rios MS, Chow CC, Cockram CS, Jacobs K, Mijovic C, Bain SC, Barnett AH, Vandewalle CL, Schuit F, Gorus FK, Tosi R, Pozzilli P, Todd JA (1996) The CTLA-4 gene region of chromosome 2q33 is linked to, and associated with type 1 diabetes. Belgian Diabetes Registry. Hum Mol Genet 5:1075–1080 [DOI] [PubMed] [Google Scholar]
- Noble JA, Valdes AM, Bugawan TL, Apple RJ, Thomson G, Erlich HA (2002) The HLA class I A locus affects susceptibility to type 1 diabetes. Hum Immunol 63:657–664 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noble JA, Valdes AM, Cook M, Klitz W, Thomson G, Erlich HA (1996) The role of HLA class II genes in insulin-dependent diabetes mellitus: molecular analysis of 180 Caucasian, multiplex families. Am J Hum Genet 59:1134–1148 [PMC free article] [PubMed] [Google Scholar]
- Noble JA, Valdes AM, Thomson G, Erlich HA (2000) The HLA class II locus DPB1 can influence susceptibility to type 1 diabetes. Diabetes 49:121–125 [DOI] [PubMed] [Google Scholar]
- Noguchi E, Shibasaki M, Arinami T, Takeda K, Yokouchi Y, Kawashima T, Yanagi H, Matsui A, Hamaguchi H (1998) Association of asthma and the interleukin-4 promoter gene in Japanese. Clin Exp Allergy 28:449–453 [DOI] [PubMed] [Google Scholar]
- Pociot F, McDermott MF (2002) Genetics of type 1 diabetes mellitus. Genes Immun 3:235–249 [DOI] [PubMed] [Google Scholar]
- Risch N (1987) Assessing the role of HLA-linked and unlinked determinants of disease. Am J Hum Genet 40:1–14 [PMC free article] [PubMed] [Google Scholar]
- Sandford AJ, Chagani T, Zhu S, Weir TD, Bai TR, Spinelli JJ, Fitzgerald JM, Behbehani NA, Tan WC, Pare PD (2000) Polymorphisms in the IL4, IL4RA, and FCERIB genes and asthma severity. J Allergy Clin Immunol 106:135–140 [DOI] [PubMed] [Google Scholar]
- Slatkin MW, Excoffier L (1996) Testing for linkage disequilibrium in genotypic data using the EM algorithm. Heredity 76:377–383 [DOI] [PubMed] [Google Scholar]
- Tisch R, McDevitt H (1996) Insulin-dependent diabetes mellitus. Cell 85:291–297 [DOI] [PubMed] [Google Scholar]
- Van der Pouw Kraan TC, van Veen A, Boeije LC, van Tuyl SA, de Groot ER, Stapel SO, Bakker A, Verweij CL, Aarden LA, van der Zee JS (1999) An IL-13 promoter polymorphism associated with increased risk of allergic asthma. Genes Immun 1:61–65 [DOI] [PubMed] [Google Scholar]
- Zaccone, P, Phillips J, Conget I, Gomis R, Haskins K, Minty A, Bendtzen K, Cooke A, Nicoletti F (1999) Interleukin-13 prevents autoimmune diabetes in NOD mice. Diabetes 48:1522–1528 [DOI] [PubMed] [Google Scholar]