Abstract
In Tetrahymena, as in other ciliates, development of the somatic macronucleus during conjugation involves extensive and reproducible rearrangements of the germ line genome, including chromosome fragmentation and excision of internal eliminated sequences (IESs). The molecular mechanisms controlling these events are poorly understood. To investigate the role that histone acetylation may play in the regulation of these processes, we treated Tetrahymena cells during conjugation with the histone deacetylase inhibitor trichostatin A (TSA). We show that TSA treatment induces developmental arrests in the early stages of conjugation but does not significantly affect the progression of conjugation once the mitotic divisions of the zygotic nucleus have occurred. Progeny produced from TSA-treated cells were examined for effects on IES excision and chromosome breakage. We found that TSA treatment caused partial inhibition of excision of five out of the six IESs analyzed but did not affect chromosome breakage at four different sites. TSA treatment greatly delayed in some cells and inhibited in most the excision events in the developing macronucleus. It also led to loss of the specialized subnuclear localization of the chromodomain protein Pdd1p that is normally associated with DNA elimination. We propose a model in which underacetylated nucleosomes mark germ line-limited sequences for excision.
The eukaryotic genome is packaged into repeated nucleosomal units that are folded in high-order chromatin fibers. The dynamics of chromatin structure plays a fundamental role in many aspects of genomic function (reviewed in references 34 and 53). Posttranslational modifications of the N-terminal tails of all core histones provide the nucleosome core particle with an enormous capacity for variability. The importance of chromatin remodeling to the regulation of gene expression has become clear partly due to the identification of several histone-modifying enzymes. In particular, the characterization of two competing enzymatic activities that regulate overall levels of histone acetylation, those of histone acetyltransferases and histone deacetylases, has established histone acetylation as a key player in modulating transcription factor access to chromatin. Determining whether histone acetylation might also regulate other DNA-templated processes such as replication, repair, and recombination is of great interest. Several recent studies have revealed that histone acetylation is crucial for the events that accompany V(D)J recombination in mammals (35, 40, 41).
The ciliated protozoa Tetrahymena thermophila provides a model system to address the question of how chromatin remodeling can regulate genome-wide recombination events. This organism undergoes massive rearrangements of its somatic genome during the sexual process of conjugation. The chromosomes are broken at about 200 sites, and about 6,000 DNA elements (called internal eliminated sequences, or IESs) are eliminated, resulting in the loss of 15% of the germ line genome (reviewed in references 15 and 54). IESs are dispersed throughout the genome and consist of single-copy and moderately repetitive sequences ranging in size from hundreds to thousands of base pairs. Most of them possess short direct repeats at their ends, one of which is maintained in the somatic genome after excision. Although excision occurs with precision and reproducibility, no consensus sequences have been identified at or near the boundaries of the eliminated elements. However, cis-acting sequences controlling the excision process have been revealed for two elements that have been extensively studied, the M and R elements (9, 19, 20). The cis determinants are located a short distance outside the ends of each element, but remarkably, their exact sequences are different for each one. Two trans-acting factors involved in this process contain chromodomains (Pdd1p [14, 37] and Pdd3p [43]). This motif is commonly found in chromosomal proteins that help establish or maintain specialized chromatin structures in other organisms (reviewed in reference 29). This suggests a connection between the formation of a specialized chromatin structure and programmed DNA excision in Tetrahymena. We imagine a scenario in which chromatin associated with germ line-limited sequences is remodeled during development, allowing the excision machinery to distinguish them from those destined for the somatic nucleus.
Histone acetylation levels are dramatically different between the germ line and the somatic nuclei in vegetative Tetrahymena cells. High levels of acetylation are found with histones isolated from the somatic nucleus, while little if any acetylation is observed with histones from the germ line nucleus during vegetative growth (50). Histone acetylation is developmentally regulated. Both histone acetylase and histone deacetylase activities are being modulated during the course of macronuclear development (12), raising the possibility that a temporal and functional relationship may exist between histone acetylation and genome rearrangements. To investigate the role of histone acetylation in DNA rearrangements in Tetrahymena, we treated Tetrahymena cells during conjugation with trichostatin A (TSA) (58, 59), the histone deacetylase inhibitor that has been largely used to increase the level of histone acetylation in living cells. We describe the effects of TSA treatment on the nuclear events of conjugation and on DNA rearrangements. We show that treatment with TSA caused failure of DNA deletion but not of chromosome breakage. The data suggest a model in which underacetylated nucleosomes mark germ line-specific sequences for excision.
MATERIALS AND METHODS
Strains and culture conditions.
All growth and manipulations of Tetrahymena were performed as described elsewhere (2). T. thermophila inbred B strains CU427 (Chx/Chx [VI, cy-s]), CU428 (Mpr/Mpr [VII, mp-s]), and B2086 (II) were used as wild-type strains in all experiments and were kindly provided by Peter Bruns (Howard Hughes Institute).
For green fluorescent protein (GFP) localization of Pdd1p, a construct (pBC-33) was made in the GFP vector pCGF-1 (M.-C. Yao and C.-H. Yao, unpublished data) consisting of the micronuclear rDNA vector (20) containing the GFP coding sequence controlled by flanking sequences of the Tetrahymena PDD1 gene (R. Coyne and M.-C. Yao, unpublished data). The entire PDD1 open reading frame was cloned in-frame at the 3′ end of the GFP coding sequence. Upon transformation with pBC-33, the ribosomal DNA vector and flanking sequences are processed, amplified, and maintained as ribosomal DNA minichromosomes (20). One such Tetrahymena transformant was mated with strain CU428. At various times during conjugation, mating cells in 1% methylcellulose were examined by fluorescence microscopy.
TSA treatment.
A stock solution of TSA (Sigma) at 1 mg/ml in 100% dimethyl sulfoxide was kept at −20°C. Just before use, TSA was diluted in 10 mM Tris-HCl at pH 7.4 and added at various times to synchronously mating wild-type cells (∼105 pairs/ml) to a final concentration of 1 μg/ml. Addition of dimethyl sulfoxide without TSA (referred to as −TSA) at the same concentration was performed in parallel as a control. Normal mating in 10 mM Tris-HCl at pH 7.4 (referred to as untreated cells) was also performed according to published methods (2). Prestarved cells mixed in the presence of 1 μg of TSA/ml failed to form pairs. In the absence of TSA, about 95% of the cells paired within the first hour after mixing. The addition of TSA was carried out on cells that had already paired, beginning at 3 h after mixing. Synchrony and progression through conjugation were assessed by fluorescence microscopy after fixation of cells in saturated HgCl2-95% ethanol (2:1) or in 70% ethanol and staining with 4 μg of DAPI (4′,6′-diamidino-2-phenylindole)/ml. After incubation, individual cells were isolated and transferred into ∼30-μl drops of SPP medium (1% proteose peptone, 0.2% dextrose, 0.1% yeast extract, 0.003% sequestrene) on a petri dish and were allowed to complete conjugation and grow at 30°C. After 3 days, the number of drops with viable cells was scored (viability). They were replica-plated into drops of medium containing cycloheximide (25 μg/ml) or 6-methylpurine (15 μg/ml). After 3 days, successful mating resulted in progeny showing resistance to both drugs. The production of progeny was calculated as the percentage of viable cells that are resistant to both drugs. Whole-cell DNAs were then isolated from pools of ∼100 progeny lines and analyzed by Southern blotting. In some cases, single cells were isolated from progeny lines and grown for DNA analysis.
Whole-cell DNA isolation.
Ten milliliter cultures of vegetatively growing (5 × 105 to 106 cells/ml) or mating cells (∼2 × 105 cells/ml) were centrifuged. Cell pellets were resuspended in 0.5 ml of freshly made lysis solution (0.5 M EDTA, 10 mM Tris [pH 9.5], 1% sodium dodecyl sulfate [SDS], 0.5 mg proteinase K/ml [Merck]). The lysates were incubated at 55°C overnight. Addition of 0.5 ml of 20% polyethylene glycol 8000-1.2 M NaCl-20 mM EDTA and chilling on ice for 1 h was followed by centrifugation at 15,000 × g for 15 min and two successive washes of the pellet with 70% ethanol. The DNA pellet was resuspended in 400 μl of Tris-EDTA buffer (10 mM Tris-HCl, 1 mM EDTA [pH7.4]). DNA was treated with RNase at 37°C for 1 h after the addition of 100 μl of 1.5 M sodium acetate-0.1 M EDTA-50 mM Tris-HCl (pH 8.0)-0.45 mg of RNase/ml. After phenol chloroform extraction, DNA was precipitated with 100% ethanol and resuspended in 150 μl of Tris-EDTA buffer.
Southern blot analysis.
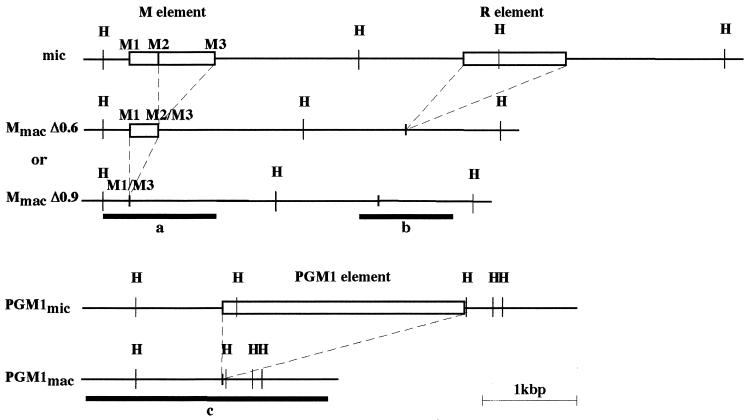
DNAs were digested with restriction enzymes under the conditions indicated by the suppliers. Fragmented DNAs were separated by electrophoresis on 0.8 to 1% agarose gels. DNA fragments were transferred from agarose gels to Hybond N+ membranes (Amersham) in 0.4 N NaOH after depurination in 0.25 N HCl. Hybridization was carried out in 7% SDS-0.5 M sodium phosphate-1% bovine serum albumin-1 mM EDTA (pH 7.2) at 65°C. Membranes were washed three times in 0.2× SSC (1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate)-0.5% SDS for 1 h at 65°C prior to autoradiography. Specific DNA fragments used as hybridization probes were radiolabeled using [α-32P]dATP (3,000 Ci/mmol) and random hexamers (Gibco/BRL) as previously described (18). Probes used were as follows: probe a was a HindIII-XbaI restriction fragment containing 1.2 kbp of macronuclear DNA from the M element (57), probe b was a AccI-EcoRI restriction fragment containing ∼1 kbp of macronuclear DNA from the R element (57), and probe c consists of a 2.5-kbp fragment containing the entire coding sequence for the PGM1 gene (13) that was amplified from Tetrahymena genomic DNA by PCR and cloned into pCRII-TOPO (Invitrogen) (see Fig. 3).
FIG. 3.
Maps of the micronuclear and macronuclear versions of the genomic region containing the M and R elements (top panel) and of the genomic region containing the PGM1 element (bottom panel). Micronuclear specific elements are represented by open boxes. The M element is excised from the micronuclear genome in two alternative forms of 0.6kbp (MmacΔ0.6) and 0.9 kbp (MmacΔ0.9). The positions and lengths of probes a, b, and c are shown. HindIII (H) restriction sites are indicated.
PCR analysis of total genomic DNA.
Two oligonucleotides (5′ AGCTTAAACAAATGCCATATTGAG 3′ and 5′ GTGGGGAGGGAGAAGATTCAAAC 3′) located 240 bp from the 5′ boundary and 25 bp from the 3′ boundary of the M element, respectively, were used to perform PCR amplification at different time points during conjugation. PCRs were carried out in 0.2 ml polypropylene tubes (USA Scientific) with reaction mixtures (25 μl) containing ∼100 ng of whole-cell DNA, 1× PCR buffer (Gibco/BRL), 0.2 mM of deoxynucleoside triphosphates, 0.5 μM of primer, and 2.5 U of Taq polymerase (Gibco/BRL). Amplifications were done in a Perkin-Elmer GeneAmp PCR system apparatus as follows: 94°C for 4 min followed by 30 cycles of 30 s at 94°C, 30 s at 55°C, and 30 s at 72°C and a final termination cycle of 7 min at 72°C.
RESULTS
Effects of TSA treatment on conjugating cells.
Since the effects of TSA treatment on the life cycle of Tetrahymena have never been reported, we investigated the process of conjugation in the presence of TSA. Vegetative Tetrahymena cells contain two types of nuclei within the same cytoplasm: the somatic macronucleus, responsible for transcription, and the germ line micronucleus, transcriptionally silent during vegetative growth. During conjugation, new macronuclei and micronuclei are generated through a series of nuclear events that are described below (Fig. 1) (22, 39). Conjugation is initiated by mixing prestarved cells of compatible mating types. After meiosis of the micronucleus, mating pairs exchange one of their haploid nuclei to form the diploid zygotic nuclei. All the prezygotic events are completed within the first 5 to 6 h after mixing. The zygotic nucleus divides twice mitotically to give rise to four nuclei; two differentiate into new macronuclei and the other two differentiate into new micronuclei. The development of the new macronucleus can be divided into three cytological stages (see Fig. 1) during which DNA rearrangements (between 12 and 14 h after mixing) (3) and DNA endoduplication also occur. The old macronucleus condenses and is eventually resorbed. The mating pair separates during this time, and one of the two micronuclei in each cell is eliminated. When cells divide after refeeding with growth medium, the two new macronuclei segregate to the daughter cells, while the micronucleus undergoes mitosis. One micronucleus and one macronucleus are present in each cell as vegetative growth resumes.
FIG. 1.
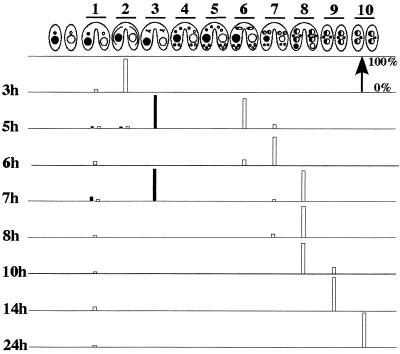
TSA treatment of conjugating cells at 3 h after mixing causes cells to arrest in metaphase of meiosis I. The developmental stages of conjugation previously described in references 22 and 39 are schematically represented as follows. Lanes: 1, cell pairing; 2, crescent stage (prophase of meiosis I); 3, metaphase of meiosis I; 4, end of meiosis II; 5, prezygotic mitosis of one of the four haploid nuclei; 6, karyogamy; 7, macronuclear development I, which is distinguished by the central location of the parental macronucleus, the anterior location of the new macronuclei, and the posterior location of the new micronuclei; 8, macronuclear development II, in which the parental macronucleus condenses and paired cells separate; 9, macronuclear development III, which begins when the parental macronucleus has been resorbed; 10, the final new macronucleus stage, in which one of the two micronuclei is eliminated and the new macronuclei have undergone DNA amplification. The percentage of cells in each cytological stage was determined after fixing and staining with DAPI. At least 200 pairs were scored for each time point by fluorescence microscopy. About 95% of the cells paired within the first hour after mixing. The white rectangles show the untreated cells. The black rectangles show the outcome for cells treated with TSA at 3 h after mixing for 2 h (until 5 h after mixing) and for 4 h (until 7 h after mixing).
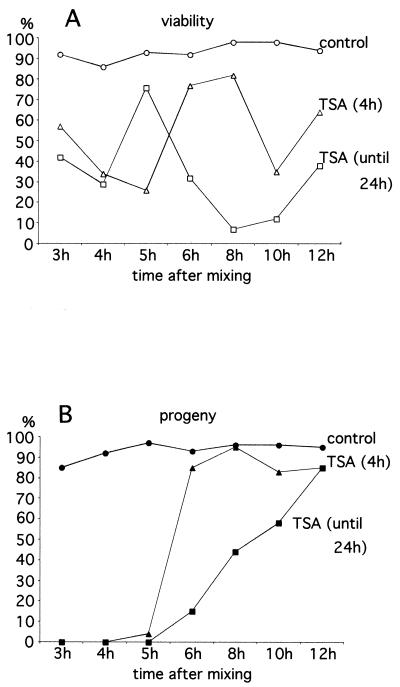
To examine the effects of TSA treatment on the nuclear events of conjugation, mating cells were incubated in the presence of TSA at 1- or 2-h intervals (beginning at 3 h and continuing until 12 h after mixing). The treatment was done either for 4 h or continued until 24 h after mixing (the later is referred to as continuous treatment). After TSA treatment, individual mating cells were cloned into fresh medium without TSA and allowed to complete conjugation and grow. Cell viability as well as progeny production were quantified from two or more separate experiments, and the results are shown in Fig. 2.
FIG. 2.
Genetic analysis of the effects of TSA treatment during conjugation. TSA was added to mating cells at various times (beginning at 3 h and continuing until 12 h after mixing) as indicated in the abscises in both graphs (A and B). The treatment was either for 4 h (triangles) or continued until 24 h after mixing (squares). Control samples are represented by circles. After treatment, individual mating pairs were cloned and the percentages of pairs giving viable cells were scored (A). The percentages of mating pairs that successfully produced sexual progeny (see Materials and Methods) are also given (B). The number of cells analyzed for each point was between 88 and 352. The results are the combined totals of two or more experiments. The discrepancy in the percentage of viability between the 4-h treatment (low viability) and the treatment until 24 h (high viability) for cells treated at 5 h after mixing may be due to bias introduced during cell cloning. At 24 h, many dying or dead cells were already immobile and thus not cloned, which raised the percentage of viable cells among those cloned.
Mating cells treated with TSA before 6 h after mixing produced fewer viable cells than the control and very few progeny in both the 4-h treatment and the continuous treatment. Consistent with these data, cytological examination of these cells never revealed new macronucleus formation. The stages of developmental arrest were complex. One striking observation came from the TSA treatment applied at 3 h after mixing and is illustrated in Fig. 1. At 3 h after mixing, the majority of the cells (90%) were in the prophase of meiosis I (crescent stage). Incubation with TSA caused cells to arrest in the metaphase of meiosis I after 2 h of treatment (87%), while control cells had already completed meiosis. Even after 4 h of treatment, TSA-treated cells did not progress further (87% are still in metaphase). Mating cells incubated with TSA at 4 and 5 h after mixing were still paired in some cases and contained in most cases one or two enlarged macronuclei by the 24-hour point.
In contrast, mating cells treated with TSA after 6 h after mixing, the time at which postzygotic division had taken place in 75% of the cells (Fig. 1), showed a different outcome. Many treated cells survived and produced true progeny. Cell viability and progeny production were significantly lower after the continuous treatment than after the 4-h treatment. Cytological examination showed little difference with the untreated controls: the pairs separate, the old macronucleus disappears, and the second micronucleus is eliminated with a similar timing. The only exception is that one of the micronuclei is not eliminated in cells that are treated from 6 h to 24 h after mixing. In summary, TSA treatment has catastrophic effects on the early stages of conjugation but does not significantly affect progression of conjugation once mitotic division of the zygotic nucleus has occurred.
Excision inhibition by TSA treatment.
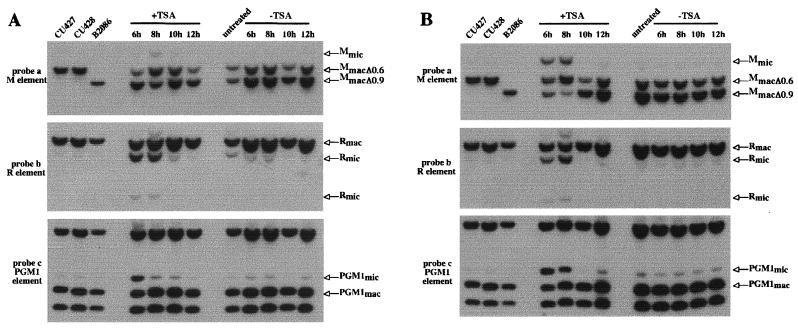
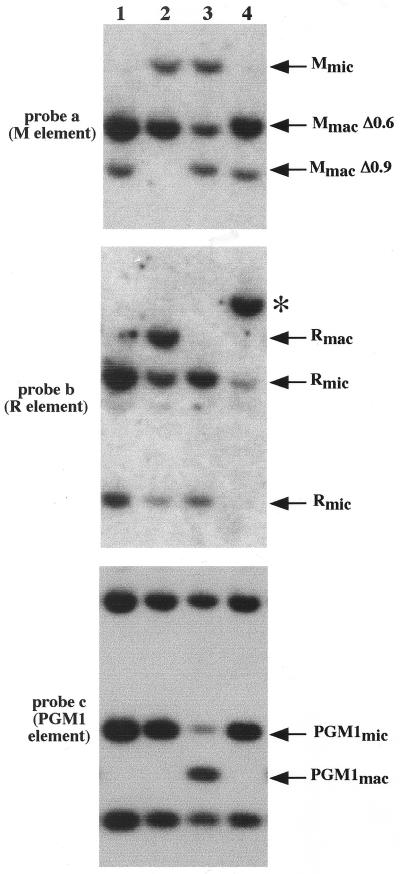
Since mating cells treated with TSA after 6 h after mixing produced progeny, we were able to examine whether the histone deacetylase inhibitor affects developmentally regulated genome rearrangements. For that purpose, we analyzed the new macronuclear genome of the sexual progeny produced after TSA treatment. Sexual progeny of approximately 100 mating cells treated at 6 h or later (i.e., at 6, 8, 10, and 12 h after mixing) for 4 h or continuously until 24 h were grown individually and then mixed for DNA extraction. DNA samples were first analyzed for IES excision by Southern hybridization. The results of hybridization of Southern blots with a probe specific for the M element (probe a, Fig. 3) are shown in the top panels of Fig. 4A and B. Figures 4A and B show the results from the 4-h and the continuous treatment experiments, respectively. Because the micronucleus-macronucleus ploidy is ∼1:20, most of the hybridizing DNA is macronuclear DNA. The M element is eliminated from the micronuclear genome during macronuclear development in two alternative forms (0.6 kbp and 0.9 kbp) that share a common right boundary (see Fig. 3) (3, 5). The parental strains (CU427 and CU428) used in this mating contain the macronuclear fragment corresponding to the 0.6-kbp deletion (MmacΔ0.6), whereas another standard strain (B2086) contains the 0.9-kbp deletion product in its macronuclear genome (MmacΔ0.9). The sexual progeny of untreated cells (Fig. 4A and B, top panels) showed both macronuclear fragments in roughly equal amounts, as expected (3). The sexual progeny of cells treated for 4 h with TSA gave similar results, indicating that the M element was correctly excised during macronuclear development (Fig. 4A, top panels). In contrast, the sexual progeny of cells treated continuously until 24 h after mixing (Fig. 4B, top panel) showed a significant amount (about 30%) of unrearranged DNA (Mmic). Thus, continuous TSA treatment starting at 6 or 8 h and lasting until 24 h after mixing caused some copies of the M element to be retained in the new macronuclear genome of the sexual progeny, whereas TSA treatment applied after 10 h after mixing did not.
FIG. 4.
TSA treatment impairs IES excision. Southern hybridization analysis was used to monitor the effect of TSA treatment on the excision of the M, R, and PGM1 elements. Control DNA samples were isolated from vegetative CU427, CU428, and B2086 strains. Experimental DNA samples were isolated from the progeny of cells that were treated at 6 h, 8 h, 10 h, and 12 h after mixing with TSA (+TSA) and without TSA (−TSA) as indicated. Each sample represents a pool of about 100 progeny lines. Total DNA was digested with HindIII. The positions of the DNA fragments corresponding to the unrearranged forms (Mmic, Rmic, and PGM1mic) and the rearranged forms (Mmac, Rmac, and PGM1mac) are indicated by arrows. Among the different HindIII macronuclear fragments revealed by probe c, only the macronuclear fragment containing the PGM1 element is indicated by the arrow as PGM1mac. Conjugating cells were treated for 4 h (A) or continuously until 24 h after mixing (B). The same blot was successively hybridized with probes a, b, and c (Fig. 3) in both panels A and B.
To determine the effect of TSA on the excision of other IESs, the same blots were stripped and successively rehybridized with two other probes. Probe b was specific for the region containing the 1.1-kbp R element located ∼2.5 kbp away from the M element (3, 4), and probe c was specific for the region containing the 2.5-kbp element located within an intron of the PGM1 gene located in a different genomic region (13) (Fig. 3). Both the micronuclear (Rmic) and the macronuclear (Rmac) forms were found in the sexual progeny of cells treated with TSA from 6 to 10 h and from 8 to 12 h after mixing, indicating that the excision of the R element was partially inhibited (Fig. 4A, middle panel). The excision of the PGM1 element was affected only in the sexual progeny of cells treated between 6 and 10 h after mixing (bottom panel of Fig. 4A). Thus, the 4-h TSA treatment partially inhibited the excision of both these elements, even though it had no effect on the excision of the M element. Continuous treatment with TSA beginning from 6 to 8 h after mixing partially affected the R and PGM1 deletion elements (Fig. 4B, middle and bottom panels), as was the case with the M element. In all cases, the progeny of cells treated with TSA at 10 h and 12 h were not affected.
Three other IESs were analyzed similarly (data not shown). The excision of two (24, 33) IESs was also impaired when cells were treated before the 10-h stage. The excision of the third element analyzed (23) was not affected. Since this element is located within an intron of a gene of unknown function, it is possible that its excision is essential for the cells to grow, which would prevent us from recovering cells that had retained the element. However, analysis of DNA from cells isolated at the 24-h point (before selection for growth) showed that the excision was not affected (data not shown). Thus, the excision of this element was not sensitive to TSA treatment.
In summary, treatment of mating cells with TSA caused partial failure of excision for five out of the six IESs analyzed in this study. This blockage depends on the time at which the treatment has been applied to mating cells (see below). In none of the samples analyzed was the inhibition of excision complete for any IESs. Similar experiments using increasing concentrations of TSA (up to a tenfold increase) did not show an increased defect of excision in the sexual progeny (data not shown). The partial failure of excision could be attributable to the fact that we were unable to treat cells with TSA earlier than 6 h after mixing without arresting development. Alternatively, there could be other deacetylase activities that are insensitive to TSA that play a role in the process (8, 25).
Since pools of 100 progeny were used in this analysis, there could be some heterogeneity among them, with some cells showing complete failure of excision and other cells no failure of excision. To assess this possibility, we isolated individual cells from some pools that showed partial failure of excision for the M, R, and PGM1 elements. DNAs of four such subclones derived from cells treated from 6 h until 24 h were analyzed by using Southern blotting (Fig. 5). Of these lines, some indeed showed complete failure of excision for the R and PGM1 elements (Fig. 5, lanes 1 and 3 for R and lanes 1, 2, and 4 for PGM1). In one cell line in which most copies of the R and PGM1 elements were not deleted, the M element deletion was not affected (Fig. 5, lane 1). This suggests that TSA treatment causes differential failure of excision for each IES within the same developing macronucleus.
FIG. 5.
Effect of TSA treatment on IES excision in individual progeny. The excision of the M, R, and PGM1 elements was analyzed by Southern hybridization in four individual cell lines (lanes 1 to 4) isolated from the sexual progeny of cells treated with TSA from 6 h until 24 h after mixing. Total DNA was digested with HindIII. The same blot was successively hybridized with probes a, b, and c (Fig. 3). The positions of the DNA fragments corresponding to the unrearranged and the rearranged forms of each element are indicated by the arrows. An aberrant form of rearrangement revealed by probe b is marked by an asterisk.
TSA treatment does not affect chromosome breakage.
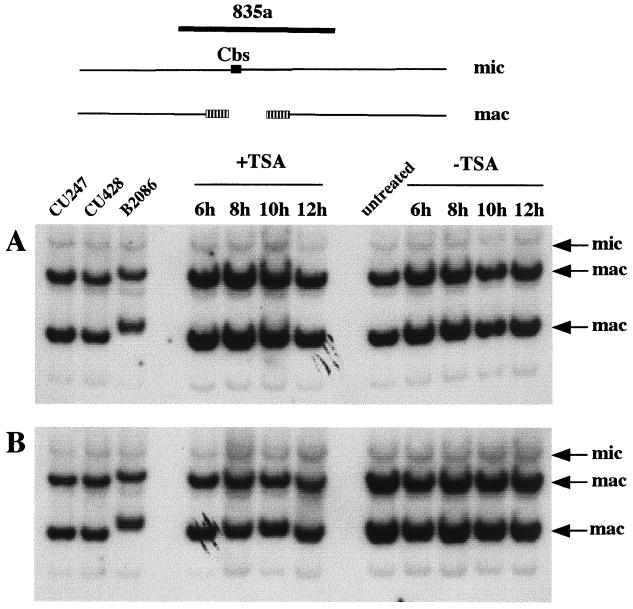
So far, we have investigated the role of TSA treatment on IES excision. To test the specificity of this effect, we examined the occurrence of the other major type of DNA rearrangement, chromosome breakage, using the same DNA samples. During macronuclear development, chromosome breakage occurs at sites containing the 15-bp chromosome breakage sequence Cbs (55, 56). Telomeres are then added to both free ends about 5 to 20 bp from the ends of Cbs (17). Consequently, a Cbs and 10 to 40 bp of DNA surrounding it are eliminated. Southern blots were hybridized with several probes that contain a Cbs and its macronuclear flanking sequences (56). Probe 835a revealed two macronuclear fragments that contain telomeres (major bands) as well as its micronuclear counterpart (a minor band), as shown in the control samples of Fig. 6. Whatever the duration of the treatment (4 h or continuous treatment [Fig. 6A and B, respectively]), the sexual progeny of TSA-treated cells showed the same pattern of hybridization as the sexual progeny of the untreated cells. We analyzed three other Cbs sites (56) and did not detect any failure in chromosome breakage. Analysis of developing macronuclei at the 24-h point also did not show any defect in chromosome breakage (data not shown), indicating that the absence of effect is likely not due to selection for growth. We therefore conclude that the process of chromosome breakage is not sensitive to TSA, which highlights the specificity of TSA treatment on the excision process.
FIG. 6.
TSA treatment does not affect chromosome breakage. Southern hybridization analysis was used to monitor chromosome breakage in sexual progeny of cells treated with TSA (+TSA) or without TSA (−TSA) for 4 h (panel A) or until 24 h (panel B). On the schematic representation of chromosome breakage, the 15-bp chromosome breakage site, Cbs, is drawn as a black box. The two macronuclear chromosomes generated after breakage are shown with hatched boxes designating telomeric repeats. All samples were digested with EcoRI. DNA samples in panel A are the same as in Fig. 4A, and DNA samples in panel B are the same as in Fig. 4B. Both blots were hybridized with probe 835a (56). The precise map and sequence of that genomic region have not been determined.
Excision events are delayed in the presence of TSA.
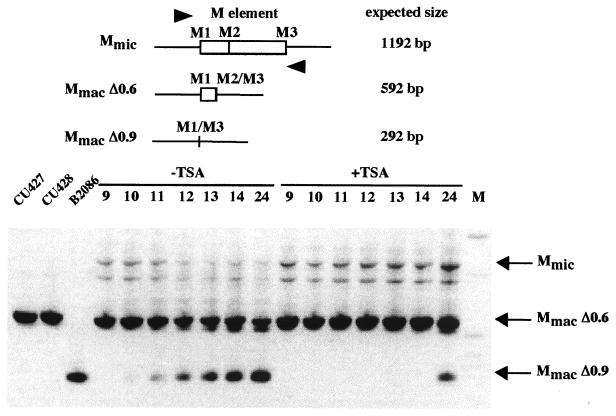
To better understand how TSA treatment affects IES excision, we looked at the excision process directly during macronuclear development. As described above, the failure of excision depends on the time of the addition of TSA to mating cells. It has been previously reported that the excision of the M and R elements normally occurs between 12 and 14 h after mixing (3). We added TSA at 6 h after mixing, and DNA was isolated from the mating cultures at 9, 10, 11, 12, 13, 14, and 24 h after mixing. To distinguish the new macronuclear junctions generated by excision from the sequences of the parental macronucleus that are also present in these cells, we took advantage of the alternative excision of the M element. The parental strains used for the mating (CU427 and CU428) contained only the 0.6-kbp deletion product in the macronucleus (MmacΔ0.6) (Fig. 7). However, both forms, MmacΔ0.6 and MmacΔ0.9, were produced in the new macronucleus of their sexual progeny (3). This feature allowed us to determine the timing of the M element excision by monitoring the appearance of the 0.9-kbp deletion product (MmacΔ0.9). We performed PCR on the DNA samples by using two oligonucleotides flanking the M element that allow the amplification of Mmic, MmacΔ0.6, and MmacΔ0.9. In untreated cells, MmacΔ0.9 was detectable as early as 10 h after mixing and increased in abundance over time (Fig. 7). This is consistent with our observation that partial blockage of excision occurs only when TSA is applied before the 10-h stage.
FIG. 7.
Timing of excision of the M element in conjugating cells with and without TSA treatment. A schematic diagram of the M element is shown on the top. The oligonucleotides used for PCR are represented by arrowheads. Expected sizes of the PCR products are indicated. PCR products amplified from DNA samples of conjugating cells were run on a 1.2% agarose gel and stained with ethidium bromide. The first three lanes show the results for PCR products amplified from control vegetative cells of CU427, CU428, and B2086 strains. Indicated time points (9, 10, 11, 12, 13, 14, and 24) refer to the times in hours after mixing at which DNAs were extracted from mass mating. TSA was added at 6 h after mixing. M is the 1-kb size marker from Gibco/BRL. Two faint bands were detected in all PCR products amplified from DNA samples of conjugating cells. The uppermost band corresponds to the expected size for Mmic.
With TSA treatment, in contrast, no excision of the M element was observed at 10 or 14 h after mixing. However, at 24 h after mixing, PCR product corresponding to MmacΔ0.9 was detected, indicating that some excision of the M element had occurred. This does not appear to have been due to weakening of TSA during the 18-h incubation (from 6 to 24 h). To test for drug efficacy, we used the supernatant of similarly treated mating cultures at 24 h to treat 3-h mating cells. This treatment induced the same developmental arrest in metaphase of meiosis as efficiently as freshly prepared TSA.
Even though some excision has occurred during TSA treatment, the amount of PCR product corresponding to Mmic was clearly more abundant in the TSA-treated cells than in the untreated cells at 24 h after mixing. This indicates that the developing macronuclei at the 24-h point still contained unrearranged forms of the M element, which is consistent with the partial blockage observed from the progeny analysis. Thus, both the timing and the efficiency of excision were greatly affected by TSA treatment.
Pdd1p localization in the presence of TSA.
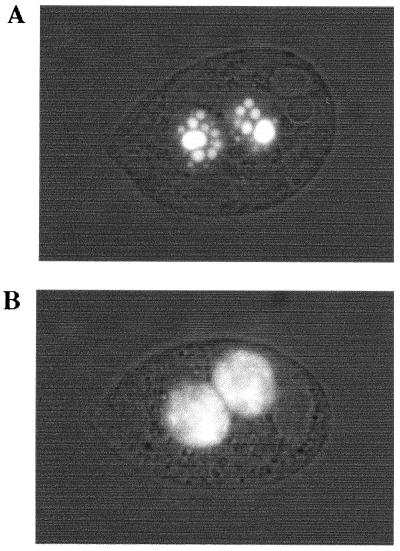
Several abundant polypeptides are specifically expressed and targeted to the developing macronuclei at the time when genome rearrangements occur (38, 43, 47). The most abundant of these, Pdd1p, is a chromodomain protein that associates with the eliminated DNA in electron-dense intranuclear structures (37, 47) and is required for the excision process (14). To test the possibility that TSA treatment alters the ability of Pdd1p to properly interact with chromatin at the eliminated sequences, we examined the expression and localization of Pdd1p during TSA treatment. By using GFP-tagged Pdd1p (R. Coyne, D. Chalker, and M.-C. Yao, unpublished data), we are able to follow Pdd1p localization in living cells. Consistent with previously reported Pdd1p immunolocalization (38), the untreated cells initially showed uniform distribution of GFP in the developing macronucleus. As conjugation progressed, the GFP became punctate, culminating with the appearance of large spherical structures at 14 h after mixing that contain micronuclear-limited sequences (Fig. 8A). In contrast, the GFP was uniformly distributed in the developing macronucleus at 14 h after mixing in TSA-treated cells (Fig. 8B). Quantitation using a fluorescence microscope showed that 92/110 of the untreated cells had a punctate pattern, whereas 0/200 of the TSA-treated cells had a similar pattern at 14 h after mixing. Thus, TSA treatment did not seem to inhibit Pdd1p expression but resulted in loss of the special Pdd1p localization that is correlated with DNA elimination. However, at 24 h after mixing, GFP had a punctate pattern in a fraction of cells treated with TSA (29/111), but not in any cells without TSA (0/100). Thus, the subnuclear structures associated with Pdd1p and DNA deletion did not form at the normal time, although they could still form at a later time in a fraction of cells. We conclude that TSA treatment leads to the inhibition of excision and the loss of the subnuclear localization of Pdd1p in most cells. In some cells it delays the timing of IES excision and, concomitantly, the formation of the Pdd1p-associated nuclear structures.
FIG. 8.
Pdd1p localization in conjugating cells incubated without (A) and with TSA (B). Cells expressing Pdd1p fused to GFP were mated with wild-type cells, and TSA was added at 6 h after mixing. At 14 h after mixing, cells are examined under a fluorescence microscope.
DISCUSSION
The role of histone deacetylation in genome rearrangements in Tetrahymena has been investigated here by using trichostatin A, a potent and specific inhibitor of histone deacetylase. This drug has been widely used to study the role of histone acetylation in gene expression. It has been shown in some cases and assumed in the others that TSA treatment increases the overall level of histone acetylation by inhibiting histone deacetylase activities (58, 59). The present study demonstrates that histone deacetylation plays an important role in the regulation of the developmentally programmed DNA deletion process in Tetrahymena. Incubation with TSA causes failure of excision of several IESs in the macronuclear genome of the sexual progeny of the treated cells. This effect is specific, since TSA treatment does not affect chromosome breakage at the four sites analyzed. It has previously been demonstrated that the M and R elements can be maintained in the macronuclear genome (10). Here we show that Tetrahymena cells containing several IESs in their macronuclear genome are viable and able to grow vegetatively. The excision inhibition induced by TSA treatment is partial, since only a fraction of the macronuclear chromosomes contain the micronuclear-specific sequences. However, we have obtained individual cell lines in which the failure of excision can be complete for some IESs. Since the excision of five IESs out of six analyzed was affected by the histone deacetylase inhibitor, it raises the possibility that the excision of many or most of the IESs that make up as much as 15% of the germ line genome is under the control of histone deacetylation.
Effects of the deacetylase inhibitor TSA on the nuclear events of conjugation.
Inhibiting histone deacetylases with TSA greatly perturbed the nuclear events in early stages of conjugation. Conjugating cells treated before the stage of the zygotic nuclear divisions failed to develop a new macronucleus and as a consequence did not produce progeny. The developmental arrest of cells in metaphase of meiosis suggests that without proper signals the conjugants do not proceed beyond a specific stage, probably a developmental checkpoint, and that to do so would require deacetylase activity. We identified a stage after which developmental progression becomes insensitive to TSA. This time point (6 h) correlates with the completion of the second postzygotic nuclear division, which is the last nuclear division of the developmental program. Past this time point, the final nuclear events of conjugation proceed normally in the presence of TSA; the differentiation of the new macronucleus and the new micronuclei take place, and the parental macronucleus is eliminated as well as one of the two micronuclei.
Other drug treatments have been shown to block macronuclear development. Nocodazole is an antimicrotubule drug that prevents macronuclear differentiation when applied before the formation of the zygotic nucleus (30, 32). Cycloheximide and actinomycin D, inhibitors of protein and RNA synthesis, respectively, are also capable of arresting development when applied before the second postzygotic division (31, 51). Interestingly, actinomycin D and cycloheximide treatments cause the same developmental arrest in metaphase of meiosis as TSA (31). In all treatments, the second zygotic division is the point past which development can no longer be prevented. What is remarkable is the fact that postzygotic events of the developmental program are able to proceed in the presence of TSA, whereas the genomic rearrangements in the developing macronucleus do not occur normally. In a similar manner, brief actinomycin D treatment of conjugating cells can block the excision process without dramatic effects on the formation of a new macronucleus (11). The nuclear events can therefore be dissociated from the rearrangement program.
A connection between histone acetylation and cell differentiation has long been known. TSA treatment promotes cell line differentiation (60) and restricts cell transformation (48). In Xenopus (1), inhibiting histone deacetylation with TSA during development greatly perturbs the differentiation program during gastrulation. It is not clear how TSA treatment causes these effects. Studies in both Schizosaccharomyces pombe (16) and mammalian cell cultures (49) indicate that TSA treatment leads to defects in chromosome segregation in mitosis. The effects of TSA treatment can thus be explained both by the role of histone acetylation in transcription regulation and by the control of the acetylation state of histones that is necessary for the maintenance of genome integrity.
A role for histone deacetylation in DNA excision.
TSA treatment of conjugating cells indicated that histone deacetylation is necessary for efficient excision of five different IESs. Interestingly, histone acetylation has been shown to play an important role in the regulation of another developmentally programmed DNA rearrangement, the V(D)J recombination of the genes encoding T cell receptors and immunoglobulins (reviewed in reference 46). TSA has been shown to enhance V(D)J recombination (40), and acetylation of histone H3 is known to accompany specific recombination events (41). More recently it has been demonstrated that histone acetylation acts in concert with the ATP-dependent remodeling factor SWI/SNF to increase the initial recombination reaction in vitro (35). Histone acetylation accompanies the activation of V(D)J recombination, while histone deacetylation is required for efficient IES excision. The detailed mechanisms by which histone acetylation regulates genome rearrangements seem to be clearly distinct in these two instances.
How might histone acetylation inhibit IES excision? Increased levels of histone acetylation could perturb the expression of genes that are involved in the excision process and consequently cause failure of excision. At this point we cannot completely rule out the possibility that the effect of TSA is indirect. However, the fact that the 4-h TSA treatment can block the excision of several IESs without having a severe effect on progeny production argues against a general effect of TSA on the expression of genes essential for development.
We favor another explanation, in which the increased histone acetylation affects the nucleosomes that are associated with IESs, directly causing inhibition of excision. One might expect that histone acetylation levels would not increase with TSA treatment in regions of the micronuclear genome that are never subject to acetylation. Inhibiting histone deacetylase activities with TSA treatment would not cause any alterations of the nucleosomes associated with IESs located in these regions. The one IES whose excision was insensitive to TSA in our experiment may well be one such example. Our data showed that histone deacetylation between 6 and 10 h of conjugation was required for efficient excision. This is just before the time IESs are actually excised from the developing macronucleus, as shown in our PCR assay for the M element (Fig. 7). We propose that IESs are associated normally with underacetylated histones, at least transiently during macronuclear development. This regulated deacetylation of histones would establish a mark that distinguishes the sequences to be eliminated from the macronucleus-destined sequences. Specific patterns of histone acetylation are maintained in different regions of eukaryotic genomes. Histones in heterochromatic regions in widely divergent species are consistently underacetylated. In mammalian metaphase chromosomes, histones H3 and H4 are underacetylated at all lysines in both constitutive heterochromatin and the facultative heterochromatin of the inactive X chromosome (7, 28). Underacetylation of histones H3 and H4 has also been implicated in the formation of heterochromatin in both Schizosaccharomyces pombe and Saccharomyces cerevisiae (reviewed in reference 21). Underacetylated histones may be a transient landmark of the chromatin associated with IESs in Tetrahymena at the time of their elimination. This analogy is further supported by the formation of electron-dense nuclear structures resembling heterochromatin within the developing macronucleus that contain the eliminated sequences at the time of their excision (37, 47). In another ciliate, Euplotes crassus, eliminated DNA may also form heterochromatin-like structures in the developing macronucleus. Changes in nucleosome spacing for the Tec transposons of E. crassus have been detected at the beginning of the macronuclear development (27). Moreover, the Tec elements show an unusual chromatin structure, as analyzed by micrococcal nuclease digestion, in contrast to the classical ladder of nucleosomes observed for macronuclear-destined sequences (26).
In our model, the underacetylated nucleosomes would tether the proteins that carry out the excision process. In cells treated with TSA, histone deacetylase activity is inhibited and increasing acetylation of associated chromatin would thus prevent the recruitment of the excision machinery. This is in good agreement with the fact that the localization of the chromodomain protein Pdd1p is greatly affected by TSA treatment. In that respect, it is interesting that exposure to TSA leads to the delocalization of heterochromatin-associated proteins in Schizosaccharomyces pombe (16) as well as in mammalian cells (49). Recent reports have indicated that methylation of lysine 9 of histone H3 is crucial in establishing a potent binding site for heterochromatin proteins (6, 36). Moreover, acetylation of lysine 9 of histone H3 inhibits its methylation by the methyltransferase SUV39H (45). One interpretation of our data would be that the inhibition of deacetylation prevents the methylation of histone H3 lysine 9, which in turn leads to the loss of Pdd1p association with the eliminated sequences and consequently to the failure of excision.
The eliminated sequences in Tetrahymena can be 1 kbp or shorter in length. Just a few modified nucleosomes would cover the entire length of these small IESs. In contrast with the histone modification patterns that distinguish euchromatin and heterochromatin domains over large regions (such as the distinct histone methylation pattern specific for the 20-kbp mating-type locus in Schizosaccharomyces pombe) (44), the chromatin remodeling involved in IES excision would be much more localized. The deacetylase activity that we have revealed in this study remains to be characterized. A histone deacetylase gene homologue to the yeast RPD3 has been isolated in Tetrahymena (52). It would be interesting to know whether it plays a role in IES excision. One important question is how the deacetylase is specifically recruited to the eliminated sequences. The fact that germ line limited sequences are transcribed between 3.5 and 8 h after mixing (11) may suggest that transcription of these sequences plays a part in the initiation of chromatin remodeling. It is possible to envision that chromatin opening associated with transcription allows the deacetylase activity to be targeted to specific locations in the genome.
Our finding that IES excision is regulated by histone deacetylation could be relevant to the intriguing epigenetic regulation of this process exerted through the maternal macronucleus. The excision of the M and R elements can be inhibited during conjugation, when these elements are introduced into the maternal macronucleus prior to mating (10). The molecular basis for this homology-dependent maternal effect is not yet elucidated. It will be of great interest to explore the possibility that histone modifications play a role in the epigenetic regulation of IES excision. Pairing interactions between molecules produced from the maternal macronucleus and unrearranged homologous sequences have been postulated to account for the sequence specificity of the phenomenon (reviewed in references 42 and 54). We speculate that these pairing interactions inhibit the excision events by interfering with histone modifications, thus altering the proper packaging of nucleosomes associated with the sequence to be eliminated.
Acknowledgments
We thank Douglas Chalker for critical reading of the manuscript.
This work was supported by grant GM26210 to M.-C. Yao from the National Institutes of Health. S.D. was a recipient of fellowships from the Association pour la Recherche sur le Cancer and Human Frontier Science Program.
REFERENCES
- 1.Almouzni, G., S. Khochbin, S. Dimitrov, and A. P. Wolffe. 1994. Histone acetylation influences both gene expression and development of Xenopus laevis. Dev. Biol. 165:654-669. [DOI] [PubMed] [Google Scholar]
- 2.Asai, D. J., and J. D. Forney. 2000. Tetrahymena thermophila, vol. 62. Academic Press, San Diego, Calif.
- 3.Austerberry, C. F., C. D. Allis, and M. C. Yao. 1984. Specific DNA rearrangements in synchronously developing nuclei of Tetrahymena. Proc. Natl. Acad. Sci. USA 81:7383-7387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Austerberry, C. F., and M. C. Yao. 1987. Nucleotide sequence structure and consistency of a developmentally regulated DNA deletion in Tetrahymena thermophila. Mol. Cell. Biol. 7:435-443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Austerberry, C. F., and M. C. Yao. 1988. Sequence structures of two developmentally regulated, alternative DNA deletion junctions in Tetrahymena thermophila. Mol. Cell. Biol. 8:3947-3950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bannister, A. J., P. Zegerman, J. F. Partridge, E. A. Miska, J. O. Thomas, R. C. Allshire, and T. Kouzarides. 2001. Selective recognition of methylated lysine 9 on histone H3 by the HP1 chromo domain. Nature 410:120-124. [DOI] [PubMed] [Google Scholar]
- 7.Belyaev, N., A. M. Keohane, and B. M. Turne. 1996. Differential underacetylation of histones H2A, H3 and H4 on the inactive X chromosome in human female cells. Hum. Genet. 97:573-578. [DOI] [PubMed] [Google Scholar]
- 8.Carmen, A. A., P. Griffin, J. Calaycay, S. Rundlett, Y. Suka, and M. Grunstein. 1999. Yeast HOS3 forms a novel trichostatin A-insensitive homodimer with intrinsic histone deacetylase activity. Proc. Natl. Acad. Sci. USA 96:12356-12361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chalker, D., A. La Terza, A. Wilson, C. Kroenke, and M. Yao. 1999. Flanking regulatory sequences of the Tetrahymena R deletion element determine the boundaries of DNA rearrangement. Mol. Cell. Biol. 19:5631-5641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chalker, D. L., and M.-C. Yao. 1996. Non-Mendelian, heritable blocks to DNA rearrangement are induced by loading the somatic nucleus of Tetrahymena thermophila with germ line-limited DNA. Mol. Cell. Biol. 16:3658-3667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chalker, D. L., and M.-C. Yao. 2001. Nongenic, bidirectional transcription precedes and may promote developmental DNA deletion in Tetrahymena thermophila. Genes Dev. 15:1287-1298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chicoine, L. G., and C. D. Allis. 1986. Regulation of histone acetylation during macronuclear differentiation in Tetrahymena: evidence for control at the level of acetylation and deacetylation. Dev. Biol. 116:477-485. [DOI] [PubMed] [Google Scholar]
- 13.Chilcoat, N. D., and A. P. Turkewitz. 1997. In vivo analysis of the major exocytosis-sensitive phosphoprotein in Tetrahymena. J. Cell Biol. 139:1197-1207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Coyne, R., M. Nikiforov, J. Smothers, C. Allis, and M. Yao. 1999. Parental expression of the chromodomain protein Pdd1p is required for completion of programmed DNA elimination and nuclear differentiation. Mol. Cell 4:865-872. [DOI] [PubMed] [Google Scholar]
- 15.Coyne, R. S., D. L. Chalker, and M.-C. Yao. 1996. Genome downsizing during ciliate development: nuclear division of labor through chromosome restructuring. Annu. Rev. Genet. 30:557-578. [DOI] [PubMed] [Google Scholar]
- 16.Ekwall, K., T. Olsson, B. M. Turner, G. Cranston, and R. C. Allshire. 1997. Transient inhibition of histone deacetylation alters the structural and functional imprint at fission yeast centromeres. Cell 91:1021-1032. [DOI] [PubMed] [Google Scholar]
- 17.Fan, Q., and M.-C. Yao. 1996. New telomere formation coupled with site-specific chromosome breakage in Tetrahymena thermophila. Mol. Cell. Biol. 16:1267-1274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Feinberg, A. P., and B. Vogelstein. 1983. A technique for radiolabeling DNA restriction endonuclease fragments to high specific activity. Anal. Biochem. 132:6-13. [DOI] [PubMed] [Google Scholar]
- 19.Godiska, R., C. James, and M. C. Yao. 1993. A distant 10-bp sequence specifies the boundaries of a programmed DNA deletion in Tetrahymena. Genes Dev. 7:2357-2365. [DOI] [PubMed] [Google Scholar]
- 20.Godiska, R., and M. C. Yao. 1990. A programmed site-specific DNA rearrangement in Tetrahymena thermophila requires flanking polypurine tracts. Cell 61:1237-1246. [DOI] [PubMed] [Google Scholar]
- 21.Grunstein, M. 1998. Yeast heterochromatin: regulation of its assembly and inheritance by histones. Cell 93:325-328. [DOI] [PubMed] [Google Scholar]
- 22.Harrison, G. S., and K. M. Karrer. 1985. DNA synthesis, methylation and degradation during conjugation in Tetrahymena thermophila. Nucleic Acids Res. 13:73-87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Heinonen, T. Y., and R. E. Pearlman. 1994. A germ line-specific sequence element in an intron in Tetrahymena thermophila. J. Biol. Chem. 269:17428-17433. [PubMed] [Google Scholar]
- 24.Huvos, P. E., M. Wu, and M. A. Gorovsky. 1998. A developmentally eliminated sequence in the flanking region of the histone H1 gene in Tetrahymena thermophila contains short repeats. J. Eukaryot. Microbiol. 45:189-197. [DOI] [PubMed] [Google Scholar]
- 25.Imai, S., C. Armstrong, M. Kaeberlein, and L. Guarente. 2000. Transcriptional silencing and longevity protein Sir2 is an NAD-dependent histone deacetylase. Nature 403:795-800. [DOI] [PubMed] [Google Scholar]
- 26.Jahn, C. L. 1999. Differentiation of chromatin during DNA elimination in Euplotes crassus. Mol. Biol. Cell 10:4217-4230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Jahn, C. L., Z. Ling, C. M. Tebeau, and L. A. Klobutcher. 1997. An unusual histone H3 specific for early macronuclear development in Euplotes crassus. Proc. Natl. Acad. Sci. USA 94:1332-1337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Jeppesen, P., and B. M. Turner. 1993. The inactive X-chromosome in female mammals is distinguished by a lack of histone H4 acetylation, a cytogenetic marker for gene expression. Cell 74:281-289. [DOI] [PubMed] [Google Scholar]
- 29.Jones, D. O., I. G. Cowell, and P. B. Singh. 2000. Mammalian chromodomain proteins: their role in genome organisation and expression. Bioessays 22:124-137. [DOI] [PubMed] [Google Scholar]
- 30.Kaczanowski, A., J. Gaertig, and J. Kubiak. 1985. Effect of the antitubulin drug nocodazole on meiosis and postmeiotic development in Tetrahymena thermophila. Induction of achiasmatic meiosis. Exp. Cell Res. 158:244-256. [DOI] [PubMed] [Google Scholar]
- 31.Kaczanowski, A., and J. Kaczanowska. 1996. Induction of blocks in nuclear divisions and overcondensation of meiotic chromosomes with cycloheximide during conjugation of Tetrahymena thermophila. J. Eukaryot. Microbiol. 43:380-388. [DOI] [PubMed] [Google Scholar]
- 32.Kaczanowski, A., M. Ramel, J. Kaczanowska, and D. Wheatley. 1991. Macronuclear differentiation in conjugating pairs of Tetrahymena treated with the antitubulin drug nocodazole. Exp. Cell Res. 195:330-337. [DOI] [PubMed] [Google Scholar]
- 33.Katoh, M., M. Hirono, T. Takemasa, M. Kimura, and Y. Watanabe. 1993. A micronucleus-specific sequence exists in the 5′-upstream region of calmodulin gene in Tetrahymena thermophila. Nucleic Acids Res. 21:2409-2414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kornberg, R. D., and Y. Lorch. 1999. Twenty-five years of the nucleosome, fundamental particle of the eukaryote chromosome. Cell 98:285-294. [DOI] [PubMed] [Google Scholar]
- 35.Kwon, J., K. Morshead, J. Guyon, R. Kingston, and M. Oettinger. 2000. Histone acetylation and hSWI/SNF remodeling act in concert to stimulate V(D)J cleavage of nucleosomal DNA. Mol. Cell 6:1037-1048. [DOI] [PubMed] [Google Scholar]
- 36.Lachner, M., D. O'Carroll, S. Rea, K. Mechtler, and T. Jenuwein. 2001. Methylation of histone H3 lysine 9 creates a binding site for HP1 proteins. Nature 410:116-120. [DOI] [PubMed] [Google Scholar]
- 37.Madireddi, M. T., R. S. Coyne, J. F. Smothers, K. M. Mickey, M.-C. Yao, and C. D. Allis. 1996. Pdd1p, a novel chromodomain-containing protein, links heterochromatin assembly and DNA elimination in Tetrahymena. Cell 87:75-84. [DOI] [PubMed] [Google Scholar]
- 38.Madireddi, M. T., M. Davis, and D. Allis. 1994. Identification of a novel polypeptide involved in the formation of DNA-containing vesicles during macronuclear development in Tetrahymena. Dev. Biol. 165:418-431. [DOI] [PubMed] [Google Scholar]
- 39.Martindale, D. W., C. D. Allis, and P. J. Bruns. 1982. Conjugation in Tetrahymena thermophila. A temporal analysis of cytological stages. Exp. Cell Res. 140:227-236. [DOI] [PubMed] [Google Scholar]
- 40.McBlane, F., and J. Boyes. 2000. Stimulation of V(D)J recombination by histone acetylation. Curr. Biol. 10:483-486. [DOI] [PubMed] [Google Scholar]
- 41.McMurry, M. T., and M. S. Krangel. 2000. A role for histone acetylation in the developmental regulation of VDJ recombination. Science 287:495-498. [DOI] [PubMed] [Google Scholar]
- 42.Meyer, E., and O. Garnier. 2002. Non-Mendelian inheritance and homology-dependent effects in ciliates. Adv. Genet. 46:305-338. [DOI] [PubMed] [Google Scholar]
- 43.Nikiforov, M., M. Gorovsky, and C. Allis. 2000. A novel chromodomain protein, Pdd3p, associates with internal eliminated sequences during macronuclear development in Tetrahymena thermophila. Mol. Cell. Biol. 20:4128-4134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Noma, K., C. D. Allis, and S. I. S. Grewal. 2001. Transitions in distinct histone H3 methylation patterns at the heterochromatin domain boundaries. Science 293:1150-1155. [DOI] [PubMed] [Google Scholar]
- 45.Rea, S., F. Eisenhaber, D. O'Carroll, B. D. Strahl, Z. W. Sun, M. Schmid, S. Opravil, K. Mechtler, C. P. Ponting, C. D. Allis, and T. Jenuwein. 2000. Regulation of chromatin structure by site-specific histone H3 methyltransferases. Nature 406:593-599. [DOI] [PubMed] [Google Scholar]
- 46.Roth, D. B., and S. Y. Roth. 2000. Unequal access: regulating V(D)J recombination through chromatin remodeling. Cell 103:699-702. [DOI] [PubMed] [Google Scholar]
- 47.Smothers, J. F., C. A. Mizzen, M. M. Tubbert, R. G. Cook, and C. D. Allis. 1997. Pdd1p associates with germline-restricted chromatin and a second novel anlagen-enriched protein in developmentally programmed DNA elimination structures. Development 124:4537-4545. [DOI] [PubMed] [Google Scholar]
- 48.Sugita, K., K. Koizumi, and H. Yoshida. 1992. Morphological reversion of sis-transformed NIH3T3 cells by trichostatin A. Cancer Res. 52:168-172. [PubMed] [Google Scholar]
- 49.Taddei, A., C. Maison, D. Roche, and G. Almouzni. 2001. Reversible disruption of pericentric heterochromatin and centromere function by inhibiting deacetylases. Nat. Cell Biol. 3:114-120. [DOI] [PubMed] [Google Scholar]
- 50.Vavra, K. J., C. D. Allis, and M. A. Gorovsky. 1982. Regulation of histone acetylation in Tetrahymena macro- and micronuclei. J. Biol. Chem. 257:2591-2598. [PubMed] [Google Scholar]
- 51.Ward, J. G., and G. Herrick. 1996. Effects of the transcription inhibitor actinomycin D on postzygotic development of Tetrahymena thermophila conjugants. Dev. Biol. 173:174-184. [DOI] [PubMed] [Google Scholar]
- 52.Wiley, E. A., R. Ohba, M.-C. Yao, and C. D. Allis. 2000. Developmentally regulated rpd3p homolog specific to the transcriptionally active macronucleus of vegetative Tetrahymena thermophila. Mol. Cell. Biol. 20:8319-8328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Wu, J., and M. Grunstein. 2000. 25 years after the nucleosome model: chromatin modifications. Trends Biochem. Sci. 25:619-623. [DOI] [PubMed] [Google Scholar]
- 54.Yao, M.-C., S. Duharcourt, and D. Chalker. 2002. Genome-wide rearrangements of DNA in ciliates, p. 730-758. In R. C. N. Craig, M. Gellert, and A. Lambowitz (ed.), Mobile DNA II. ASM Press, Washington D.C.
- 55.Yao, M.-C., C.-H. Yao, and B. Monks. 1990. The controlling sequence for site-specific chromosome breakage in Tetrahymena. Cell 63:763-772. [DOI] [PubMed] [Google Scholar]
- 56.Yao, M.-C., K. Zheng, and C.-H. Yao. 1987. A conserved nucleotide sequence at the sites of developmentally regulated chromosomal breakage in Tetrahymena. Cell 48:779-788. [DOI] [PubMed] [Google Scholar]
- 57.Yao, M. C., J. Choi, S. Yokoyama, C. F. Austerberry, and C. H. Yao. 1984. DNA elimination in Tetrahymena: a developmental process involving extensive breakage and rejoining of DNA at defined sites. Cell 36:433-440. [DOI] [PubMed] [Google Scholar]
- 58.Yoshida, M., S. Horinouchi, and T. Beppu. 1995. Trichostatin A and trapoxin: novel chemical probes for the role of histone acetylation in chromatin structure and function. Bioessays 17:423-430. [DOI] [PubMed] [Google Scholar]
- 59.Yoshida, M., M. Kijima, M. Akita, and T. Beppu. 1990. Potent and specific inhibition of mammalian histone deacetylase both in vivo and in vitro by trichostatin A. J. Biol. Chem. 265:17174-17179. [PubMed] [Google Scholar]
- 60.Yoshida, M., S. Nomura, and T. Beppu. 1987. Effects of trichostatins on differentiation of murine erythroleukemia cells. Cancer Res. 47:3688-3691. [PubMed] [Google Scholar]