Abstract
A common haplotype spanning 250 kb in the cytokine gene cluster on chromosome 5q31 has recently been reported to be strongly associated with Crohn disease (CD) in Canadian families. We have replicated this finding by both the transmission-disequilibrium test (TDT) (P=.016) and in a case-control association study (P=.008) in a large European cohort of patients with CD, although the increase in disease risk was small (odds ratio 1.49 for homozygotes, 95% CI 1.11–2.0). No association was detected in families or individuals with ulcerative colitis (UC). Stratification of offspring with CD in the TDT sample by mutation status in the CD susceptibility gene CARD15 showed that the association with the 5q31 risk haplotype was present only in offspring with at least one of the known CARD15 disease susceptibility alleles (P=.044). The 5q31 risk haplotype frequency was 53.1% in unrelated individuals with CD who had one or two CARD15 mutations versus 43.7% in control subjects (P=.0001) but was not significantly elevated in individuals with CD who had no CARD15 mutations (45.4%, P=.41). Kaplan-Meier survival analysis of age at disease onset showed a significantly earlier onset in homozygotes for the 5q31 risk haplotype (P=.0019). These findings suggest that genetic variants at the 5q31 (IBD5) locus may hasten the onset of Crohn disease and cooperate with CARD15 in disease causation.
Crohn disease (CD [MIM 266600]) and ulcerative colitis (UC [MIM 191390]) are two forms of chronic inflammatory bowel disease (IBD) with distinct clinical features. CD affects any part of the gastrointestinal tract, whereas in UC the inflammation is confined to the colon and rectum. Genetic linkage studies identified a susceptibility locus for CD on chromosome 16 (Hugot et al. 1996), and subsequent positional cloning and candidate gene studies revealed mutations in the NOD2 gene (now known as CARD15) at this locus that were strongly associated with CD (Hampe et al. 2001; Hugot et al. 2001; Ogura et al. 2001). A genomewide search in Canadian families found evidence of linkage to CD on chromosome 5q31 (Rioux et al. 2000), and linkage disequilibrium (LD) mapping of this locus detected association of a common haplotype in the 5q31 cytokine gene cluster with CD (Rioux et al. 2001). This haplotype contained 11 SNPs that were in almost complete LD with each other and were strongly associated with disease. The recent report (Dahlman et al. 2002) of a lack of confirmation of a published association between a genetic variant in the IL12B gene and type 1 diabetes (Morahan et al. 2001) exemplifies the importance of independent replication of such findings in complex disorders. We therefore sought to replicate the association of the 5q31 locus with CD in a large sample of European patients. We also investigated whether this association extends to UC, since common genes may contribute to susceptibility to both of these IBD subphenotypes (Watts and Satsangi 2002).
Two SNPs (C2063G and C2198G) that lie ∼67 kb apart on the common disease-associated haplotype were genotyped in 267 unrelated individuals and confirmed to be in strong LD (Δ=0.87; D′=0.91). C2063G, which is a nonsynonymous change in a GENSCAN predicted gene (Rioux et al. 2001), was then genotyped in a set of 427 British and 138 German families (Hampe et al. 1999) containing a total of 511 offspring with CD and 320 with UC. All genotyping was performed using the TaqMan biallelic discrimination system (Livak et al. 1995) on an ABI 7700 DNA analyzer. Allelic transmission distortion to these two disease phenotypes was assessed by the transmission-disequilibrium test (TDT) (Spielman et al. 1993), implemented using TRANSMIT (Clayton 1999). An association test using transmission to all affected offspring was performed by a bootstrap simulation across families (TRANSMIT). A χ2 test confirmed that genotypes were in Hardy-Weinberg equilibrium. Excess transmission of the G allele of the C2063G SNP to affected offspring was observed in CD (P=.016 (table 1) but not in UC. The SNP was then genotyped in an independent set of unrelated British individuals with CD (n=684) and UC (n=388) (Cuthbert et al. 2002) and in 701 British control subjects. The frequency of the G allele in patients with CD was significantly higher than in controls (P=.008) but was not increased in patients with UC (table 1). The CD odds ratios (ORs) for the CG and GG genotypes were 1.19 (95% CI 0.93–1.52) and 1.49 (95% CI 1.11–2.01), respectively.
Table 1.
TDT and Case-Control Analyses for the C2063G SNP at 5q31
|
Value for Analysis |
||||||||||
| TDT |
Cases and Controls |
|||||||||
| Phenotype | NTDTa | Obsb | Expc | PTDTd | NCCe | CC | CG | GG | Freq(G) | PCCf |
| CD | 511 | 484 | 458.9 | .016 | 684 | 190 | 327 | 167 | 48.3% | .008 |
| UC | 320 | 249 | 263.3 | .063 | 388 | 146 | 172 | 70 | 40.2% | NSg |
| Control | … | … | … | … | 701 | 231 | 334 | 136 | 43.2% | … |
NTDT = number of affected offspring.
Obs = observed number of transmissions of G allele to affected offspring.
Exp = expected number of transmissions of G allele to affected offspring.
PTDT = P value from TDT association of allele G.
NCC = number of case and control subjects.
PCC = P value from test for difference in frequency of allele G in cases and controls, assessed with a χ2 test.
NS = not significant (P>.05).
In view of the strong contribution of mutations in the CARD15 gene to the risk of CD, we investigated the possibility of interaction between this gene and the 5q31 locus in CD. Both the TDT and the case-control sample were genotyped for the C2063G SNP and for the three SNPs in the CARD15 gene (R702W, G908R, and 1007fs) that are independently associated with disease risk (Hugot et al. 2001; Cuthbert et al. 2002). In the TDT sample, stratification of offspring with CD by their CARD15 mutation status showed some evidence of association in offspring carrying at least one CARD15 disease-susceptibility allele (DSA) (n=203; P=.044) but no association in offspring without the three mutations (n=259; P=.20). No significant differences in the distribution of CARD15 and 5q31 genotypes were observed between familial and sporadically ascertained patients with CD (P=.7). A single individual with CD was therefore randomly selected from each family, and these were combined with the sporadically ascertained patients with CD, providing a total of 943 unrelated patients with CD and 676 control subjects. These patients with CD and control subjects were stratified according to whether they had no, one, or two CARD15 DSAs, and the frequency of the C2063G SNP was determined in these three groups (table 2). By use of a χ2 test of independence of ordinal data, the distribution of C2063G genotypes was shown to be associated with the number of CARD15 mutations in patients with CD (P=.001) but not in control subjects (P>.5). Individuals with CD who had one or two CARD15 DSAs had a significantly higher frequency of the 2063G allele than those with none (53.1% vs. 45.4%; P=.0018). The frequency of 2063G in all unrelated individuals with CD (48.0%) was significantly higher than in control subjects (43.7%; P=.016). The association of this haplotype with CD was much stronger in affected individuals with one or two CARD15 DSAs (53.1% vs. 43.7% in control subjects; P=.0001) but was not significant in case subjects with CD who had no CARD15 DSAs (45.4% vs. 43.7%; P=.41). There was no significant difference in G allele frequency in control subjects with different CARD15 genotypes. Logistic regression analysis by Splus v5.1 was then used to estimate ORs for each genotype and for combinations of genotypes, modeling case/control status on 5q31 and CARD15 genotypes (table 2). Risk owing to 5q31 was modeled by the number of alleles (zero, one, or two), and independent risks were calculated for CARD15 genotypes, giving an OR of 1.12 for each copy of the 5q31 G allele and ORs of 2.47 and 22.05 for CARD15 −/+ and +/+, respectively. ORs for specific 5q31/CARD15 genotypes were obtained by multiplying the baseline contributions for each locus, and 95% CIs were obtained directly from logistic regression parameter estimates. CARD15 contributed most of the disease risk (P<10-10); the increased risk owing to 5q31 genotypes in individuals with CD carrying no CARD15 DSAs was not significant. The combined population attributable risk for these two loci was 31%.
Table 2.
Case-Control Genotypes for the 5q31 and CARD15 Loci, with Disease Risk Estimated by Logistic Regression[Note]
|
Controls (N=676)a |
CD Cases (N=943)a |
5q31/CARD15 OR (95% CI) |
|||||||||
| CARD15 Genotype | CC | CG | GG | Freq(G) | CC | CG | GG | Freq(G) | CC | CG | GG |
| −/− | 186 | 282 | 111 | 43.5% | 190 | 296 | 133 | 45.4% | 1 | 1.12 (.97–1.29) | 1.25 (.94–1.67) |
| −/+ | 33 | 39 | 22 | 44.1% | 59 | 123 | 69 | 52.0% | 2.47 (1.90–3.22) | 2.77 (2.06–3.71) | 3.10 (2.12–4.53) |
| +/+ | 0 | 2 | 1 | … | 11 | 41 | 21 | 56.8% | 22.05 (7.30–66.67) | 24.70 (8.13–75.06) | 27.67 (8.89–86.04) |
Note.— CARD15 genotypes are defined by the presence or absence of three CD DSAs: −/− (wild type), −/+ (heterozygous for mutation), and +/+ (homozygous or compound heterozygous for mutation).
N = number of case and control subjects.
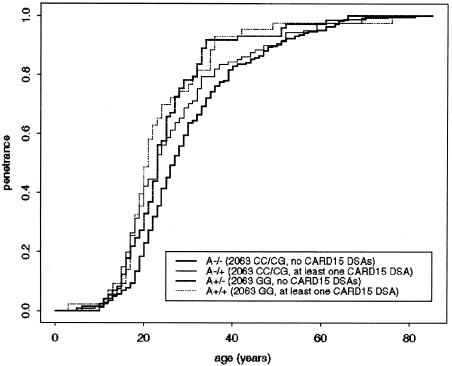
The strongest evidence for linkage to 5q31 has been found in families with early-onset CD (Rioux et al. 2000). We therefore considered the effects of CARD15 and 5q31 loci on age at disease onset in 500 independent individuals with CD. Survival curves (years until age at CD onset) were calculated using Kaplan-Meier survival analysis (Splus v5.1) and were compared using a Mantel-Haenszel test (fig. 1). A significantly earlier age at onset occurred in the 2063 GG patients with CD than in CC/CG patients with CD (P=.0019); an earlier age at onset in patients with CD carrying at least one CARD15 DSA was marginally significant (P=.048). The age at disease onset in patients with CD with the genotype A−/− (2063 CC/CG, no CARD15 DSAs) was therefore compared with each of the two-locus early-onset high-risk genotypes, A−/+ (CC/CG, at least one CARD15 DSA), A+/− (GG, no CARD15 DSAs), and A+/+ (GG, at least one CARD15 DSA) (table 3). A significantly earlier age at onset occurred in both 2063 GG patients with CD who also carried at least one CARD15 DSA (A+/+; median age 21 years; P=.003) and those who did not carry a CARD15 DSA (A+/−; median age 23 years; P=.002), compared with the baseline genotype (A−/−; median age 27 years). The age at onset in 2063 GG patients with CD who also carried at least one CARD15 DSA (A−/+) was not significantly different from those without a CARD15 DSA. In 2063 CC/CG patients with CD, the effect of CARD15 on age at onset was only marginally significant (P=.058). Disease-onset risk ratios, as defined by the ratio of the relative disease-onset rates (observed/expected number of cases) for the risk and baseline genotypes (table 3), were calculated. The relative disease onset risk ratios between two-locus risk genotypes and the baseline genotype are 1.23 (A−/+), 1.49 (A+/−), and 1.59 (A+/+). By age 21 years, 58.1% of 2063 GG patients with CD who also carried at least one CARD15 DSA had developed the disease (95% CI 40.5–70.6) compared with 27.5% of patients with CD who did not carry a 5q31 or CARD15 risk genotype (95% CI 21.6–32.9). By age 36 years, 93% of individuals with the high-risk genotype at both loci had developed the disease. We have shown elsewhere that the presence of CARD15 mutations was associated with the ileal form of CD (Cuthbert et al. 2002). We therefore investigated the possibility of an association of the 5q31 haplotype with the site of disease in 551 independent individuals with CD. No association was found; the frequency of the 2063G allele was 48.7% in affected individuals with ileal disease and 48.4% in affected individuals with colon-specific disease (P=.99).
Figure 1.
Kaplan-Meyer survival curves (age until disease onset) for the baseline genotype A−/− (2063 CC/CG, no CARD15 DSAs) and CARD15/5q31 early-onset risk genotypes: A−/+ (2063 CC/CG, one or two CARD15 DSAs), A+/− (2063 GG, no CARD15 DSAs) and A+/+ (2063 GG, one or two CARD15 DSAs).
Table 3.
Disease Onset Age Distribution by CARD15/5q31 Genotypes and Comparison of Baseline and Risk Genotypes
|
Age at Onset(years) |
No. of Cases |
|||
| Disease OnsetRisk Genotype | Mean | Median (95% CI) | Observed(N) | Expecteda (Pb) |
| A−/− | 30.1 | 27 (25–29) | 244 | … |
| A−/+ | 27.1 | 23 (20–26) | 121 | 105.0 (.058) |
| A+/− | 25.2 | 23 (22–25) | 73 | 52.9 (.002) |
| A+/+ | 24.1 | 21 (19–24) | 43 | 28.6 (.003) |
Expected number of cases, under the assumption of no difference in age at disease onset between risk genotype (2063 GG or one or two CARD15 DSAs) and baseline genotype (no or one copy of 2063 G allele and no CARD15 DSAs).
P value obtained from test of difference between two survival curves (risk genotype vs. baseline genotype).
Our family- and population-based tests for association of genetic variation at the cytokine gene cluster on chromosome 5q31 and disease support the primary evidence for the existence of a CD susceptibility gene at this locus (IBD5) (Rioux et al. 2001). We found no evidence of association with UC, which was not tested in the original study. The disease risk conferred by this locus in our large sample of individuals with CD was only 1.49 in 2063G homozygotes, which is substantially less than the sixfold risk predicted by Rioux et al (2001). The low risk is consistent with the absence of significant linkage to this locus in the genome scan from the British and German populations (Hampe et al. 1999) and underlines the difficulty in detection of linkage at loci of modest effect (Risch and Merikangas 1996). The most striking properties of the 5q31 locus, however, were its significantly higher frequency in our sample of 943 patients with CD who had one or two CARD15 mutations and its association with a younger age at disease onset. The early onset is consistent with the original linkage study (Rioux et al. 2000), which showed a higher LOD score at this locus in families with at least one affected sibling with a diagnosis at age 16 years or younger. These authors did not find evidence of interaction of the 5q31 disease risk haplotype with CARD15 in 84 CD trios but pointed out that larger samples might be required to detect it (Vermeire et al. 2002). The identity of the actual disease susceptibility gene at the 5q31 locus is unknown, but our findings suggest that it may interact with CARD15 in a common pathophysiological pathway. The 5q31 locus contains the genes for the cytokines IL4, IL5, and IL13, and CARD15 appears to be involved in bacterial lipopolysaccharide-induced activation of the transcription factor NF-κB in mononuclear phagocytes (Ogura et al. 2001). Since abnormal regulation of monocyte activation by IL4 and IL13 has been described in CD (Schreiber et al. 1995), it is possible that a hypomorphic allele in one of these regulatory cytokines in combination with a mutation in CARD15 may lead to an early and persistent proinflammatory response in the gastrointestinal tract. Thus, the 5q31 locus may contain a modifier gene which hastens disease onset in some genetically susceptible individuals. Such models must remain speculative until the function of CARD15 is more precisely defined and the identity of the CD susceptibility gene at 5q31 is known. These loci have roles in other inflammatory disorders; genetic and functional studies have implicated the 5q31 cytokine gene cluster in asthma (Cookson 2002), and mutations in CARD15 have been detected in Blau syndrome, which is a Mendelian disorder characterized by granulomatous arthritis (Miceli-Richard et al. 2001). The connection between these two loci in CD may, therefore, be of broader significance in the etiology of chronic inflammatory disease.
Acknowledgments
We thank Prof. J. Lennard-Jones and Drs. S. Bridger and J. Lee, for patient ascertainment, and A. Craggs and J. Hirst, for help in obtaining blood samples. This work was supported by the Wellcome Trust, the Generation Trust, the European Union Fifth Framework Programme, and the Deutsche Forschungsgemeinschaft.
Electronic-Database Information
URLs for data presented herein are as follows:
- Online Mendelian Inheritance in Man (OMIM), http://www.ncbi.nih.gov/Omim/ (for Crohn disease and ulcerative colitis)
- TRANSMIT http://www-gene.cimr.cam.ac.uk/clayton/software/transmit.txt (for version 2.5)
References
- Clayton D (1999) A generalization of the transmission/disequilibrium test for uncertain-haplotype transmission. Am J Hum Genet 65:1170–1177 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cookson WO (2002) Asthma genetics. Chest 121:7S–13S [DOI] [PubMed] [Google Scholar]
- Cuthbert AP, Fisher SA, Mirza MM, King K, Hampe J, Croucher PJ, Mascheretti S, Sanderson J, Forbes A, Mansfield J, Schreiber S, Lewis CM, Mathew CG (2002) The contribution of NOD2 gene mutations to the risk and site of disease in inflammatory bowel disease. Gastroenterology 122:867–874 [DOI] [PubMed] [Google Scholar]
- Dahlman I, Eaves IA, Kosoy R, Morrison VA, Heward J, Gough SC, Allahabadia A, Franklyn JA, Tuomilehto J, Tuomilehto-Wolf E, Cucca F, Guja C, Ionescu-Tirgoviste C, Stevens H, Carr P, Nutland S, McKinney P, Shield JP, Wang W, Cordell HJ, Walker N, Todd JA, Concannon P (2002) Parameters for reliable results in genetic association studies in common disease. Nat Genet 30:149–150 [DOI] [PubMed] [Google Scholar]
- Hampe J, Cuthbert A, Croucher PJ, Mirza MM, Mascheretti S, Fisher S, Frenzel H, King K, Hasselmeyer A, MacPherson AJ, Bridger S, van Deventer S, Forbes A, Nikolaus S, Lennard-Jones JE, Foelsch UR, Krawczak M, Lewis C, Schreiber S, Mathew CG (2001) Association between insertion mutation in NOD2 gene and Crohn’s disease in German and British populations. Lancet 357:1925–1928 [DOI] [PubMed] [Google Scholar]
- Hampe J, Schreiber S, Shaw SH, Lau KF, Bridger S, Macpherson AJS, Cardon LR, Sakul H, Harris TJ, Buckler A, Hall J, Stokkers P, van Deventer SJJ, Nürnberg P, Mirza MM, Lee JCW, Lennard-Jones JE, Mathew CG, Curran ME (1999) A genomewide analysis provides evidence for novel linkages in inflammatory bowel disease in a large European cohort. Am J Hum Genet 64:808–816 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hugot JP, Chamaillard M, Zouali H, Lesage S, Cezard JP, Belaiche J, Almer S, Tysk C, O’Morain CA, Gassull M, Binder V, Finkel Y, Cortot A, Modigliani R, Laurent-Puig P, Gower-Rousseau C, Macry J, Colombel JF, Sahbatou M, Thomas G (2001) Association of NOD2 leucine-rich repeat variants with susceptibility to Crohn’s disease. Nature 411:599–603 [DOI] [PubMed] [Google Scholar]
- Hugot JP, Laurent-Puig P, Gower-Rousseau C, Olson JM, Lee JC, Beaugerie L, Naom I, Dupas JL, Van Gossum A, Orholm M, Bonaiti-Pellie C, Weissenbach J, Mathew CG, Lennard-Jones JE, Cortot A, Colombel JF, Thomas G (1996) Mapping of a susceptibility locus for Crohn's disease on chromosome 16. Nature 379:821–823 [DOI] [PubMed] [Google Scholar]
- Livak KJ, Marmaro J, Todd JA (1995) Towards fully automated genome-wide polymorphism screening. Nat Genet 9:341–342 [DOI] [PubMed] [Google Scholar]
- Miceli-Richard C, Lesage S, Rybojad M, Prieur AM, Manouvrier-Hanu S, Hafner R, Chamaillard M, Zouali H, Thomas G, Hugot JP (2001) CARD15 mutations in Blau syndrome. Nat Genet 29:19–20 [DOI] [PubMed] [Google Scholar]
- Morahan G, Huang D, Ymer SI, Cancilla MR, Stephen K, Dabadghao P, Werther G, Tait BD, Harrison LC, Colman PG (2001) Linkage disequilibrium of a type 1 diabetes susceptibility locus with a regulatory IL12B allele. Nat Genet 27:218–221 [DOI] [PubMed] [Google Scholar]
- Ogura Y, Bonen DK, Inohara N, Nicolae DL, Chen FF, Ramos R, Britton H, Moran T, Karaliuskas R, Duerr RH, Achkar JP, Brant SR, Bayless TM, Kirschner BS, Hanauer SB, Nunez G, Cho JH (2001) A frameshift mutation in NOD2 associated with susceptibility to Crohn’s disease. Nature 411:603–606 [DOI] [PubMed] [Google Scholar]
- Rioux JD, Daly MJ, Silverberg MS, Lindblad K, Steinhart H, Cohen Z, Delmonte T, et al (2001) Genetic variation in the 5q31 cytokine gene cluster confers susceptibility to Crohn disease. Nat Genet 29:223–228 [DOI] [PubMed] [Google Scholar]
- Rioux JD, Silverberg MS, Daly MJ, Steinhart AH, McLeod RS, Griffiths AM, Green T, Brettin TS, Stone V, Bull SB, Bitton A, Williams CN, Greenberg GR, Cohen Z, Lander ES, Hudson TJ, Siminovitch KA (2000) Genomewide search in Canadian families with inflammatory bowel disease reveals two novel susceptibility loci. Am J Hum Genet 66:1863–1870 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Risch N, Merikangas K (1996) The future of genetic studies of complex human diseases. Science 273:1516–1517 [DOI] [PubMed] [Google Scholar]
- Schreiber S, Heinig T, Panzer U, Reinking R, Bouchard A, Stahl PD, Raedler A (1995) Impaired response of activated mononuclear phagocytes to interleukin 4 in inflammatory bowel disease. Gastroenterology 108:21–33 [DOI] [PubMed] [Google Scholar]
- Spielman RS, McGinnis RE, Ewens WJ (1993) Transmission test for linkage disequilibrium: the insulin gene region and insulin-dependent diabetes mellitus (IDDM). Am J Hum Genet 52:506–516 [PMC free article] [PubMed] [Google Scholar]
- Vermeire S, Wild G, Kocher K, Cousineau J, Dufresne L, Bitton A, Langelier D, Pare P, Lapointe G, Cohen A, Daly MJ, Rioux JD (2002) CARD15 genetic variation in a Quebec population: prevalence, genotype-phenotype relationship, and haplotype structure. Am J Hum Genet 71:74–83 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watts DA, Satsangi J (2002) The genetic jigsaw of inflammatory bowel disease. Gut Suppl 50:iii31–iii36 [DOI] [PMC free article] [PubMed] [Google Scholar]