Abstract
There is an inverse relationship between serum bilirubin concentrations and risk of coronary artery disease. The strength of the association is similar to that of smoking, systolic blood pressure, and HDL cholesterol. We carried out a genomewide scan in a Framingham Heart Study. Our study sample consisted of 330 families with 1,394 sibling pairs, 681 cousin pairs, and 89 avuncular pairs. Using variance-component methods, the heritability was estimated to be 49%±6%, and the genome scan demonstrated significant evidence of linkage of serum bilirubin to chromosome 2q, with a LOD score of 3.8 at location 243 cM. The peak multipoint LOD score is located 1 cM away from the uridine diphosphate glycosyltransferase 1 (UGT1A1) gene. UGT1A1 catalyzes the conjugation of bilirubin with glucuronic acid and thus enhances bilirubin elimination; therefore, it is an important candidate gene for serum bilirubin. Gilbert syndrome, a hyperbilirubinemic syndrome, has a population frequency of 2%–19% and is mainly due to a TA insertion at the promoter region of UGT1A1. Only one other region in the genome produced a multipoint LOD score >1 (LOD = 1.3). Our findings suggest that UGT1A1 may be a major gene controlling serum bilirubin levels in the population.
Recent studies, including a Framingham Offspring Study, have shown that there is an inverse relationship between serum bilirubin concentrations and risk of coronary artery disease (CAD) (Schwertner et al. 1994; Breimer et al. 1995; Hopkins et al. 1996; Levinson 1997; Djousse et al. 2001; Hunt et al. 2001). The strength of the association with CAD was similar to that of smoking, systolic blood pressure, and HDL cholesterol (Schwertner 1998; Schwertner and Fischer 2000). Several studies indicate bilirubin is an effective antioxidant that efficiently scavenges peroxyl radicals; suppresses the oxidation of lipids and lipoproteins, especially low density lipoprotein; and thus acts against plaque formation and subsequent atherosclerosis (Wu et al. 1991, 1996; Miller et al. 1993; Lamont et al. 1997). Thus, there is increasing interest in factors that regulate levels of serum bilirubin.
Serum bilirubin is derived primarily from the degradation of hemoglobin, catalyzed by heme oxygenase, and is transported to the liver by binding to albumin. Within hepatocytes, the solubility of bilirubin is increased by the addition of one or two molecules of glucuronic acid, catalyzed by UGT1A1 (MIM 191740). Bilirubin monoglucuronide and diglucuronide metabolites are then actively transported into the bile.
In humans, the heritability of total serum bilirubin has been estimated to be 58% (Williams et al. 1991). Two segregation analyses in large population-based cohorts have suggested that there is a major gene with the rarer genotype associated with elevated bilirubin levels (Hunt et al. 1996, 2001). Very recently, the authors of a genome scan on serum bilirubin in a white population ascertained by the presence of early CAD reported a locus on chromosome 2q telomere, 2q34–37, that influences variation in serum bilirubin levels (Kronenberg et al. 2002). Here, we report the results of a serum bilirubin genome scan in unselected families, the Framingham Heart Study.
The Framingham Heart study is a population-based cohort study. In brief, the Framingham Heart Study began in 1948 with the recruitment of 5,209 residents from Framingham, MA. The subject individuals have undergone biennial examinations since the study began. In 1971, the Framingham Offspring Study (Kannel et al. 1979) was initiated. In total, there were 5,124 subjects recruited. The offspring subjects have been examined every 4 years (except the first two examinations, which have 8 years intervening). The current analysis consists of 1,702 genotyped subject individuals from the 330 largest extended families within the study.
Total serum bilirubin was measured during the first and second examinations of the offspring, using the ultramicromethod (Walters and Gerarde 1970). Our analysis was restricted to bilirubin measurements in the first examination, since far fewer individuals had serum bilirubin measured in the second examination. The correlation coefficient between the two examinations is 0.6, P<.01.
Genomic DNA was isolated from nucleated blood cells. DNA samples were sent to the Marshfield Mammalian Genotyping Service. At an average of 10 cM density, 399 microsatellite markers (Screening Set 8) (Yuan et al. 1997) covered the genome with an average marker heterozygosity of 0.77.
Variation in serum bilirubin due to known factors was tested and removed, using SOLAR, to enhance the ability of linkage analysis to detect genetically determined variation (Almasy and Blangero 1998). The covariates selected were all statistically significantly associated in a regression model and were incorporated in both the heritability estimation and the linkage analysis. These were sex, height, weight, total cholesterol, hematocrit, albumin, SGOT, smoking (no. of cigarettes/d), and alcohol intake (oz./wk). At first, BMI (instead of height and weight) was tested but was not significant in the multiple-regression model. When BMI was replaced by height and weight, both variables were highly significant. In essence, SOLAR uses the residuals for heritability estimation and two-point and multipoint linkage analysis. Further analyses were conducted using normalized deviates. SOLAR makes use of all information in pedigrees of any size and structure. Residual heritability is the proportion of total phenotypic variation due to additive genetic effects, after removing the variation attributable to covariates. Marker allele frequencies were estimated from the study participants and then used to estimate the proportion of alleles shared identical by descent (IBD) among all relative pairs. SOLAR uses a likelihood-ratio test to evaluate linkage by comparing a purely polygenic model (without consideration of genetic marker information) with a model that incorporates marker information at a specific locus (two-point analysis) or several nearby markers in a chromosome (multipoint analysis). We also repeated the variance-components linkage analysis using the normalized deviates, calculated from ranked standardized residuals in regression models conducted in SAS.
Since this method is based on the assumption of a multivariate normal distribution, violations of the assumption may result in inflated type I error rates (Allison et al. 1999; Blangero et al. 2000, 2001). To ensure a more accurate result, we used an empirical P value method implemented in SOLAR for LOD score adjustment (Blangero et al. 2000). A fully informative marker, which was not linked to the trait studied, was simulated. The IBD information for this marker was calculated, and then linkage analysis was performed. The empirical P values for the trait were obtained on the basis of the LOD score distribution generated from 10,000 replicates. This method provides robust LOD scores for data with nonnormal distributions. As a second approach to account for the nonnormality in these data, the analysis of normalized deviates was also pursued.
There were 1,702 individuals in the 330 largest extended families who were genotyped. Among them, 394 individuals belonged to the original cohort and did not have serum bilirubin measured. The total number of individuals with serum bilirubin measured was 1,612. The mean values of the phenotypes of the 1,612 (50% male) individuals are displayed in table 1. The total number of individuals with serum bilirubin measured and genotyped was 1,264, including 1,394 sibling pairs, 681 cousin pairs, and 89 avuncular pairs.
Table 1.
Characteristics of the 1,612 Framingham Offspring with Bilirubin Measured
|
Value in Subjects |
||||
| Male |
Female |
|||
| Variable | Mean ± SD | Range | Mean ± SD | Range |
| Total bilirubin (mg/dl) | 8.8 ± 3.8 | 2–35 | 7.1 ± 2.9 | 2–27 |
| Age (years) | 32.3 ± 10.4 | 11–64 | 33.3 ± 10.5 | 10–62 |
| Height (inches) | 68.9 ± 3.3 | 53–78 | 63.6 ± 2.7 | 52–71 |
| Weight (pounds) | 176.1 ± 29.4 | 54–264 | 137.1 ± 27.8 | 67–281 |
| Total cholesterol (mg/dl) | 193.5 ± 38.6 | 96–138 | 188.4 ± 37.9 | 100–346 |
| Hematocrit (%) | 45.5 ± 2.9 | 34–62 | 40.6 ± 2.8 | 28–52 |
| Albumin (mg/dl) | 48.1 ± 3.1 | 32–57 | 45.8 ± 3.3 | 26–59 |
| SGOTa (mg/dl) | 47.9 ± 20.0 | 18–221 | 40.4 ± 17.7 | 8–285 |
| Smoking (no./d) | 15.0 ± 16.4 | 0–90 | 11.0 ± 14.1 | 0–88 |
| Alcohol (oz./wk) | 4.9 ± 5.9 | 0–43 | 2.2 ± 3.0 | 0–34 |
Liver serum glutamic oxaloacetic transaminase.
The distribution of serum bilirubin was not normal, P<.01, and the estimates of skewness and kurtosis were 2.41±0.03 and 9.50±0.07, respectively, without covariates adjustment.
The heritability estimate from the serum bilirubin, after adjusting for covariates, was 49%±6%. The proportion of variance due to all final covariates is ∼11%.
For two-point linkage analysis, LOD scores of 1 or higher were observed for markers on chromosomes 4, 5, and 7 and for three adjacent markers on chromosome 2 (table 2).
Table 2.
Markers with Two-Point LOD Score of 1 or Higher
| Marker | Chromosome | Location(cM) | LOD Score |
| D2S1363 | 2 | 227 | 1.7 |
| D2S427 | 2 | 237 | 1.7 |
| 112yd2 | 2 | 261 | 1.5 |
| D4S1647 | 4 | 105 | 1.2 |
| D5S820 | 5 | 160 | 1.1 |
| GATA118G10 | 7 | 79 | 1.4 |
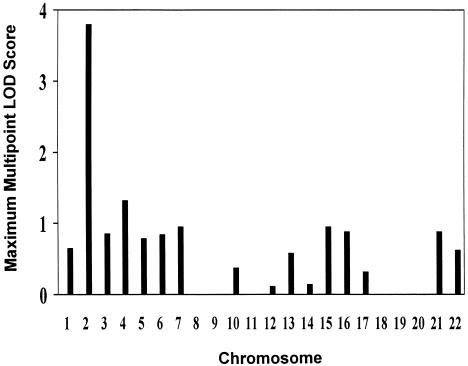
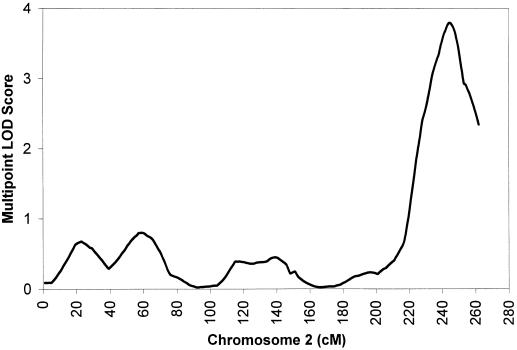
The highest multipoint LOD scores for each of the 22 autosomes are presented in figure 1. For multipoint analysis, a maximum LOD score of 3.8 (P=.00001) was observed on the chromosome 2q telomere, with the peak location in 243 cM (fig. 2) with comprehensive human genetic maps (Broman et al. 1998). Using normalized deviates also produced very similar results, with a maximum LOD score of 3.9 located in 244 cM. We also carried out a regular LOD score analysis without adjusting for nonnormality; a maximum LOD score of 5.7 was obtained at the same location, 2q 243 cM. Except for this, there was only one region on chromosome 4 (96 cM) in the genome that had a maximum multipoint LOD score ⩾1 (LOD=1.3).
Figure 1.
Maximum multipoint LOD scores for serum bilirubin for the 22 autosomes. X-axis values are the number of 22 autosomes. Y-axis values are maximum multipoint LOD scores. Linkage analyses were conducted using multivariable residuals, adjusted for sex, height, weight, total cholesterol, hematocrit, albumin, SGOT, smoking, and alcohol intake.
Figure 2.
Multipoint LOD scores for serum bilirubin on chromosome 2. X-axis values are cM. Y-axis values are multipoint LOD scores. Linkage analyses were conducted using multivariable residuals, adjusted for sex, height, weight, total cholesterol, hematocrit, albumin, SGOT, smoking, and alcohol intake.
Peak evidence for linkage on chromosome 2 occurred in the region bounded by markers D2S427 and GATA178G09. The LOD-one supporting interval (the region corresponding to maximum LOD score minus 1) for the location of serum bilirubin QTL spans a 22-cM interval flanked by markers D2S1363 and 112yd4, which are 34 cM apart. The results provided substantial evidence for a serum bilirubin QTL on 2q telomere.
Our results are consistent with a recently reported genome scan, conducted in a population enriched for early-onset cardiovascular disease, in which a locus with a multipoint LOD score of 3.01 was identified in this region (Kronenberg et al. 2002). Thus, our finding complement and extend the provocative findings of Kronenberg et al., conducted in a subset of families selected for disease. Kronenberg et al. also provided evidence for loci on chromosome 9, 10, and 18 that may possibly influence serum bilirubin levels. None of the loci on chromosome 9, 10, and 18 overlap with our findings. In addition to coming from a different study population, the different results may be due to differences in linkage analysis methods. Our study was conducted using empirical P value and normalized deviate methods to adjust LOD score for nonnormality of data, whereas Kronenberg et al. used both parametric and nonparametric linkage methods without adjustment for nonnormality. Without adjustment for nonnormality, our results showed LOD=5.7 at 2q, 243 cM.
Within the chromosome region of 2q telomere lies an important candidate gene, UGT1A1. This gene is ∼5 Mb (∼5 cM) telomeric from marker D2S427. The peak maximum LOD score in this area is located 6 cM telomeric from marker D2S427. Therefore, UGT1A1 and the peak maximum LOD score almost overlap. To date, no other genes known to regulate serum bilirubin metabolism have been found in this region.
Uridine diphosphoglucuronate glucuronosyltransferases (UGTs) are a family of enzymes that conjugate various substrates with glucuronic acid and enhance their elimination through the bile. UGT1A1 is the physiologically relevant isoform and significantly contributes to bilirubin glucuronidation. Mutations in UGT1A1 lead to complete or partial inactivation of the enzyme, causing three nonhemolytic unconjugated hyperbilirubinemia syndromes, type I and type II Crigler-Najjar syndromes and Gilbert syndrome. Crigler-Najjar syndromes are rare and lethal forms of autosomal inherited diseases, with a complete lack of bilirubin UGT activity (type I) or a severe deficiency of the enzyme (type II). Type I and type II Crigler-Najjar syndromes are mainly associated with missense mutations in the coding region of the UGT1A1 gene and have a frequency of 1 in 1 million births. Gilbert syndrome, a more prevalent (2%–19% in population studies) and essentially benign condition, is principally caused by a TA insertion at the TATAA promoter region upstream of the UGT1A1 exon. The gene frequency of this mutation among the white population is extraordinarily high, 0.34–0.40 (Bosma et al. 1995; Monaghan et al. 1996; Beutler et al. 1998; Morsche et al. 2001) and is very close to the estimated major gene allele frequency from the two segregation analyses (Hunt et al. 1996, 2001).
Combining the results of previous association and segregation studies, the results of this genome scan, and the physiological function of the gene, we speculate that UGT1A1 may be a major gene controlling serum bilirubin levels.
Two segregation studies indicated that a major gene may protect ∼12% of the population against CAD, and most individuals with CAD lacked the protective effect from this gene associated with bilirubin levels (Hunt et al. 1996, 2001). If ⩾12% of the population have a strong protective effect against CAD owing to the UGT1A1 gene, this would have significant population implications in future CAD prevention. Given the substantial magnitude of variability in bilirubin explained by the chromosome 2 region of linkage, our data support further study of the role of the UGT1A1 gene variants in bilirubin determination and in risk for CAD and related cardiovascular diseases.
In conclusion, our results provide evidence that a major gene with a large effect influencing bilirubin levels exists on chromosome 2q telomere. We have identified a chromosomal region implicated in several Mendelian hyperbilirubinemic disorders, and within this region resides an important candidate gene, UGT1A1. Further research is warranted to examine the linkage between the UGT 1A1 gene and serum bilirubin levels, as well as association studies to examine for associations of polymorphisms in the UGT1A1 gene with serum bilirubin and CAD.
Electronic-Database Information
URLs for data presented herein are as follows:
- Mammalian Genotyping Service, http://research.marshfieldclinic.org/genetics/
- Online Mendelian Inheritance in Man (OMIM), http://www.ncbi.nlm.nih.gov/Omim/ (for UGT1A1) [PubMed]
References
- Allison DB, Neale MC, Zannolli R, Schork NJ, Amos CI, Blangero J (1999) Testing the robustness of the likelihood-ratio test in a variance-component quantitative-trait loci-mapping procedure. Am J Hum Genet 65:531–544 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Almasy L, Blangero J (1998) Multipoint quantitative-trait linkage analysis in general pedigrees. Am J Hum Genet 62:1198–1211 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beutler E, Gelbart T, Demina A (1998) Racial variability in the UDP-glucuronosyltransferase 1 (UGT1A1) promoter: a balanced polymorphism for regulation of bilirubin metabolism. Proc Natl Acad Sci USA 95:8170–8174 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blangero J, Williams JT, Almasy L (2000) Robust LOD scores for variance component-based linkage analysis. Genet Epidemiol 19:S8–S14 [DOI] [PubMed] [Google Scholar]
- ——— (2001) Variance component methods for detecting complex trait loci. Adv Genet 42:151–181 [DOI] [PubMed] [Google Scholar]
- Bosma PJ, Chowdhury JR, Bakker C, Gantla S, de Boer A, Oostra BA, Lindhout D, Tytgat GNJ, Jansen PLM, Oude Elferick RPJ, Chowdhury NR (1995) The genetic basis of the reduced expression of bilirubin UDP-glucuronosyltransferase 1 in Gilbert’s syndrome. N Engl J Med 333:1171–1175 [DOI] [PubMed] [Google Scholar]
- Breimer LH, Wannamethee G, Ebrahim S, Shaper AG (1995) Serum bilirubin and risk of ischemic heart disease in middle-aged British men. Clin Chem 41:1504–1508 [PubMed] [Google Scholar]
- Broman KW, Murray JC, Sheffield VC, White RL, Weber JL (1998) Comprehensive human genetic maps: individual and sex-specific variation in recombination. Am J Hum Genet 63:861–869 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Djousse L, Levy D, Cupples LA, Evans JC, D’Agostino RB, Ellison RC (2001) Total serum bilirubin and risk of cardiovascular disease in the Framingham offspring study. Am J Cardiol 87:1196–1200 [DOI] [PubMed] [Google Scholar]
- Hopkins PN, Wu LL, Hunt SC, James BC, Vincent GM, Williams RR (1996) Higher serum bilirubin is associated with decreased risk for early familial coronary artery disease. Arterioscler Thromb Vasc Biol 16:250–255 [DOI] [PubMed] [Google Scholar]
- Hunt SC, Kronenberg F, Eckfeld JH, Hopkins PN, Myers RH, Heiss G (2001) Association of plasma bilirubin with coronary heart disease and segregation of bilirubin as a major gene trait: the NHLBI family heart study. Atherosclerosis 154:747–754 [DOI] [PubMed] [Google Scholar]
- Hunt SC, Wu LL, Hopkins PN, Williams RR (1996) Evidence for a major gene elevating serum bilirubin concentration in Utah pedigrees. Arterioscler Thromb Vasc Biol 16:912–917 [DOI] [PubMed] [Google Scholar]
- Kannel WB, Feinleib M, McNamara PM, Garrison RJ, Castelli WP (1979) An investigation of coronary heart disease in families: the Framingham offspring study. Am J Epidemiol 110:281–290 [DOI] [PubMed] [Google Scholar]
- Kronenberg F, Coon H, Gutin A, Abkevich V, Samuels ME, Ballinger DG, Hopkins PN, Hunt SC (2002) A genome scan for loci influencing anti-atherogenic serum bilirubin levels. Europ J Hum Genet 10:539–546 [DOI] [PubMed] [Google Scholar]
- Lamont J, Campbell J, FitzGerld P (1997) Measurement of individuals versus total antioxidants. Clin Chem 43:852–853 [PubMed] [Google Scholar]
- Levinson SS (1997) Relationship between bilirubin, apolipoprotein B, and coronary artery disease. Ann Clin Lab Sci 27:185–192 [PubMed] [Google Scholar]
- Miller NG, Rice-Evens C, Davies MJ, Gopinathan V, Milner A (1993) A novel method for measuring antioxidant capacity and its application to monitoring the antioxidant status in premature neonates. Clin Sci 84:407–412 [DOI] [PubMed] [Google Scholar]
- Monaghan G, Ryan M, Seddon R, Burchell B (1996) Genetic variation in bilirubin UDP-glucuronosyltranferase gene promoter and Gilbert syndrome. Lancet 347:578–581 [DOI] [PubMed] [Google Scholar]
- Morsche RH, Zusterzeel PL, Raijmakers MT, Roes EM, Steegers EA, Peters WH (2001) Polymorphism in the promoter region of the bilirubin UDP-glucuronosyltransferase (Gilbert’s syndrome) in healthy Dutch subjects. Hepatology 33:765 [DOI] [PubMed] [Google Scholar]
- Schwertner HA (1998) Association of smoking and low serum bilirubin antioxidant concentrations. Atherosclerosis 136:383–387 [DOI] [PubMed] [Google Scholar]
- Schwertner HA, Fischer JR Jr (2000) Comparison of various lipid, lipoprotein, and bilirubin combinations as risk factors for predicting coronary artery disease. Atherosclerosis 150: 381–387 [DOI] [PubMed] [Google Scholar]
- Schwertner HA, Jackson WG, Tolan G (1994) Association of low serum concentration of bilirubin with increased risk of coronary artery disease. Clin Chem 40:18–23 [PubMed] [Google Scholar]
- Walters MI, Gerarde HW (1970) An ultramicromethod for the determination of conjugated and total bilirubin in serum or plasma. Microchem J 15:231–243 [Google Scholar]
- Williams RR, Hasstedt SJ, Hunt SC, Wu LL, Hopkins PN, Berry TD, Stults BM, Barlow GK, Kuida H (1991) Genetic traits related to hypertension and electrolyte metabolism. Hypertension 17:I69–I73 [DOI] [PubMed] [Google Scholar]
- Wu TW, Fung KP, Wu J, Yang CC, Weisel RD (1996) Antioxidation of human low density lipoprotein by unconjugated and conjugated bilirubins. Biochem Pharmacol 51:859–862 [DOI] [PubMed] [Google Scholar]
- Wu TW, Wu J, Li RK, Mickle D, Carey D (1991) Albumin-bound bilirubins protect human ventricular myocytes against oxyradical damage. Biochem Cell Biol 69:683–688 [DOI] [PubMed] [Google Scholar]
- Yuan B, Vaske D, Weber JL, Beck J, Sheffield VC (1997) Improved set of short-tandem-repeat polymorphisms for screening the human genome. Am J Hum Genet 60:459–460 [PMC free article] [PubMed] [Google Scholar]