Abstract
Acrocapitofemoral dysplasia is a recently delineated autosomal recessive skeletal dysplasia, characterized clinically by short stature with short limbs and radiographically by cone-shaped epiphyses, mainly in hands and hips. Genomewide homozygosity mapping in two consanguineous families linked the locus to 2q35-q36 with a maximum two-point LOD score of 8.02 at marker D2S2248. Two recombination events defined the minimal critical region between markers D2S2248 and D2S2151 (3.74 cM). Using a candidate-gene approach, we identified two missense mutations in the amino-terminal signaling domain of the gene encoding Indian hedgehog (IHH). Both affected individuals of family 1 are homozygous for a 137C→T transition (P46L), and the three patients in family 2 are homozygous for a 569T→C transition (V190A). The two mutant amino acids are strongly conserved and predicted to be located outside the region where brachydactyly type A-1 mutations are clustered.
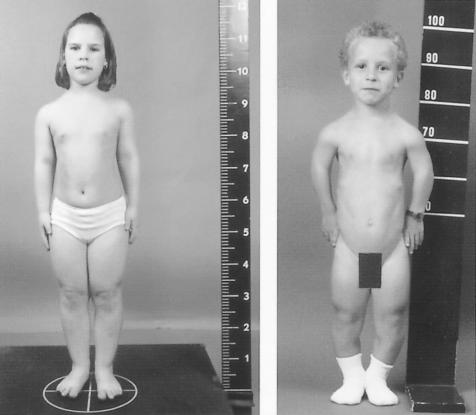
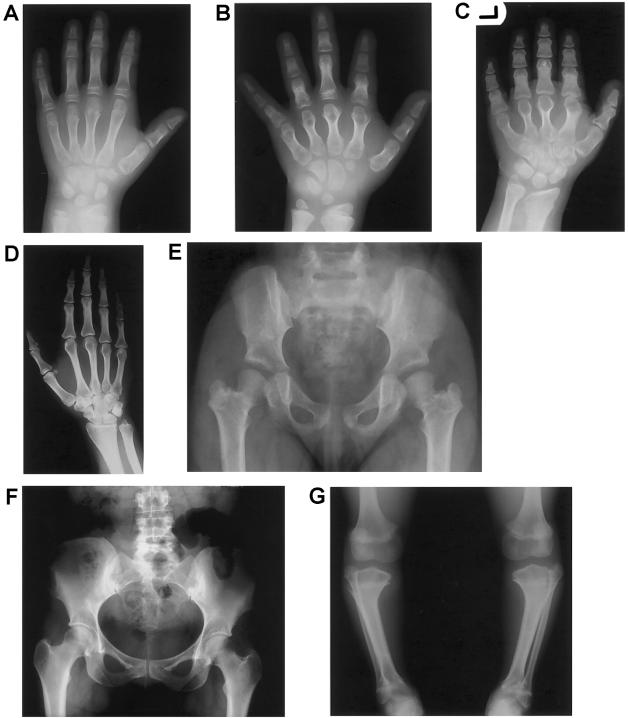
Recently, we delineated a new autosomal recessive skeletal dysplasia in two consanguineous families of Belgian and Dutch origin (Mortier et al. 2003). The clinical phenotype is characterized by short stature of variable degree with short limbs and brachydactyly, relatively large head, and narrow thorax with pectus deformities (fig. 1). Affected individuals do not exhibit associated congenital anomalies and are of normal intelligence. Radiographically, cone-shaped epiphyses (CSE) or a similar epiphyseal configuration are observed in the hands, the proximal part of the femur, and, to a variable degree, at the shoulders, knees, and ankles (fig. 2). These CSE appear early in childhood and disappear with the premature fusion of the growth plate before puberty, resulting in shortening of the involved skeletal components. This process leads, in the hips, to a characteristic egg-shaped femoral head attached to a very short femoral neck and, in the hands, to shortening of the tubular bones, especially the middle phalanges. Since all affected individuals show these striking and characteristic abnormalities in hands and hips, we named the disorder acrocapitofemoral dysplasia (ACFD).
Figure 1.
Clinical variability illustrated by two affected individuals. Left panel, Patient VII-4 at age 10.5 years. Note mild disproportionate short stature with brachydactyly. Right panel, Short-limb dwarfism with narrow thorax and genu varum in patient IX-2 at age 9 years.
Figure 2.
Most characteristic radiographic abnormalities in the affected individuals. A–C, Radiographs of the left hand. A, Cone-shaped epiphyses in the thumbs and the middle and distal phalanges of digits II–V of patient VII-4 at age 9.5 years. B, Shortening of the tubular bones more pronounced in patient IX-2 at age 11 years. Note the remnants of cone-shaped epiphyses and the fused “tear-drop” metacarpal epiphyses. C, Severe brachydactyly in patient IX-1 at age 14 years. Note complete epimetaphyseal fusion of metacarpals and phalanges. D, Radiograph of the right hand of parent VI-1, showing no obvious signs of BDA1. E, Radiograph of the pelvis in patient VII-4 at age 6 years, showing coxa vara with egg-shaped femoral head and very short femoral neck after epimetaphyseal fusion. F, Normal radiograph of the pelvis in parent VI-1, with no evidence of a premature fusion of the proximal femoral growth plate (normal femoral head and neck). G, Radiograph of the lower limbs in patient IX-2 at age 11 years. Varus deformity of the tibiae with proximal and distal premature epimetaphyseal fusion is most likely the result of cone-shaped epiphyses. Note the voluminous distal femoral epiphyses and the proximal overgrowth of the fibulae.
To unravel the genetic basis of this skeletal dysplasia, we performed homozygosity mapping in both consanguineous families, using 400 genetic markers with an average spacing of 10 cM (ABI PRISM Linkage Mapping Set version 2) (Lander and Botstein 1987). Initially, the genomewide screening was performed on four affected individuals (VII-1, VII-4, IX-2, and IX-5) from both families (fig. 3). A fifth patient (IX-1), later identified, was subsequently used in the analysis. Appropriate informed consent was obtained from all human subjects involved in the study. Alleles were scored using the Genescan and Genemapper software (Applied Biosystems). After the initial screening, the region on 2q35-q36 was further investigated, because four affected individuals were homozygous by descent for two consecutive markers, D2S325 and D2S2382. Analysis with additional, closely spaced markers refined the candidate region to between D2S2382 and D2S163, a distance of about 5 cM (fig. 3). A recombination event between loci D2S2382 and D2S2248 in patient IX-2 (fig. 3B) defined the proximal boundary of the genetic interval. A recombination event between loci D2S2151 and D2S163 in individual IX-4 (fig. 3B) defined the distal boundary of the candidate region. The MLINK program of the LINKAGE software package (Lathrop and Lalouel 1984) was used to calculate two-point LOD scores between the disease phenotype and each of the markers, assuming a recessive disease with an allele frequency of 0.001 and marker allele frequencies of 0.125. This resulted in a maximum LOD score of 8.02 at θ=0 for marker D2S2248 (table 1). A search on the human genome resources of the National Center for Biotechnology Information disclosed several candidate genes within the linked region on chromosome 2q35-q36: IGFBP2 and IGFBP5 (both encoding an insulin-like growth-factor–binding protein), IHH (encoding Indian hedgehog), and STK36 (human homologue of the Drosophila melanogaster gene fused). We selected IHH as the strongest candidate because this gene is expressed in the growth plate (Kindblom et al. 2002), dominant mutations have been identified in brachydactyly type A1 (BDA1 [MIM 112500]) (Gao et al. 2001; McCready et al. 2002), and Ihh-null mice exhibit a short-limb dwarfism phenotype (St-Jacques et al. 1999).
Figure 3.
Pedigrees of (Belgian) family 1 (A) and (Dutch) family 2 (B). The genotypes of five markers within the linked region on chromosome 2 are shown. The haplotype cosegregating with the disorder is indicated with a black bar.
Table 1.
Two-Point LOD Scores for Linkage of the ACFD Locus to Chromosome 2q35-q36
| Combined LOD Score at θ= |
Family 1 |
Family 2 |
Families 1 and 2 |
|||||||||
| Marker | Positiona | .00 | .01 | .05 | .10 | .30 | Zmax | θ | Zmax | θ | Zmax | θ |
| D2S2382 | 213.49 | −∞ | 7.01 | 6.81 | 5.96 | 2.03 | 3.24 | 0 | 3.89 | .05 | 7.09 | .02 |
| D2S2248 | 214.71 | 8.02 | 7.80 | 6.92 | 5.789 | 1.66 | 1.90 | 0 | 6.12 | 0 | 8.02 | 0 |
| D2S1371 | 215.25 | 6.32 | 6.11 | 5.28 | 4.26 | .97 | 1.40 | 0 | 4.92 | 0 | 6.32 | 0 |
| D2S2151 | 218.45 | 5.02 | 4.85 | 4.17 | 3.34 | .74 | .63 | 0 | 4.40 | 0 | 5.02 | 0 |
| D2S163 | 218.45 | −∞ | 10.85 | 10.62 | 9.72 | 4.98 | 2.02 | 0 | 8.89 | .01 | 10.92 | .02 |
The position is given in centiMorgans according to the Marshfield map.
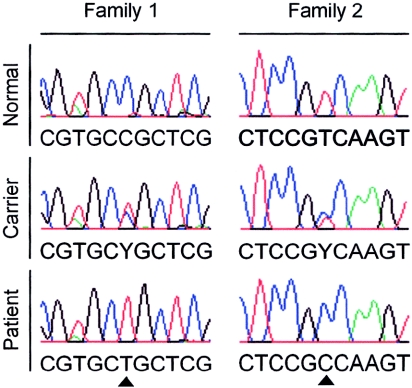
Sequence analysis of Indian hedgehog (IHH [GenBank accession number Q14623]) in the affected individuals revealed two distinct nucleotide changes. In the two affected individuals in family 1, a homozygous 137C→T transition, predicted to result in a P46L substitution, was observed (fig. 4). In the three affected individuals in family 2, a homozygous 569T→C transition, predicted to result in a V190A substitution, was identified (fig. 4). Both substitutions are located in the amino-terminal domain of the Indian hedgehog protein, which is responsible for local and long-range signaling (Porter et al. 1995). Heterozygosity for the 137C→T and 569T→C mutation was observed in the unaffected parents in family 1 and 2, respectively. Both sequence changes were absent in 75 unrelated control subjects (150 chromosomes), confirming that they do not represent common polymorphisms. Finally, alignment of the protein sequence of homologues of Indian hedgehog in several species showed that both proline at position 46 and valine at position 190 are strongly conserved residues (fig. 5).
Figure 4.
Sequence analysis of IHH in both families. The normal (top), heterozygous (middle), and homozygous mutant (bottom) sequences are shown. The 137C→T mutation is presented on the left, and the 569T→C mutation is shown on the right (triangles).
Figure 5.
Cross-species alignment of the amino-terminal signaling domain of several Hedgehog homologues: HH Drome, IHH Mouse, IHH Brare, DHH Mouse, DHH2 Xenla, TWHH Brare, and AmphiHH (GenBank accession numbers Q02936, P97812, Q98862, Q61488, Q91611, Q90419, and CAA74169.1). Amino acids matching the HH Drome sequence are represented as dots. ACFD mutations are indicated with a gray bar and BDA1 mutations with an asterisk.
Indian hedgehog is one of the three mammalian homologues of the D. melanogaster segment polarity gene, Hedgehog (Jeong and McMahon 2002). The Hedgehog gene family encodes a group of secreted signaling molecules that are essential for growth and patterning of many different body parts of vertebrate and invertebrate embryos (Ingham and McMahon 2001). These molecules are processed after translation by internal cleavage to generate an amino-terminal fragment and a carboxy-terminal fragment with the subsequent addition of cholesterol at the C-terminus of the amino-terminal fragment. This amino-terminal fragment has all the known signaling activity, whereas the carboxy-terminal fragment is responsible for the autoproteolytic process (Lee et al. 1994; Porter et al. 1995).
The precise regulation of proliferation and hypertrophic differentiation of chondrocytes in the growth plate is critical for balancing longitudinal growth and ossification of bones (Erlebacher et al. 1995). Several lines of evidence indicate that Ihh signaling controls growth of bones by coordinating chondrocyte proliferation and differentiation (Vortkamp et al. 1996; Long et al. 2001). The regulation of differentiation is indirectly mediated through the production of parathyroid hormone–related peptide (PTHrP). By stimulating PTHrP production in the periarticular growth plate, Ihh delays the switch from proliferation to differentiation. In this way, Ihh determines the rate and site in the growth plate at which hypertrophic differentiation occurs (Karp et al. 2000). We postulate that the recessive mutations, identified in our patients with ACFD, cause an increased rate of chondrocyte differentiation by diminishing Indian hedgehog signaling in the growth plate. This increased rate of differentiation is radiographically reflected by the appearance of CSE and subsequent premature closure of the growth plate, with shortening of the bone as a consequence. The cone-shaped configuration of the epiphyses may indicate that this increased rate of differentiation is most pronounced or starts earlier at the center of the physis. There is no evidence for a concentration gradient of IHH and/or PTHrP within a certain zone of the growth plate that could explain an earlier fusion at the center. However, in normal conditions, the knee growth plates seem to close earlier in their central portion than in their peripheral portion (Sasaki et al. 2002). Of note are also the presence of CSE in other skeletal dysplasias affecting the balance between chondrocyte proliferation and differentiation in the growth plate (e.g., cartilage hair hypoplasia [MIM 250250]) and the appearance of CSE after infantile meningococcemia (Patriquin et al. 1981; Grogan et al. 1989; Giedion 1998).
Radiographic evaluation of the heterozygous parents of patients VII-1, VII-4 (family 1), and IX-2 (family 2) does not reveal obvious signs of BDA1 (fig. 2D). None of these parents show absent or rudimentary middle phalanges or middle phalanges fused with the distal phalanges. Mild shortening of middle phalanges II–IV (up to 2.5 SD below the mean) and severe shortening of the middle phalanx V (up to 5 SD below the mean) are observed only in the parents of patients VII-1 and VII-4. The parents of patient IX-2 show only relative shortening (>2 SD below the mean) of the metacarpals and proximal phalanges (data not shown). In addition, both parents of patients VII-1 and VII-4 (family 1) show normal radiographs of the pelvis (fig. 2F).
A similar situation, where heterozygotes of the recessive mutations do not exhibit the dominant phenotype, has been observed in brachydactyly type B (BDB [MIM 113000]) and Robinow syndrome (MIM 268310). Heterozygous mutations flanking the intracellular tyrosine kinase domain of ROR2 cause BDB (Schwabe et al. 2000). Homozygous ROR2 mutations scattered throughout the entire coding region cause Robinow syndrome. However, heterozygous carriers of Robinow syndrome do not seem to have BDB (Afzal et al. 2000; Schwabe et al. 2000; van Bokhoven et al. 2000). A possible explanation may be that Robinow mutations are loss-of-function mutations with no phenotypic consequences in carriers, whereas BDB mutations exert a dominant effect. In contrast to these observations, loss-of-function mutations of GDF5 cause brachydactyly type C (BDC [MIM 113100]) in the heterozygous state and Grebe dysplasia (MIM 200700), an autosomal recessive acromesomelic dysplasia, in the homozygous state, with radiographic features of BDC in the heterozygous carriers of Grebe mutations (Costa et al. 1998; Everman et al. 2002; Faiyaz-Ul-Haque et al. 2002).
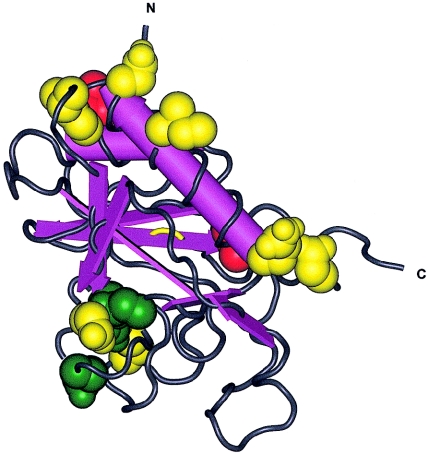
These studies together with our data indicate that the phenotypic outcome of mutations in genes involved in brachydactyly is not only dependent on the homozygous versus heterozygous state but also on the location and nature of the mutation. The mutations in IHH, causing either BDA1 or ACFD, are both missense mutations. However, their precise locations within the amino-terminal signaling domain differ. To compare the locations of different Hedgehog mutations, the crystal structure of the amino-terminal domain of mouse Shh (1VHH [Molecular Modeling DataBase accession number 3860]) can be used because of the high similarity between human IHH and mouse Shh (GenBank accession number Q62226) (Hall et al. 1995) (fig. 6). The mutations reported in BDA1 (fig. 6, green) are predicted to be clustered together on one side of the protein, whereas the ACFD (fig. 6, red) mutations seem to be located at the other side, in regions where mutations in SHH (GenBank accession number Q15465) have been reported to cause holoprosencephaly (HPE3 [MIM 142945]) (fig. 6, yellow). In addition, the P46L mutation is located in a region important for binding of the sonic hedgehog protein to its receptor Patched (Fuse et al. 1999). In conclusion, this study indicates that mutations in IHH can cause not only BDA1 but also a more severe skeletal dysplasia with short-limb dwarfism (ACFD). Additional in vitro and in vivo experiments are necessary to investigate the consequences of the different (BDA1 and ACFD) mutations on Indian hedgehog signaling in the growth plate.
Figure 6.
Tertiary structure of the amino-terminal signaling domain of mouse Shh with the indication of the corresponding location of the different missense mutations in human IHH and SHH (Human Gene Mutation Database). ACFD mutations (IHH) are represented in red, BDA1 mutations (IHH) in green, and HPE3 mutations (SHH) in yellow. The cluster in the middle (bottom) contains the four BDA1 mutations (E95K, D100E, D100N, and E131K) and two HPE3 mutations (D93V and H140P). The cluster at the carboxy-terminal end (middle right) contains one ACFD mutation (V190A) and two HPE3 mutations (Q105H and E193Q). The cluster at the amino-terminal end (top) contains one ACFD mutation (P46L) and four HPE3 mutations (I116F, N120K, W122G, and W122R).
Acknowledgments
We are indebted to the families for their interest and cooperation. Special thanks to M. L. Duyts for her excellent technical assistance in preparing the radiographic illustrations. We are grateful to Mike Briggs for critical review of the manuscript. This study is supported, in part, by the Fund for Scientific Research, Flanders, with a mandate “fundamental clinical research” to G.M., and by the Fifth Framework of the specific research and technological development program “Quality of Life and Management of Living Resources” of the European Commission (Contract QLG1-CT-2001-02188) to J.H., A.D.P., and G.M.
Electronic-Database Information
Accession numbers and URLs for data presented herein are as follows:
- GenBank, http://ncbi.nlm.nih.gov/Genbank/ (for HH Drome [accession number Q02936], IHH Mouse [accession number P97812], IHH Brare [accession number Q98862], DHH Mouse [accession number Q61488], DHH2 Xenla [accession number Q91611], TWHH Brare [accession number Q90419], AmphiHH [accession number CAA74169.1], IHH Human [accession number Q14623], SHH Human [accession number Q15465], and SHH Mouse [accession number Q62226])
- Human Gene Mutation Database, http://archive.uwcm.ac.uk/uwcm/mg/hgmd0.html
- Marshfield Map, http://research.marshfieldclinic.org/genetics/Map_Markers/maps/IndexMapFrames.html
- Molecular Modeling DataBase, http://ncbi.nlm.nih.gov/Structure/ (for 1VHH [accession number 3860])
- Online Mendelian Inheritance in Man (OMIM), http://www.ncbi.nlm.nih.gov/Omim/ (for BDA1, BDB, BDC, Robinow syndrome, Grebe dysplasia, and HPE3)
References
- Afzal AR, Rajab A, Fenske CD, Oldridge M, Elanko N, Ternes-Pereira E, Tuysuz B, Murday VA, Patton MA, Wilkie AO, Jeffery S (2000) Recessive Robinow syndrome, allelic to dominant brachydactyly type B, is caused by mutation of ROR2. Nat Genet 25:419–422 [DOI] [PubMed] [Google Scholar]
- Costa T, Ramsby G, Cassia F, Peters KR, Soares J, Correa J, Quelce-Salgado A, Tsipouras P (1998) Grebe syndrome: clinical and radiographic findings in affected individuals and heterozygous carriers. Am J Med Genet 75:523–529 [DOI] [PubMed] [Google Scholar]
- Erlebacher A, Filvaroff EH, Gitelman SE, Derynck R (1995) Toward a molecular understanding of skeletal development. Cell 80:371–378 [DOI] [PubMed] [Google Scholar]
- Everman, DB, Bartels CF, Yang Y, Yanamandra N, Goodman FR, Mendoza-Londono JR, Savarirayan R, White SM, Graham JM Jr, Gale RP, Svarch E, Newman WG, Kleckers AR, Francomano CA, Govindaiah V, Singh L, Morrison S, Thomas JT, Warman ML (2002) The mutational spectrum of brachydactyly type C. Am J Med Genet 112:291–296 [DOI] [PubMed] [Google Scholar]
- Faiyaz-Ul-Haque M, Ahmad W, Wahab A, Haque S, Azim AC, Zaidi SH, Teebi AS, Ahmad M, Cohn DH, Siddique T, Tsui LC (2002) Frameshift mutation in the cartilage-derived morphogenetic protein 1 (CDMP1) gene and severe acromesomelic chondrodysplasia resembling Grebe-type chondrodysplasia. Am J Med Genet 111:31–37 [DOI] [PubMed] [Google Scholar]
- Fuse N, Maiti T, Wang B, Porter JA, Hall TM, Leahy DJ, Beachy PA (1999) Sonic hedgehog protein signals not as a hydrolytic enzyme but as an apparent ligand for patched. Proc Natl Acad Sci USA 96:10992–10999 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao B, Guo J, She C, Shu A, Yang M, Tan Z, Yang X, Guo S, Feng G, He L (2001) Mutations in IHH, encoding Indian hedgehog, cause brachydactyly type A-1. Nat Genet 28:386–388 [DOI] [PubMed] [Google Scholar]
- Giedion A (1998) Phalangeal cone-shaped epiphyses of the hand: their natural history, diagnostic sensitivity, and specificity in cartilage hair hypoplasia and the trichorhinophalangeal syndromes I and II. Pediatr Radiol 28:751–758 [DOI] [PubMed] [Google Scholar]
- Grogan DP, Love SM, Ogden JA, Millar EA, Johnson LO (1989) Chondro-osseous growth abnormalities after meningococcemia: a clinical and histopathological study. J Bone Joint Surg Am 71:920–928 [PubMed] [Google Scholar]
- Hall TM, Porter JA, Beachy PA, Leahy DJ (1995) A potential catalytic site revealed by the 1.7-A crystal structure of the amino-terminal signalling domain of Sonic hedgehog. Nature 378:212–216 [DOI] [PubMed] [Google Scholar]
- Ingham PW, McMahon AP (2001) Hedgehog signaling in animal development: paradigms and principles. Genes Dev 15:3059–3087 [DOI] [PubMed] [Google Scholar]
- Jeong J, McMahon AP (2002) Cholesterol modification of hedgehog family proteins. J Clin Invest 110:591–596 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karp SJ, Schipani E, St-Jacques B, Hunzelman J, Kronenberg H, McMahon APBR (2000) Indian hedgehog coordinates endochondral bone growth and morphogenesis via parathyroid hormone related-protein-dependent and -independent pathways. Development 127:543–548 [DOI] [PubMed] [Google Scholar]
- Kindblom JM, Nilsson O, Hurme T, Ohlsson C, Savendahl L (2002) Expression and localization of Indian hedgehog (Ihh) and parathyroid hormone related protein (PTHrP) in the human growth plate during pubertal development. J Endocrinol 174:R1–R6 [DOI] [PubMed] [Google Scholar]
- Lander ES, Botstein D (1987) Homozygosity mapping: a way to map human recessive traits with the DNA of inbred children. Science 236:1567–1570 [DOI] [PubMed] [Google Scholar]
- Lathrop GM, Lalouel JM (1984) Easy calculations of LOD scores and genetic risks on small computers. Am J Hum Genet 36:460–465 [PMC free article] [PubMed] [Google Scholar]
- Lee JJ, Ekker SC, von Kessler DP, Porter JA, Sun BI, Beachy PA (1994) Autoproteolysis in hedgehog protein biogenesis. Science 266:1528–1537 [DOI] [PubMed] [Google Scholar]
- Long F, Zhang XM, Karp S, Yang Y, McMahon AP (2001) Genetic manipulation of hedgehog signaling in the endochondral skeleton reveals a direct role in the regulation of chondrocyte proliferation. Development 128:5099–5108 [DOI] [PubMed] [Google Scholar]
- McCready ME, Sweeney E, Fryer AE, Donnai D, Baig A, Racacho L, Warman ML, Hunter AG, Bulman DE (2002) A novel mutation in the IHH gene causes brachydactyly type A1: a 95-year-old mystery resolved. Hum Genet 111:368–375 [DOI] [PubMed] [Google Scholar]
- Mortier GR, Kramer PPG, Giedion A, Beemer FA (2003) Acrocapitofemoral dysplasia: an autosomal recessive skeletal dysplasia with cone-shaped epiphyses in hands and hips. J Med Genet 40:201–207 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patriquin HB, Trias A, Jecquier S, Marton D (1981) Late sequelae of infantile meningococcemia in growing bones of children. Radiology 141:77–82 [DOI] [PubMed] [Google Scholar]
- Porter JA, von Kessler DP, Ekker SC, Young KE, Lee JJ, Moses K, Beachy PA (1995) The product of hedgehog autoproteolytic cleavage active in local and long-range signalling. Nature 374:363–366 [DOI] [PubMed] [Google Scholar]
- Sasaki T, Ishibashi Y, Okamura Y, Toh S, Sasaki T (2002) MRI evaluation of growth plate closure rate and pattern in the normal knee joint. J Knee Surg 15:72–76 [PubMed] [Google Scholar]
- Schwabe GC, Tinschert S, Buschow C, Meinecke P, Wolff G, Gillessen-Kaesbach G, Oldridge M, Wilkie AOM, Kömec R, Mundlos S (2000) Distinct mutations in the receptor tyrosine kinase gene ROR2 cause brachydactyly type B. Am J Hum Genet 67:822–831 [DOI] [PMC free article] [PubMed] [Google Scholar]
- St-Jacques B, Hammerschmidt M, McMahon AP (1999) Indian hedgehog signaling regulates proliferation and differentiation of chondrocytes and is essential for bone formation. Genes Dev 13:2072–2086 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Bokhoven H, Celli J, Kayserili H, van Beusekom E, Balci S, Brussel W, Skovby F, Kerr B, Percin EF, Akarsu N, Brunner HG (2000) Mutation of the gene encoding the ROR2 tyrosine kinase causes autosomal recessive Robinow syndrome. Nat Genet 25:423–426 [DOI] [PubMed] [Google Scholar]
- Vortkamp A, Lee K, Lanske B, Segre GV, Kronenberg HM, Tabin CJBR (1996) Regulation of rate of cartilage differentiation by Indian hedgehog and PTH-related protein. Science 273:613–622 [DOI] [PubMed] [Google Scholar]