Abstract
Atrioventricular septal defects (AVSD) are common cardiovascular malformations, occurring in 3.5/10,000 births. Although frequently associated with trisomy 21, autosomal dominant AVSD has also been described. Recently we identified and characterized the cell adhesion molecule CRELD1 (previously known as “cirrin”) as a candidate gene for the AVSD2 locus mapping to chromosome 3p25. Analysis of the CRELD1 gene from individuals with non–trisomy 21–associated AVSD identified heterozygous missense mutations in nearly 6% of this population, including mutations in isolated AVSD and AVSD associated with heterotaxy syndrome. CRELD1 is the first human gene to be implicated in the pathogenesis of isolated AVSD and AVSD in the context of heterotaxy, which provides an important step in unraveling the pathogenesis of AVSD.
Congenital heart malformations are the most common form of birth defect, occurring in ∼1% of live births. Atrioventricular septal defects (AVSD) are a subset of malformations that constitute ⩾7.4% of all recognized congenital heart defects (Ferencz et al. 1997). The formation of the atrioventricular septa and valves from progenitor cardiac structures called “endocardial cushions” is required to produce the normal four-chambered heart (Eisenberg and Markwald 1995). During atrioventricular valvuloseptal morphogenesis, the endocardial cushions expand as they are infiltrated by extracellular matrix secreted from the surrounding myocardium. The cushions then fuse and are remodeled to form the atrioventricular valves and septa (Markwald et al. 1981; Eisenberg and Markwald 1995). Failure of this process results in AVSD, with the degree of severity dependent on the stage at which the developmental failure occurs (Wessels and Markwald 2000).
AVSD are a spectrum of cardiac malformations that result in a persistent common atrioventricular canal (Jacobs et al. 2000). The complete form of AVSD involves underdevelopment of the lower part of the atrial septum and the upper part of the ventricular septum. There is also a single common atrioventricular valve that results in unrestricted circulatory communication between the atria and ventricles. A less severe form, known as “partial AVSD” or “ostium primum atrial septal defect,” has a deficiency of the atrial septum. Defects of the inlet muscular ventricular septum and isolated cleft mitral valve are also considered to be part of the AVSD clinical spectrum. Complete AVSD are clinically apparent at birth, whereas less severe forms, such as an isolated cleft mitral valve or small defects of the atrial or ventricular septa, may go undetected.
AVSD are most frequently associated with trisomy 21, but there is considerable evidence for genetic heterogeneity. There are several other syndromes that can have AVSD as a clinical finding, including Ivemark syndrome (MIM 208530), 3p− syndrome (MIM 606217), autosomal heterotaxy syndrome (MIM 605376), Ellis-van Creveld syndrome (MIM 225500), CHARGE syndrome (MIM 214800), and Kaufman-McKusick syndrome (MIM 236700). In addition to syndromic AVSD, several large families with isolated AVSD inherited in an autosomal dominant pattern with incomplete penetrance and variable expression have been described (O’Nuallain et al. 1977; Emanuel et al. 1983; Wilson et al. 1993; Cousineau et al. 1994; Kumar et al. 1994; Amati et al. 1995). Study of one large family resulted in the identification of the AVSD1 locus on chromosome 1p31-p21 (MIM 606215) by use of a combination of DNA pooling and shared segment analysis in a high-density genome screen (Sheffield et al. 1997). Although the responsible gene has not yet been identified, that study demonstrated the existence of a congenital heart defect susceptibility gene inherited as an autosomal dominant trait with incomplete penetrance.
A second AVSD locus, AVSD2 (MIM 606217), was defined through analysis of chromosomal breakpoints in 3p− syndrome, which results from a deletion of 3p25-pter (Phipps et al. 1994; Drumheller et al. 1996; Green et al. 2000). The features of this syndrome include developmental delay, microcephaly, ptosis, telecanthus, and micrognathia. Variable features that are presumably due to differences in proximal breakpoints include postaxial polydactyly, renal and gastrointestinal abnormalities, cleft palate, and congenital heart defects. Congenital heart defects occur in approximately one-third of individuals with 3p− syndrome and are typically AVSD (Green et al. 2000). Complete AVSD has been described in at least one individual (Ramer et al. 1989). Ventricular septal defects and atrial septal defects have also been associated with 3p− syndrome (Phipps et al. 1994).
From these data, it is clear that there is a substantial amount of genetic heterogeneity, incomplete penetrance, and variable expression associated with AVSD, increasing the difficulty of finding the susceptibility genes. In fact, the majority of AVSD not related to trisomy 21 occur as sporadic cases of isolated AVSD (Digilio et al. 1999). The relatively high incidence of sporadic versus familial cases indicates that isolated AVSD is usually genetically complex, with multiple factors contributing to susceptibility and variability of expression.
We recently identified a novel cell adhesion molecule, CRELD1 (GenBank accession number AF452623), as a candidate for the AVSD2 locus, on the basis of its mapping to chromosome 3p25 and its expression in the developing heart (Rupp et al. 2002). To test the hypothesis that mutation of CRELD1 is associated with non–trisomy 21–related AVSD, we analyzed a group of subjects with complete or partial AVSD for CRELD1 mutations. Subjects for this study were recruited through the Oregon Registry of Congenital Heart Defects or by an approved protocol for the investigation of congenital heart disease through The Children’s Hospital of Philadelphia. All studies were done with informed consent and conformed to institutional guidelines. We screened 50 unrelated subjects with AVSD and no known cytogenetic abnormalities. The specific phenotypes are listed in table 1. With the exception of the group with heterotaxy, the subjects did not have extracardiac abnormalities. Samples for DNA sequencing were prepared by PCR amplification of fragments encompassing each of the 10 major coding exons of CRELD1, including ⩾50 bp of each flanking intron.
Table 1.
Clinical and Molecular Findings in Patients with AVSD
| Phenotype | No. of Subjects with Nucleotide Change | Nucleotide Change and Position | Amino Acid Change |
| Complete AVSD (n=13) | 0 | ||
| Partial AVSD (ostium primum ASD) (n=22) | 2 | C4148T, exon 9 | T311I |
| C4201T, exon 9 | R329C | ||
| AVSD and hypoplastic arch (n=1) | 0 | ||
| AVSD and COAa (n=2) | 0 | ||
| Partial AVSD and DORVb (n=1) | 0 | ||
| Partial AVSD and heterotaxy (n=11) | 1 | G1566A, exon 3 | R107H |
COA = coarctation of the aorta.
DORV = double-outlet right ventricle.
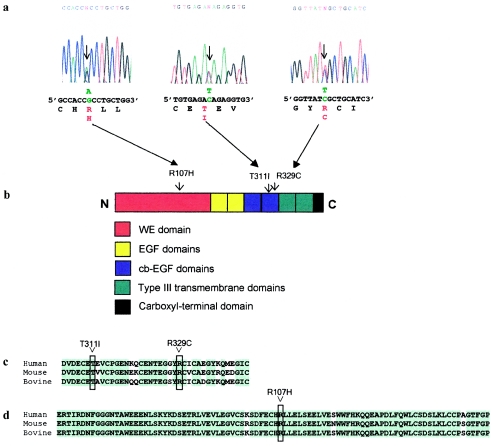
A single-base change, C4201T, was identified in one individual (subject 1) with an isolated partial AVSD (fig. 1a). The mutation results in a substitution of cysteine for arginine at amino acid 329 (R329C) in the second cb-EGF domain (fig. 1b). The addition of a free cysteine residue to a cb-EGF domain is predicted to disrupt the disulfide-bonding pattern for that domain. Another heterozygous C→T transition in exon 9, C4148T, was detected in an unrelated individual (subject 2), also with an isolated partial AVSD (fig. 1a). The C4148T change results in a substitution of isoleucine for threonine at amino acid 311 (T311I) and in the second cb-EGF domain (fig. 1b). These alterations were not detected in 400 normal chromosomes from a North American white control population, which is representative of the subjects’ racial background.
Figure 1.
a, Sequence analyses identifying missense mutations in CRELD1. The arrows on each electropherogram indicate the variant nucleotides, with the wild-type sequence shown below. The altered nucleotides are shown in green. The single-letter–amino acid translation is under the first base of each codon. The altered amino acid residues are in red. Sequences from the complementary strands showed the same heterozygous changes. b, Diagrammatic representation of CRELD1 protein. The approximate positions of the amino acid changes are indicated above the diagram of CRELD1, with arrowheads pointing to the position of the substituted amino acid. c, Alignment of the sequence for the cb-EGF domain from the human, mouse, and bovine CRELD1 genes. d, Alignment of partial sequence of the WE domain from the human, mouse, and bovine CRELD1 genes. Amino acids that are identical among the three species are highlighted in green. The amino acid residues changed by the missense mutations in humans are boxed.
A third heterozygous missense mutation was detected in an individual with a partial AVSD and evidence of heterotaxy syndrome. In addition to the partial AVSD, this individual had dextrocardia, right ventricle aorta with pulmonary atresia, and a right aortic arch. No extracardiac abnormalities were detected. The single-base G→A substitution, G1566A, is in exon 3 and results in a substitution of histidine for arginine (R107H) in the highly conserved WE domain (fig. 1a and 1b). This subject is of mixed Hispanic and African American descent. The base substitution was not detected in 200 chromosomes from an African American control population or in 100 chromosomes from a Hispanic control population.
The R329C, T311I, and R107H mutations affect highly conserved residues and result in nonconservative changes in amino acid sequence (fig. 1c and 1d). In each case, the gene was sequenced in its entirety, and no additional mutations were found, indicating that these mutations act in a dominant fashion. In addition, the range of phenotypes associated with mutations in the present study demonstrates considerable variability of expression for CRELD1 mutations.
Family members of subject 1 were available for study. Analysis of genomic DNA from the first-degree relatives showed inheritance of the R329C mutation (allele T4201) from the father. A brother and sister of the subject also carry the T4201 allele, although none of these family members reported having a heart defect. Two-dimensional echocardiography and color flow Doppler studies were performed on the parents and carrier sister of the proband. In each case, the studies showed that these individuals had structurally normal hearts with normal ventricular function and no evidence of congenital heart disease (data not shown). As has previously been observed in AVSD, these findings are consistent with incomplete penetrance (Pierpont et al. 2000). Given the low penetrance observed in this family, we propose that CRELD1 is an AVSD-susceptibility gene and that CRELD1 mutations increase the risk of developing a heart defect, rather than being directly causative.
The two mutations associated with isolated partial AVSD occurred in a cb-EGF domain (fig. 2). This type of domain is prevalent in many extracellular proteins, and heterozygous mutations affecting cb-EGF domains have been associated with a number of genetic disorders. Of particular interest, arginine-to-cysteine and threonine-to-isoleucine substitutions in cb-EGF domains have been noted in hemophilia B, Marfan syndrome, familial hypercholesterolemia, and heterotaxy due to CFC1 mutations (Krawczak and Cooper 1997; Collod-Beroud et al. 1998; Bamford et al. 2000).
Figure 2.
Diagrammatic representation of the cb-EGF domain harboring the T311I and R329C mutations. Conserved amino acid residues defining cb-EGF domains are indicated by the single-letter–amino acid code. Black lines show the disulfide-bonding pattern for the conserved cysteine residues. The positions of mutated amino acid residues are indicated by black circles; the β-hydroxylated asparagine residue is indicated by an asterisk.
It was anticipated that the introduction of a free cysteine by the R329C mutation would disrupt protein structure. To determine if the R329C substitution altered characteristics of CRELD1, we expressed the wild-type or mutant allele with a carboxyl-terminal FLAG-epitope tag in stably transfected HEK 293 cells and visualized the protein by western blot analysis. Mobility of the C329 mutant protein was slightly retarded, compared with that of wild-type CRELD1 on SDS-PAGE in the presence of reducing agents (fig. 3a). The amino acid substitution alone does not explain an apparent difference in molecular weight but could result in altered posttranslational modification or a change in protein conformation. Digestion with N-glycosidase F prior to SDS-PAGE increased the mobility of both the wild-type and C329 protein but did not alter the difference in mobility between the two molecules (fig. 3a). The data demonstrate that CRELD1 is N-glycosylated, as was predicted by the presence of two N-glycosylation consensus sites (Rupp et al. 2002), but also that the altered mobility is not due to exposure of additional N-glycosylation sites. Digestion with a mixture of glycohydrolases did not resolve the difference in mobility between the normal and mutant molecule, indicating that the discrepancy is not due to alteration of any common form of glycosylation (fig. 3b).
Figure 3.
a, Western blot showing the mobility shift of the C329 mutant protein, compared with wild-type (wt) CRELD1 without PNGase F digestion. Duplicate samples are loaded in alternating lanes. Digestion of the proteins with PNGase F results in a slight increase in mobility, compared with undigested protein, but does not resolve the difference in mobility between wt and C329 CRELD1. b, Western blot showing digestion of wt and 329C CRELD1 with PNGase F alone (+) or a mixture of glycohydrolases (PNGase F, sialidase A, endo-O-glycosidase, β(1,4)galactosidase, and glucosaminidase) that cleave all glycosidase linkages (++), compared with undigested protein (−). Note that digestion with additional glycohydrolases does not further shift the protein band, compared with digestion with PNGase F, indicating that CRELD1 is N-glycosylated but does not have other glycosylation sites to account for the decreased mobility of C329, compared with wt protein.
cb-EGF domains are posttranslationally modified to contain a β-hydroxylated asparagine or aspartate residue in the calcium-binding region. Treatment of stably transfected cells with 10 μM 2,2′-dipyridyl, an inhibitor of β-hydroxylase, for 24 h did not alter the mobility of the C329 protein, indicating that a change in β-hydroxylation was not the cause of the shift in mobility (data not shown). Further analysis will be required to determine the cause of the altered mobility of the C329 mutant protein.
We conclude that the missense changes described here represent disease-related mutations of CRELD1 in these subjects. None of the mutations were found in a minimum of 100 control chromosomes, indicating a specific association with AVSD. The mutations we have identified to date are all associated with partial AVSD, occurring in nearly 9% of that population. It is possible that CRELD1 mutations are not associated with complete AVSD, although analysis of a larger complete AVSD population is required. CRELD1 is the first human gene to be directly associated with AVSD. The cell adhesion molecule DSCAM has been identified as a candidate gene for trisomy 21–related AVSD (Barlow et al. 2001), but, to date, no DSCAM mutations have been reported. The identification of mutations in CRELD1 associated with AVSD highlight the importance of cell adhesion in valvuloseptal morphogenesis. Furthermore, this study suggests that isolated AVSD may share a common genetic etiology with AVSD associated with heterotaxy syndrome, although study of additional heterotaxy subjects is needed for confirmation. This is analogous to CFC1 mutations, where loss of function has been associated with left-right laterality defects with heart malformations (Bamford et al. 2000), as well as isolated transposition of the great arteries or double-outlet right ventricle (Goldmuntz et al. 2002). Our study shows that CRELD1 plays an important role in normal and abnormal valvuloseptal morphogenesis and that mutations that affect CRELD1 structure predispose to AVSD. Identification of a gene involved in the pathogenesis of AVSD provides new insight into a biochemical pathway that is important in the correct formation of the atrioventricular valves and septa.
Acknowledgments
We thank Dr. Robert Glanville, for technical advice, and Kathryn Lesowski and Darcie Babcock, for technical support. This research was supported in part by Research Grant 1-FY02-765 from the March of Dimes Birth Defects Foundation (to C.L.M.), a grant from the Medical Research Foundation/Oregon Health Sciences Foundation (to C.L.M.), and Public Health Service grant 5 M01 RR00334.
Electronic-Database Information
Accession number and URLs for data presented herein are as follows:
- GenBank, http://www.ncbi.nlm.nih.gov/GenBank/ (for the previously published cDNA sequence for human CRELD1 [accession number AF452623])
- Online Mendelian Inheritance in Man (OMIM), http://www.ncbi.nlm.nih.gov/Omim/ (for Ivemark syndrome, autosomal heterotaxy syndrome, 3p− syndrome, Ellis-van Creveld syndrome, CHARGE syndrome, Kaufman-McKusick syndrome, AVSD1, AVSD2, and CRELD1)
References
- Amati F, Mari A, Mingarelli R, Gennarelli M, Digilio MC, Giannotti A, Marino B, Novelli G, Dallapiccola B (1995) Two pedigrees of autosomal dominant atrioventricular canal defect (AVCD): exclusion from the critical region on 8p. Am J Med Genet 57:483–488 [DOI] [PubMed] [Google Scholar]
- Bamford RN, Roessler E, Burdine RD, Saplakoglu U, dela Cruz J, Splitt M, Goodship JA, Towbin J, Bowers P, Ferrero GB, Marino B, Schier AF, Shen MM, Muenke M, Casey B (2000) Loss-of-function mutations in the EGF-CFC gene CFC1 are associated with human left-right laterality defects. Nat Genet 26:365–369 [DOI] [PubMed] [Google Scholar]
- Barlow GM, Chen XN, Shi ZY, Lyons GE, Kurnit DM, Celle L, Spinner NB, Zackai E, Pettenati MJ, Van Riper AJ, Vekemans MJ, Mjaatvedt CH, Korenberg JR (2001) Down syndrome congenital heart disease: a narrowed region and a candidate gene. Genet Med 3:91–101 [DOI] [PubMed] [Google Scholar]
- Collod-Beroud G, Beroud C, Ades L, Black C, Boxer M, Brocks DJH, Holman KJ, de Paepe A, Francke U, Grau U, Hayward C, Klein HG, Liu WG, Nuytinck L, Peltonen L, Perez ABA, Rantamaki T, Junien C, Boileau C (1998) Marfan database (3rd ed): new mutations and new routines for the software. Nucleic Acids Research 26:229–233 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cousineau AJ, Lauer RM, Pierpont ME, Burns TL, Ardinger RH, Patil SR, Sheffield VC (1994) Linkage analysis of autosomal dominant atrioventricular canal defects: exclusion of chromosome 21. Hum Genet 93:103–108 [DOI] [PubMed] [Google Scholar]
- Digilio MC, Marino B, Toscano A, Giannotti A, Dallapiccola B (1999) Atrioventricular canal defect without Down syndrome: a heterogeneous malformation. Am J Med Genet 85:140–146 [PubMed] [Google Scholar]
- Drumheller T, McGillivray C, Behrner D, MacLeod P, McFadden DE, Roberson J, Venditti C, Chorney D, Chorney M, Smith DI (1996) Precise localisation of 3p25 breakpoints in four patients with the 3p− syndrome. J Med Genet 33:842–847 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eisenberg LM, Markwald RR (1995) Molecular regulation of atrioventricular valvuloseptal morphogenesis. Circ Res 77:1–6 [DOI] [PubMed] [Google Scholar]
- Emanuel R, Somerville J, Inns A, Withers R (1983) Evidence of congenital heart disease in the offspring of parents with atrioventricular defects. Br Heart J 49:144–147 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferencz C, Loffredo CA, Rubin JD, Magee CA (eds) (1997) Perspectives in pediatric cardiology. Vol 4, Futura Publishing Company, Mount Kisco, NY [Google Scholar]
- Goldmuntz E, Bamford R, Karkera JD, dela Cruz J, Roessler E, Muenke M (2002) CFC1 mutations in patients with transposition of the great arteries and double-outlet right ventricle. Am J Hum Genet 70:776–780 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Green EK, Priestley MD, Waters J, Maliszewska C, Latif F, Maher ER (2000) Detailed mapping of a congenital heart disease gene in chromosome 3p25. J Med Genet 37:581–587 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacobs JP, Burke RP, Quintessenza JA, Mavroudis C (2000) Congenital heart surgery nomenclature and database project: atrioventricular canal defect. Ann Thorac Surg 69:S36–S43 [DOI] [PubMed] [Google Scholar]
- Krawczak M, Cooper DN (1997) The mutation database. Trends in Genetics 13:121–122 [DOI] [PubMed] [Google Scholar]
- Kumar A, Williams CA, Victorica BE (1994) Familial atrioventricular septal defect: possible genetic mechanisms. Br Heart J 71:79–81 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Markwald RR, Krook JM, Kitten GT, Runyan RB (1981) Endocardial cushion tissue development: structural analyses on the attachment of extracellular matrix to migrating mesenchymal cell surfaces. Scanning Electron Microscopy 261–274 [PubMed] [Google Scholar]
- O’Nuallain S, Hall JG, Stamm SJ (1977) Autosomal dominant inheritance of endocardial cushion defect. Birth Defects Orig Artic Ser 13:143–147 [PubMed] [Google Scholar]
- Phipps ME, Latif F, Prowse A, Payne SJ, Dietz-Band J, Leversha M, Affara NA, Moore AT, Tolmie J, Schinzel A, Lerman MI, Ferguson-Smith MA, Maher ER (1994) Molecular genetic analysis of the 3p− syndrome. Hum Molec Genet 3:903–908 [DOI] [PubMed] [Google Scholar]
- Pierpont MEM, Markwald RR, Lin AE (2000) Genetic aspects of atrioventricular septal defects. Am J Med Genet 97:289–296 [PubMed] [Google Scholar]
- Ramer JC, Ladda RL, Frankel C (1989) Two infants with del(3)(p25pter) and a review of previously reported cases. Am J Med Genet 33:108–112 [DOI] [PubMed] [Google Scholar]
- Rupp PA, Fouad GT, Egelston CA, Reifsteck CA, Olson SB, Knosp WM, Glanville RW, Thornburg KL, Robinson SW, Maslen CL (2002) Identification, genomic organization and mRNA expression of CRELD1, the founding member of a unique family of matricellular proteins. Gene 293:47–57 [DOI] [PubMed] [Google Scholar]
- Sheffield VC, Pierpont ME, Nishimura D, Beek JS, Burns TL, Berg MA, Stone EM, Patil SR, Lauer RM (1997) Identification of a complex congenital heart defect susceptibility locus by using DNA pooling and shared segment analysis. Hum Molec Genet 6:117–121 [DOI] [PubMed] [Google Scholar]
- Wessels A, Markwald R (2000) Cardiac morphogenesis and dysmorphogenesis. I. Normal development. Methods Mol Biol 136:239–259 [DOI] [PubMed] [Google Scholar]
- Wilson L, Curtis A, Korenberg JR, Schipper RD, Allan L, Chenevix-Trench G, Stephenson A, Goodship J, Burn J (1993) A large, dominant pedigree of atrioventricular septal defect (AVSD): exclusion from the Down syndrome critical region on chromosome 21. Am J Hum Genet 53:1262–1268 [PMC free article] [PubMed] [Google Scholar]