Abstract
CBS1 from Magnaporthe grisea is a structural and functional homolog of the cystathionine β-synthase (CBS) gene from Saccharomyces cerevisiae. Our studies indicated that M. grisea can utilize homocysteine and methionine through a CBS-independent pathway. The results also revealed responses of M. grisea to homocysteine that are reminiscent of human homocystinuria.
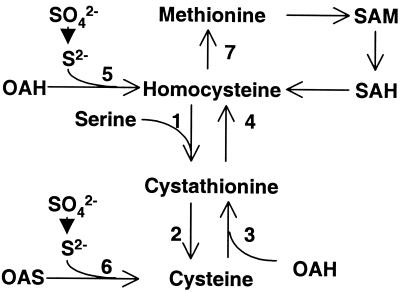
In the filamentous fungi Aspergillus nidulans and Neurospora crassa, inorganic sulfur is assimilated directly into either homocysteine (Fig. 1, enzyme 5) or cysteine (Fig. 1, enzyme 6) (15). The transsulfuration pathways allow the interconversion of homocysteine and cysteine, with the intermediary formation of cystathionine (Fig. 1) (15). Cystathionine β-synthase (CBS) catalyzes the formation of cystathionine from homocysteine and serine (Fig. 1, enzyme 1). Cysteine is synthesized from cystathionine in a reaction catalyzed by cystathionine γ-lyase (Fig. 1, enzyme 2). There is only one existing transsulfuration pathway in mammals, i.e., from homocysteine to cysteine (6). In A. nidulans, N. crassa, and the yeast Saccharomyces cerevisiae, an opposite transsulfuration pathway is present, allowing the conversion of cysteine to homocysteine (Fig. 1, enzymes 3 and 4) (5, 15). In addition, there is no evidence that enzymes 3 and 4 can catalyze the reverse reaction from homocysteine to cysteine. CBS has been conserved in eukaryotic evolution (12) and is directly involved in the removal of homocysteine from the methionine cycle. In humans, CBS deficiency results in an elevated level of circulating homocysteine (homocystinuria), which is a risk factor for a number of neurological defects and vascular diseases (17). This disorder is commonly caused by recessive mutations in the human CBS gene (17).
FIG. 1.
Transsulfuration pathways in the filamentous fungi A. nidulans and N. crassa. OAH, O-acetyl-homoserine; OAS, O-acetyl-serine; SAM, S-adenosyl-methionine; SAH, S-adenosyl-homocysteine. Enzymes: 1, CBS; 2, cystathionine γ-lyase; 3, cystathionine γ-synthase; 4, cystathionine β-lyase; 5, homocysteine synthase; 6, CYS; 7, methionine synthase.
A genome-wide effort (7) has been initiated to study gene functions in Magnaporthe grisea, a filamentous fungus that causes diseases in rice and other cereal crops (18). As part of this effort, a cosmid clone from an M. grisea (strain Guy11) (13) genomic library (7) was shotgun sequenced as described previously (8). BLASTX searches (1) of the sequence against the National Center for Biotechnology Information nonredundant protein database (27 June 2001) identified a putative gene, CBS1, encoding a CBS-like protein. The coding sequence (GenBank accession number AF422799) is interrupted by an intron of 71 bp, a finding which was confirmed by comparison to a cDNA sequence.
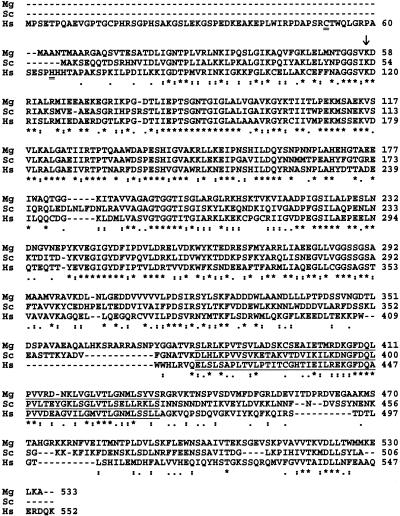
The deduced gene product of CBS1 shares extensive homology with CBS proteins from S. cerevisiae (46% identity) and humans (45% identity) (Fig. 2). CBS is a pyridoxal phosphate (PLP)-dependent enzyme (10). In human CBS, Lys119 is the PLP binding residue (11), and this residue is conserved in M. grisea and S. cerevisiae (Fig. 2). Similarly, the human CBS domain, comprising residues 417 to 470 (2), can be identified in the S. cerevisiae and M. grisea proteins (Fig. 2) by hidden Markov model searches against the Pfam database (P score = 1.8 × 10−14; 18 February 2002). CBS domains are also present in a wide range of unrelated proteins (2). The region containing the human CBS domain is involved in regulation by S-adenosyl-l-methionine (9). The Cys52 and His65 residues that axially coordinate the iron in the heme group of human CBS (16) are not conserved in M. grisea and S. cerevisiae (Fig. 2). In fact, S. cerevisiae CBS was recently found to be a nonheme protein (10, 14). It is therefore likely that the M. grisea enzyme does not contain a heme group, either. In S. cerevisiae, the biosynthesis of cysteine occurs exclusively through the CBS pathway, and CBS null mutants are cysteine auxotrophs (5). We demonstrated that the introduction of an expression plasmid containing M. grisea CBS1 rescued the growth defect of a CBS-deficient S. cerevisiae strain in the absence of cysteine (data not shown). These findings indicate that M. grisea CBS1 is a structural and functional homolog of the S. cerevisiae CBS gene.
FIG. 2.
Amino acid alignment of M. grisea CBS1 (Mg) with S. cerevisiae CBS (Sc) (GenBank accession number AAC37401) and human CBS (Hs) (GenBank accession number A55760). Sequences were aligned by using the Clustal W program and Lasergene software (DNASTAR, Madison, Wis.). Gaps in the alignment are indicated by dashes. Asterisks indicate identical residues in all sequences. Colons indicate conservative substitutions. Dots indicate semiconserved substitutions. Lys119 in human CBS is involved in PLP binding and is conserved in M. grisea and S. cerevisiae (arrow). The Cys52 and His65 residues that axially coordinate the iron in the heme group in human CBS are doubly underlined. The CBS domains in the C termini of the sequences are underlined.
CBS1 is a single-copy gene in M. grisea, as revealed by genomic Southern analysis (data not shown). We performed in silico hybridization (TBLASTN searches) (1) of the CBS1 amino acid sequence against our internal M. grisea unigene database and the complete N. crassa genome database (version 2; Whitehead Institute, MIT Center for Genome Research; www-genome.wi.nit.edu/annotation/fungi/neurospora). There was no evidence of a second gene encoding CBS in either filamentous fungus. The closest matches from both databases were sequences encoding cysteine synthase (CYS)-like proteins. CBS and CYS are related proteins, and both require PLP as a cofactor. The amino acid sequence identity between M. grisea CBS1 and A. nidulans CYS (GenBank accession number P50867, the only annotated filamentous fungal CYS in National Center for Biotechnology Information nr as of 18 October 2001) is 38%. However, CYS proteins are considerably shorter (∼300 amino acids) than CBS proteins (>500 amino acids) (5). There are no indications that CYS proteins from different species exhibit CBS activities. Based on these findings, we conclude that CBS1 is the only gene encoding CBS in M. grisea.
CBS1 was deleted from M. grisea by replacement with a modified hygromycin phosophotransferase gene (4) as described previously (21). The null (cbs1) mutants were found to retain virulence on rice (data not shown). In addition, the cbs1 mutants were not auxotrophic and were able to utilize inorganic sulfate, cysteine, cystathionine, homocysteine, or methionine as a sole sulfur source (data not shown). In the absence of inorganic sulfur sources, the pathway through CBS (Fig. 1) is the only known route for cysteine biosynthesis in filamentous fungi and other microbes (16) (Kegg metabolic pathways) (http://www.genome.ad.jp/kegg/metabolism.html) (8 January 2002). Thus, homocysteine and methionine can be utilized via a CBS-independent pathway in M. grisea cbs1 mutants. Similarly, Schizosaccharomyces pombe, which lacks CBS naturally, is able to convert methionine to cysteine (3). Alternatively, homocysteine or methionine may be degraded through unknown pathways and the resulting sulfide ion may be assimilated by M. grisea.
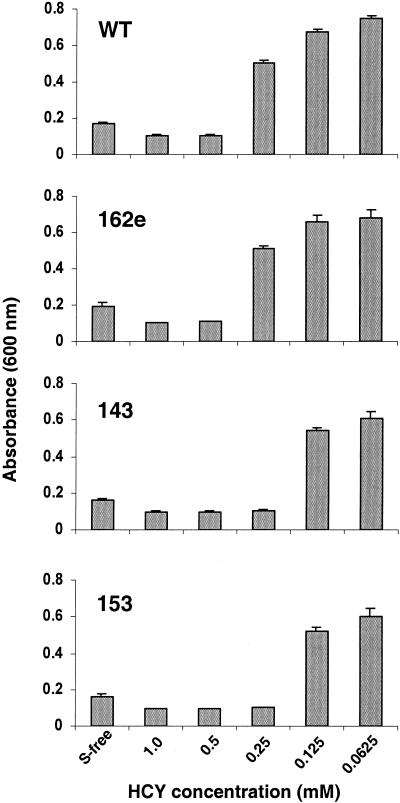
Our growth studies revealed that homocysteine is toxic to M. grisea. Spore suspensions were inoculated into minimal medium (MM) (18) containing different concentrations of homocysteine as described previously (7). Fungal growth was monitored as an increase in absorbance at 600 nm. Complete inhibition of growth was observed when the wild-type (WT) strain was grown in homocysteine at a concentration of 0.5 mM or higher (Fig. 3). The cbs1 mutants were hypersensitive to exogenous homocysteine. For example, the growth of the cbs1 mutants was completely inhibited at a concentration of 0.25 mM (Fig. 3). Inhibitory effects on growth were not evident with cysteine, cystathionine, or methionine at all the tested concentrations (data not shown). Toxicity of homocysteine for fungal growth has not been described elsewhere. However, humans with homocystinuria have been known to develop different clinical phenotypes caused by elevated levels of circulating homocysteine (17). This disorder is most frequently a consequence of CBS deficiencies. It is possible that M. grisea and human CBS proteins share a common physiological function as a detoxification mechanism for homocysteine.
FIG. 3.
Inhibition of growth of M. grisea strains by homocysteine. The fungal strains were grown in MM containing different concentrations of homocysteine (HCY) as a sole sulfur source. Mycelial growth was monitored by measuring the absorbance at 600 nm 7 days after inoculation. Growth inhibition is reflected by an absorbance value lower than that obtained in sulfur-free (S-free) medium. The WT strain and an ectopic transformant (162e) were inhibited at ≥0.5 mM homocysteine. cbs1 mutants 143 and 153 showed increased sensitivity to homocysteine. Data are reported as means (columns) and standard deviations (error bars).
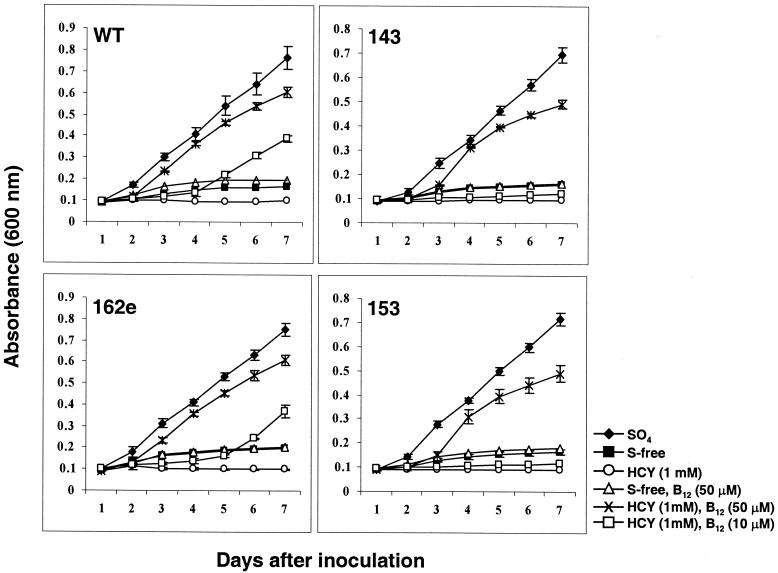
Vitamin or coenzyme treatments of homocystinuria patients serve to enhance pathways that remove excess circulating homocysteine (19). Interestingly, we demonstrated that the addition of vitamin B12 relieved the toxicity of homocysteine for M. grisea. As shown in Fig. 4, supplementation with vitamin B12 at 50 μM allowed both the WT and cbs1 mutant strains to grow in MM containing 1 mM homocysteine. The vitamin B12 response appeared to be concentration dependent. Thus, the growth of WT and ectopic strains in the presence of homocysteine was moderately restored when 10 μM vitamin B12 was supplied (Fig. 4). The growth of the cbs1 mutants, which were more sensitive to homocysteine, remained inhibited at 10 μM vitamin B12 (Fig. 4). In the human body, homocysteine either can be remethylated to methionine by methionine synthase or can undergo transsulfuration reactions via CBS to form cysteine (6). The remethylation of homocysteine to methionine by methionine synthase is dependent on vitamin B12, betaine, and folate, while the CBS-catalyzed reaction requires vitamin B6 as a coenzyme (20). In M. grisea, methionine synthase (Fig. 1, enzyme 7) activities likely were enhanced with vitamin B12 supplementation to remove homocysteine.
FIG. 4.
Effect of vitamin B12 on homocysteine sensitivity in M. grisea strains. Vitamin B12 was added to MM containing homocysteine (HCY) as the sole sulfur source. Complete inhibition of growth was observed for all strains growing in the presence of 1 mM homocysteine. Mycelial growth was restored in all strains when 50 μM vitamin B12 was added to the medium. At 10 μM vitamin B12, the growth of the WT strain and an ectopic transformant (162e) was delayed and partially restored. cbs1 mutants 143 and 153, which are hypersensitive to homocysteine, did not respond to vitamin B12 at this concentration. Note that vitamin B12 added to sulfur-free (S-free) medium did not support growth. Fungal strains growing in MM containing inorganic sulfate (SO4) as the sole sulfur source were used as positive controls. Data are reported as means (as indicated) and standard deviations (error bars).
In conclusion, our studies of CBS1 in M. grisea indicate that homocysteine and methionine can be utilized by the fungus through pathways that are independent of CBS. In addition, our results reveal similarities between M. grisea and humans with regard to sensitivities to homocysteine and responsiveness to vitamin B12 supplementation. The fungus may be exploited as a system to screen for therapeutic agents to relieve homocysteine toxicity.
Acknowledgments
We thank members of the DNA Technology Group at Paradigm Genetics, Inc., for their sequencing efforts. We also thank Jeffrey Shuster, Matthew Tanzer, Todd M. DeZwaan, Weiwen Zhang, and Keith Allen, Paradigm Genetics, Inc., and the anonymous reviewers for critical reviews and helpful suggestions.
REFERENCES
- 1.Altschul, S. F., T. L. Madden, A. A. Schaffer, J. Zhang, Z. Zhang, W. Miller, and D. J. Lipman. 1997. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 25:3389-3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bateman, A. 1997. The structure of a domain common to archaebacteria and the homocystinuria disease protein. Trends Biochem. Sci. 22:12-13. [DOI] [PubMed] [Google Scholar]
- 3.Brzywczy, J., and A. Paszewski. 1994. Sulfur amino acid metabolism in Schizosaccharomyces pombe: occurrence of two O-acetylhomoserine sulfhydrylases and the lack of the reverse transulfuration pathway. FEMS Microbiol. Lett. 121:171-174. [DOI] [PubMed] [Google Scholar]
- 4.Carroll, A. N., J. A. Sweigard, and B. Valent. 1994. Improved vectors for selecting resistance to hygromycin. Fungal Genet. Newsl. 41:22. [Google Scholar]
- 5.Cherest, H., D. Thomas, and Y. Surdin-Kerjan. 1993. Cysteine biosynthesis in Saccharomyces cerevisiae occurs through the transsulfuration pathway which has been built up by enzyme recruitment. J. Bacteriol. 175:5366-5374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Finkelstein, J. D., and J. J. Martin. 2000. Homocysteine. Int. J. Biochem. Cell Biol. 32:385-389. [DOI] [PubMed] [Google Scholar]
- 7.Hamer, L., K. Adachi, V. Montenegro-Chamorro, M. M. Tanzer, S. K. Mahanty, C. Lo, R. W. Tarpey, A. R. Skalchunes, R. W. Heiniger, S. A. Frank, B. A. Darveaux, D. J. Lampe, T. M. Slater, L. Ramamurthy, T. M. DeZwaan, G. H. Nelson, J. R. Shuster, J. Woessner, and J. Hamer. 2001. Gene discovery and gene function assignment in filamentous fungi. Proc. Natl. Acad. Sci. USA 98:5110-5115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hamer, L., H. Pan, K. Adachi, M. J. Orbach, A. Page, L. Ramamurthy, and J. P. Woessner. 2001. Regions of microsynteny in Magnaporthe grisea and Neurospora crassa. Fungal Genet. Biol. 33:137-143. [DOI] [PubMed] [Google Scholar]
- 9.Janos̆ík, M., V. Kery, M. Gaustadnes, K. N. Maclean, and J. P. Kraus. 2001. Regulation of human cystathionine β-synthase by S-adenosyl-l-methionine: evidence for two catalytically active conformations involving an autoinhibitory domain in the C-terminal region. Biochemistry 49:10625-10633. [DOI] [PubMed] [Google Scholar]
- 10.Jhee, K.-H., P. McPhie, and E. W. Miles. 2000. Yeast cystathionine β-synthase is a pyridoxal phosphate enzyme, but unlike the human enzyme, is not a heme protein. J. Biol. Chem. 275:11541-11544. [DOI] [PubMed] [Google Scholar]
- 11.Kery, V., G. Bukovska, and J. P. Kraus. 1994. Transsulfuration depends on heme in addition to pyridoxal 5′-phosphate. Cystathionine β-synthase is a heme protein. J. Biol. Chem. 269:25283-25288. [PubMed] [Google Scholar]
- 12.Kruger, W. D., and D. R. Cox. 1994. A yeast system for expression of human cystathionine β-synthase: structural and functional conservation of the human and yeast genes. Proc. Natl. Acad. Sci. USA 91:6614-6618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Leung, H., E. S. Borromeo, M. A. Bernado, and J. L. Notteghem. 1999. Genetic analysis of virulence in rice blast fungus Magnaporthe grisea. Phytopathology 78:1227-1233. [Google Scholar]
- 14.Maclean, K. N., M. Janos̆ík, J. Oliveriusová, V. Kery, and J. P. Kraus. 2000. Transsulfuration in Saccharomyces cerevisiae is not dependent on heme: purification and characterization of recombinant yeast cystathionine β-synthase. J. Inorg. Biochem. 81:161-171. [DOI] [PubMed] [Google Scholar]
- 15.Marzluf, G. A. 1997. Molecular genetics of sulfur assimilation in filamentous fungi and yeast. Annu. Rev. Microbiol. 51:73-96. [DOI] [PubMed] [Google Scholar]
- 16.Meier, M., M. Janos̆ík, V. Kery, J. P. Kraus, and P. Burhard. 2001. Structure of human cystathionine β-synthase: a unique pyridoxal 5′-phosphate-dependent heme protein. EMBO J. 20:3910-3916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Mudd, S. H., H. L. Levy, and J. P. Kraus. 2001. Disorders of transsulfuration, p. 2007-2056. In C. R. Scriver, A. Beaudet, W. Sly, and D. Valle (ed.) The metabolic basis of inherited diseases. McGraw-Hill Book Co., New York, N.Y.
- 18.Talbot, N. J., D. J. Ebbole, and J. E. Hamer. 1993. Identification and characterization of MPG1, a gene involved in pathogenicity from the rice blast fungus Magnaporthe grisea. Plant Cell 5:1575-1590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Walter, J. H., J. E. Wraith, and F. J. White. 1998. Strategies for the treatment of cystathionine beta-synthase deficiency: the experience of the Willink Biochemical Genetics Unit over the past 30 years. Eur. J. Pediatr. 157(Suppl. 2):S71-S76. [DOI] [PubMed] [Google Scholar]
- 20.Wilcken, D. E. L., and B. Wilcken. 1997. The natural history of vascular disease in homocystinuria and the effects of treatment. J. Inherit. Metab. Dis. 20:295-300. [DOI] [PubMed] [Google Scholar]
- 21.Wu, S. C., K. S. Ham, A. G. Darvill, and P. Albersheim. 1997. Deletion of two endo-β-1,4-xylanase genes reveals additional isozymes secreted by the rice blast fungus. Mol. Plant-Microbe Interact. 10:700-708. [Google Scholar]