Abstract
Mitochondrial (mt) impairment, particularly within complex I of the electron transport system, has been implicated in the pathogenesis of Parkinson disease (PD). More than half of mitochondrially encoded polypeptides form part of the reduced nicotinamide adenine dinucleotide dehydrogenase (NADH) complex I enzyme. To test the hypothesis that mtDNA variation contributes to PD expression, we genotyped 10 single-nucleotide polymorphisms (SNPs) that define the European mtDNA haplogroups in 609 white patients with PD and 340 unaffected white control subjects. Overall, individuals classified as haplogroup J (odds ratio [OR] 0.55; 95% confidence interval [CI] 0.34–0.91; P=.02) or K (OR 0.52; 95% CI 0.30–0.90; P=.02) demonstrated a significant decrease in risk of PD versus individuals carrying the most common haplogroup, H. Furthermore, a specific SNP that defines these two haplogroups, 10398G, is strongly associated with this protective effect (OR 0.53; 95% CI 0.39–0.73; P=.0001). SNP 10398G causes a nonconservative amino acid change from threonine to alanine within the NADH dehydrogenase 3 (ND3) of complex I. After stratification by sex, this decrease in risk appeared stronger in women than in men (OR 0.43; 95% CI 0.27–0.71; P=.0009). In addition, SNP 9055A of ATP6 demonstrated a protective effect for women (OR 0.45; 95% CI 0.22–0.93; P=.03). Our results suggest that ND3 is an important factor in PD susceptibility among white individuals and could help explain the role of complex I in PD expression.
Introduction
Selective degeneration of dopaminergic neurons within the substantia nigra (SN) manifests in the clinical signs of Parkinson disease (PD [MIM 168600]), including resting tremor, rigidity, gait disturbances, and bradykinesia. The etiology of idiopathic PD is currently undefined; however, it is now largely accepted that genetic susceptibility factors exist and may interact with environmental exposures, leading to the development of this late-onset degenerative disease.
Several reports suggest that mitochondrial (mt) dysfunction may be involved in the expression of PD (Greenamyre et al. 1999, 2001). The activity of complex I, which is the first complex of the electron transport chain, has been reported to be reduced within SN tissue and platelets of patients with PD, an observation suggestive of a systemic defect (Parker et al. 1989; Schapira et al. 1989, 1990). Furthermore, biochemical analyses, using cytoplasmic hybrid cells (cybrids), have demonstrated moderate impairment of complex I in patients with sporadic PD, indicating that mtDNA aberrations may cause the deficit in energy production (Swerdlow et al. 1996; Gu et al. 1998). In these cybrid systems, several other pathological events were observed, including excessive generation of reactive oxygen species (ROS), which leads to oxidative stress (Swerdlow et al. 1996; Cassarino et al. 1997), increased expression of antioxidative proteins (bcl-2 and bcl-XL), signaling a reaction to cell stress (Veech et al. 2000) and the creation of abnormal mitochondria morphology with protein inclusion bodies (Trimmer et al. 2000). In addition, animal models have shown that environmental toxins that inhibit complex I activity cause clinical manifestations of PD (Betarbet et al. 2002). A seminal study by Betarbet and colleagues (2000) provided compelling evidence of this causal relationship in rat model experiments that involved chronic exposure to the pesticide rotenone. In the same study, Betarbet and colleagues showed that inhibition of complex I by rotenone caused the selective degeneration of SN dopaminergic neurons, accumulation of ubiquitin, and α-synuclein–positive cytoplasmic inclusions, as well as development of tremor and rigidity within subjects. Furthermore, the pathological findings were replicated in an in vitro model that provided evidence of molecular mechanisms that may lead to gradual neuronal cell death that is reminiscent of PD (Sherer et al. 2002). Accordingly, these studies support the hypothesis that changes in complex I could contribute to a primary pathogenic susceptibility mechanism in PD.
Interestingly, seven of the polypeptides encoded by the mt genome are also known to code for protein subunits of complex I. We hypothesize that genetic polymorphisms within the mt genome could act as susceptibility factors and contribute to the expression of PD, either directly or through interactions with nuclear encoded genes or environmental toxins. To test the hypothesis that mt variation contributes to this expression, we genotyped predefined European mtDNA haplogroups (Torroni et al. 1996) in a large study of white patients with PD and white unaffected control subjects (table 1).
Table 1.
Characterization of European Haplogroups
| SNP |
||||||||||
| Haplogroup | 1719 | 4580 | 7028 | 8251 | 9055 | 10398 | 12308 | 13368 | 13708 | 16391 |
| H | … | … | C | … | … | A | … | … | … | … |
| I | A | … | T | A | … | G | … | … | … | A |
| J | … | … | T | … | … | G | … | … | A | … |
| K | … | … | T | … | A | G | G | … | … | … |
| T | … | … | T | … | … | A | … | A | … | … |
| U | … | … | T | … | … | A | G | … | … | … |
| V | … | A | T | … | … | A | … | … | … | … |
| W | … | … | T | A | … | A | … | … | … | … |
| X | A | … | T | … | … | A | … | … | … | … |
Subjects and Methods
Sample
A total of 609 unrelated patients of European ancestry who had PD were included in the present study. Cases were ascertained through the Morris K. Udall Parkinson’s Disease Center of Excellence, Duke Center for Human Genetics (DCHG), and from the DCHG/GlaxoSmithKline Parkinson’s Disease Genetics Collaboration. The 340 white control subjects were collected from spouses of patients with Alzheimer disease ascertained through the Joseph and Kathleen Bryan Alzheimer’s Disease Research Center. Control subjects had no significant signs of cognitive or neurological impairment when enrolled in the study. Mean ± SD age at onset (AAO) in affected individuals in the sample is 62 years ± 12. AAO is self-reported by the patient with PD and is defined as the age at which the affected individual first noticed one of the cardinal signs of PD. Mean ± SD age at examination (AAE) of patients with PD is 66 years ± 12, and mean ± SD AAE of control subjects is 69 years ± 9. AAE was defined as the age at which the affected or unaffected participant was clinically examined by study personnel. The overall sample consists of 57% males and 43% females. The PD case group is composed of 63% men and 37% women, whereas the control group consists of 44% men and 56% women. Written consent was obtained from all participants, in accordance with protocols approved by the institutional review board at each contributing center. A board-certified neurologist specializing in movement disorders or a physician assistant experienced in neurological disorders examined individuals, and rigorous clinical criteria were used for diagnosis of PD. All patients with PD had at least two principal signs of PD (resting tremor, bradykinesia, or rigidity) and no clinical features of any other parkinsonian syndrome.
Classification of Haplogroups
Ten SNPs within coding genes and the control region were chosen for genotyping, according to the previous work of Torroni et al. (1996). SNPs within RFLP sites were identified so that the allelic discrimination method Taqman could be employed (table 1). By comparing the complete, revised Cambridge genomic sequence (Andrews et al. 1999) with the Japanese (Anderson et al. 1981), Swedish (Arnason et al. 1996), and African (Horai et al. 1995) reference sequence genomes, we were able to identify the nucleotide change within each restriction site (GenBank). All primer and probe sequences are available upon request.
SNP Genotyping
Genomic DNA was isolated from whole blood samples by the DCHG DNA banking core facility, using Puregene (Gentra Systems). High-throughput genotyping was established using the 5′ nuclease allelic discrimination Taqman assay in a 384-well format on the ABI Prism 7900HT Sequence Detection System (Applied Biosystems). In each chamber of the 384-well sample plates, 20 ng of DNA was distributed using a Hydra HTS Workstation microdispensing system (Robbins Scientific). Probes and primers for each SNP were designed using ABI Prism Primer Express software, version 2.0 (Applied Biosystems). All probes designed with a black-hole quencher reporter were generated by Integrated DNA Technologies, and all minor-groove–binding Taqman probes were manufactured by Applied Biosystems.
To each well, 5 μl of master mix (0.2 U/μl Taqman Universal PCR Master Mix; 0.9 ng/μl of each forward and reverse primer; and 0.2 ng/μl of each probe) was dispensed by a MultiProbe2 204DT (Packard Instruments). The amplification reaction was conducted on a 384-well ABI Dual GeneAmp PCR System 9700, utilizing the following program: 50°C for 2 min, 95°C for 10 min, 95°C for 15 s, and 62°C for 1 min, repeated for 40 cycles; samples were held at 4°C after cycling completion. Data were generated on an ABI Prism 7900HT Sequence Detection System (SDS) and were analyzed using the associated SDS, version 2.0, software.
The few samples that fell outside SNP clusters were sequenced for genotyping. Sequencing primers were designed using the Vector NTI Suite 6 software package (InforMax) and the Primer3 Web site. DNA sequencing was conducted on an ABI Prism 3100 Genetic Analyzer (Applied Biosystems). Sequencing analysis was performed using the ABI Prism Sequencing Analysis software, version 3.7, and Sequencher software, version 4.0.5. In addition to the positive control samples, four negative control samples were assayed per plate. For quality control, samples from 24 individuals were duplicated on each 384-well plate. Technicians performing the SNP genotyping were blinded to the duplications. In addition, two DNA samples from the CEPH were sequenced for each SNP, were tested eight times per plate, and were also used as blind internal controls. All quality-control samples were compared in the Duke Center for Human Genetics Data Coordinating Center. Data were stored and managed by the PEDIGENE system (Haynes et al. 1995).
Statistical Analysis
All statistical analyses were performed using SAS software, release 8.1 (SAS Institute). Statistical significance was declared at α = .05. A t test was conducted to evaluate differences in AAE between case and control subjects, with a significant difference found (P=.0001). To assess differences in distribution of sex between cases and controls, we used a χ2 test and found a significant difference (P=.0001). Therefore, to adjust for potential confounding, we used AAE and sex as covariates in the analyses. We performed unconditional logistic regression to generate ORs with their associated 95% CIs, to assess odds of carrying each mt SNP in PD cases compared with controls. In addition, we used unconditional logistic regression to simultaneously assess odds of PD cases carrying specific haplogroups. Since haplogroup carrier status was a categorical independent variable with more than two categories, there are multiple ways to assign the reference group: each haplogroup can be compared with a common haplogroup, or each haplogroup can be compared with all other haplogroups pooled into one group. An advantage of using a common haplogroup as the reference is that it is more homogeneous than a pooling of different haplogroups, and it means that each haplogroup is compared with the same reference group for consistency. We performed the analysis, using both approaches for comparison. First, H was chosen as a reference group, since it is found at the highest frequency (40%–50%) among European populations. We also tested for association of a specific haplogroup (e.g., K) relative to all other haplogroups, by pooling frequencies of non-K. This is conceptually the same as the binary SNP allele comparison. P values reported for SNPs and haplogroups are based on the Wald χ2 statistic for the particular SNP or haplogroup and are not adjusted for multiple testing.
Results
All nine major European haplogroups were observed in our sample and did not differ significantly from a previous study of a similar North American control population (Torroni et al. 1994) (table 2). In addition, a nearly identical percentage of individuals (8.2% in control subjects and 8.5% in patients with PD) did not fit into these nine predefined haplogroups and were classified as “others.” This group most likely consists of rare European haplogroups (e.g., R or Z) or the historical admixture known to exist in the North American white population (Finnila et al. 2000; Richards et al. 2000). Therefore, comparison of overall population haplogroups suggests that the control population was well matched to our PD cases, and it supports an absence of significant substructure (see the “Discussion” section).
Table 2.
Haplogroup Counts and Frequencies Overall
| PD(N=609) |
Control(N=340) |
Total(N=949) |
||||
| Haplogroup | n | % | n | % | n | % |
| H | 273 | 44.8 | 134 | 39.4 | 407 | 42.9 |
| I | 20 | 3.3 | 11 | 3.2 | 31 | 3.3 |
| J | 43 | 7.1 | 38 | 11.2 | 81 | 8.5 |
| K | 34 | 5.6 | 32 | 9.4 | 66 | 6.9 |
| T | 53 | 8.7 | 36 | 10.6 | 89 | 9.4 |
| U | 94 | 15.4 | 41 | 12.1 | 135 | 14.2 |
| V | 24 | 3.9 | 10 | 2.9 | 36 | 3.6 |
| W | 8 | 1.3 | 5 | 1.5 | 13 | 1.4 |
| X | 8 | 1.3 | 5 | 1.5 | 13 | 1.4 |
| Other | 52 | 8.5 | 28 | 8.2 | 80 | 8.4 |
Evaluation of genotyping results, by use of the Taqman method, revealed 100% match of all duplications. Although heteroplasmy was not specifically tested, we did not observe the occurrence of multiple mtDNA copies (wild type or mutant) in any individual sequenced (N=125).
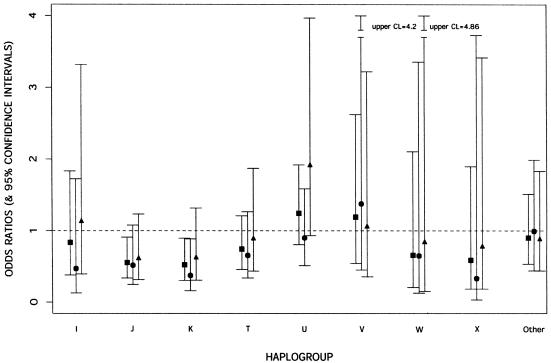
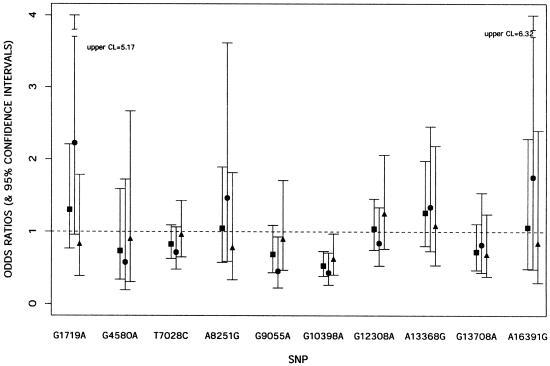
The frequencies of both haplogroup J (OR 0.55; 95% CI 0.34–0.91; P=.02) and haplogroup K (OR 0.52; 95% CI 0.31–0.90; P=.02) were lower in PD cases than in controls when each group was compared with the common haplogroup H (table 3 fig. 1). A similar finding (P=.03) was revealed when each haplogroup was analyzed by comparing it with all other haplogroups pooled together (data not shown). In a comparison designed to determine what made these two haplogroups (J and K) uniquely different from the other haplogroups tested, one SNP located at position 10398 was identified. We therefore tested this SNP independently and found that the difference in 10398G allele frequency between patients with PD and control subjects was highly significant (OR 0.53; 95% CI 0.39– 0.73; P=.0001). The 10398G allele causes a nonconservative amino acid change from threonine (Thr) (hydrophilic) to alanine (Ala) (hydrophobic) within the NADH dehydrogenase 3 (ND3) gene, which is a subunit of complex I. Further stratification of the data set by sex revealed that the 10398G effect appeared to be stronger in women (OR 0.43; 95% CI 0.27–0.71; P=.0009) than in men (OR 0.62; 95% CI 0.41– 0.97; P=.04). Moreover, this analysis showed that SNP 9055A, found within the ATP6 gene, has a mild protective effect only among women (OR 0.46; 95% CI 0.22–0.91; P=0.03) (table 3; fig. 2). In addition, we found that SNP allele 13708A, located within the NADH dehydrogenase 5 (ND5) gene, is protective in the group ⩾70 years of age (OR 0.27; 95% CI 0.09–0.77; P=.01). All other SNPs used for constructing the haplotypes failed to reach significance for association in the overall and stratified data sets.
Table 3.
OR of mt Haplogroups and SNPs Overall[Note]
| 95% CI |
||||
| mt Variant | OR | LowerBoundary | UpperBoundary | P |
| Haplogroup: | ||||
| I | .83 | .38 | 1.83 | .65 |
| J | .55 | .34 | .91 | .02 |
| K | .52 | .30 | .90 | .02 |
| T | .74 | .46 | 1.21 | .23 |
| U | 1.24 | .81 | 1.92 | .33 |
| V | 1.19 | .54 | 2.62 | .67 |
| W | .67 | .20 | 2.11 | .48 |
| X | .59 | .18 | 1.90 | .37 |
| Other | .90 | .53 | 1.51 | .69 |
| SNP: | ||||
| 1719GA | 1.30 | .77 | 2.21 | .33 |
| 4580GA | .74 | .34 | 1.59 | .44 |
| 7028TC | .83 | .63 | 1.09 | .18 |
| 8251GA | 1.05 | .58 | 1.89 | .88 |
| 9055GA | .69 | .44 | 1.09 | .11 |
| 10398GA | .53 | .39 | .73 | .0001 |
| 12308AG | 1.04 | .75 | 1.45 | .80 |
| 13368AG | 1.26 | .80 | 1.98 | .31 |
| 13708GA | .72 | .47 | 1.11 | .14 |
| 16391AG | 1.06 | .49 | 2.29 | .88 |
Total sample consists of 949 total individuals, of whom 609 are patients with PD. OR for each haplogroup was established by comparison with reference haplogroup H.
Figure 1.
OR of individual mt haplogroup for risk of PD. ▪ indicates results for both sexes combined; ● denotes results for women; ▴ denotes results for men. Bars indicate 95% CIs. CL = confidence limit.
Figure 2.
OR results of each SNP for risk of PD. ▪ indicates results for both sexes combined; ● denotes results for women; ▴ denotes results for men. Bars indicate 95% CIs. CL = confidence limit.
Discussion
The mechanism(s) of mt dysfunction observed experimentally in PD has not yet been explained. Here, we present evidence for the involvement of specific inherited mt haplogroups and SNPs in conferring both risk of and protection from the common form of PD.
In association studies, population stratification could lead to false associations between gene markers and disease. However, we were careful to match cases and controls for ethnic background during clinical evaluation, and specific mt haplogroups matched well with previous studies of North American white populations (table 2) (Torroni and Wallace 1994). Given that the “European” haplogroup frequencies have been well documented, the similarity of haplogroup proportions in our control population to those in other studies supports the conclusion that this control and PD group are representative of the general white population. Furthermore, a recent study regarding population subdivision has suggested that carefully matched case-control samples in U.S. and European populations, like those presented here, are “unlikely to contain levels of structure that would result in significantly inflated numbers of false-positive associations” (Ardlie et al. 2002). The etiology of the apparent protective effect of the J and K haplogroups and the 10398G polymorphism contained within them is not yet known, although the association suggests a significant biological role. Indeed, the protective effect of the J haplogroup is interesting, given the published association between J and longevity in northern Italian men (De Benedictis et al. 1999, 2000). Conversely, haplogroup J has also been associated with increased risk for expression of a maternally inherited blindness disorder (Leber hereditary optic neuropathy) (Brown et al. 2002).
Both associated polymorphisms (10398G and 13708A) cause nonconservative amino acid changes from Thr to Ala within ND3 and from Ala to Thr within ND5. These subunits are two of the seven mt-encoded peptides making up the 43 enzymatic subunits of complex I.
Several possible mechanisms for the apparent protective effect of the associated alleles are possible. Given the biological role of complex I, it is conceivable that the strongly associated 10398G allele may increase the performance of complex I within the brain and other tissues in individuals derived from J and K haplogroup lineages. This may be specifically advantageous to the functioning of the complex I in a normal state or under abnormal oxidative stress conditions. Individuals who inherit alternative genotypes that are not protective (or are mildly deleterious) may have inadequate capability for energy metabolism when under cellular stress and thus be at greater risk for developing PD.
Alternatively, the associated 10398 SNP may act as a surrogate marker for a causative variation. For instance, a variation may reside in an mt gene and be in linkage disequilibrium with 10398. One other haplogroup (I) carrying 10398G did not give the same significant findings as haplogroups J and K. Although I, J, and K all share the 10398G polymorphism, their evolutionary relationships are clearly divergent in a phylogeny of mt lineages whereby an A→G substitution has reoccurred at that position several times throughout human evolution (Herrnstadt et al. 2002). Haplogroup I may lack the apparent protective effect, because of the presence of other polymorphisms that are not shared with J and K. However, the frequency of I is relatively low (3%), which reduced the power of the analysis to detect association, even if it was present.
An additional explanation for the pathological mechanism of the 10398 polymorphism is that the 10398A allele may cause a greater ROS production than 10398G. Complex I normally produces ROS during cellular activity; however, when the complex is compromised or inhibited, generation of ROS is enhanced, leading to oxidative stress (Orth and Schapira 2002). Lipids, proteins, and DNA within SN tissue are particularly susceptible to oxidative damage (Jenner and Olanow 1998). It has been proposed that excessive oxidative stress over time may be one of the mechanisms responsible for the degeneration of SN neurons and the concomitant accumulation of fibrillar proteins (Betarbet et al. 2000; Sherer et al. 2002). Therefore, the toxic effects resulting from oxidative injury caused by mtDNA polymorphisms plus environmental exposure to pesticides (rotenone or other xenotoxins) could act synergistically to cause PD.
Our data demonstrated that the apparent protective effect of the 10398G allele was stronger among the women (P=.0009) than among the men (P=.04). Furthermore, SNP allele 9055A, which partly defines haplogroup K, was found to decrease PD risk only in women. These findings are interesting, given the results from multiple clinical studies showing that incidence of PD among men is higher than that among women (male:female ratio 1.5–2.5:1.0) (Tanner and Goldman 1996; Swerdlow et al. 2001). Therefore, our results suggest a possible rationale for the higher susceptibility observed among women.
Interestingly, a previous study also demonstrated that the 10398G polymorphism was found at an increased frequency in a control population when compared with patients with PD (Simon et al. 2000). Although this difference was statistically significant (P=.001), the authors correctly dismissed their findings, because their control group was not ethnically matched to the white cases. Allele 10398G is found in only 26% of white individuals, whereas it is found at greater frequencies in Asians (66%) and sub-Saharan Africans (91%–96%) (Torroni and Wallace 1994; Wallace et al. 1999). Since their control group was ethnically mixed, the frequency difference could have resulted from this known difference in population substructure. This contrasts with our study, in which case-control samples were ethnically matched. Therefore, additional population studies will be needed to confirm the protective effect of 10398G allele in white populations.
However, the findings of Simon et al. (2000) raise the intriguing observation that the allele frequency of 10398A (risk allele) among populations appears to correspond to the trends in PD prevalence worldwide, in that disease is believed to be most common in white individuals, who have a higher frequency of this risk allele (10398A), than in members of other major ethnic groups (Muthane et al. 2001).
Concerning the applicability of the results in other general, ethnically mixed populations, if the 10398G allele is the major functional contributor to the observed association effect, then we would expect the effect to be similar in all individuals carrying this allele. However, we cannot rule out potential interactions with other white mt or nuclear genes, and our finding therefore applies only to white individuals from Europe and North America. Further studies in other ethnic groups will be needed to evaluate the protective effect of 10398G.
Although there is a growing body of evidence to support the role of mtDNA in the pathogenesis of PD, few gains have been made in the discovery of causative or risk alleles. Our findings may help elucidate the mechanism behind the mt dysfunction found in patients with PD, as well as contribute to the understanding of why a higher disease prevalence is reported in white individuals and in men. In addition, these data provide the basis for the exploration of gene-gene or gene-environment interactions that may affect pathogenesis of PD. Future biochemical studies will be needed to confirm the functional significance of these associations.
Acknowledgments
We are grateful to the families who participated in this study. We also thank the personnel at the Center for Human Genetics, Institute for Genome Sciences and Policy, Duke University Medical Center, for excellent clinical, technical, and administrative support. This research was supported, in part, by NIH program project grants 2 P50 NS39764-02 and P01 NS26630, National Institute on Aging grant 1R01-AG-20135-01, the McKnight Foundation (McKnight Neuroscience of Brain Disorders Award), and GlaxoSmithKline. J.M.vdW. is supported by a postdoctoral fellowship award from the American Parkinson’s Disease Association.
Electronic-Database Information
Accession numbers and URLs for data presented herein are as follows:
- GenBank, http://www.ncbi.nlm.nih.gov/Genbank/ (for mt reference sequences: Cambridge [accession number NC001807], revised Cambridge [accession number J01415], Japanese [accession number AB055387], Swedish [accession number X93334], and African [accession number D38112])
- Online Mendelian Inheritance in Man (OMIM), http://www.ncbi.nlm.nih.gov/Omim/
- Primer3 Web site, http://www-genome.wi.mit.edu/cgi-bin/primer/primer3_www.cgi
References
- Anderson S, Bankier AT, Barrell BG, de Bruijn MH, Coulson AR, Drouin J, Eperon IC, Nierlich DP, Roe BA, Sanger F, Schreier PH, Smith AJ, Staden R, Young IG (1981) Sequence and organization of the human mitochondrial genome. Nature 290:457–465 [DOI] [PubMed] [Google Scholar]
- Andrews RM, Kubacka I, Chinnery PF, Lightowlers RN, Turnbull DM, Howell N (1999) Reanalysis and revision of the Cambridge reference sequence for human mitochondrial DNA. Nat Genet 23:147 [DOI] [PubMed] [Google Scholar]
- Ardlie KG, Lunetta KL, Seielstad M (2002) Testing for population subdivision and association in four case-control studies. Am J Hum Genet 71:304–311 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arnason U, Xu X, Gullberg A (1996) Comparison between the complete mitochondrial DNA sequences of Homo and the common chimpanzee based on nonchimeric sequences. J Mol Evol 42:145–152 [DOI] [PubMed] [Google Scholar]
- Betarbet R, Sherer TB, Greenamyre JT (2002) Animal models of Parkinson’s disease. Bioessays 24:308–318 [DOI] [PubMed] [Google Scholar]
- Betarbet R, Sherer TB, MacKenzie G, Garcia-Osuna M, Panov AV, Greenamyre JT (2000) Chronic systemic pesticide exposure reproduces features of Parkinson’s disease. Nat Neurosci 3:1301–1306 [DOI] [PubMed] [Google Scholar]
- Brown MD, Starikovskaya E, Derbeneva O, Hosseini S, Allen JC, Mikhailovskaya IE, Sukernik RI, Wallace DC (2002) The role of mtDNA background in disease expression: a new primary LHON mutation associated with western eurasian haplogroup J. Hum Genet 110:130–138 [DOI] [PubMed] [Google Scholar]
- Cassarino DS, Fall CP, Swerdlow RH, Smith TS, Halvorsen EM, Miller SW, Parks JP, Parker WD Jr, Bennett JP Jr (1997) Elevated reactive oxygen species and antioxidant enzyme activities in animal and cellular models of Parkinson’s disease. Biochim Biophys Acta 1362:77–86 [DOI] [PubMed] [Google Scholar]
- De Benedictis G, Carrieri G, Garasto S, Rose G, Varcasia O, Bonafe M, Franceschi C, Jazwinski SM (2000) Does a retrograde response in human aging and longevity exist? Exp Gerontol 35:795–801 [DOI] [PubMed] [Google Scholar]
- De Benedictis G, Rose G, Carrieri G, De Luca M, Falcone E, Passarino G, Bonafe M, Monti D, Baggio G, Bertolini S, Mari D, Mattace R, Franceschi C (1999) Mitochondrial DNA inherited variants are associated with successful aging and longevity in humans. FASEB J 13:1532–1536 [DOI] [PubMed] [Google Scholar]
- Finnila S, Hassinen IE, Ala-Kokko L, Majamaa K (2000) Phylogenetic network of the mtDNA haplogroup U in northern Finland based on sequence analysis of the complete coding region by conformation-sensitive gel electrophoresis. Am J Hum Genet 66:1017–1026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greenamyre JT, MacKenzie G, Peng TI, Stephans SE (1999) Mitochondrial dysfunction in Parkinson’s disease. Biochem Soc Symp 66:85–97 [DOI] [PubMed] [Google Scholar]
- Greenamyre JT, Sherer TB, Betarbet R, Panov AV (2001) Complex I and Parkinson’s disease. IUBMB Life 52:135–141 [DOI] [PubMed] [Google Scholar]
- Gu M, Cooper JM, Taanman JW, Schapira AHV (1998) Mitochondrial DNA transmission of the mitochondrial defect in Parkinson’s disease. Ann Neurol 44:177–186 [DOI] [PubMed] [Google Scholar]
- Haynes C, Speer MC, Peedin M, Roses AD, Haines JL, Vance JM, Pericak-Vance MA (1995) PEDIGENE: a comprehensive data management system to facilitate efficient and rapid disease gene mapping. Am J Hum Genet Suppl 57:A193 [Google Scholar]
- Herrnstadt C, Elson JL, Fahy E, Preston G, Turnbull DM, Anderson C, Ghosh SS, Olefsky JM, Beal MF, Davis RE, Howell N (2002) Reduced-median-network analysis of complete mitochondrial DNA coding-region sequences for the major African, Asian, and European haplogroups. Am J Hum Genet 70:1152–1171 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horai S, Hayasaka K, Kondo R, Tsugane K, Takahata N (1995) Recent African origin of modern humans revealed by complete sequences of hominoid mitochondrial DNAs. Proc Natl Acad Sci USA 92:532–536 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jenner P, Olanow CW (1998) Understanding cell death in Parkinson’s disease. Ann Neurol 44:S72-S84 [DOI] [PubMed] [Google Scholar]
- Muthane U, Jain S, Gururaj G (2001) Hunting genes in Parkinson’s disease from the roots. Med Hypotheses 57:51–55 [DOI] [PubMed] [Google Scholar]
- Orth M, Schapira AH (2002) Mitochondrial involvement in Parkinson’s disease. Neurochem Int 40:533–541 [DOI] [PubMed] [Google Scholar]
- Parker WD Jr, Boyson SJ, Parks JK (1989) Abnormalities of the electron transport chain in idiopathic Parkinson’s disease. Ann Neurol 26:719–723 [DOI] [PubMed] [Google Scholar]
- Richards M, Macaulay V, Hickey E, Vega E, Sykes B, Guida V, Rengo C, et al (2000) Tracing European founder lineages in the Near Eastern mtDNA pool. Am J Hum Genet 67:1251–1276 [PMC free article] [PubMed] [Google Scholar]
- Schapira AH, Cooper JM, Dexter D, Clark JB, Jenner P, Marsden CD (1990) Mitochondrial complex I deficiency in Parkinson’s disease. J Neurochem 54:823–827 [DOI] [PubMed] [Google Scholar]
- Schapira AH, Cooper JM, Dexter D, Jenner P, Clark JB, Marsden CD (1989) Mitochondrial complex I deficiency in Parkinson’s disease. Lancet 1:1269 [DOI] [PubMed] [Google Scholar]
- Sherer TB, Betarbet R, Stout AK, Lund S, Baptista M, Panov AV, Cookson MR, Greenamyre JT (2002) An in vitro model of Parkinson’s disease: linking mitochondrial impairment to altered α-synuclein metabolism and oxidative damage. J Neurosci 22:7006–7015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simon DK, Mayeux R, Marder K, Kowall NW, Beal MF, Johns DR (2000) Mitochondrial DNA mutations in complex I and tRNA genes in Parkinson’s disease. Neurology 54:703–709 [DOI] [PubMed] [Google Scholar]
- Swerdlow RH, Parker WD, Currie LJ, Bennett JP, Harrison MB, Trugman JM, Wooten GF (2001) Gender ratio differences between Parkinson’s disease patients and their affected relatives. Parkinsonism Relat Disord 7:129–133 [DOI] [PubMed] [Google Scholar]
- Swerdlow RH, Parks JK, Miller SW, Tuttle JB, Trimmer PA, Sheehan JP, Bennett JPJ, Davis RE, Parker WD (1996) Origin and functional consequences of the complex I defect in Parkinson’s disease. Ann Neurol 40:663–671 [DOI] [PubMed] [Google Scholar]
- Tanner CM, Goldman SM (1996) Epidemiology of Parkinson’s disease. Neurol Clin 14:317–335 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Torroni A, Huoponen K, Francalacci P, Petrozzi M, Morelli L, Scozzari R, Obinu D, Savontaus M-L, Wallace DC (1996) Classification of European mtDNAs from an analysis of three European populations. Genetics 144:1835–1850 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Torroni A, Lott MT, Cabell MF, Chen YS, Lavergne L, Wallace DC (1994) mtDNA and the origin of Caucasians: identification of ancient Caucasian-specific haplogroups, one of which is prone to a recurrent somatic duplication in the D-loop region. Am J Hum Genet 55:760–776 [PMC free article] [PubMed] [Google Scholar]
- Torroni A, Wallace DC (1994) Mitochondrial DNA variation in human populations and implications for detection of mitochondrial DNA mutations of pathological significance. J Bioenerg Biomembr 26:261–271 [DOI] [PubMed] [Google Scholar]
- Trimmer PA, Swerdlow RH, Parks JK, Keeney P, Bennett JP Jr, Miller SW, Davis RE, Parker WD Jr (2000) Abnormal mitochondrial morphology in sporadic Parkinson’s and Alzheimer’s disease cybrid cell lines. Exp Neurol 162:37–50 [DOI] [PubMed] [Google Scholar]
- Veech GA, Dennis J, Keeney PM, Fall CP, Swerdlow RH, Parker WD Jr, Bennett JP Jr (2000) Disrupted mitochondrial electron transport function increases expression of anti-apoptotic bcl-2 and bcl-XL proteins in SH-SY5Y neuroblastoma and in Parkinson disease cybrid cells through oxidative stress. J Neurosci Res 61: 693–700 [DOI] [PubMed] [Google Scholar]
- Wallace DC, Brown MD, Lott MT (1999) Mitochondrial DNA variation in human evolution and disease. Gene 238:211–230 [DOI] [PubMed] [Google Scholar]