Abstract
Retrotransposition of L1 LINEs (long interspersed elements) continues to sculpt the human genome. However, because recent insertions are dimorphic, they are not fully represented in sequence databases. Here, we have developed a system, termed “ATLAS” (amplification typing of L1 active subfamilies), that enables the selective amplification and display of DNA fragments containing the termini of human-specific L1s and their respective flanking sequences. We demonstrate that ATLAS is robust and that the resultant display patterns are highly reproducible, segregate in Centre d'Etude du Polymorphisme Humain pedigrees, and provide an individual-specific fingerprint. ATLAS also allows the identification of L1s that are absent from current genome databases, and we show that some of these L1s can retrotranspose at high frequencies in cultured human cells. Finally, we demonstrate that ATLAS also can identify single-nucleotide polymorphisms within a subset of older, primate-specific L1s. Thus, ATLAS provides a simple, high-throughput means to assess genetic variation associated with L1 retrotransposons.
Introduction
L1 LINEs (long interspersed elements) are abundant non–long-terminal-repeat retrotransposons that comprise ∼17% of human DNA (Lander et al. 2001). The vast majority of L1s retrotransposed in the distant past and continued to accumulate mutations at the pseudogene rate. Thus, these ancient L1 insertions generally are fixed with respect to presence in human populations and are retrotransposition defective (Smit et al. 1995; Lander et al. 2001). In contrast, the average human genome contains ∼60–100 retrotransposition-competent L1s (RC-L1s) (Sassaman et al. 1997; Moran and Gilbert 2002), and many of them are dimorphic, indicating that they have retrotransposed since the origin of our species (Sheen et al. 2000; Ovchinnikov et al. 2001; Myers et al. 2002).
RC-L1s are 6.0 kb in length and contain a 5′ UTR, two nonoverlapping ORFs (ORF1 and ORF2), and a 3′ UTR that ends in a poly(A) tail (Scott et al. 1987; Dombroski et al. 1991). ORF1 encodes a 40-kDa nucleic acid binding protein (ORF1p) (Holmes et al. 1994; Hohjoh and Singer 1996; Hohjoh and Singer 1997), whereas ORF2 encodes a protein (ORF2p) with both endonuclease and reverse-transcriptase activities (Mathias et al. 1991; Feng et al. 1996). Both ORF1p and ORF2p are required for retrotransposition (Moran et al. 1996), which likely occurs by a mechanism termed “target-site–primed reverse transcription” (Luan et al. 1993; Luan and Eickbush 1995; Feng et al. 1996; Moran et al 1996).
L1 retrotransposition continues to have an impact on the human genome. To date, 14 de novo insertions have been identified that have resulted in either a genetic disorder or a novel polymorphism (for review, see Moran 1999; Moran and Gilbert 2002). Sequence analysis has demonstrated that 13 insertions have an ACA trinucleotide at positions 5930–5932 of their 3′ UTR (relative to L1 retrotransposable element 1) (Dombroski et al. 1991) (fig. 1a), which is diagnostic for the youngest human L1 subfamily (i.e., the Ta subfamily) (Skowronski et al. 1988). The remaining insertion contains an ACG trinucleotide at this position within its 3′ UTR, which is diagnostic for the slightly older, pre-Ta subfamily (Kazazian et al. 1988; Boissinot et al. 2000; Lander et al. 2001). The development of a cultured-cell retrotransposition assay subsequently confirmed that the Ta subfamily comprises the majority of human RC-L1s (Moran et al. 1996; Sassaman et al. 1997; Kimberland et al. 1999).
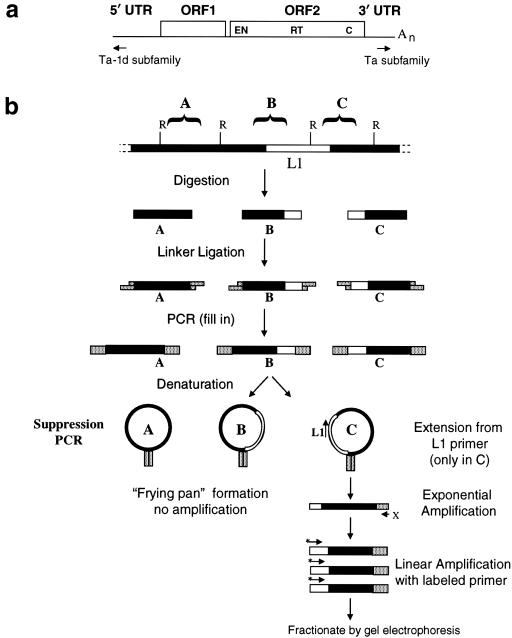
Figure 1.
Principle of ATLAS. a, Overview of LI structure. A schematic of a full-length Ta-1d L1 retrotransposon indicates the position of 5′ and 3′ UTRs, ORFs, and Ta-1d–specific and Ta-subfamily–specific primers. b, Principle of ATLAS, as depicted by a flow chart of the ATLAS procedure. Digestion of genomic DNA and ligation to a GC-rich oligonucleotide linker create a library of fragments flanked by defined sequences (A–C) (top). The denatured single-stranded DNA can self-anneal within the common linker sequences to form a frying pan–shaped intermediate that is inefficiently amplified (A and B). By contrast, subfamily-specific L1 primers can anneal within the frying pan (C), leading to extension products containing a single linker, which subsequently can undergo exponential amplification (bottom). Black bars indicate genomic DNA sequence, white bars indicate L1 sequence, and speckled bars indicate linker sequences. Restriction sites (R) that are compatible with the linker and the L1-specific (L1) and linker-specific (X) primers are shown.
The L1 Ta subfamily is specific to humans (Boissinot et al. 2000) and consists of ∼535 members that amplified during the past 2 million years (Lander et al. 2001; Myers et al. 2002). Approximately 160 Ta-subfamily L1s are full length, and phylogenetic analyses have demonstrated that they can be divided into two groups (the older is termed “Ta-0,” and the younger is termed “Ta-1”) on the basis of the presence of diagnostic nucleotides at positions 5536 and 5539 of ORF2 (Boissinot et al. 2000). In addition, full-length Ta-1 L1s can be subclassified on the basis of the presence (Ta-1nd) or absence (Ta-1d) of a single guanosine nucleotide at position 72 of their respective 5′ UTRs (Boissinot et al. 2000) (fig. 1a). Because the allele frequency of an individual L1 generally is correlated with its age, Ta-subfamily L1s consistently comprise most of the dimorphic L1s in the human genome. For example, ∼30% of Ta-0 L1s are dimorphic, whereas ∼56% of Ta-1 L1s are dimorphic (Boissinot et al. 2000). Thus, although the Ta subfamily comprises only ∼0.1% of human L1s, its members represent a valuable source of identical-by-descent markers that can be used in population studies (Sheen et al. 2000; Ovchinnikov et al. 2001; Myers et al. 2002).
PCR-based procedures previously have been developed to identify dimorphic retrotransposon insertions in human DNA. For example, L1 display, which utilizes PCR primers specific to subfamilies of L1s, has been successful in the identification of candidate dimorphic L1 insertions (Sheen et al. 2000; Ovchinnikov et al. 2001). Similarly, suppression PCR (Lavrentieva et al. 1999) has been combined with both differential and subtractive hybridization, to identify human-specific endogenous retrovirus insertions (Buzdin et al. 2002; Mamedov et al. 2002). However, though effective, both of these methods are labor intensive and are not readily amenable to high-throughput analysis.
Here, we report the development of a system, termed “ATLAS” (amplification typing of L1 active subfamilies), that combines the principles employed in suppression PCR and L1 display. We demonstrate that ATLAS enables the selective amplification and display of human-specific L1s and that the resultant display patterns are highly reproducible, segregate in CEPH pedigrees, and are unique to a given individual. Moreover, ATLAS enables the identification of RC-L1s that are absent from current genome databases and has allowed the discovery of SNPs associated with a subset of older, primate-specific L1s. Thus, ATLAS is a powerful system to assess genetic variation associated with L1 retrotransposons.
Material and Methods
Oligonucleotides
The oligonucleotides used in the present study (table S1 [online only, at the authors' Web site]) were purchased from Invitrogen and were purified by ethanol precipitation prior to quantification by UV spectrophotometry. The linker primers were purified by high-performance liquid chromatography.
A Database of Young Full-Length L1 Insertions
BLAST searches (Altschul et al. 1990) of nonredundant human genomic databases with a full-length RC-L1 (L1.3 [GenBank accession number L19088]) (Dombroski et al. 1993; Sassaman et al. 1997) were performed to identify sequence contigs that contain full-length Ta-subset L1s. The FASTA alignment algorithm (Pearson and Lipman 1988) was used to identify sequences with >98% sequence identity, over ⩾5,922 colinear bases (allowing a maximum 5′ truncation of 100 bp) when aligned with L1.3 (GenBank accession number L19088). The database was rendered nonredundant by exhaustive pairwise comparisons utilizing the full-length L1 and 1,000 bp of its 5′ and 3′ flanking sequences. Sequences displaying >95% identity within either flank were discarded. Each sequence was annotated with respect to typical L1 structural hallmarks (i.e., insertion at a consensus endonuclease cleavage site, presence and position of short direct target-site duplications [TSDs], simple or composite poly(A) tail structures, ORF status, and subfamily diagnostic SNP status). For L1s present in contigs that are not of “finished” sequence quality, completion status was monitored regularly. Summary statistics for the database (available at the authors' Web site) are illustrated in table 1. The 256 L1 sequences satisfying these criteria were aligned using the Clustal W algorithm (Thompson et al. 1994), and the multiple alignment was manually refined using the Seaview editor (Galtier et al. 1996), at the Human Genome Mapping Project Resource Centre. For the full L1 sequence alignment, see the supplementary data available online, at the authors' Web site.
Table 1.
Summary Statistics of a Full-Length Human-Specific L1 Database[Note]
| L1 Subfamily | Totala | TSDsIdentifiedb | TSDsUnidentifiedc | ORF1/ORF2Intactd | ORF1/ORF2 Interruptede |
| Ta | 144 | 137 | 7 | 64 | 80 |
| Pre-Ta | 86 | 83 | 3 | 27 | 59 |
| Non-Ta | 22 | 19 | 3 | 3 | 19 |
| Other | 4 |
4 |
0 |
2 |
2 |
| Total | 256 | 243 | 13 | 96 | 160 |
The full database is available online, at the authors' Web site.
Total number of L1s, from each subfamily, meeting the selection criteria.
Number of L1s that are flanked by TSDs located within 5 kb of flanking DNA.
Number of L1s that are not flanked by TSDs.
Number of L1s from each respective subfamily with intact ORFs.
Number of L1s in which either ORF1 and/or ORF2 contain chain-termination mutations.
DNA Isolation and Sequence Analysis
Plasmid DNAs were purified on Qiagen midi prep columns. BigDye Terminator Cycle Sequencing (ABI Prism) was performed on an Applied Biosystems DNA sequencer (ABI 377 or ABI 3700) at the University of Michigan Sequencing Core facilities.
PCR Containment Procedures
To maintain PCR hygiene, we prepared and manipulated all reagents in a Class II flow hood (Forma) with UV (354 nm) decontamination facilities. The hood was housed outside the laboratory, and caution was taken to exclude L1-containing PCR products and L1-containing clones from the hood and its immediate environment. All manipulations were conducted with equipment that regularly was UV decontaminated.
Suppression Library Construction and Amplification
Genomic DNA (600 ng) was digested to completion with 20 U of AseI (NEB) overnight at 37°C. Heating at 65°C for 20 min inactivated the enzyme, and 100 ng of the digested DNA was ligated to a 40-fold molar excess of the annealed suppression linker (RBMSL2) (table S1 [online only, at the authors' Web site]). The linkers were annealed by heating equal volumes (20 μM) of RBMSL2 and RBD3 at 65°C for 10 min and allowing them to slowly cool to room temperature. Ligations were performed overnight at 14°C–16°C in 1 × NEB3 (50 mM of Tris HCl [pH 7.9], 10 mM of MgCl2, 100 mM of NaCl, and 1 mM of dithiothreitol) supplemented with 1 mM of ATP and 400 U of T4 DNA ligase. Heating at 65°C for 20 min inactivated the ligase and melted away the “dummy” RBD3 oligonucleotide (see table S1 [online only, at the authors' Web site]). Excess linkers and DNA fragments that were too short (i.e., <100 bp) to contain flanking genomic sequences were removed using the Qiaquick PCR purification system (Qiagen). Approximately 2.5–5 ng of ligated genomic DNA then was amplified in a 15-μl reaction volume containing 1.25 μM of either RB5PA2 (5′ ATLAS) or RB3PA1 (3′ ATLAS), 1.25 μM of RBX4 (linker-specific primer), 0.5 U of Taq DNA polymerase (Sigma), and 1 × PCR buffer (50 mM of Tris HCl, 12 mM of NH4SO4, 5 mM of MgCl2, 7.4 mM of 2-mercaptoethanol, 125 μg/ml of BSA, and 1.1 mM of dNTPs). Amplification was performed, in a Perkin-Elmer 9600, under the following cycling conditions: 1 cycle at 96°C for 2 min; followed by 30 cycles at 96°C for 30 s, 64°C for 30 s, and 72°C for 1 min; and a final extension at 72°C for 10 min.
In addition to AseI, library ligations were optimized for use with AciI-, MseI-, and TaqI-digested genomic DNA (with appropriately modified linkers). Whereas all figures (1–6) present data derived from AseI-generated libraries, some of the ATLAS clones subjected to sequence analysis were derived from genomic DNA digested with these other enzymes (see table S2 [online only, at the authors' Web site]).
Figure 6.
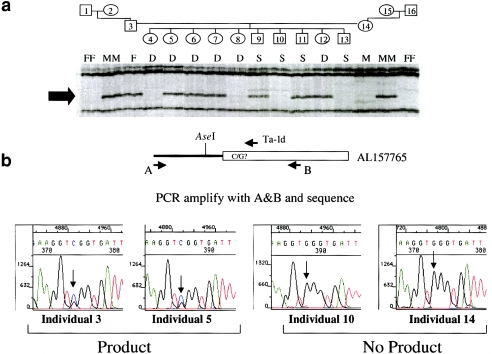
Use of ATLAS to identify SNPs within older L1s. a, Segregation of a dimorphic 5′ (Ta-1d) ATLAS product in CEPH family 1331. A dimorphic 5′ ATLAS display product (see “D” in fig. 5) is shown. b, Identification, through sequencing, of a cosegregating SNP at the L1 primer site. Primers placed outside the AseI restriction site and Ta-1d L1 primer-annealing site enabled PCR amplification of the product from each individual (1–16) in CEPH family 1331. Direct sequencing of the products revealed that an ATLAS display product occurred only when an individual inherited an allele, of the L1, containing an exact match to the Ta-1d primer-annealing site. Individuals 3 and 5 are heterozygous for the matched/mismatched sequence (arrows indicate the double G/C peak). In contrast, individuals 10 and 14 are homozygous for the mismatched allele (arrows indicate the single G peak).
Optimization of Suppression PCR
To maximally enrich for human-specific L1s, we optimized the suppression PCR. Since the suppression effect is temperature dependent, we could not simply raise the primer-annealing temperature to optimize the reaction. Thus, we successively 5′ truncated the L1-specific primers used in ATLAS, to determine the minimum primer length required in order to selectively amplify a locus-specific product that resided either upstream (5′ ATLAS) or downstream (3′ ATLAS) of a known Ta-1d L1. The desired 5′ product was amplified only when RB5PA, RB5PA1, or RB5PA2 was used as a primer (data not shown). Similarly, the desired 3′ product was amplified only when either RB3PA or RB3PA1 was used as a primer (data not shown). Thus, all subsequent 5′ ATLAS reactions were conducted with RB5PA2, and all 3′ ATLAS reactions were conducted with RB3PA1. We also selected the linker primer (RBX4) that amplified the desired product most efficiently, and it was used in all subsequent ATLAS reactions. (For primer details, see table S1 [online only, at the authors' Web site].)
Optimization reactions were fractionated on 2% agarose gels, and the products were transferred, by Southern blotting, to nylon filters (Nytran; Schleicher and Schuell). DNA was fixed to the membrane by baking at 80°C for 2 h. The filters were hybridized to α-32P–labeled (Rediprime II; Amersham Pharmacia) unique sequence probes derived from the empty allele of the locus. Hybridization reactions (at 65°C) were performed overnight in 7% SDS, 1 mM of EDTA (pH 8.0), and 0.5 M of disodium phosphate/sodium dihydrogen phosphate buffer (pH 7.2) (Church and Gilbert 1984). Filters were washed at 65°C, twice in 0.2 × SSC (300 mM NaCl and 30 mM sodium citrate) and 0.5% SDS for 15 min and once in 0.1 × SSC (150 mM NaCl and 15 mM sodium citrate) and 0.1% SDS for 30 min. Radioactive signals were visualized by autoradiography. Notably, this procedure enabled the detection of single-copy sequences from 2.5–5 ng of input genomic DNA, an enrichment of ⩾2,000-fold (relative to a genomic Southern blot input of 10 μg).
Labeling and Display of ATLAS Products
Seventy-five picomoles of the original amplification primer (RB5PA2 or RB3PA1) was labeled in a 20-μl reaction volume containing 50 μCi of γ-33P ATP and 10 U of T4 DNA kinase at 37°C for 60 min. Treatment at 65°C for 20 min inactivated the kinase, and the unincorporated radioisotope was removed using Sephadex G-25 spin columns (Amersham Pharmacia). Aliquots of the labeled primer (0.9–1.9 pmol) were subsequently used in linear amplifications, to generate the ATLAS display products. Labeling reactions were conducted in 9-μl reaction volumes containing 10 mM of Tris HCl (pH 8.3), 50 mM of KCl, 1.5 mM of MgCl2, 250 mM of dNTPs, 0.2 U of Taq DNA polymerase (Sigma), and 1 μl of the primary PCR product. Linear amplification was performed using the following cycling conditions: 1 cycle at 96°C for 2 min; followed by 60 cycles at 96°C for 30 s, 64°C for 30 s, 72°C for 90 s; and a final extension at 72°C for 10 min. Five microliters of the resultant products were mixed with 5 μl of sequencing stop mix (95% formamide, 20 mM EDTA, 0.05 % bromophenol blue, and 0.05% xylene cyanol FF), and the mixture was denatured at 85°C for 4 min. Two microliters of the denatured products were resolved on 5.0% Long Ranger (Flowgen), 1 × glycerol–tolerant (Amersham Pharmacia) polyacrylamide gels containing 50% urea, and the dried gel was visualized by autoradiography.
Recovery and Cloning of ATLAS Products
Alignment between the dried gel and the autoradiograph containing the ATLAS display products was achieved using radioactive (33P) ink spots. ATLAS products were excised from the gel by cutting through the film and gel, using sterile scalpel blades. Gel fragments were placed in 20–40 μl of buffer (1 mM Tris HCl [pH 8.0]) and were frozen at −20°C. The gel was reautoradiographed to ensure that the correct product was excised, and the thawed gel fragments subsequently were reamplified by adding 1 μl of the eluate to 20-μl PCRs. The resultant products were separated on a 2% agarose gel, were purified using the Qiaquick gel extraction kit (Qiagen), and were cloned using the pGEM-T easy kit (Promega). DNAs from clones containing insertions were sequenced using the T7 or SP6 oligonucleotide primers. Clone sequence identifiers, as well as their corresponding accession numbers, are given in table S2 (online only, at the authors' Web site).
Confirmation of Dimorphic Status
DNA sequences flanking dimorphic 5′ ATLAS display products were identified within the December 2001 freeze of the human genome working draft (HGWD) sequence by using the BLAT server (Kent 2002), at the UCSC Genome Bioinformatics Web site. Genomic sequence data were used to design PCR primers 5′ of the AseI site and 3′ of the predicted L1 insertion site. Two PCRs were used to determine the insertion status of a given L1 in DNAs from the 10 individuals used in the original ATLAS analysis. Cycling conditions were as follows: 1 cycle at 96°C for 2 min; followed by 30 cycles at 96°C for 30 s, 64°C for 30 s, and 72°C for 90 s; and a final extension at 72°C for 10 min.
Linkage Analysis
Dimorphic full-length L1 insertions identified by 5′ ATLAS in CEPH family 1331 were located in the HGWD sequence by using BLAT (Kent 2002), and flanking STR markers were acquired using the NCBI Map Viewer human genome utility (see the Entrez Genome View Web site). Segregation data for these loci were downloaded from the CEPH-Généthon Web site. L1 insertion status was coded as a dominant trait, and evidence for single-point linkage to each STR marker was tested using the Fastlink program (Cottingham et al. 1993), at the Human Genome Mapping Project Resource Centre.
Amplification and Cloning of Full-Length L1 Retrotransposons
Fifty to five hundred nanograms of genomic DNA from an individual carrying a given insertion was subjected to long-range PCR amplification with the Expand Long Template PCR kit (Roche), using primers complementary to sequences that flank the L1. Conditions and cycling parameters were performed as recommended by the kit manufacturer. PCR products were purified and cloned into the pGEM-T easy cloning vector (Promega). The cloned L1s were restricted with NotI and BstZ17I, and the resultant fragment was used to replace the corresponding fragment from pJM102/L1.3. The presence of a full-length L1 in pCEP4 was verified by restriction mapping and by sequencing of the 5′ and 3′ ends of the L1 insert.
L1 Retrotransposition Assay
HeLa cells were grown at 37°C in an atmosphere containing 7% carbon dioxide and 100% humidity in Dulbecco’s modified Eagle medium (DMEM) lacking sodium pyruvate (Gibco BRL). DMEM was supplemented with 10% fetal calf serum and 1 × penicillin-streptomycin-L-glutamine (100 × stock; Gibco BRL). Cell passage was performed using standard techniques, and retrotransposition was monitored using the transient retrotransposition assay (Wei et al. 2000, 2001).
Results
Principle of ATLAS
We initially sought to develop a system that would allow us to selectively discriminate human-specific L1s from the vast majority of older L1s present in the genome. To accomplish this goal, we combined the principles employed in L1 display (Sheen et al. 2000) and suppression PCR (Lavrentieva et al. 1999; Buzdin et al. 2002; Mamedov et al. 2002), to develop ATLAS (as outlined in fig. 1b). First, genomic DNA is digested to completion with a restriction endonuclease (denoted by “R” in fig. 1b), to generate a subset of DNA fragments that contain the 5′ and 3′ termini of L1s, as well as their immediate flanking genomic sequences. The digested DNA is ligated to a GC-rich oligonucleotide linker, creating a library of small DNA fragments (typically 100–1,000 bp), which are used as templates in PCRs containing both L1- and linker-specific oligonucleotide primers. Intramolecular annealing of the terminal-linker sequences suppresses the amplification of non–L1-containing templates by forming “frying pan”–shaped intermediates. This suppression effect (Lavrentieva et al. 1999) can be relieved if an L1-specific primer anneals within the loop of the frying pan (fig. 1b, right-hand side). The resultant amplicons contain only a single terminal linker and therefore can undergo exponential amplification in subsequent cycles of PCR, leading to an enrichment of L1 termini and their immediate flanking sequences. Then, aliquots of the primary PCR products are used as templates in linear PCRs containing a 33P-labeled subfamily-specific L1 oligonucleotide. Fractionation of the radiolabeled extension products on denaturing polyacrylamide gels, followed by autoradiography, results in a bar code–like display of fragments containing L1s and their immediate flanking sequences.
Results from a Typical ATLAS Experiment
A typical ATLAS experiment conducted with AseI-digested genomic DNA from three unrelated individuals is shown in figure 2. A primer (RB5PA2) specific for the Ta-1d diagnostic SNP in the L1 5′ UTR was used to generate the 5′ display products, whereas a primer (RB3PA1) specific for the Ta-subfamily sequence present in the L1 3′ UTR was used to generate the 3′ display products (for primer details, see table S1 [online only, at the authors' Web site]). The presence of a band indicates that an individual is either homozygous or hetero-/hemizygous for the presence of an L1 insertion. The absence of a band indicates that an individual either lacks the L1 insertion or contains a sequence-specific variant within either the oligonucleotide primer-annealing site or the restriction site used in the construction of the ATLAS library.
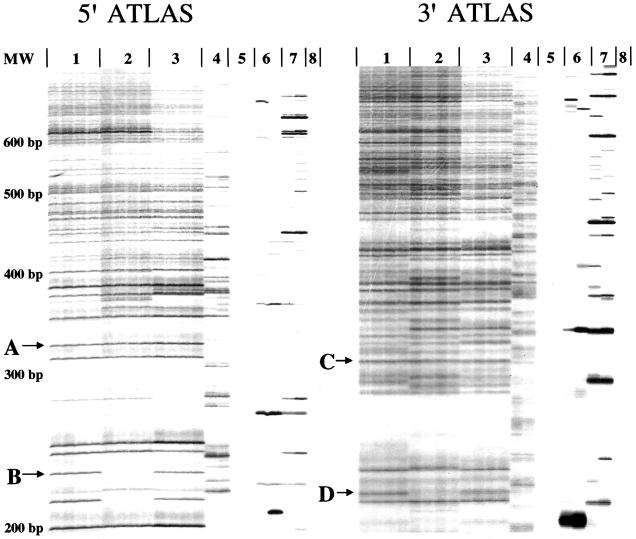
Figure 2.
Representative ATLAS array. Two independent ATLAS display patterns from the DNAs from three unrelated individuals (lanes 1–3) were generated with primers specific to either the 5′ UTR L1 Ta-1d subfamily (left) or the 3′ UTR L1 Ta subfamily (right). The libraries were labeled in duplicate, resulting in four display lanes per individual. Lane 4 reactions were conducted in the absence of restriction digestion; lane 5 reactions were conducted in the absence of genomic DNA; lane 6 reactions were conducted in the absence of T4 DNA ligase; lane 7 reactions were conducted in the absence of linker; and lane 8 reactions were conducted in the absence of primary amplification products. “A” and “C” indicate invariant bands, whereas “B” and “D” indicate variant bands. Size standards are indicated to the left of the gel.
Comparison of the display products between individuals revealed variant and invariant bands. Invariant bands (see “A” and “C” in fig. 2) indicate candidate L1 insertions that are present in all three individuals. By contrast, variant bands (see “B” and “D” in fig. 2) indicate candidate dimorphic L1 insertions. As expected, the 3′ display products were more numerous than the 5′ display products, because ∼60% of young L1s are 5′ truncated and lack the L1 5′ UTR (Boissinot et al. 2000). Band broadening also is observed in the 3′ display products, which likely results from “stuttering” at the L1 poly(A) tail during amplification. Notably, we detected more invariant bands than anticipated in the 5′ display, but the reason for this result was elucidated in subsequent experiments (see the subsection “5′ ATLAS Can Identify Genetic Variation Associated with a Subset of Older, Primate-Specific L1s,” below).
As controls for reaction specificity, we demonstrated that the ATLAS display products are dependent on the presence of input DNA (fig. 2, lanes 5 and 8), restriction-enzyme digestion (fig. 2, lane 4), ligation (fig. 2, lane 6), and the presence of the linker (fig. 2, lane 7). Product characterization revealed that the reproducible banding pattern observed in the absence of the restriction enzyme results from inter-L1 PCR amplification (data not shown). By contrast, the banding patterns observed in the absence of either ligase (fig. 2, lane 6) or linkers (fig. 2, lane 7) are stochastic and likely result from rare mispriming events in the primary PCR. Importantly, reproducible ATLAS patterns were generated only when all the components (genomic DNA, restriction enzyme, ligase, and linkers) were present in the reactions.
ATLAS Is Reproducible and Robust
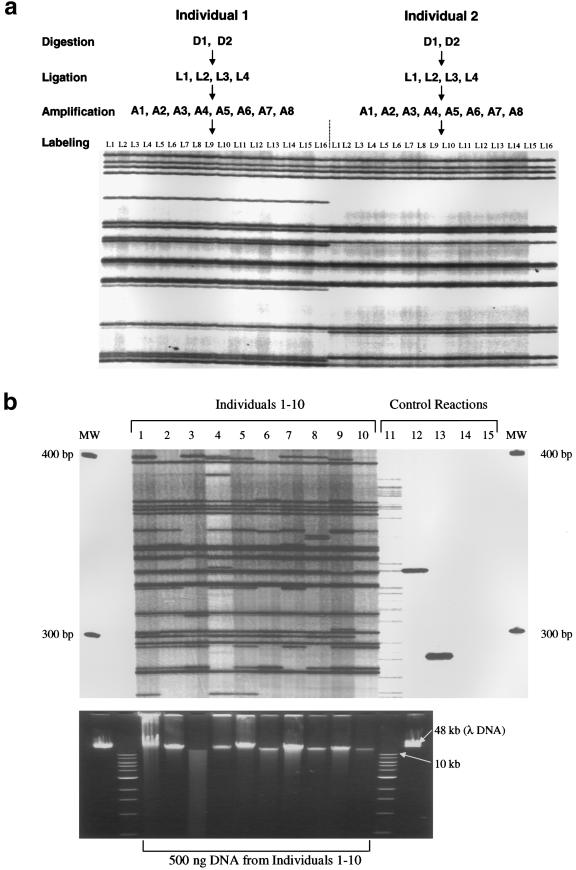
To test whether ATLAS was reproducible, we used two independent AseI-digested genomic DNA samples from two unrelated individuals as templates in individual ATLAS reactions (fig. 3a). At each step during the ATLAS procedure (ligation, suppression PCR, and labeling PCR), reactions were prepared in duplicate. The resultant ATLAS display patterns for each respective DNA are identical (fig. 3a), indicating that ATLAS is highly reproducible.
Figure 3.
ATLAS's reproducibility and robustness. a, High reproducibility. Genomic DNA samples (D1 and D2) from two unrelated individuals were digested, in duplicate, with AseI, and each digestion was ligated, in duplicate, to a compatible linker (L1–L4). Each of the purified ligation reactions (A1–A8) was amplified in duplicate, and the resultant primary amplifications (L1–L16) were radiolabeled in duplicate. The resultant ATLAS display patterns are shown for each of the 16 replicates. b, Robustness. Top, 5′ ATLAS performed, in duplicate, on 10 unrelated individuals (lanes 1–10). Controls (lanes 11–15) are the same as in figure 2. Bottom, Genomic DNAs (∼500 ng), used to generate the above 5′ ATLAS display patterns, resolved on a 0.8% agarose gel that contained ethidium bromide. Note the significant amount of degradation in the DNA sample from individual 3. MW = molecular weight markers; λ DNA = genomic DNA from bacteriophage lambda.
Restriction-based library-construction methods also can be sensitive to DNA quality, since the effective copy number of a “target” sequence is dependent on the mean DNA fragment size. We hypothesized that the utilization of suppression PCR over small distances (100–1,000 bp) should minimize the effect that DNA quality has on the ATLAS display pattern. To test this, we performed duplicate ATLAS reactions on genomic DNAs of differing quality from 10 unrelated individuals (fig. 3b). Despite considerable variation in DNA quality (e.g., see fig. 3b, sample 3), the display patterns are quantitatively and qualitatively comparable, indicating that ATLAS is robust.
3′ ATLAS Selectively Amplifies Human-Specific L1s
To determine whether ATLAS truly enriches for human-specific L1 insertions, we cloned and sequenced 19 dimorphic 3′ ATLAS display products that were absent from at least 1 of the 10 DNAs examined in figure 3b. Each product had the predicted structure because it contained the 3′ end of a Ta-subfamily L1, a poly(A) tail, and flanking genomic DNA (tables 2 and S2 [online only, at the authors' Web site]).
Table 2.
Characterization of ATLAS Display Products[Note]
| Reactiona | Total ProductsCharacterized(L1Hs Elements)b | Flanks Localizedin HGWDc | Localized L1Hs Elementsd | Primate-SpecificL1se | Novel DimorphicL1Hs Elementsf |
| 3′ ATLAS | 19 (19) | 8 | 8 | 0 | 3 |
| 5′ ATLAS | 56 (23) | 47 | 23 | 24 | 15 |
Derived from table S2 (online only, at the authors' Web site).
Type of ATLAS reaction, 5′ (Ta-1d) specific or 3′ (Ta) specific.
Total number of ATLAS products characterized; the number of human-specific L1 (L1Hs) elements is shown in parentheses.
Number of characterized products for which the flanking genomic sequences could be localized in the HGWD.
Number of localized insertions that were identified as human specific.
Number of localized insertions that were identified as primate specific.
Number of localized human-specific L1 insertions that were novel (i.e., absent from the HGWD).
Next, we used BLAT to map the location of each of the flanking sequences in the HGWD. In 5 of 19 instances, the 3′ flanking sequences were located downstream of a known Ta-subfamily L1. In 3 of 19 instances, there was no L1 upstream of flanking sequence, suggesting that the preintegration site (i.e., the sequence lacking the L1 insertion) actually is present in the HGWD. Finally, in 11 of 19 instances, we were unable to unambiguously map the 3′ flanking sequences, because (1) they were absent from the HGWD (in five cases); (2) they were composed of high-copy-number repeats (in three cases: two L1s were flanked by α-satellite DNA, and one L1 was flanked by an Alu element); or (3) they were too short to be assigned, because the restriction site used to create the 3′ display library was located within 10 bp of the L1 poly(A) tail (in three cases). Thus, our data indicate that 3′ ATLAS selectively displays Ta-subfamily L1s and that it enables the identification of human-specific L1 insertions that are absent from the HGWD (also see the subsection “5′ ATLAS Identifies Human-Specific L1s,” below).
5′ ATLAS Identifies Human-Specific L1s
To assess the fidelity of 5′ ATLAS, we cloned and sequenced 56 dimorphic display products and mapped their respective 5′ flanking sequences to the HGWD. As above (see the subsection “3′ ATLAS Selectively Amplifies Human-Specific L1s”), in 9 of 56 instances, we were unable to unambiguously map the 5′ flanking sequences, because (1) they were absent from the HGWD (in three cases); (2) they were composed of high-copy-number repeats (in two cases: one L1 was flanked by α-satellite DNA, and another L1 was flanked by human satellite II DNA); or (3) they were too short to be assigned, because the restriction site used to create the 5′ display library was located within 10 bp of the L1 terminus (in four cases). The L1 sequences present in these nine clones show high identity to L1.3, indicating that they likely are derived from Ta-1d L1s.
In 47 of 56 instances, the flanking sequences could be unambiguously identified in the HGWD, and, in 23 instances, the display products had the predicted structure and contained the 5′ end of a Ta-1d L1 and its flanking genomic DNA. Of those L1s, 8 were present in the HGWD, whereas 15 were not (tables 2 and S2 [online only, at the authors' Web site]). The other 24 of these 47 cases are described later (see the subsection “5′ ATLAS Can Identify Genetic Variation Associated with a Subset of Older, Primate-Specific L1s,” below).
Independent Verification of the Dimorphic Status of Human-Specific L1s Identified by 5′ ATLAS
To verify that 5′ ATLAS truly identifies dimorphic Ta-1d L1s, we used conventional PCR analyses to independently assess the presence/absence status of 9 of the 23 full-length L1s identified above (see the subsection “5′ ATLAS Identifies Human-Specific L1s”) (table 3). Amplification using 5′ and 3′ flanking primers or a 3′ flanking primer and a Ta-subfamily specific primer (fig. 4a) enabled the discrimination of all possible insertion states in two PCRs. Typing of the nine loci in 10 unrelated individuals confirmed the ATLAS results (fig. 4b and data not shown).
Table 3.
Characterization of Dimorphic Full-Length L1s Identified by ATLAS
| AccessionNumbera | Chromosomal Locationb | Presencein HGWDc | Identity to L1.3d(%) | Retrotransposition Activitye | Insertion Allele Frequencyf | Population Distributiong |
| AC005885 | 12q24.32 | + | 99.7 | ND | .63 | U |
| AC044907 | 15q25.2 | + | 99.6 | ND | .43 | U |
| AC009414 | 2p22.2 | − | 100.0 | − | .32 | U |
| AL121819 | 14q23.1 | − | 99.3 | ++++ | .24 | U |
| AC008826h | 5p13.3 | − | 99.8 | −/+ | .16 | U |
| AL358779 | 9q31.3 | − | 99.1 | ++++ | .09 | U |
| AC011092 | 11p15.3 | − | 99.2 | − | .07 | AS/NE/SA |
| AC087224 | 18q21.2 | − | 99.2 | −/+ | .01 | NE |
| AL021407i | 6p22.3 | − | 99.3 | ++++ | .0 | P |
GenBank accession number of each genomic locus.
Chromosomal location of each L1 as determined using the HGWD BLAT server.
Whether the L1 is present (+) or absent (−) in the HGWD.
Relative retrotransposition activity as compared to L1.3. ++++ = L1s that retrotransposed at ∼50%–100% the level of L1.3; − = L1s that do not retrotranspose; −/+ = L1s that show marginal activity and retrotransposed at <1% the level of L1.3; ND = not tested.
Insertion allele frequency of the respective filled sites in 90 unrelated individuals from four worldwide populations.
Population distribution of the L1s. U = ubiquitous; AS = restricted to Asians; NE = northern Europeans; SA = South Americans.
The AL021407 insertion apparently is private to the individual in whom it was discovered.
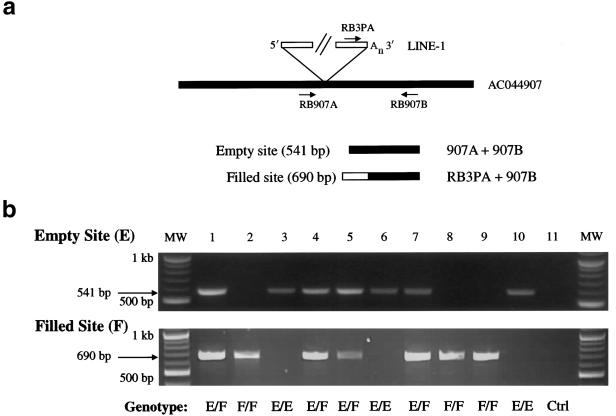
Figure 4.
Dimorphic Ta-1d L1s, identified by 5′ ATLAS. a, Independent assay to assess the dimorphic status of Ta-1d L1s. PCR typing assays were developed to confirm the presence (filled)/absence (empty) status of Ta-1d L1s identified in the 5′ ATLAS reactions shown in panel b. Empty sites, lacking the L1 insertion, can be amplified with primers flanking the insertion site (e.g., RB907A and RB907B). Filled sites, containing the L1 insertion, can be amplified with a Ta-subfamily–specific L1 3′ UTR primer and a 3′ flanking primer (e.g., RB3PA and RB907B). b, Results of PCR typing experiments. Top, Amplification of empty site through PCR typing by use of the RB907A and RB907B primers. Bottom, Amplification of filled site through PCR typing by use of the RB3PA and RB907B primers. Note that individual DNA samples (1–10) can be homozygous for the presence of the L1 (F/F; lanes 2, 8, and 9), heterozygous (EF; lanes 1, 4, 5, and 7), or homozygous for the absence of the L1 (E/E; lanes 3, 6, and 10). The 11th reaction (Ctrl) is a control PCR with genomic DNA omitted. MW = molecular weight markers.
Geographic Distribution of Human-Specific L1s Identified by 5′ ATLAS
To determine their prevalence and distribution, we used PCRs to type the nine dimorphic full-length L1 insertions identified above (see the subsection “Independent Verification of the Dimorphic Status of Human-Specific L1s Identified by 5′ ATLAS”) in 90 randomly selected individuals representative of worldwide human populations. The panel comprises genomic DNAs from 30 Africans/African Americans, 20 Asians, 20 northern Europeans, and 20 South Americans. Each locus was tested for deviation from Hardy-Weinberg equilibrium (HWE) by a permutation-test method (Guo and Thompson 1992), using the Arlequin population genetics software suite (Schneider et al. 2000). After a Bonferroni correction, none of the loci showed a significant deviation in any population, indicating that they are in HWE.
As expected, the allele frequency of an L1 identified by 5′ ATLAS was correlated with the occurrence of the insertion in the HGWD (table 3). Furthermore—consistent with the recent origin of low-allele-frequency insertions—two L1s (ACO11092 and AC087224) were restricted to particular populations, whereas a third L1 (AL021407) represented a private polymorphism, which was present only in a single individual (table 3). Thus, as with 3′ ATLAS, we conclude that 5′ ATLAS enables the amplification of human-specific L1 insertions and that some of those L1s are absent from the HGWD.
Some Low-Insertion-Frequency Polymorphic Ta-1d L1s Are Retrotransposition Competent
We hypothesized that some of the newly identified Ta-1d full-length L1s should remain retrotransposition competent. To test this hypothesis, we examined seven L1s for their ability to retrotranspose in cultured human HeLa cells. Three L1s (AL358779, AL121819, and AL021407) (table S2 [online only, at the authors' Web site]) retrotransposed at levels similar to L1.3 (GenBank accession number L19088), a known active element (table 3), whereas two other L1s (AC008826 and AC087224) reproducibly retrotransposed at extremely low (∼0.1%) levels. Thus, ATLAS allows the identification of RC-L1s that are absent from the HGWD. Moreover, since AL021407 was present only in a single individual (table 3; also see the subsection “Geographic Distribution of Human-Specific L1s Identified by 5′ ATLAS,” above), our data are consistent with previous studies, which have showed that recent mutagenic L1 insertions retain the ability to retrotranspose (Naas et al. 1998; Kimberland et al. 1999).
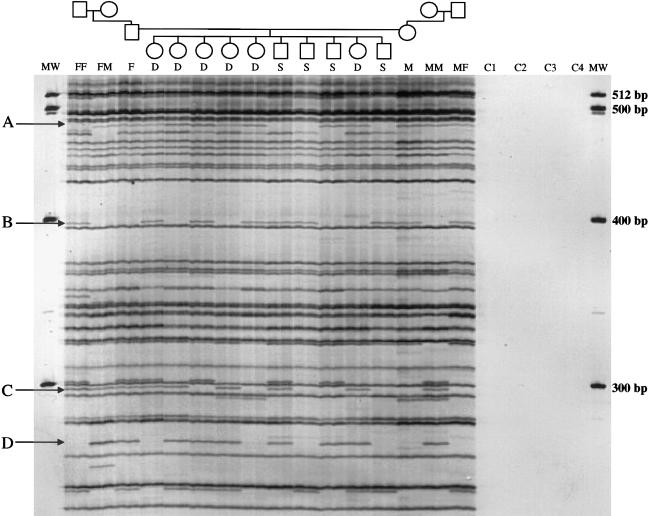
5′ ATLAS Banding Patterns Segregate in Families
In principle, ATLAS display patterns should exhibit Mendelian segregation. To test this hypothesis, we subjected genomic DNAs from a large three-generation CEPH family (paternal and maternal grandparents, parents, and 10 children) to 5′ ATLAS, and 13 loci were analyzed with respect to band presence or absence. In all cases, the dimorphic bands showed stable transmission throughout the pedigree (in fig. 5, four examples are indicated by “A”–“D”). Notably, even though only 13 loci were examined for segregation, each of the 10 children had unique display patterns, indicating that ATLAS generates individual-specific “fingerprints.” Finally, in one instance (AC015617, a novel insertion at 18q22.3), we established that the locus showed significant linkage (LOD score >3.0) to three flanking Généthon STR markers (data not shown), indicating that the presence/absence pattern observed is not due to an independently segregating paralogous locus.
Figure 5.
ATLAS products' segregation in families. 5′ Ta-1d–specific ATLAS was performed on a three-generation CEPH pedigree (1331). FF = paternal grandfather; FM = paternal grandmother; F = father; S/D = son/daughter; M = mother; MF = maternal grandfather; MM = maternal grandmother. “C1” denotes a control sample in which water was substituted for genomic DNA at digestion; “C2” denotes a control sample in which water was substituted for library DNA; and “C3” and “C4” denote control samples in which water was substituted for the primary PCR products in the labeling reaction. Arrows (“A”–“D”) indicate dimorphic array products. MW = molecular weight markers.
5′ ATLAS Can Identify Genetic Variation Associated with a Subset of Older, Primate-Specific L1s
In 24 of 47 instances, the dimorphic 5′ ATLAS display products contained older, primate-specific full-length L1s (from the L1PA2, L1PA3, L1PA4, L1PA7, L1PA8, or L1P subfamilies [Smit et al. 1995]) and their respective 5′ flanking DNA sequences. This result was unexpected, since previous studies would predict that these older L1s should be fixed with respect to presence in human DNA. Thus, we hypothesized that sequence-specific variants or SNPs could explain why these display products appeared to be dimorphic.
To demonstrate conclusively that 5′ ATLAS could selectively amplify a subset of older L1s, we analyzed nine invariant 5′ display products that were present in all 10 DNAs examined in figure 3b. Sequence analysis revealed that each invariant band originated from a member of an older, primate-specific L1 subfamily (L1PA2, L1PA3, L1PA4, L1PA7, and L1P) that contained an exact match to the Ta-1d primer sequence (data not shown). Thus, 5′ ATLAS can identify a subset of older L1s that fortuitously harbor a Ta-1d primer-binding site.
We next PCR amplified an apparently dimorphic older L1 (AL157765) from the DNAs of each individual represented in the CEPH family and analyzed its sequence in detail (figs. 6a and 6b). Individuals exhibiting a display product had an allele, of the older L1, that contained a SNP, resulting in a perfect match to the Ta-1d primer (this SNP also is present in dbSNP [rs2503417]). In contrast, individuals lacking the display product contained two alleles, of the older L1, that lack that SNP (fig. 6b). Thus, our data demonstrate that 5′ ATLAS is exquisitely sensitive and can discriminate between L1 alleles that differ by 1 bp. Notably, similar results were obtained for two other L1s (AL133391 and AC066611), and additional product characterization revealed that the apparent dimorphism in AC093527 was due to a 1-bp insertion/deletion polymorphism within the amplified product. Moreover, we conclude that the apparent dimorphism observed in AC011302 likely is due to a deletion or mutation that eliminates one of the primer-annealing sites, because the L1 could be amplified only from individuals harboring the ATLAS display product (see table S2 [online only, at the authors' Web site]).
We next wished to determine why the other 19 older L1s appeared to be dimorphic. As above, in seven instances, analysis of the HGWD revealed that the older L1 contained at least one nucleotide difference from the 5′ Ta-1d primer used in the ATLAS reaction. However, in 12 instances, the L1 in the HGWD matched to the Ta-1d primer-annealing site and is flanked by the restriction site used in library construction. Moreover, sequence analysis showed that these 12 L1s were not mosaic elements that consisted of the 5′ end of a young L1 and the 3′ end of an older L1 (data not shown).
To determine the reason for the apparent dimorphism associated with these 12 older L1s, we attempted to amplify the candidate loci by using PCR primers that flanked both the restriction site used in library construction and the Ta-1d primer-annealing site (e.g., see the placement of primers A and B in fig. 6a). Six loci were amplified efficiently. In two instances, we could not design locus-specific primers, because the 5′ flanking sequence was embedded in repeated DNA. In four instances, the designed primers failed to give specific amplification of the L1, despite extensive optimization (data not shown).
PCR and sequencing analysis demonstrated that four loci (AC022884, AL360002, AC087714, and AL583822) were present in the DNAs from all 10 individuals examined (i.e., even those who apparently lacked the L1). However, the sizes of those products were smaller than expected, indicating that the cloned L1 did not represent the dimorphic display product but instead was derived from a faster-migrating invariant product that appeared at the expected position on the display gel. Fortunately, this limitation of the gel-resolution system can be overcome by simple modifications of the ATLAS procedure, as we showed by cloning the authentic dimorphic L1s from two of the above samples (for details, see the “Discussion” section).
Finally, in two instances (AL160032 and AC93265), the display products showed dimorphism between peripheral-blood DNA (band absent) and tumor DNA (band present) derived from anonymous patients with Wilms tumor. Direct sequencing of the PCR products confirmed that both sets of DNA had an identical L1 of the predicted size. However, because the original ATLAS library was made using AciI, which is sensitive to CpG methylation status, the apparent dimorphism likely is due to hypomethylation of the L1 promoter in tumor DNA (Alves et al. 1996; Florl et al. 1999; Takai et al. 2000). Thus, unexpectedly, our initial analysis suggests that ATLAS can detect epigenetic changes associated with L1s.
Discussion
We have combined the principles employed in L1 display (Sheen et al. 2000) and suppression PCR (Lavrentieva et al. 1999; Buzdin et al. 2002; Mamedov et al. 2002), to develop ATLAS, and we have demonstrated that this improved system provides a rapid and robust means to assess genetic variation associated with human L1s. ATLAS can selectively identify dozens of human-specific L1s in a single reaction and is superior to L1 display, which allows for the acquisition of only a few dimorphic L1s per experiment and requires the use of multiple primers, low-resolution agarose gel electrophoresis, and Southern blotting, to detect dimorphic insertions (Sheen et al. 2000). Moreover, we predict that simple technical modification, such as the use of fluorescent-labeled primers and the separation of the ATLAS display products on automated DNA sequencers, will enable high-throughput identification of more dimorphic L1s.
In the course of developing ATLAS, we compiled an extensive database (available online, at the authors' Web site) of full-length human-specific L1s from the HGWD. These L1s were multiply aligned to assess the variation that exists around known subfamily-specific sequence variants, allowing us to design primers that would selectively amplify the majority of Ta-subfamily L1s present in the HGWD (the alignment is also available online, at the authors' Web site). Interestingly, we identified several pre-Ta–subfamily and non-Ta–subfamily L1s that contain intact ORFs (table 1), some of which remain retrotransposition competent (Brouha et al., in press). Clearly, modifications of the L1-specific primers used in the ATLAS protocol should allow for an assessment of the variation associated with pre-Ta and other primate-specific L1 subfamilies.
We have showed that ATLAS identified 18 recent human-specific L1 insertions that are absent from current genome databases. Indeed, the ease with which we isolated low-allele-frequency L1 insertions (in three cases; see the “Results” section) suggests that young L1s are far more common in human populations than previously suspected. Thus, although the HGWD provides a valuable resource to identify human-specific L1s, it only provides a “snapshot” of extant L1 diversity.
Our data also demonstrate that a subset of primate-specific L1s share fortuitous sequence identity with the Ta-1d–specific primer-annealing site. As expected, many of these L1s were identified as invariant 5′ ATLAS display products; however, we did identify 24 older L1s that appeared to be dimorphic. In 14 instances, detailed analyses of the display products revealed the reason for the apparent dimorphism. In 12 cases, the older L1s accumulated base-substitution polymorphisms within the Ta-1d primer-annealing site (in 10 cases), contained size polymorphisms within the amplified product (in 1 case), or likely contained a deletion or mutation resulting in the loss of one of the primer-annealing sites (in 1 case). Similarly, in two other instances, the apparent dimorphism likely reflects methylation differences in the L1 promoter that exist between the peripheral-blood and tumor DNA samples from patients with Wilms tumor. Thus, we conclude that these 14 older L1s truly are fixed with respect to presence and that their apparent dimorphism results from DNA mutations or epigenetic differences associated with the L1s.
In four instances, we did not clone the actual dimorphic L1 but instead mistakenly cloned a minor product that contained a fixed, anomalously migrating older L1. Thus, we must conclude that the rapid-acquisition strategy used to characterize display products occasionally can result in product misidentification. Fortunately, this minor problem, which is due to a limitation of the gel-resolution system, can be overcome easily by using DNA fingerprinting to rapidly identify the “majority product” present in multiple, independent clones derived from a given display product. Alternatively, clones can be obtained from the filled or empty sites of different individuals; L1s that are present only at the filled position would represent the authentic dimorphic product. Indeed, in samples in which AL360002 and AC022884 originally were identified as anomalously migrating display products, the above strategies successfully led to the identification of AL512410 and AL155765 as the authentic dimorphic products (see table S2 [online only, at the authors' Web site]).
In a final six instances, we could not independently reamplify the older L1; thus, we could not determine the actual reason for the apparent dimorphism. Parsimony would dictate that virtually all of these older L1s also are fixed with respect to presence and that their apparent dimorphism is due to either DNA mutations associated with the L1s or product misidentification (as seen above). However, it is possible that a subset of older L1s appear to be dimorphic because of gene-conversion events, between different L1 subfamilies, that could effectively eliminate primer-annealing sites (Kass et al. 1995; Saxton et al. 1998; Myers et al. 2002). Alternatively, it is formally possible that dimorphisms result from the existence of deletion alleles, which encompass the location of the display product, although such instances likely would be rare (Edwards et al. 1992). Notably, either of the above explanations may account for the results observed with AC0110302 (see the “Results” section and table S2 [online only, at the authors' Web site]). Nonetheless, taken together, our data demonstrate that ATLAS is an exquisitely sensitive method for the identification of genetic variation associated with L1s.
ATLAS now provides a genomic tool to learn even more about L1 biology. For example, it can be used to conduct high-throughput screens for human-specific dimorphic L1s in geographically diverse populations. Such screens will enable the identification of identical-by-descent markers of a known ancestral state that will be useful tools in the study of human population genetics and demography. Similarly, our initial analyses suggest that epigenetic variation associated with L1s can be ascertained by using enzymes, in the construction of ATLAS libraries, that differentially cleave methylated and unmethylated genomic DNA.
Finally, the reproducibility and individual-specific nature of ATLAS display patterns suggests that ATLAS could be employed to study de novo L1 retrotransposition in small pools of sperm-derived DNA. Similarly, the individual-specific fingerprints that are generated suggest that ATLAS may be applicable to forensic situations in which only small amounts of degraded DNA are available for identification purposes. Indeed, the large number of independently segregating markers and the rarity of reversion make the probability of misidentification extremely small.
In closing, ATLAS provides a powerful means to identify genetic variation associated with L1 retrotransposons. Indeed, our data indicate that, in human populations, there exist numerous L1s that are absent from current sequence databases. The ability to rapidly identify these L1s will lead to a greater appreciation of how they continue to sculpt our genome.
Acknowledgments
Dr. Mark Batzer, Dr. Thomas Glaser, and Prof. Sir Alec Jeffreys, F.R.S., kindly provided DNAs used in these studies. We thank members of the University of Michigan DNA Sequencing Core, for help with sequencing; Dr. Timothy Bestor, Dr. Thomas Glover, Dr. Stephen B. Gruber, Prof. Haig Kazazian Jr., Dr. Jeffrey Long, and other members of the Moran laboratory, for critically evaluating the manuscript; Dr. Arian Smit and Dr. Victor Pollara, for helping us to gain early access to L1-sequence–containing contigs isolated from the Golden Path; Mr. Colin Veal, for assistance with the linkage analysis; and Dr. Celia May, for Web site construction. R.M.B. is supported by a Wellcome Trust International Prize Travelling Fellowship (058305/B/99Z). J.V.M. is supported, in part, by grants from the W. M. Keck Foundation and the National Institutes of Health (GM60518). The University of Michigan Cancer Center helped to defray some of the costs of DNA sequencing.
Electronic-Database Information
The accession number and URLs for data presented herein are as follows:
- Authors' Web Site, http://www.le.ac.uk/ge/ajj/LINE1/ (for supplementary tables S1 and S2, as well as full-length L1 database and full-length L1 alignment)
- BLAST, http://www.ncbi.nlm.nih.gov/BLAST/
- CEPH-Généthon Web Site, http://www.cephb.fr/ceph-genethon-map.html
- dbSNP Home Page, http://www.ncbi.nlm.nih.gov/SNP/ (for rs2503417)
- Entrez Genome View, http://www.ncbi.nlm.nih.gov/mapview/map_search.cgi? (for NCBI Map Viewer)
- GenBank, http://www.ncbi.nlm.nih.gov/Genbank/ (for L1.3 [accession number L19088])
- Human Genome Mapping Project Resource Center, http://www.hgmp.mrc.ac.uk/ (for BLAST/FASTA, Clustal W, Seaview, and Fastlink)
- UCSC Genome Bioinformatics (“Golden Path”), http://genome.ucsc.edu/ (for BLAT)
References
- Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ (1990) Basic local alignment search tool. J Mol Biol 215:403–410 [DOI] [PubMed] [Google Scholar]
- Alves G, Tatro A, Fanning T (1996) Differential methylation of human LINE-1 retrotransposons in malignant cells. Gene 176:39–44 [DOI] [PubMed] [Google Scholar]
- Boissinot S, Chevret P, Furano AV (2000) L1 (LINE-1) retrotransposon evolution and amplification in recent human history. Mol Biol Evol 17:915–928 [DOI] [PubMed] [Google Scholar]
- Brouha B, Schustak J, Badge RM, Lutz-Prigge S, Farley AH, Moran JV, Kazazian HH Jr. Hot L1s account for the bulk of retrotransposition in the human population. Proc Natl Acad Sci USA (in press) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buzdin A, Khodosevich K, Mamedov I, Vinogradova T, Lebedev Y, Hunsmann G, Sverdlov E (2002) A technique for genome-wide identification of differences in the interspersed repeats integrations between closely related genomes and its application to detection of human-specific integrations of HERV-K LTRs. Genomics 79:413–422 [DOI] [PubMed] [Google Scholar]
- Church GM, Gilbert W (1984) Genomic sequencing. Proc Natl Acad Sci USA 81:1991–1995 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cottingham RW Jr, Idury RM, Schäffer AA (1993) Faster sequential genetic linkage computations. Am J Hum Genet 53:252–263 [PMC free article] [PubMed] [Google Scholar]
- Dombroski BA, Mathias SL, Nanthakumar E, Scott AF, Kazazian HH Jr (1991) Isolation of an active human transposable element. Science 254:1805–1808 [DOI] [PubMed] [Google Scholar]
- Dombroski BA, Scott AF, Kazazian HH Jr (1993) Two additional potential retrotransposons isolated from a human L1 subfamily that contains an active retrotransposable element. Proc Natl Acad Sci USA 90:6513–6517 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edwards MC, Gibbs RA (1992) A human dimorphism resulting from loss of an Alu. Genomics 14:590–597 [DOI] [PubMed] [Google Scholar]
- Feng Q, Moran JV, Kazazian HH Jr, Boeke JD (1996) Human L1 retrotransposon encodes a conserved endonuclease required for retrotransposition. Cell 87:905–916 [DOI] [PubMed] [Google Scholar]
- Florl AR, Lower R, Schmitz-Drager BJ, Schulz WA (1999) DNA methylation and expression of LINE-1 and HERV-K provirus sequences in urothelial and renal cell carcinomas. Br J Cancer 80:1312–1321 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galtier N, Gouy M, Gautier C (1996) SEAVIEW and PHYLO_WIN: two graphic tools for sequence alignment and molecular phylogeny. Comput Appl Biosci 12:543–548 [DOI] [PubMed] [Google Scholar]
- Guo SW, Thompson EA (1992) A Monte Carlo method for combined segregation and linkage analysis. Am J Hum Genet 51:1111–1126 [PMC free article] [PubMed] [Google Scholar]
- Hohjoh H, Singer MF (1996) Cytoplasmic ribonucleoprotein complexes containing human LINE-1 protein and RNA. EMBO J 15:630–639 [PMC free article] [PubMed] [Google Scholar]
- ——— (1997) Sequence-specific single-strand RNA binding protein encoded by the human LINE-1 retrotransposon. EMBO J 16:6034–6043 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holmes SE, Dombroski BA, Krebs CM, Boehm CD, Kazazian HH Jr (1994) A new retrotransposable human L1 element from the LRE2 locus on chromosome 1q produces a chimaeric insertion. Nat Genet 7:143–148 [DOI] [PubMed] [Google Scholar]
- Kass DH, Batzer MA, Deininger PL (1995) Gene conversion as a secondary mechanism of short interspersed element (SINE) evolution. Mol Cell Biol 15:19–25 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kazazian HH Jr, Wong C, Youssoufian H, Scott AF, Phillips DG, Antonarakis SE (1988) Haemophilia A resulting from de novo insertion of L1 sequences represents a novel mechanism for mutation in man. Nature 332:164–166 [DOI] [PubMed] [Google Scholar]
- Kent WJ (2002) BLAT—the BLAST-like alignment tool. Genome Res 12:656–664 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kimberland ML, Divoky V, Prchal J, Schwahn U, Berger W, Kazazian HH Jr (1999) Full-length human L1 insertions retain the capacity for high frequency retrotransposition in cultured cells. Hum Mol Genet 8:1557–1560 [DOI] [PubMed] [Google Scholar]
- Lander ES, Linton LM, Birren B, Nusbaum C, Zody MC, Baldwin J, Devon K, et al (2001) Initial sequencing and analysis of the human genome. Nature 409:860–921 [DOI] [PubMed] [Google Scholar]
- Lavrentieva I, Broude NE, Lebedev Y, Gottesman II, Lukyanov SA, Smith CL, Sverdlov ED (1999) High polymorphism level of genomic sequences flanking insertion sites of human endogenous retroviral long terminal repeats. FEBS Lett 443:341–347 [DOI] [PubMed] [Google Scholar]
- Luan DD, Eickbush TH (1995) RNA template requirements for target DNA-primed reverse transcription by the R2 retrotransposable element. Mol Cell Biol 15:3882–3891 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luan DD, Korman MH, Jakubczak JL, Eickbush TH (1993) Reverse transcription of R2Bm RNA is primed by a nick at the chromosomal target site: a mechanism for non-LTR retrotransposition. Cell 72:595–605 [DOI] [PubMed] [Google Scholar]
- Mamedov I, Batrak A, Buzdin A, Arzumanyan E, Lebedev Y, Sverdlov ED (2002) Genome-wide comparison of differences in the integration sites of interspersed repeats between closely related genomes. Nucleic Acids Res 30:e71 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mathias SL, Scott AF, Kazazian HH Jr, Boeke JD, Gabriel A (1991) Reverse transcriptase encoded by a human transposable element. Science 254:1808–1810 [DOI] [PubMed] [Google Scholar]
- Moran JV (1999) Human L1 retrotransposition: insights and peculiarities learned from a cultured cell retrotransposition assay. Genetica 107:39–51 [PubMed] [Google Scholar]
- Moran JV, Gilbert N (2002) Mammalian LINE-1 retrotransposons and related elements. In: Craig N, Craggie R, Gellert M, Lambowitz A (eds) Mobile DNA II. ASM Press, Washington, DC, pp 836–869 [Google Scholar]
- Moran JV, Holmes SE, Naas TP, DeBerardinis RJ, Boeke JD, Kazazian HH Jr (1996) High frequency retrotransposition in cultured mammalian cells. Cell 87:917–927 [DOI] [PubMed] [Google Scholar]
- Myers JS, Vincent BJ, Udall H, Watkins WS, Morrish TA, Kilroy GE, Swergold GD, Henke J, Henke L, Moran JV, Jorde LB, Batzer MA (2002) A comprehensive analysis of recently integrated human Ta L1 elements. Am J Hum Genet 71:312–326 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naas TP, DeBerardinis RJ, Moran JV, Ostertag EM, Kingsmore SF, Seldin MF, Hayashizaki Y, Martin SL, Kazazian HH Jr (1998) An actively retrotransposing, novel subfamily of mouse L1 elements. EMBO J 17:590–597 (erratum 20:2608 [2001]) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ovchinnikov I, Troxel AB, Swergold GD (2001) Genomic characterization of recent human LINE-1 insertions: evidence supporting random insertion. Genome Res 11:2050–2058 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pearson WR, Lipman DJ (1988) Improved tools for biological sequence comparison. Proc Natl Acad Sci USA 85:2444–2448 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sassaman DM, Dombroski BA, Moran JV, Kimberland ML, Naas TP, DeBerardinis RJ, Gabriel A, Swergold GD, Kazazian HH Jr (1997) Many human L1 elements are capable of retrotransposition. Nat Genet 16:37–43 [DOI] [PubMed] [Google Scholar]
- Saxton JA, Martin SL (1998) Recombination between subtypes creates a mosaic lineage of LINE-1 that is expressed and actively retrotransposing in the mouse genome. J Mol Biol 280:611–622 [DOI] [PubMed] [Google Scholar]
- Schneider S, Roessli D, Excoffier L (2000) Arlequin, version 2.000: a software for population genetics data analysis. Genetics and Biometry Laboratory, University of Geneva, Geneva [Google Scholar]
- Scott AF, Schmeckpeper BJ, Abdelrazik M, Comey CT, O’Hara B, Rossiter JP, Cooley T, Heath P, Smith KD, Margolet L (1987) Origin of the human L1 elements: proposed progenitor genes deduced from a consensus DNA sequence. Genomics 1:113–125 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheen F, Sherry ST, Risch GM, Robichaux M, Nasidze I, Stoneking M, Batzer MA, Swergold GD (2000) Reading between the LINEs: human genomic variation induced by LINE-1 retrotransposition. Genome Res 10:1496–1508 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skowronski J, Fanning TG, Singer MF (1988) Unit-length LINE-1 transcripts in human teratocarcinoma cells. Mol Cell Biol 8:1385–1397 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smit AF, Toth G, Riggs AD, Jurka J (1995) Ancestral, mammalian-wide subfamilies of LINE-1 repetitive sequences. J Mol Biol 246:401–417 [DOI] [PubMed] [Google Scholar]
- Takai D, Yagi Y, Habib N, Sugimura T, Ushijima T (2000) Hypomethylation of LINE1 retrotransposon in human hepatocellular carcinomas, but not in surrounding liver cirrhosis. Jpn J Clin Oncol 30:306–309 [DOI] [PubMed] [Google Scholar]
- Thompson JD, Higgins DG, Gibson TJ (1994) CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res 22:4673–4680 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei W, Gilbert N, Ooi SL, Lawler JF, Ostertag EM, Kazazian HH Jr, Boeke JD, Moran JV (2001) Human L1 retrotransposition: cis preference versus trans complementation. Mol Cell Biol 21:1429–1439 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei W, Morrish TA, Alisch RS, Moran JV (2000) A transient assay reveals that cultured human cells can accommodate multiple LINE-1 retrotransposition events. Anal Biochem 284:435–438 [DOI] [PubMed] [Google Scholar]