Abstract
We report a nonepisodic autosomal dominant (AD) spinocerebellar ataxia (SCA) not caused by a nucleotide repeat expansion that is, to our knowledge, the first such SCA. The AD SCAs currently comprise a group of ⩾16 genetically distinct neurodegenerative conditions, all characterized by progressive incoordination of gait and limbs and by speech and eye-movement disturbances. Six of the nine SCAs for which the genes are known result from CAG expansions that encode polyglutamine tracts. Noncoding CAG, CTG, and ATTCT expansions are responsible for three other SCAs. Approximately 30% of families with SCA do not have linkage to the known loci. We recently mapped the locus for an AD SCA in a family (AT08) to chromosome 19q13.4-qter. A particularly compelling candidate gene, PRKCG, encodes protein kinase C γ (PKCγ), a member of a family of serine/threonine kinases. The entire coding region of PRKCG was sequenced in an affected member of family AT08 and in a group of 39 unrelated patients with ataxia not attributable to trinucleotide expansions. Three different nonconservative missense mutations in highly conserved residues in C1, the cysteine-rich region of the protein, were found in family AT08, another familial case, and a sporadic case. The mutations cosegregated with disease in both families. Structural modeling predicts that two of these amino acid substitutions would severely abrogate the zinc-binding or phorbol ester–binding capabilities of the protein. Immunohistochemical studies on cerebellar tissue from an affected member of family AT08 demonstrated reduced staining for both PKCγ and ataxin 1 in Purkinje cells, whereas staining for calbindin was preserved. These results strongly support a new mechanism for neuronal cell dysfunction and death in hereditary ataxias and suggest that there may be a common pathway for PKCγ-related and polyglutamine-related neurodegeneration.
Introduction
The nonepisodic autosomal dominant (AD) spinocerebellar ataxias (SCAs) share the clinical features of progressive incoordination of gait, dysarthria, dysphagia, and limb dysmetria, which are often associated with abnormal eye movements. Additional features, such as mental retardation, retinopathy, peripheral neuropathy, or myoclonus, distinguish some subtypes of the hereditary ataxias (Rosenberg 1995; Mariotti and DiDonato 2001; also see “Hereditary Ataxia Overview,” by T.D.B., in GeneReviews, available at the GeneTests Home Page). The clinical presentation reflects degeneration of the cerebellar cortex and various other regions of the CNS. The incidence of the disease is ∼1–5/100,000, with an average age at onset in the 3rd decade of life.
Trinucleotide repeat expansions, (CAG)n, that encode polyglutamine tracts are responsible for SCA1, SCA2, SCA3, SCA6, SCA7, and SCA17, whereas expanded CTG, ATTCT, and noncoding CAG repeats are responsible for SCA8, SCA10, and SCA12, respectively. In North American populations, ∼30% of families with SCA do not have linkage to the known loci (Moseley et al. 1998). We recently mapped the locus for an AD SCA, without other defining clinical features, in a family (AT08) to a 22-cM region on chromosome 19q13.4-qter (Brkanac et al. 2002). A maximum two-point LOD score of 2.57 was obtained for marker D19S254 at recombination fraction 0, and a multipoint LOD score of 4.72 was obtained at D19S926. Among the >300 genes and ESTs in the minimal region listed in the National Center for Biotechnology Information (NCBI) database on December 20, 2001 (for the NCBI Map Viewer, see the Entrez Genome View Web site), one particularly compelling candidate gene was PRKCG (MIM 179680), encoding protein kinase C γ (PKCγ).
PKCγ is a member of the conventional subgroup of a serine/threonine kinase family (Coussens et al. 1986; Knopf et al. 1986) that plays a role in such diverse processes as signal transduction, cell proliferation and differentiation, synaptic transmission, and tumor promotion (e.g., see Tanaka and Nishizuka 1994; Zeidman et al. 1999; Newton 2001). The PKC isozymes differ in their tissue distributions and responsiveness to calcium and phospholipids (e.g., see Tanaka and Nishizuka 1994; Ho et al. 2001). The amino-terminal regulatory domain of PKCγ contains two cysteine-rich regions (Cys1 and Cys2), collectively termed “C1” (each of which interacts with two zinc ions and provides high-affinity diacylglycerol [DAG]/phorbol ester binding), and a Ca2+-sensitive region, termed “C2” (figs. 1A and 1B) (for review, see Newton 2001). The carboxyl-terminal catalytic domain contains kinase and substrate-recognition regions.
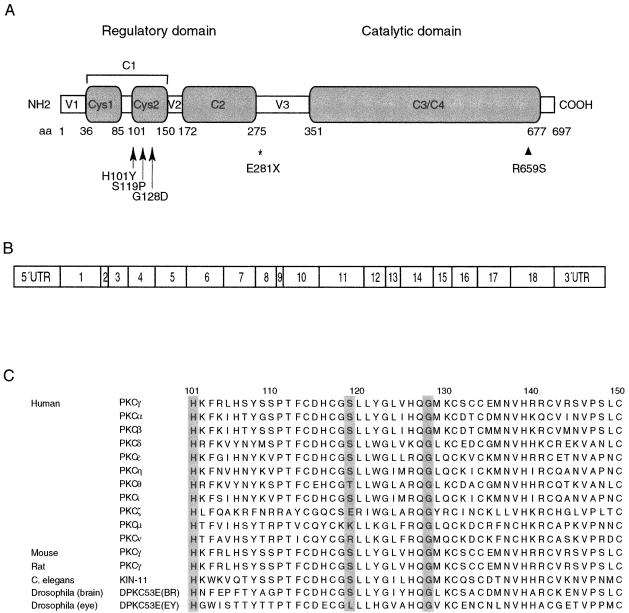
Figure 1.
Organization of PRKCG and corresponding protein. A, Functional domains, showing the conserved (C) and variable (V) regions. The amino acid (aa) boundaries of the regions are shown as predicted in the SMART database. The mutations found in the present study are indicated by arrows. The position of a reported mutation in retinitis pigmentosa (Al-Maghtheh et al. 1998) is indicated by an arrowhead, and the relative position of the rat agu mutation (Craig et al. 2001) is indicated by an asterisk. B, Exonic organization. C, Amino acid sequence alignment in the Cys2 region, showing striking evolutionary conservation of PKCγ and other isozymes of PKC at three mutation sites found in the present study (shown in gray).
Tissue-expression patterns and rodent models strongly suggested PKCγ as a candidate for the chromosome 19q13.4-qter ataxia locus. In vertebrates, the expression of PKCγ is high in brain and spinal cord (Saito et al. 1988), with particularly high expression in Purkinje cells of the cerebellar cortex (Barmack et al. 2000). PKCγ-knockout mice have mildly abnormal gait and incoordination (Abeliovich et al. 1993; C. Chen et al. 1995) associated with abnormal persistence of multiple-climbing-fiber innervation onto Purkinje cells (C. Chen et al. 1995). The spontaneously arising nonsense mutation in PKCγ in agu rats produces a recessive parkinsonian-like movement disorder accompanied by progressively more impaired, staggering gait (Craig et al. 2001). Transgenic mice that overexpress ataxin 1 containing an expanded polyglutamine tract develop pathological changes in Purkinje cells and a progressive ataxia similar to that seen in patients with SCA1 (Burright et al. 1995; Clark et al. 1997). In these SCA1 transgenic mice, PKCγ protein concentration is decreased and mislocalized from the Purkinje cell dendritic membrane into cytoplasmic vacuoles (Skinner et al. 2001). For these reasons, we evaluated PRKCG for DNA sequence alterations in family AT08.
Subjects and Methods
Patients
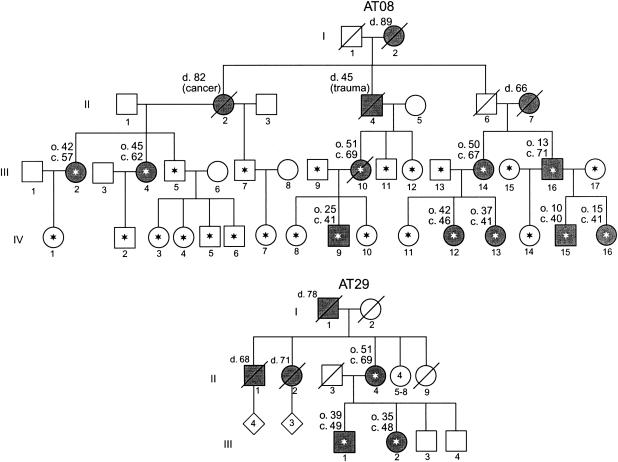
Subjects and families referred to the Medical Genetics Clinic at the University of Washington Medical Center or seen in the Neurology Clinic at the Puget Sound Veterans Affairs Hospital were evaluated by one of us (T.D.B.) and were included in the study if they met a clinical diagnosis of unexplained cerebellar ataxia. Subjects gave informed consent for blood draw and DNA studies under protocols approved by the University of Washington institutional review board. Families AT08 and AT29 have a relatively uncomplicated form of cerebellar ataxia and no apparent decrease of life span. Additional clinical details of family AT08 can be found elsewhere (Brkanac et al. 2002). Pedigrees of family AT08 and another multiplex family in which PRKCG mutations were found are shown in figure 2.
Figure 2.
Pedigrees of two multiplex kindreds in which PRKCG mutations were identified. Asterisks denote subjects who provided blood samples. Symbols representing clinically affected individuals are blackened, and deceased individuals are indicated by a slash mark. Age at death (d.), approximate age at onset of symptoms (o.), and current age (c.) are shown, in years, above and to the left of the relevant symbols.
Mutation/Polymorphism Detection
DNA was isolated from peripheral blood, and the 18 exons and splice junctions of the PRKCG gene were PCR amplified with AmpliGold Taq (Applied Biosystems), using the following nine intronic primer pairs (Integrated DNA Technologies): exons 1 and 2, forward (ctgcctttggctcttcct) and reverse (cagcctccaccctcctga) (1,016 bp); exon 3, forward (cgctctctctttccaatttt) and reverse (gaggaggagaaccaggtgt) (203 bp); exons 4 and 5, forward (caaggcaggaggaaaagata) and reverse (acaagtgccttgggtcag) (542 bp); exon 6, forward (gcttggaactcttgattgct) and reverse (ccactaggaccctcagatca) (310 bp); exons 7–9, forward (acctccagcaccaaggat) and reverse (atgtgtggggaattgaagac) (1,059 bp); exons 10 and 11, forward (ttgggagcatttccttatcg) and reverse (ctcgccctaaactcagaatc) (825 bp); exons 12–14, forward (gtctgatagttggcggtggt) and reverse (cagtgccaagctcacctg) (896 bp); exons 15 and 16, forward (gggaagagcttgtgctgaaa) and reverse (tcaggaatgggagcattttt) (1,880 bp); and exons 17 and 18, forward (ttctctgggtctacctgtcc) and reverse (ttagtggtgtggtctctgga) (932 bp). The amplification protocol included an initial denaturation at 95°C for 5 min; 32 cycles of 94°C for 30 s, 56°C (or 54°C in some fragments) for 45 s, and 72°C for 60 s; and a final extension at 72°C for 10 min. PCR-amplified fragments were cleaned by exonuclease I and shrimp alkaline phosphatase digestion using ExoSAP-IT (USB Corporation). Direct DNA sequencing of the purified fragments was performed as described elsewhere (D. H. Chen et al. 2002) with either the same forward primers as in the PCR amplifications or the following primers within the amplimers: exon 2 (ctggattcctgggtctgaag), exons 8 and 9 (cttccaatgtctttgcctct), exons 13 and 14 (atccagccactgaccttct), exon 16 (ggcatccgagataggaaatg), and exon 18 (cagacaccatgaagcatgaata). For confirmation of the sequence alterations, exon 4 was also sequenced in reverse.
Radioisotope dideoxy sequencing with two bases (wild-type C and mutant T) was performed to evaluate the cosegregation of ataxia and the mutation in family AT08 and to screen 96 normal control individuals for a possible polymorphism. The forward primer for exon 4 was end-labeled with [γ32]P by a T4 kinase reaction, and sequencing was performed with the AmpliCycle Sequencing Kit (Applied Biosystems). The sequencing products were electrophoresed at 50°C on 6% polyacrylamide gels containing 7 M urea.
The single-nucleotide substitutions 355T→C and 383G→A altered the restriction endonuclease digestion patterns of HaeIII and MwoI, respectively. RFLP analyses with these enzymes (New England Biolabs) were performed on 260-bp exon 4 fragments (PCR amplified with the forward primer given above and the reverse primer atttcccggaacccagac) from 96 normal control individuals, under the conditions suggested by the manufacturer. Restriction fragments were separated on 3% agarose gels. HaeIII digestion generated fragments of 11, 25, 36, and 188 bp in wild type and 11, 25, 36, 58, and 130 bp in 355T→C mutants. MwoI digestion generated fragments of 55, 59, 72, and 74 bp in wild type and 55, 59, and 146 bp in 383G→A mutants. An additional 96 normal control individuals were evaluated for all three sequence changes in exon 4 by fluorescence-labeling sequencing methods.
Protein-Structure Modeling
Residues 100–153 of the nuclear magnetic resonance (NMR) structure of PKCγ (i.e., only the ordered residues; see Xu et al. 1997) were used for the molecular dynamics (MD) simulations. Initial mutant structures were created with the Insight97 program (Accelrys) by replacing the original residues with the best-fitting rotamers. The protein structures were inserted into a box of TIP3P waters with boundaries ⩾10 Å away from any protein atom. Simulations were performed with the Amber program (Pearlman et al. 1995), using the Cornell95 force field (Cornell et al. 1995), a constant dielectric ɛ=1.0. Zinc ions were modeled using the cationic dummy method of Pang et al. (2000). The particle-mesh Ewald method provided a proper treatment of the long-range electrostatic forces, and a cutoff of 10 Å was applied to van der Waals forces. Bonds were constrained with the Shake algorithm. Each solvated protein was energy minimized (5,500 cycles) and was then heated to 298 K (50 ps) and equilibrated (150 ps). This was followed by a 1.0-ns MD simulation (NVE ensemble; 1-fs time steps), to study the behavior of the proteins.
Immunohistochemistry
Microtome sections (5 μm) were cut from paraffin-embedded sections of cerebellar tissue. For enhancement of the immune reaction, the slides were pretreated by boiling in Na citrate for both ataxin 1 and ubiquitin staining and by protease digestion for PKCγ staining. Standard avidin-biotin complex immunochemical staining was performed. Mouse monoclonal anti-PKCγ (ZyMed Laboratories), rabbit polyclonal anti–ataxin 1 (ZyMed Laboratories), rabbit anti-calbindin (Sigma-Aldrich), and rabbit anti-ubiquitin (East Acres Biologicals) were used at dilutions of 1:20, 1:50, 1:2,000, and 1:8,000, respectively.
Results
Identification of PRKCG Mutations in Patients with Ataxia
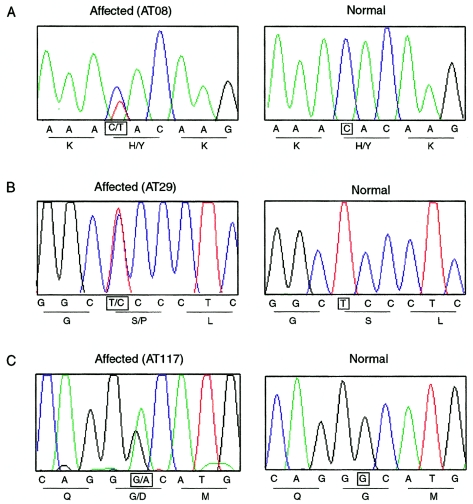
The entire coding region of PRKCG was sequenced in genomic DNA from an affected individual (IV-13) in family AT08 (fig. 2). A C→T transition in nucleotide 301, counted from the start codon in mRNA (GenBank accession number NM_002739), was detected in exon 4, predicting substitution of hydrophilic tyrosine for hydrophobic histidine at position 101 (H101Y) (fig. 3A). All individuals from whom DNA samples were obtained were screened for the mutation. The mutation segregated with disease in all nine other affected members of the family. Two clinically unaffected at-risk individuals in their 20s also inherited the mutation (not indicated on the pedigree, for consideration of confidentiality). The remaining 13 at-risk individuals and 4 spouses had the wild-type sequence. This histidine residue, in the Cys2 region, is evolutionarily conserved in all mammals and invertebrates studied and in all Cys2 regions in the PKC family (fig. 1C). The C→T nucleotide change was not found in 192 normal control individuals (384 chromosomes). In comparing the residue numbers in the literature, we noticed that there is a discrepancy between the numbering shown for rat PKCγ by Knopf et al. (1986) and the NCBI database (GenBank accession number NM_002739). Knopf et al. inserted a space at position 14 in the rat molecule to allow alignment of the residues of several PKC isoforms. Because some authors referred directly or indirectly to the article by Knopf et al., beyond residue 13, their numbering is one higher for the rat and human PKCγ proteins. We used the numbering given in the NCBI database.
Figure 3.
Sequence chromatograms for portions of PRKCG exon 4, showing heterozygous mutations in affected individuals from three families with SCA as compared to control individuals. A, A C→T transition in nucleotide 301 in family AT08. B, A T→C transition in nucleotide 355 in family AT29. C, A G→A transition in nucleotide 383 in family AT117. The relevant nucleotides are highlighted by squares. Predicted amino acids are also shown under the corresponding nucleotide sequence.
PRKCG was then sequenced in 39 additional subjects with ataxia, 27 with positive family histories and 12 with apparently sporadic disease. All subjects had previously tested negative for expansions in the genes for SCA1, SCA2, SCA3, and SCA6. Twenty-seven subjects had also tested negative for abnormal alleles of SCA7 and SCA8. Two other mutations in exon 4 were detected that affect highly conserved residues in the Cys2 region (figs. 3B and 3C). A T→C transition in nucleotide 355, predicting a serine-to-proline substitution at residue 119 (S119P), was found in an affected woman and her affected son and daughter (mean age at onset 42 years, range 35–51 years; family AT29 in fig. 2). A G→A transition in nucleotide 383, predicting a glycine-to-aspartate substitution at residue 128 (G128D), was found in a 55-year-old man with onset in his early 20s who had no family history of ataxia (family AT117); his father and mother died at ages 83 and 54 years, respectively. The serine and glycine residues are also conserved in all mammalian Cys2 regions and most PKC family members (fig. 1C). One hundred ninety-two control samples (384 chromosomes) were tested, and none exhibited either of these two single-nucleotide changes.
Solution-Structure Modeling
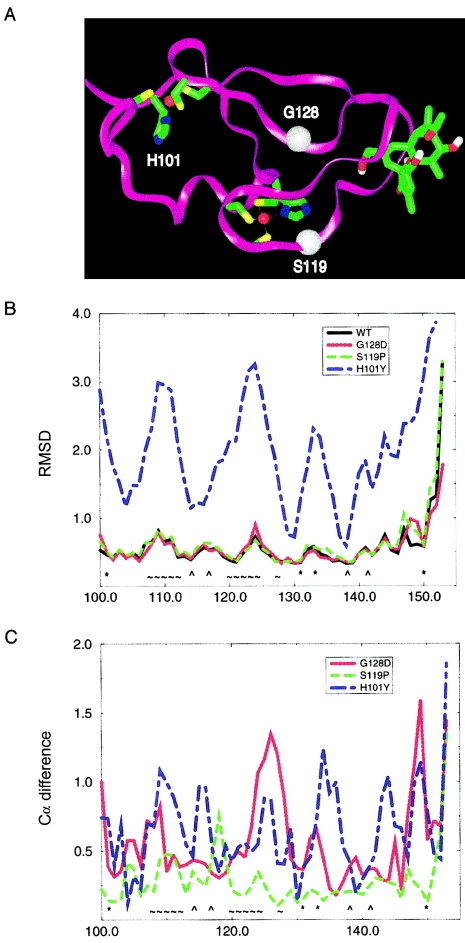
Residues involved in zinc and phorbol ester binding in the C1 domain of rat PKCγ and PKCδ were previously identified by in vitro binding assays (Quest et al. 1994; Kazanietz et al. 1995; Xu et al. 1997) and were confirmed by NMR spectroscopy (Xu et al. 1997) and x-ray crystallography (Zhang et al. 1995). We compared the molecular structure of each of the three mutants to the wild-type structure through MD simulations in water at room temperature for the Cys2 region of PKCγ (fig. 4). Because the phorbol ester binding site is partially collapsed in the PKCγ NMR structure when it is devoid of a ligand (Xu et al. 1997), the MD simulations were repeated with the following modification: the conformation of this binding site was altered into the conformation seen in the crystal structure of the homologous PKCδ phorbol acetate complex (Zhang et al. 1995). Phorbol acetate fit perfectly in this new PKCγ conformation. This shortcut was applied because the transition between these two conformational states typically takes longer than 1 μs. The results of both sets of simulations were virtually identical; however, only the second set is appropriate for the evaluation of phorbol ester binding. For conciseness, only the results of the second set of simulations are shown in figure 4. Our solution-structure modeling predicts that two of the three mutations detected in patients with ataxia would have a dire effect on the function of the PKCγ protein. The H101Y protein conformation fluctuated wildly during the simulation, with protein backbone atoms moving >3.0 Å, especially in the Zn1 and phorbol ester binding sites, whereas the wild type and the two other mutant proteins were stable (fig. 4B). Our modeling predictions are consistent with previous in vitro functional studies of the Cys2 region of protein kinase C. A deletion study (Quest et al. 1994) of the phorbol binding domain of rat PKCγ identified this residue (which was denoted as “His102”) as critical for both zinc coordination and phorbol ester binding. Substitution of glycine for histidine at the analogous position in rat PKCδ completely abolished phorbol binding (Kazanietz et al. 1995). To our knowledge, residues 119 and 128 have not been studied previously in mutational analyses. Although residue 128 is not within the phorbol ester binding site, our structural modeling predicted that the substitution of aspartate for glycine indirectly changes the shape of the binding site, with a maximum shift of >1.0 Å for Leu124 (fig. 4C). Hence, it is likely that the G128D mutation reduces the binding affinity of phorbol esters. Only a local change in protein backbone position was observed for the substitution of proline for serine at residue 119. Thus, there is no obvious structural explanation for the deleterious effects of this mutation. However, the observation that Ser119 is conserved in PKCγ in all mammals studied, as well as in PKCγ homologues in Caenorhabditis elegans and Drosophila brain and most other PKC isoforms, argues for the functional importance of this residue (fig. 1C).
Figure 4.
MD studies of wild-type and mutant Cys2 regions of PKCγ. A,Three-dimensional structure of the Cys2 region of wild-type PKCγ with phorbol acetate bound. H101 is a ligand in the first zinc binding site; zinc is shown in red. S119 is close to the second zinc binding site, as well as the phorbol acetate binding site. G128 is also in the vicinity of the phorbol acetate binding site; Cα positions of S119 and G128 are indicated by white spheres. B, Fluctuations of protein backbone coordinates per residue, expressed as root-mean-square deviation (RMSD) (in Å) for wild type and three mutants. Note the instability of the H101Y mutant in comparison to wild type and the two other mutants. C, Differences in average structures, calculated as distance between Cα positions (in Å) per residue between wild type and each of the three mutants. Note the global shift of the structure in the H101Y mutant and the shifts in the phorbol ester binding site in the G128D mutant. WT = wild type; * = first zinc binding site; ⁁ = second zinc binding site; ∼ = phorbol binding site.
Immunohistochemical Studies
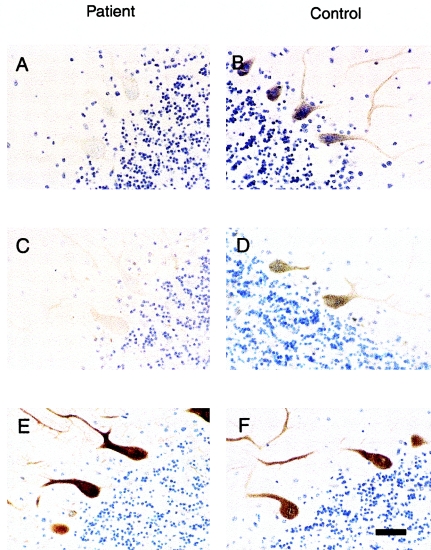
We examined PKCγ immunoreactivity in cerebellar tissue from an affected person (II-7) in family AT08 who died of a ruptured left-middle cerebral artery aneurysm at age 66 years (fig. 2). Limited postmortem brain was available, and there was no information on the time between death and fixation of tissues. There was selective marked loss of Purkinje cells in the cerebellum (Brkanac et al. 2002). Immunohistochemical studies showed that the residual Purkinje cells all stained very poorly for PKCγ relative to control cerebellum, although there was cell-to-cell variability (figs. 5A and 5B). Because PKCγ has been reported to be reduced in Purkinje cells in SCA1 transgenic mice (Skinner et al. 2001), we were interested in determining the expression of ataxin 1 in the cerebellum of the patient with mutant PKCγ. The majority of Purkinje cells showed lack of staining for ataxin 1, and only a minority retained faint cytoplasmic staining (figs. 5C and 5D). Staining was normal for calbindin, a protein that is highly expressed in Purkinje cells (figs. 5E and 5F), demonstrating that the observations with PKCγ and ataxin 1 were unlikely to be nonspecific sequelae of cell dysfunction and death. A hallmark pathological feature of several SCAs is the aggregation of the expanded protein in ubiquitin-positive nuclear inclusions; such inclusions were not detected in the patient whom we studied (not shown).
Figure 5.
Reduced immunoreactivity for PKCγ and ataxin 1 in an ataxic patient from family AT08 who carries the H101Y mutation. A and B, PKCγ immunohistochemistry. The scattered surviving Purkinje cells in a postmortem cerebellar section showed reduced but variable staining for antibody to PKCγ (A), compared with intense staining in membrane and dendrites from an age-matched control (B). C and D, Ataxin 1 immunohistochemistry. The majority of Purkinje cells from the same ataxic patient showed no immunostaining for ataxin 1, but a minority of cells were weakly positive. Control Purkinje cells were intensely stained in cytoplasm (D). E and F, Calbindin immunohistochemistry. Staining for calbindin was similar in the ataxic patient and the control individual, demonstrating that the loss of PKCγ and ataxin 1 did not represent nonspecific decreased protein synthesis. Scale bar = 75 μm.
Discussion
We present here evidence supporting a new mechanism for nonepisodic AD hereditary ataxia, apparently unrelated to a nucleotide repeat expansion. The missense mutations (H101Y, S119P, and G128D) in the Cys2 region of the C1 domain of PKCγ appear to cause a disorder that, on clinical grounds, is indistinguishable from other “uncomplicated” SCAs. The patients with mutations in PRKCG whom we studied displayed an adult-onset cerebellar ataxia without any differentiating features, such as cognitive decline, visual or other sensory loss, axial myoclonus, or peripheral neuropathy. The mean age at onset in the 14 persons with ataxia was 33 years (range 10–51 years), and there was no evidence for a shortened life span. Examination of the two pedigrees in figure 2 shows a trend for earlier age at onset in the youngest generation. However, this observation may be an artifact. With increased awareness of the disease in the families, the age at diagnosis in the younger generation may be more accurate than the recollected age at onset in the older generation. Full sequencing of the coding region and UTRs of PRKCG did not reveal a trinucleotide repeat tract. Although it is theoretically possible that there is a repeat expansion in an intron, it is highly unlikely that an undetected expansion is present in each of the three mutant alleles.
In the 40 clinical cases studied that were not attributed to SCA1, SCA2, SCA3, and SCA6, mutations in PKCγ are responsible for 7.5% of cases (3/40; 2 of 28 families and 1 of 12 sporadic cases). PKCγ is within the minimal region for SCA14, a disorder described in a single Japanese family and characterized by early-onset axial myoclonus (Yamashita et al. 2000). Although the phenotype of the families that we studied does not include this sign, it is possible that a mutation in a different portion of PKCγ is responsible for SCA14. As additional families are studied, a better clinical picture of the range of the complete phenotype of this type of ataxia will be developed, and genotype-phenotype correlations can then be evaluated.
There are substantial differences in the phenotypes conferred by different PKCγ mutations in humans, mice, and rats. In a candidate-gene analysis for retinitis pigmentosum type 11 (RP11), a neurodegenerative retinal disorder that maps to chromosome 19q13.4, an identical mutation, R659S, in the catalytic domain was reported in four unrelated individuals with familial retinitis pigmentosa (Al-Maghtheh et al. 1998). This alteration, affecting a conserved residue, segregated with disease in the two families available for study. However, no mutation was found in six other families with RP11 (Al-Maghtheh et al. 1998; Dryja et al. 1999). None of the PKCγ-mutation carriers in the families reported here has experienced unusual visual loss, and an electroretinogram performed as part of a previous clinical evaluation of the affected sporadic case (in family AT117) was normal.
The phenotype of the recessive agu rat resembles that of SCA only in clumsy gait (Craig et al. 2001). None of the affected individuals in our study show features of parkinsonism, such as tremor, rigidity, or bradykinesia. The agu mutation results in a protein truncated to 280 amino acids, missing the entire catalytic domain of PKCγ. The amino-terminal regulatory portion of PKCγ was not studied to determine if it could be detected in neurons or if it aggregated improperly. Although the loss of PKCγ kinase activity likely contributes to this neurological phenotype, the differences in the phenotypes of the knockout mouse and the homozygous agu-mutant rat suggest a role for a dysfunctional amino-terminal moiety of the protein.
It is interesting that the neurological phenotype of the heterozygous mutation in the patients described in the present article appears to be more severe than that of the knockout mouse (Abeliovich et al. 1993; C. Chen et al. 1995). This observation makes it unlikely that the enzyme is rate limiting or that the intracellular concentration of PKCγ is critical for some downstream process. Although the phenotypic difference may be an artifact of the different life spans of the two species, an alternative hypothesis is that the presence of the mutant protein produces more-toxic effects on the cells than does the absence of the protein. Perhaps the mutant proteins have a dominant negative effect by competing with the wild-type protein for attachment sites on the membrane or for substrate binding. PKCγ translocates between the plasma membrane and the cytosol during its activation cycle (Newton 2001). This process involves binding of DAG and Zn2+. The N-terminal region of PKCγ regulates the C-terminal kinase activity. Therefore, disruption of zinc and DAG binding by H101Y and G128D may indirectly alter the enzyme’s phosphorylation function. It is also possible that the mutations exert their toxic effect by increasing the intracellular concentration of Zn2+(Koh 2001). Our modeling study did not identify an effect of S119P on zinc or DAG binding sites. Perhaps the substitution of proline for serine affects the molecule's solubility, a property that cannot be predicted by solution-structure modeling. In addition, the clustering of all three mutations in exon 4 may suggest a common mechanism for pathogenicity; for example, this region of PKCγ may be critical for interaction with another functional protein.
Current understanding of the pathogeneses of polyglutamine-expansion diseases is evolving. For SCA1, it appears that the presence of mutant ataxin 1 in the nucleus is necessary and sufficient for development of disease and neuronal cell death (Klement et al. 1998). Although a hallmark pathological feature of SCA1 is aggregation of the expanded protein along with components of the ubiquitin-conjugation system in nuclear inclusions, neither aggregation nor sequestration of the protein is required. The observation that PKCγ is reduced and abnormally localized in Purkinje cells of SCA1 transgenic mice (Skinner et al. 2001) raises the possibility that it plays an essential role in the development and/or progression of this disorder. Our observation of decreased expression of ataxin 1 in Purkinje cells containing a mutation in PKCγ suggests that there may be a common pathway for neurodegeneration of these and other types of SCA.
Acknowledgments
We are grateful to the many family members who participated in these studies. Randy Small assisted with the immunohistochemical evaluations. The research was supported, in part, by funds from the Department of Veterans Affairs (support to T.D.B., H.L., W.H.R., and J.W.), National Institute of Environmental Health Sciences grant P30ES07033 (to C.L.M.J.V.), the Mary Gates Endowment for Students (to L.B. and P.J.C.), a grant from the National Aeronautics and Space Administration to the Washington Space Grant program (to L.B. and P.J.C.), P. Clementz, and J. Cook.
Note added in manuscript.—After this article was accepted for publication, an article describing a missense mutation in the fibroblast growth factor 14 gene in a single family with an ataxia syndrome was published by van Swieten et al. (2003).
Electronic-Database Information
The accession number and URLs for data presented herein are as follows:
- Entrez Genome View, http://www.ncbi.nlm.nih.gov/mapview/map_search.cgi?chr=hum_chr.inf (for NCBI Map Viewer)
- GenBank, http://www.ncbi.nlm.nih.gov/Genbank/ (for PRKCG mRNA sequence [accession number NM_002739])
- GeneTests Home Page, http://www.geneclinics.org/ or http://www.genetests.org/ (for “Hereditary Ataxia Overview,” by T.D.B., in GeneReviews)
- Online Mendelian Inheritance in Man (OMIM), http://www.ncbi.nlm.nih.gov/Omim/ (for PRKCG) [PubMed]
- SMART, http://smart.embl-heidelberg.de/ (for domain prediction within PKCγ)
References
- Abeliovich A, Paylor R, Chen C, Kim JJ, Wehner JM, Tonegawa S (1993) PKC γ mutant mice exhibit mild deficits in spatial and contextual learning. Cell 75:1263–1271 [DOI] [PubMed] [Google Scholar]
- Al-Maghtheh M, Vithana EN, Inglehearn CF, Moore T, Bird AC, Bhattacharya SS (1998) Segregation of a PRKCG mutation in two RP11 families. Am J Hum Genet 62:1248–1252 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barmack NH, Qian Z, Yoshimura J (2000) Regional and cellular distribution of protein kinase C in rat cerebellar Purkinje cells. J Comp Neurol 427:235–254 [DOI] [PubMed] [Google Scholar]
- Brkanac Z, Bylenok L, Fernandez M, Matsushita M, Lipe H, Wolff, J, Nochlin D, Raskind WH, Bird TD (2002) A new dominant spinocerebellar ataxia is linked to chromosome 19q13.4. Arch Neurol 59:1291–1295 [DOI] [PubMed] [Google Scholar]
- Burright EN, Clark HB, Servadio A, Matilla T, Feddersen RM, Yunis WS, Duvick LA, Zoghbi HY, Orr HT (1995) SCA1 transgenic mice: a model for neurodegeneration caused by an expanded CAG trinucleotide repeat. Cell 82:937–948 [DOI] [PubMed] [Google Scholar]
- Chen C, Kano M, Abeliovich A, Chen L, Bao S, Kim JJ, Hashimoto K, Thompson RF, Tonegawa S (1995) Impaired motor coordination correlates with persistent multiple climbing fiber innervation in PKCγ mutant mice. Cell 83:1233–1242 [DOI] [PubMed] [Google Scholar]
- Chen DH, Lipe HP, Qin Z, Bird TD (2002) Cerebral cavernous malformation: novel mutation in a Chinese family and evidence for heterogeneity. J Neurol Sci 196:91–96 [DOI] [PubMed] [Google Scholar]
- Clark HB, Burright EN, Yunis WA, Larson S, Wilcox C, Hartman B, Matilla A, Zoghbi HY, Orr HT (1997) Purkinje cell expression of a mutant allele of SCA1 in transgenic mice leads to disparate effects on motor behaviors, followed by progressive cerebellar dysfunction and histological alterations. J Neurosci 17:7385–7395 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cornell WD, Cieplak P, Bayly CI, Gould IR, Merz KM, Ferguson DM, Spellmeyer DC, Fox T, Caldwell JW, Kollman PA (1995) A second generation force field for the simulation of proteins, nucleic acids, and organic molecules. J Am Chem Soc 117:5179–5197 [Google Scholar]
- Coussens L, Parker PJ, Rhee L, Yang-Feng TL, Chen E, Waterfield MD, Francke U, Ullrich A (1986) Multiple, distinct forms of bovine and human protein kinase C suggest diversity in cellular signaling pathways. Science 233:859–866 [DOI] [PubMed] [Google Scholar]
- Craig NJ, Durán Alonso MB, Hawker KL, Shiels P, Glencorse TA, Campbell JM, Bennett NK, Canham M, Donals D, Gardiner M, Gilmore DP, MacDonald RJ, Maitland K, McCallion AS, Russell D, Payne AP, Sutcliffe RG, Davies RW (2001) A candidate gene for human neurodegenerative disorders: a rat PKCγ mutation causes a parkinsonian syndrome. Nat Neurosci 4:1061–1062 [DOI] [PubMed] [Google Scholar]
- Dryja TP, McEvoy J, McGee TL, Berson EL (1999) No mutations in the coding region of the PRKCG gene in three families with retinitis pigmentosa linked to the RP11 locus on chromosome 19q. Am J Hum Genet 65:926–928 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ho C, Slater SJ, Stagliano B, Stubbs CD (2001) The C1 domain of protein kinase C as a lipid bilayer surface sensing module. Biochemistry 40:10334–10341 [DOI] [PubMed] [Google Scholar]
- Kazanietz MG, Wang S, Milnes GWA, Lewin NE, Liu HL, Blumberg PM (1995) Residues in the second cysteine-rich region of protein kinase C δ relevant to phorbol ester binding as revealed by site-directed mutagenesis. J Biol Chem 270:21852–21859 [DOI] [PubMed] [Google Scholar]
- Klement IA, Skinner PJ, Kaytor MD, Yi H, Hersch SM, Clark HB, Zoghbi H, Orr HT (1998) Ataxin-1 nuclear localization and aggregation: role in polyglutamine-induced disease in SCA1 transgenic mice. Cell 95:41–53 [DOI] [PubMed] [Google Scholar]
- Knopf J, Lee M, Sultzman L, Kriz R, Loomis C, Hewick R, Bell R (1986) Cloning and expression of multiple protein kinase C cDNAs. Cell 46:491–502 [DOI] [PubMed] [Google Scholar]
- Koh JY (2001) Zinc and disease of the brain. Mol Neurobiol 24:99–106 [DOI] [PubMed] [Google Scholar]
- Mariotti C, DiDonato S (2001) Cerebellar/spinocerebellar syndromes. Neurol Sci 22 Suppl 2:S88–S92 [DOI] [PubMed] [Google Scholar]
- Moseley ML, Benzow KA, Schut LJ, Bird TD, Gomez CM, Barkhaus PE, Blindauer KA, Labuda M, Pandolfo M, Koob MD, Ranum LP (1998) Incidence of dominant spinocerebellar and Friedreich triplet repeats among 361 ataxia families. Neurology 51:1666–1671 [DOI] [PubMed] [Google Scholar]
- Newton AC (2001) Protein kinase C: structural and spatial regulation by phosphorylation, cofactors, and macromolecular interactions. Chem Rev 101:2353–2364 [DOI] [PubMed] [Google Scholar]
- Pang YP, Xu K, Yazal JE, Prendergas FG (2000) Successful molecular dynamics simulation of the zinc-bound farnesyltransferase using the cationic dummy atom approach. Protein Sci 9:1857–1865 (erratum 9:2583) [PMC free article] [PubMed] [Google Scholar]
- Pearlman DA, Case DA, Caldwell JW, Ross WS, Cheatham TE, Debolt S, Ferguson D, Seibel G, Kollman PA (1995) AMBER, a package of computer-programs for applying molecular mechanics, normal-mode analysis, molecular-dynamics and free-energy calculations to simulate the structural and energetic properties of molecules. Comput Phys Commun 91:1–41 [Google Scholar]
- Quest AFG, Bardes ESG, Bell RM (1994) A phorbol ester binding domain of protein kinase Cγ: deletion analysis of the Cys2 domain defines a minimal 43–amino acid peptide. J Biol Chem 269:2961–2970 [PubMed] [Google Scholar]
- Rosenberg RN (1995) Autosomal dominant cerebellar phenotypes: the genotype has settled the issue. Neurology 45:1–5 [DOI] [PubMed] [Google Scholar]
- Saito N, Kikkawa U, Nishizuka Y, Tanaka C (1988) Distribution of protein kinase C-like immunoreactive neurons in rat brain. J Neurosci 8:369–382 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skinner PJ, Vierra-Green CA, Clark HB, Zoghbi HY, Orr HT (2001) Altered trafficking of membrane proteins in Purkinje cells of SCA1 transgenic mice. Am J Pathol 159:905–913 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka C, Nishizuka Y (1994) The protein kinase C family for neuronal signaling. Annu Rev Neurosci 17:551–567 [DOI] [PubMed] [Google Scholar]
- van Swieten JC, Brusse E, de Graaf BM, Krieger E, van de Graaf R, de Koning I, Maat-Kievit A, Leegwater P, Dooijes D, Oostra BA, Heutink P (2003) A mutation in the fibroblast growth factor 14 gene is associated with autosomal dominant cerebral ataxia. Am J Hum Genet 72:191–199 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu RX, Pawelczyk T, Xia T-H, Brown SC (1997) NMR structure of a protein kinase C-γ phorbol-binding domain and study of protein-lipid micelle interactions. Biochemistry 36:10709–10717 [DOI] [PubMed] [Google Scholar]
- Yamashita I, Sasaki H, Yabe I, Fukazawa T, Nogoshi S, Komeichi K, Takada A, Shiraishi K, Takiyama Y, Nishizawa M, Kaneko J, Tanaka H, Tsuji S, Tashiro K (2000) A novel locus for dominant cerebellar ataxia (SCA14) maps to a 10.2-cM interval flanked by D19S206 and D19S605 on chromosome 19q13.4-qter. Ann Neurol 48:156–163 [DOI] [PubMed] [Google Scholar]
- Zeidman R, Petterssson L, Sailaja PR, Truedsson E, Fagerstrom S, Pahlman S, Larsson C (1999) Novel and classical protein kinase C isoforms have different functions in proliferation, survival and differentiation of neuroblastoma cells. Int J Cancer 81:494–501 [DOI] [PubMed] [Google Scholar]
- Zhang G, Kazanietz MG, Blumberg PM, Hurley JH (1995) Crystal structure of the Cys2 activator-binding domain of protein kinase C δ in complex with phorbol ester. Cell 81:917–924 [DOI] [PubMed] [Google Scholar]