Abstract
Several theoretical studies have suggested that large samples of randomly ascertained siblings can be used to ascertain phenotypically extreme individuals and thereby increase power to detect genetic linkage in complex traits. Here, we report a genetic linkage scan using extremely discordant and concordant sibling pairs, selected from 34,580 sibling pairs in the southwest of England who completed a personality questionnaire. We performed a genomewide scan for quantitative-trait loci (QTLs) that influence variation in the personality trait of neuroticism, or emotional stability, and we established genomewide empirical significance thresholds by simulation. The maximum pointwise P values, expressed as the negative logarithm (base 10), were found on 1q (3.95), 4q (3.84), 7p (3.90), 12q (4.74), and 13q (3.81). These five loci met or exceeded the 5% genomewide significance threshold of 3.8 (negative logarithm of the P value). QTLs on chromosomes 1, 12, and 13 are likely to be female specific. One locus, on chromosome 1, is syntenic with that reported from QTL mapping of rodent emotionality, an animal model of neuroticism, suggesting that some animal and human QTLs influencing emotional stability may be homologous.
Introduction
It is commonly believed that human personalities vary enormously, but it also clear that the characterization of personality differences is challenging. Psychologists now agree that most of the variation can be explained by a small number of personality factors, including neuroticism (a measure of emotional stability), which manifests at one extreme as anxiety, depression, moodiness, low self-esteem, and diffidence (Loehlin and Nichols 1976; Zuckerman et al. 1988; Digman 1990; Deary and Matthews 1993; Cloninger 1994). It is perhaps not surprising that a number of studies have described a relationship between high scores on measures of neuroticism and major depressive disorder (Zeiss and Lewinsohn 1988; Hirschfeld et al. 1989; Duggan et al. 1990, 1995; Roy 1990); knowing more about the etiology of neuroticism may advance our understanding not only of the biology of personality but also of one of the most common mental disorders.
In common with other personality factors, genetic effects are known to account for a substantial proportion of the variation in neuroticism: the additive genetic variance is estimated to be 27%–31%, and nonadditive effects are estimated to be 14%–17% (Loehlin and Nichols 1976; Floderus-Myrhed et al. 1980; Rose et al. 1988; Eaves et al. 1989, 1998, 1999; Loehlin 1992; Lake et al. 2000). Heritability in liability to neuroticism is comparable to other complex traits that have been the subject of genome scans to identify susceptibility loci, and the genetic architecture of neuroticism is unlikely to be unusual: it is reasonable to suppose that a large number of genes each make a small contribution to the genetic variation, rendering their detection extremely difficult.
Quantitative genetic analysis of personality has also revealed a complex relationship between sex and neuroticism. There is a consistent finding of higher mean neuroticism scores in females, but, with one exception (Viken et al. 1994), studies report no difference in heritability between males and females. Three studies find evidence of sex-specific genetic factors for neuroticism (Eaves et al. 1989; Martin et al. 2000; Fanous et al. 2002), although one did not (Lake et al. 2000).
Attempts to uncover the genetic basis of complex human traits, such as neuroticism, have frequently foundered because of the small phenotypic effect attributable to each locus and the consequent need for very large sample sizes in genetic linkage studies—on the order of tens of thousands of randomly selected sibships. However, assessment of neuroticism by postal questionnaire has made it possible to obtain samples of sufficient size (Boomsma et al. 2000; Kirk et al. 2000; Martin et al. 2000; Sham et al. 2000); furthermore, the amount of genotyping required to detect a locus in such a large sample can be considerably reduced, while maintaining statistical power, by selecting those sibling pairs most likely to show deviation from an expected proportion of allele sharing (Carey and Williamson 1991; Cardon and Fulker 1994; Eaves and Meyer 1994; Risch and Zhang 1995, 1996; Heo et al. 2002). Genetically informative pairs for linkage are concordant for either extremely high or extremely low scores, or they are discordant, with one member of the pair having an extremely low score and the other having an extremely high score. Each of the three groups—concordant high, concordant low, or discordant—has different power to detect loci, depending on the genetic model. When the genetic model underlying neuroticism is unknown, selecting all three groups maximizes the chance of detecting a genetic effect (Heo et al. 2002). Here, we describe the successful identification of genetic loci influencing variation in neuroticism, detected by linkage analysis of extremely concordant and discordant sibling pairs selected from a sample of 34,580 sibling pairs in 20,427 independent sibships (Martin et al. 2000).
Subjects and Methods
Sample Ascertainment
We contacted all men and women between 30 and 50 years of age who were listed in general practitioner registers in the southwest of England. Of those contacted, 88,141 individuals agreed to return the full 90-item revised Eysenck Personality Questionnaire (EPQ) (Eysenck and Eysenck 1975). Most of the sibships (15,259/20,427 [75%]) consisted only of the proband and one additional sibling, but larger sibships—4,146, 838, 151, and 23 sibships of sizes 3, 4, 5, and 6, respectively—were also identified. The total sample thus consists of 34,580 sibling pairs in 20,427 independent sibships. A full description of the study design and EPQ results has been published elsewhere (Martin et al. 2000). Research protocols and all procedures used in the study were approved by ethical review panels in the United Kingdom.
We selected the most genetically informative pairs as follows. After transforming the scores (as described below, in the “Phenotypes” subsection), we regressed the scores on age and sex and standardized the residuals. Then, the residuals were ranked and centered for each sibling, around a mean of 0. We took the product of the mean-centered ranks for each sibling pair and selected the highest and lowest 2.5% of the rank products.
Phenotypes
The neuroticism scale of the revised EPQ consists of 23 questions scored on a two-point scale (Eysenck and Eysenck 1975). We used an arcsine transformation of the raw neuroticism scores to remove association between the mean and variance of the measure (Eaves et al. 1989). Twin studies have found a highly significant quadratic regression of absolute intrapair differences on the pair means of MZ twins, suggesting a gene-by-environment interaction (Eaves et al. 1989). The error variance is highest for intermediate values and lowest for extremes, indicating that the interaction may be due mainly to a correlation between the true score and the measurement error. The angular transformation removes this correlation and is preferred over other transformations in genetic studies (Martin and Jardine 1986). The angular transformation is formulated as 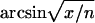 , where x is the number of items scored positively by a subject on a particular scale and n is the maximum possible score on that scale (n=23). After transformation, we regressed the transformed neuroticism scores on age and sex, and we computed standardized residual scores, which were used in subsequent linkage mapping.
, where x is the number of items scored positively by a subject on a particular scale and n is the maximum possible score on that scale (n=23). After transformation, we regressed the transformed neuroticism scores on age and sex, and we computed standardized residual scores, which were used in subsequent linkage mapping.
DNA Extraction and Preparation
Cytosoft cytology brushes were used to collect mouth swabs (Medical Packaging). DNA was extracted using BioRad Instagene Matrix, following the manufacturer’s instructions. The final DNA volume of 500 μl was diluted 40-fold for subsequent PCRs. DNA from completed families was aliquoted into 96-well plates ready for PCR amplification.
Microsatellite Genotyping
We genotyped 388 highly polymorphic markers that span all 22 autosomes. All markers came from the ABI Prism LMS2-MD10 panels (Applied Biosystems). PCR primers were labeled with 6-FAM, HEX, or TET phosphoramidite (Applied Biosystems). PCRs were performed in 96-well Costar plates in a 10-μl volume, with 40 ng of template genomic DNA, on PTC-225 thermocyclers (MJ Research). Pooled products were run on a 3700 sequencer (Applied Biosystems), and the results were analyzed by means of Genescan (version 2.0) and Genotyper (version 2.1) software, to derive allele sizes (Reed et al. 1994).
Error Checking
A number of quality control tests were performed. At the level of genotyping, each 96-well genotyping plate contained a reference individual (CEPH standard 1347-02; Coriell Institute) for quality control, plate identity, and orientation. After a genotyping run on the automated sequencing machine, manually scored genotypes and associated ABI trace files were loaded into a relational database that contained all phenotypes and family relationships. This allowed us to run error-checking procedures on the genetic data, assess raw data files, and edit genotypes when necessary, all with the same software package.
Pedigrees containing two or more typed individuals were examined for genotyping errors by use of Sibmed (Douglas et al. 2000), the error-checking option in Merlin (Abecasis et al. 2002), and Pedcheck (O'Connell and Weeks 1998). For Sibmed analysis, a prescribed false-positive rate of <0.001 was set, given a prior genotyping error rate of 0.01. Marker haplotypes were generated using Genehunter 2.0 to identify any chromosomes showing an excessive number of recombination events (Kruglyak et al. 1996). Family relationships were examined by identity-by-descent (IBD)–based methods implemented in Relative (Goring and Ott 1997) and Relpair (Epstein et al. 2000).
Individuals with suspected errors that could not be explained as errors in family relationships were regenotyped. As a further method for the detection of error, we identified pairs of individuals whose mean IBD (proportion of alleles shared IBD), across the genome, was >0.55 or <0.45. These individuals were regenotyped at all loci. Genotype trace files of inconsistent duplicate genotypes were manually reevaluated and were compared with other family members, to determine the source of the error. If no explanation could be found for the error, then genotypes for that marker for the whole family were discarded.
Statistical Analysis
Maximum-likelihood estimates of IBD posterior probabilities were estimated for each sibling pair by using the Merlin computer program (Abecasis et al. 2002). We define D2 and S2 as the squared sibling-pair trait difference and squared sibling-pair trait sum, respectively. Several authors have pointed out that D2 and S2 are independent (when the phenotype is normally distributed) and that a combination of both is more efficient than the use of either (Fulker and Cherny 1996; Amos et al. 1997; Wright 1997; Drigalenko 1998; Forrest 2001), and there have been a number of suggestions about how best to combine the phenotypes (Xu et al. 2000; Forrest 2001; Sham and Purcell 2001; Visscher and Hopper 2001; Sham et al. 2002). We used the approach suggested by Visscher and Hopper (2001), who propose regressing both D2 and S2 on 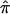 , the estimated proportion of genes IBD at the marker locus, where
, the estimated proportion of genes IBD at the marker locus, where 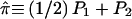 and Pj is the posterior probability that the pair shares j alleles IBD. The coefficients from the two regressions are weighted by the inverse of their variance (Visscher and Hopper 2001) and are combined to give a single measure.
and Pj is the posterior probability that the pair shares j alleles IBD. The coefficients from the two regressions are weighted by the inverse of their variance (Visscher and Hopper 2001) and are combined to give a single measure.
For the test of deviation from the expected amount of allele sharing, we first grouped all pairs as either concordant or discordant and calculated the mean proportion of alleles shared IBD at each position along the genome. We calculated the difference from the expected value (0.5) as either 0.5 subtracted from the mean IBD, for the concordant pairs, or the mean IBD subtracted from 0.5, for the discordant pairs. In this way, we obtained a consistent sign for both types of pair. We then conducted a t test by dividing this difference by the sample estimate of the SEM.
Regression and allele-sharing tests were performed using Perl code written for this purpose. The code is freely available at Jonathan Flint's Web site.
Results
Sample Characteristics
We have previously conducted a population-based study of personality, in the southwest of England, in which 34,580 sibling pairs completed the EPQ (Martin et al. 2000). Descriptive statistics for the transformed neuroticism scale in this sample are given in table 1. The data were evaluated by factorial analysis of variance, to assess mean differences as a function of sex and age. Of the total sample, 52,249 (59.3%) were female, and 35,892 (40.7%) were male. All scales showed mean differences for sex, as well as significant main effects for age. The sibling correlation in the sample is 0.171 for neuroticism.
Table 1.
Descriptive Statistics for the Neuroticism Scale of the EPQ in 12,836 Female Probands, 7,579 Male Probands, 12,581 Female Siblings, and 7,834 Male Siblings[Note]
|
Mean (SD) |
|||||
| Respondent Group | Total | Males | Females | Sex tStatistic | P |
| Probandsa | .79 (−.3) | .71 (−.3) | .83 (−.3) | 29.91 | <.001 |
| Siblings | .77 (−.3) | .69 (−.3) | .83 (−.3) | 31.688 | <.001 |
Note.— The neuroticism scale is an angular transformation of the raw neuroticism score.
Respondents in the initial cohort whose sibling(s) also responded to the questionnaire.
Following a procedure described in the “Subjects and Methods” section, we selected 408 discordant pairs, 414 concordant-low–scoring pairs and 410 concordant-high–scoring pairs, giving a total of 1,232 independent pairs. We wrote to both family members and, when possible, parents, asking them to send us a cheek swab. In total, we wrote to 3,160 people.
We obtained responses from 2,491 individuals, comprising 807 families. Although the overall response rate was high (78%), we obtained only 629 independent pairs (51% of those requested), because only one sibling in a family replied. We received swabs from both parents for 302 families and from one parent for 335 families, but we could use parental samples for only 207 of the families that had returned two extreme-scoring siblings. These parental samples were included in the genome scan. We obtained 204 discordant pairs, 224 concordant-low pairs, and 201 concordant-high pairs who were suitable for genotyping.
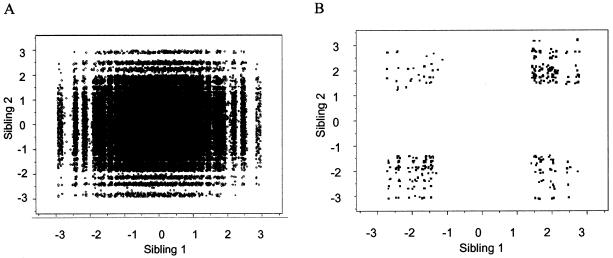
Examination of genotype data with Merlin, Relative, and Relpair resulted in exclusion of 68 pedigrees, on the grounds of demonstrable nonpaternity, half-paternity, or inadequate genotype data for the assessment of family relationships. Twenty-seven pedigrees were discarded from the extremely discordant group, whereas 19 in the concordant-low group and 22 in the concordant-high group were discarded. The final pedigree data set consisted of 182 discordant pairs (44% of those requested), 205 concordant-low pairs (49% of those requested), and 174 concordant-high pairs (42% of those requested). Figure 1 shows the distribution of the extreme-scoring sibling pairs compared to the distribution of the total sample.
Figure 1.
Scatterplots of the distribution of neuroticism scores for each sibling pair. The scale is a standardized residual of the transformed sex- and age-regressed neuroticism scores. A, Distribution of entire sample. B, Distribution of selected sample.
Genotype Characteristics
We obtained usable data from 382 markers (98% of markers), with a mean marker spacing of 10.2 cM and a mean marker heterozygosity of 77.1%. Inheritance checking was limited by the relatively small number of parents in our sample, but the inclusion of rigorous quality controls—in particular, the regenotyping of dubious genotypes—meant that the success rate for completed genotypes in the sample was 82%. Overall, 27% of genotypes were duplicated. Inconsistencies could not be resolved in 2.1% of suspected errors, or ∼0.5% of the total genotypes generated in the sample.
Regression Analysis
To find evidence for linkage, we used a regression-based approach. Regressing a score that combines the squared difference and squared sum of each sibling pair’s phenotype has power to detect genetic effects that is approximately equivalent to a variance-components analysis under normality (Sham and Purcell 2001; Visscher and Hopper 2001; Sham et al. 2002); such regression has the advantage that it is more robust to departures from normality that are due to scaling artifacts (Allison et al. 1999, 2000), and its speed of computation makes it easier to implement nonparametric tests of significance.
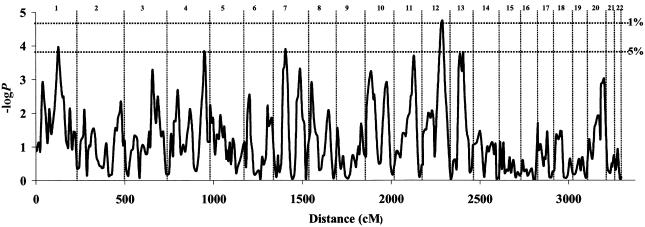
Figure 2 shows the regression results evaluated at every 5 cM across the genome. The statistic shown is the negative logarithm (base 10) of the P value (hereafter referred to as the “−logP” value) obtained from the regression. We calculated a t statistic by using the method of Visscher and Hopper (2001); equivalent results were obtained using the method of Sham and Purcell (2001) and Sham et al. (2002), which weights the squared sums and differences by the sibling correlation (0.171 in our sample) (results not shown).
Figure 2.
Multipoint linkage analysis of the genome for individual variation in neuroticism. The −logP values (vertical axis) for the Visscher-Hopper regression are shown (Visscher and Hopper 2001). The cumulative distance is given at the bottom, and chromosome numbers are given at the top. The two dotted, horizontal lines represent the empirically derived genomewide significance thresholds (5% and 1%).
Empirical Significance Thresholds
Although, under the null hypothesis, the regression method is quite robust to departures from normality, given the extreme departures from normality that were due to selection on a scale with limited range, we derived the empirical genomewide significance threshold by simulation, to protect against false positives and potentially enhance the power of the analysis.
To assess the significance of the regression analysis, we used simulations to estimate the expected number per genome scan of −logP scores equal to or greater than the observed −logP score. Simulations were performed with both Simulate (Terwilliger et al. 1993) and Merlin (Abecasis et al. 2002), using the number of alleles and frequencies in the real data set while maintaining patterns of missing genotypes and genetic distances between markers. The simulations assume no genotype-phenotype linkage and maintain the family structures and phenotypic scores. We obtained 10,000 replicates from each program, and we analyzed data from each replicate by regression, as described above (see the “Regression Analysis” subsection). Because we applied a one-tailed test of significance in our genome scan, from each simulation, we kept the maximum −logP values associated with a regression coefficient whose sign was in the expected direction (positive for the Visscher-Hopper regression [Visscher and Hopper 2001]). The distribution of the maximum −logP result from each simulated scan is used to estimate the probability that the observed value could have occurred by chance; evidence for linkage is considered significant when the observed −logP score is expected to occur less than once in 20 genome scans. Therefore, −logP values were ranked, and the value that demarcated the highest 5% was taken as the 5% significance threshold.
Both simulation programs gave the same results: the genomewide 5% significance threshold is 0.00014 (−logP 3.8), and the corresponding 1% value is 0.00002 (−logP 4.7). The genomewide 5% threshold was exceeded on five chromosome arms: 1q (3.95), 4q (3.84), 7p (3.90), 12q (4.74), and 13q (3.81). The −logP statistics for markers are given in table 2.
Table 2.
Pointwise Significance Values for Seven Loci That Influence Neuroticism
| −logP Value for |
||||
| Chromosome (Marker) | Distance(cM) | P | CompleteSample | TruncatedSamplea |
| 1 (D1S2868) | 126 | .00011 | 3.95 | 3.73 |
| 4 (D4S1539) | 176 | .00014 | 3.84 | 3.84 |
| 7 (D7S516) | 42 | .00013 | 3.90 | 4.14 |
| 8 (D8S277) | 8 | .00117 | 2.93 | 2.55 |
| 11 (D11S898) | 99 | .00020 | 3.70 | 3.59 |
| 12 (D12S346) | 105 | .00002 | 4.74 | 4.81 |
| 13 (D13S153) | 64 | .00015 | 3.81 | 3.81 |
For when extreme phenotypes have been truncated.
Test of Mean IBD Deviation
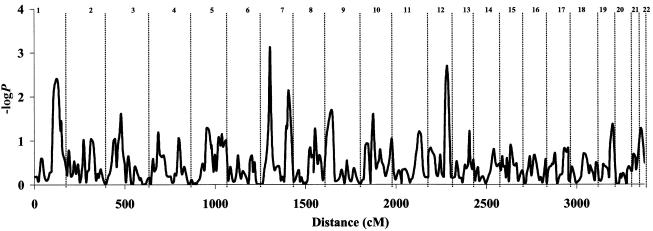
We also applied a test of deviation from the expected amount of allele sharing. At susceptibility loci, the mean proportion of alleles that are IBD is expected to be <0.5 in discordant sibling pairs but >0.5 in concordant pairs. Table 3 shows the mean IBD deviation and associated t statistics for both discordant and concordant sibling pairs at the loci mentioned above (see the “Empirical Significance Thresholds” subsection); results were consistent with expectations. To test the significance of the mean IBD deviation from the expected value of 0.5, we conducted a t test, using information combined from both groups (see the “Subjects and Methods” section). Figure 3 shows the −logP values of the t statistic, across the genome. Because the information content of the phenotypic data is reduced, this method is much less powerful than the regression method; however, −logP values >2 were achieved at three loci: 1q (2.4), 7q (3.2), and 12q (2.7).
Table 3.
Mean IBD Deviation and Associated t Statistics for Sibling Pairs, Discordant and Concordant for Extreme Neuroticism Scores
|
Discordant Pairs |
Concordant Pairs |
|||
| Chromosome (Marker) | Mean IBD | t | Mean IBD | t |
| 1 (D1S2868) | .45 | 2.11 | .53 | 2.35 |
| 4 (D4S1539) | .46 | 2.20 | .51 | .80 |
| 7 (D7S516) | .45 | 2.78 | .54 | 2.52 |
| 8 (D8S277) | .46 | 2.05 | .51 | .72 |
| 11 (D11S898) | .46 | 1.98 | .52 | 1.62 |
| 12 (D12S346) | .45 | 2.49 | .54 | 2.38 |
| 13 (D13S153) | .46 | 1.81 | .52 | 1.30 |
Figure 3.
The −logP values (vertical axis) of a t test for deviation from the expected mean IBD across the autosomal chromosomes. Distance is given at the bottom.
Sensitivity Analyses
Failure to replicate apparently robust results in linkage studies of complex traits has engendered justifiable skepticism (Risch and Botstein 1996). Consequently, we evaluated the robustness of our results in two ways:
First, to counter the potential bias of the presence of outliers (Wang et al. 1998), we truncated the most extreme phenotypes, a process that has been shown to improve the power of QTL detection (Fernández et al. 2002). We identified the 5% most extreme individuals (2.5% in each direction) and truncated their scores to the value of the least extreme (∼2.4 SDs outside the mean). Table 2 shows the −logP results from these analyses. After truncation, four loci exceeded a −logP value of 3.8, and two of these loci showed an increase in the −logP value (to 4.1, on chromosome 7, and 4.8, on chromosome 12). These findings suggest that our positive results are not merely artifacts of the undue influence of a few outliers.
Second, we considered whether our results could be false positives by examining the distribution of the t statistics that we obtained. Because we conducted one-tailed testing, significant results in the opposite tail have no biological interpretation and can be assumed to represent random deviations in the test statistic. If there were some unknown artifact inflating the variance of the test statistic, then, under the null hypothesis, the distribution of the test statistic would still be symmetrical. Therefore, if we obtained highly significant results in the predicted direction but obtained an approximately equal number of similarly significant results in the opposite direction, then the apparently significant results we obtained would be in doubt. Alternatively, if we obtained highly significant results in the predicted direction and far fewer or none of similar significance in the opposite direction, then our confidence in the apparently significant results that we obtained would be strengthened. Table 4 compares the maximum −logP values on each chromosome for the two directions. No pointwise −logP values in the wrong direction were greater than 3, compared with six in the expected direction.
Table 4.
Maximum −logP Values on Each Chromosome
| −logP Value, WhenRegression Coefficient Is in |
||
| Chromosome | BiologicallyImplausibleDirection | ExpectedDirection |
| 1 | 1.29 | 3.95 |
| 2 | 1.67 | .91 |
| 3 | 2.83 | .78 |
| 4 | .12 | 3.84 |
| 5 | 1.26 | 1.58 |
| 6 | 1.93 | .33 |
| 7 | .80 | 3.90 |
| 8 | 1.43 | 2.93 |
| 9 | 1.02 | .32 |
| 10 | .47 | 2.83 |
| 11 | .42 | 3.70 |
| 12 | .06 | 4.74 |
| 13 | .27 | 3.81 |
| 14 | .58 | .83 |
| 15 | .59 | .57 |
| 16 | 1.02 | .26 |
| 17 | .85 | .04 |
| 18 | .83 | .28 |
| 19 | 1.26 | .03 |
| 20 | .08 | 2.56 |
| 21 | .45 | .02 |
| 22 | 2.50 | .00 |
Sex Effects
Because of the reported association between sex and neuroticism (Eaves et al. 1989; Martin et al. 2000), we sought to determine whether there were sex-specific effects. Overall, there was an excess of women in the sample: 36.6% of the sample were male, and 63.4% were female. As has been observed in other studies, the sibling correlation of neuroticism between same-sex pairs (0.181 for male pairs and 0.186 for female pairs) is greater than that between opposite-sex pairs (0.157), suggesting that loci contributing to variation in neuroticism are different between men and women. We performed a number of analyses to examine whether loci had sex-specific effects.
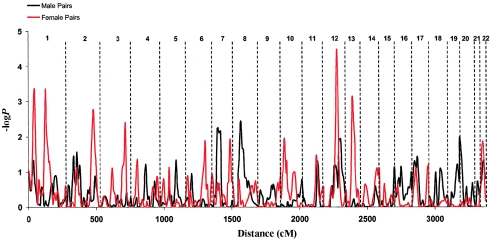
We analyzed male pairs and female pairs separately, using the same regression analysis employed in the genome scan for all subjects. The result is shown in figure 4: a red line shows the genomewide −logP values for female pairs, and a black line shows the values for male pairs. Loci on chromosomes 1 and 13 appear to be female specific, and loci on chromosomes 7 and 8 appear to be male specific. In table 5, we show the negative −logP values for the male, female, and opposite-sex pairs. At one locus, for male pairs on chromosome 1, the sign of the t statistic of the Visscher-Hopper regression (Visscher and Hopper 2001) was in the opposite direction to that expected, suggesting that the locus had no effect.
Figure 4.
Genomewide linkage analysis for individual variation in neuroticism in female-female (red line) and male-male (black line) sibling pairs. The −logP values (vertical axis) for the Visscher-Hopper regression are shown (Visscher and Hopper 2001). The cumulative distance is displayed at the bottom, and chromosome numbers are given at the top.
Table 5.
Sex-Specific Effects
|
Resultsa for |
|||
| Chromosome (Marker) | Male Pairs | Female Pairs | Opposite-Sex Pairs |
| 1 (D1S2868) | −.45 (.02) | 3.20 (2.24) | 3.98 (2.16) |
| 4 (D4S1539) | .43 (.30) | 1.42 (.40) | 2.07 (.80) |
| 7 (D7S516) | 2.09 (.98) | 1.07 (.20) | 2.44 (.86) |
| 8 (D8S277) | 2.72 (1.02) | 1.26 (.90) | 1.35 (.21) |
| 11 (D11S898) | 1.55 (.61) | 1.69 (.34) | 2.30 (.74) |
| 12 (D12S346) | .24 (.19) | 4.37 (2.12) | 3.60 (1.85) |
| 13 (D13S153) | .27 (.21) | 3.70 (1.15) | 3.25 (1.20) |
For the regression analysis of same-sex and opposite-sex pairs, −logP values are given. The sign of the regression coefficient is shown by the sign of the −logP value (negative in only one case, for male pairs on chromosome 1). The significance of the sex-specific effect is shown in parentheses, again as a −logP value, determined by simulation.
We used a simulation strategy to determine whether the genetic effect could be attributed specifically to any of the three possible pair types (male-male, female-female, and male-female). In a series of 10,000 simulations, we randomly reassigned the sex of each individual but maintained the same number of male-male, female-female, and male-female pairs. All other features of the data set (genotypes, phenotypes, and pedigree structure) remained unaltered. Sibling pairs from each simulation were separated into three groups, and the five loci that had been found to be significant in the complete data were analyzed using Visscher-Hopper regression (Visscher and Hopper 2001). The significance of the result for each of the three pair types in the original data set was determined on the basis of the percentile position it occupied in the ranked results from the corresponding simulated data sets.
In table 5, we show the significance of the result as determined by simulation (again expressed as a −logP value). Results for female pairs at loci on chromosomes 1, 12, and 13 were significant at the 1% level; results for male pairs were significant at the 1% level on chromosome 8 and just failed to meet the 1% level on chromosome 7. Consistent with a sex-specific effect, we do not observe any cases in which the results for opposite-sex pairs are more significant than those for the same-sex pairs. Note that this test does not distinguish a sex-specific effect from the effect of sex-pair type. There are no well-developed methods available that use sibling pairs to test for sex-specific effects.
Discussion
Our genetic linkage study, using a design based on extremely concordant and discordant sibling pairs, found loci, on chromosomes 1, 4, 7, and 13, that exceed a 5% genomewide significance threshold and found one locus, on chromosome 12, that exceeds a 1% threshold. We believe that these linkage results are robust for a number of reasons: (1) we have determined the empirical genomewide significance threshold (by simulation), indicating that our findings are not due to the unusual phenotypic distribution of the sample; (2) we conducted an analysis of allele sharing and found that, as expected, discordant sibling pairs have less allele sharing and concordant siblings have more allele sharing; (3) the loci remain significant when we exclude individuals in the tails of the distribution; and (4) the maximum t statistic acquired in the opposite tail of the distribution does not reach our 5% significance threshold.
Our results raise a number of issues, two of which are especially notable. First, it is probable that the five loci exceeding our 5% significance threshold are only a fraction of those contributing to variation in neuroticism. Given that we have genotyped only 2.5% of our sample, we can provide only inaccurate estimates of the effect size attributable to each locus, but those figures suggest that the five loci do not explain all the known genetic variance. On the basis of an analysis of variance of the genotyped sample, the five loci explain 23% of the phenotypic variance, which will be an overestimate of their effect in the total, unselected sample (Beavis 1998). Since the genetic variance of neuroticism is ∼40%, it is likely that other loci remain undetected. Some of these loci may well be represented by peaks on our genome scan that fail to exceed our significance threshold. Figure 2 shows that a number of loci attain a −logP value >2—notably, on chromosomes 3 (−logP 2.8, at 109 cM), 10 (−logP 2.4, at 116 cM), and 20 (−logP 2.6, at 11 cM)—as well as additional peaks on chromosomes 1 (−logP 2.1, at 37 cM) and 7 (−logP 3.6, at 124 cM).
Second, we have shown that some loci apparently act in a sex-specific manner, as had been suggested by previous quantitative genetic analyses (Eaves et al. 1989; Martin et al. 2000; Fanous et al. 2002). Indeed, there may be some loss of power when sex effects are ignored, because, when a locus has no influence on one sex, random deviations in the test statistic may result in the regression coefficients having opposite signs for the two sexes, as was seen, for example, at the locus on chromosome 1 (table 5). It is interesting that separate analysis of the same-sex pairs (fig. 4) shows suggestive evidence of additional loci on chromosomes 1 (−logP 3.31, at 37 cM), 2 (−logP 2.2, at 194 cM), and 10 (−logP 2.46, at 59 cM).
A major reason for investigating the genetic basis of neuroticism is that personality is known to be involved in both the risk of developing common psychiatric diseases and the modification of the severity of common psychiatric diseases. Analysis of twin data showed that ∼55% of the genetic liability of major depression is shared with neuroticism (Kendler et al. 1993), a figure in agreement with our sibling data (Martin et al. 2000). There is also evidence that neuroticism and some anxiety disorders have genetic factors in common (Jardine et al. 1984; Kendler et al. 1993). To show how our results compare with linkage studies of related conditions, we have listed the results of relevant reports in table 6.
Table 6.
Linkage Analysis of Psychiatric Disorders Genetically Related to Neuroticism
| Phenotype | Chromosome | Distance(cM) | LOD | NPLa | Reference |
| Anxiety susceptibility | 1 | 218 | 2.05 | Smoller et al. 2001 | |
| Panic | 1 | 260 | 2.04 | Gelernter et al. 2001 | |
| Alcoholism or depression | 1 | 120 | 4.66 | Nurnberger et al. 2001 | |
| Anorexia nervosa plus covariates | 1 | 210 | 3.46 | Devlin et al. 2002 | |
| Comorbid alcoholism and depression | 2 | 248 | 3.26 | Nurnberger et al. 2001 | |
| Anorexia nervosa plus covariates | 2 | 114 | 2.22 | Devlin et al. 2002 | |
| Recurrent major depression | 2 | 205 | 6.86 | Zubenko et al. 2002 | |
| Panic | 7 | 47 | 2.54 | Knowles et al. 1998 | |
| Panic | 7 | 63 | 2.23 | Crowe et al. 2001 | |
| Depression | 7 | 150 | 2.87 | Nurnberger et al. 2001 | |
| Harm avoidance | 8 | 17 | 3.2 | Cloninger et al. 1998 | |
| Anxiety susceptibility | 10 | 148 | 2.38 | Smoller et al. 2001 | |
| Panic | 11 | 5 | 2.01 | Gelernter et al. 2001 | |
| Harm avoidance | 11 | 194 | 1.6 | Cloninger et al. 1998 | |
| Panic/agoraphobia | 12 | 66 | 4.96 | Smoller et al. 2001 | |
| Anorexia nervosa plus covariates | 13 | 26 | 2.5 | Devlin et al. 2002 | |
| Harm avoidance | 18 | 109 | 1.6 | Cloninger et al. 1998 |
Nonparametric linkage score.
Loci, on 8p and 11q, that influence anxiety proneness, a personality factor very similar, if not identical, to neuroticism, have already been identified in a genome scan using the Tridimensional Personality Questionnaire (Cloninger et al. 1998) (LOD scores of 3.2 and 1.6, respectively; see table 6). Although neither locus achieved significance at the 5% threshold in our sample, we did obtain linkage evidence for both: −logP 2.9, on 8p, and 3.7, on 11q (table 2). Intriguingly, four studies report loci, on 1q, that influence traits genetically related to neuroticism, one of which (for depression) maps on top of the locus reported here at 120 cM (Nurnberger et al. 2001). There are also two reports of loci that influence panic disorder in the same 7p region that influences neuroticism. Furthermore, there is evidence for a depression-susceptibility locus, on 2q, that, according to one report, is female specific (Zubenko et al. 2002). Our sex-specific analyses provide some evidence in support of a similar finding on 2q for neuroticism (fig. 4).
There have been a considerable number of association studies of personality, some using the EPQ. The 1996 report (Lesch et al. 1996) of an association between variation in the serotonin transporter gene (5HTT) and neuroticism generated much interest (Ricketts et al. 1998; Murakami et al. 1999; Hu et al. 2000), but subsequent reports have been contradictory (Mazzanti et al. 1998; Kotler et al. 1999; Kumakiri et al. 1999; Greenberg et al. 2000). We detected no QTLs on chromosome 17, the location of the 5HTT gene, but it may be that the effect size attributable to the locus is too small to be detected by linkage: a recent meta-analysis of association studies of personality reported that the 5HTT gene had a marginally significant effect on neuroticism (P=.038) (Munafò et al. 2003).
The locus on chromosome 1 is intriguing not only because it may also influence vulnerability to depression but also because it may be syntenic with loci discovered in animal studies. We have demonstrated that a locus in the middle of rat chromosome 5 influences behavior in a number of tests of rodent emotionality, a model of neuroticism (Fernández-Teruel et al. 2002). The locus is syntenic with 1p in humans, but the low resolution of both human and rat mapping studies makes it impossible to say whether the same genes influence the trait in both species. In humans, we have suggestive evidence for linkage at a locus in this region: D1S218 (−logP 2.1). High-resolution mapping in the mouse detected a number of loci that influence emotionality on chromosome 1, syntenic with human chromosome 1q (Talbot et al. 1999; Mott et al. 2000). Association testing, using candidate genes discovered in the 0.8-cM region containing the mouse locus, will be able to determine whether the same genes influence neuroticism in human subjects and variation in emotionality in the mouse. The congruence of human and animal studies may provide a way to identify genes that contribute to susceptibility to a number of emotional disorders.
Acknowledgments
This work was funded by the Wellcome Trust and was supported, in part, by National Institutes of Health grants R01DK056366, R01ES09912, MH-01458, and P41RR006009.
Electronic-Database Information
The URL for data presented herein is as follows:
- Jonathan Flint's Web Site, http://www.well.ox.ac.uk/flint/software.shtml (for Perl code)
References
- Abecasis GR, Cherny SS, Cookson WO, Cardon LR (2002) Merlin—rapid analysis of dense genetic maps using sparse gene flow trees. Nat Genet 30:97–101 [DOI] [PubMed] [Google Scholar]
- Allison DB, Fernández JR, Heo M, Beasley TM (2000) Testing the robustness of the new Haseman-Elston quantitative-trait loci–mapping procedure. Am J Hum Genet 67:249–252 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allison DB, Neale MC, Zannolli R, Schork NJ, Amos CI, Blangero J (1999) Testing the robustness of the likelihood-ratio test in a variance-component quantitative-trait loci–mapping procedure. Am J Hum Genet 65:531–544 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amos CI, Krushkal J, Thiel TJ, Young A, Zhu DK, Boerwinkle E, de Andrade M (1997) Comparison of model-free linkage mapping strategies for the study of a complex trait. Genet Epidemiol 14:743–748 [DOI] [PubMed] [Google Scholar]
- Beavis WD (1998) QTL analyses: power, precision and accuracy. In: Paterson AH (ed) Molecular analysis of complex traits. CRC Press, Boca Raton, FL, pp 123–150 [Google Scholar]
- Boomsma DI, Beem AL, van den Berg M, Dolan CV, Koopmans JR, Vink JM, de Geus EJ, Slagboom PE (2000) Netherlands Twin Family Study of Anxious Depression (NETSAD). Twin Res 3:323–334 [DOI] [PubMed] [Google Scholar]
- Cardon LR, Fulker DW (1994) The power of interval mapping of quantitative trait loci, using selected sib pairs. Am J Hum Genet 55:825–833 [PMC free article] [PubMed] [Google Scholar]
- Carey G, Williamson J (1991) Linkage analysis of quantitative traits: increased power by using selected samples. Am J Hum Genet 49:786–796 [PMC free article] [PubMed] [Google Scholar]
- Cloninger CR (1994) Temperament and personality. Curr Opin Neurobiol 4:266–273 [DOI] [PubMed] [Google Scholar]
- Cloninger CR, Van Eerdewegh P, Goate A, Edenberg HJ, Blangero J, Hesselbrock V, Reich T, et al (1998) Anxiety proneness linked to epistatic loci in genome scan of human personality traits. Am J Med Genet 81:313–317 [DOI] [PubMed] [Google Scholar]
- Crowe RR, Goedken R, Samuelson S, Wilson R, Nelson J, Noyes R Jr (2001) Genomewide survey of panic disorder. Am J Med Genet 105:105–109 [PubMed] [Google Scholar]
- Deary IJ, Matthews G (1993) Personality traits are alive and well. The Psychologist 299–311 [Google Scholar]
- Devlin B, Bacanu SA, Klump KL, Bulik CM, Fichter MM, Halmi KA, Kaplan AS, Strober M, Treasure J, Woodside DB, Berrettini WH, Kaye WH (2002) Linkage analysis of anorexia nervosa incorporating behavioral covariates. Hum Mol Genet 11:689–696 [DOI] [PubMed] [Google Scholar]
- Digman JM (1990) Personality structure: emergence of the 5-factor model. Annu Rev Psychol 41:417–440 [Google Scholar]
- Douglas JA, Boehnke M, Lange K (2000) A multipoint method for detecting genotyping errors and mutations in sibling-pair linkage data. Am J Hum Genet 66:1287–1297 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drigalenko E (1998) How sib pairs reveal linkage. Am J Hum Genet 63:1242–1245 [PMC free article] [PubMed] [Google Scholar]
- Duggan CF, Lee AS, Murray RM (1990) Does personality predict long-term outcome in depression? Br J Psychiatry 157:19–24 [DOI] [PubMed] [Google Scholar]
- Duggan C, Sham P, Lee A, Minne C, Murray R (1995) Neuroticism: a vulnerability marker for depression evidence from a family study. J Affect Disord 35:139–143 [DOI] [PubMed] [Google Scholar]
- Eaves LJ, Eysenck HJ, Martin NG (eds) (1989) Genes, culture and personality: an empirical approach. Academic Press, London [Google Scholar]
- Eaves L, Heath A, Martin N, Maes H, Neale M, Kendler K, Kirk K, Corey L (1999) Comparing the biological and cultural inheritance of personality and social attitudes in the Virginia 30,000 study of twins and their relatives. Twin Res 2:62–80 [DOI] [PubMed] [Google Scholar]
- Eaves LJ, Heath AC, Neale MC, Hewitt JK, Martin NG (1998) Sex differences and non-additivity in the effects of genes on personality. Twin Res 1:131–137 [DOI] [PubMed] [Google Scholar]
- Eaves L, Meyer J (1994) Locating human quantitative trait loci: guidelines for the selection of sibling pairs for genotyping. Behav Genet 24:443–455 [DOI] [PubMed] [Google Scholar]
- Epstein MP, Duren WL, Boehnke M (2000) Improved inference of relationship for pairs of individuals. Am J Hum Genet 67:1219–1231 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eysenck HJ, Eysenck SBG (eds) (1975) Manual of the Eysenck Personality Questionnaire. Educational and Industrial Testing Service, San Diego, CA [Google Scholar]
- Fanous A, Gardner CO, Prescott CA, Cancro R, Kendler KS (2002) Neuroticism, major depression and gender: a population-based twin study. Psychol Med 32:719–728 [DOI] [PubMed] [Google Scholar]
- Fernández JR, Etzel C, Beasley TM, Shete S, Amos CI, Allison DB (2002) Improving the power of sib pair quantitative trait loci detection by phenotype Winsorization. Hum Hered 53:59–67 [DOI] [PubMed] [Google Scholar]
- Fernández-Teruel A, Escorihuela RM, Gray JA, Aguilar R, Gil L, Giménez-Llort L, Tobeña A, Bhomra A, Nicod A, Mott R, Driscoll P, Dawson GR, Flint J (2002) A quantitative trait locus influencing anxiety in the laboratory rat. Genome Res 12:618–626 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Floderus-Myrhed B, Pedersen N, Rasmuson I (1980) Assessment of heritability for personality, based on a short form of the Eysenck Personality Inventory. Behav Genet 10:153–162 [DOI] [PubMed] [Google Scholar]
- Forrest WF (2001) Weighting improves the “new Haseman-Elston” method. Hum Hered 52:47–54 [DOI] [PubMed] [Google Scholar]
- Fulker DW, Cherny SS (1996) An improved multipoint sib-pair analysis of quantitative traits. Behav Genet 26:527–532 [DOI] [PubMed] [Google Scholar]
- Gelernter J, Bonvicini K, Page G, Woods SW, Goddard AW, Kruger S, Pauls DL, Goodson S (2001) Linkage genome scan for loci predisposing to panic disorder or agoraphobia. Am J Med Genet 105:548–557 [DOI] [PubMed] [Google Scholar]
- Göring HH, Ott J (1997) Relationship estimation in affected sib pair analysis of late-onset diseases. Eur J Hum Genet 5:69–77 [PubMed] [Google Scholar]
- Greenberg BD, Li Q, Lucas FR, Hu S, Sirota LA, Benjamin J, Lesch KP, Hamer D, Murphy DL (2000) Association between the serotonin transporter promoter polymorphism and personality traits in a primarily female population sample. Am J Med Genet 96:202–216 [DOI] [PubMed] [Google Scholar]
- Heo M, Faith MS, Allison DB (2002) Power and sample sizes for linkage with extreme sampling under an oligogenic model for quantitative traits. Behav Genet 32:23–36 [DOI] [PubMed] [Google Scholar]
- Hirschfeld RMA, Klerman GL, Lavori P, Keller MB, Griffith P, Coryell W (1989) Premorbid personality assessments of first onset of major depression. Arch Gen Psychiatry 46:345–350 [DOI] [PubMed] [Google Scholar]
- Hu S, Brody CL, Fisher C, Gunzerath L, Nelson ML, Sabol SZ, Sirota LA, Marcus SE, Greenberg BD, Murphy DL, Hamer DH (2000) Interaction between the serotonin transporter gene and neuroticism in cigarette smoking behavior. Mol Psychiatry 5:181–188 [DOI] [PubMed] [Google Scholar]
- Jardine R, Martin NG, Henderson AS (1984) Genetic covariation between neuroticism and the symptoms of anxiety and depression. Genet Epidemiol 1:89–107 [DOI] [PubMed] [Google Scholar]
- Kendler KS, Neale MC, Kessler RC, Heath AC, Eaves LJ (1993) A longitudinal twin study of personality and major depression in women. Arch Gen Psychiatry 50:853–862 [DOI] [PubMed] [Google Scholar]
- Kirk KM, Birley AJ, Statham DJ, Haddon B, Lake RI, Andrews JG, Martin NG (2000) Anxiety and depression in twin and sib pairs extremely discordant and concordant for neuroticism: prodromus to a linkage study. Twin Res 3:299–309 [DOI] [PubMed] [Google Scholar]
- Knowles JA, Fyer AJ, Vieland VJ, Weissman MM, Hodge SE, Heiman GA, Haghighi F, de Jesus GM, Rassnick H, Preud'homme-Rivelli X, Austin T, Cunjak J, Mick S, Fine LD, Woodley KA, Das K, Maier W, Adams PB, Freimer NB, Klein DF, Gilliam TC (1998) Results of a genome-wide genetic screen for panic disorder. Am J Med Genet 81:139–147 [DOI] [PubMed] [Google Scholar]
- Kotler M, Cohen H, Kremer I, Mel H, Horowitz R, Ohel N, Gritsenko I, Nemanov L, Katz M, Ebstein R (1999) No association between the serotonin transporter promoter region (5-HTTLPR) and the dopamine D3 receptor (BalI D3DR) polymorphisms and heroin addiction. Mol Psychiatry 4:313–314 [DOI] [PubMed] [Google Scholar]
- Kruglyak L, Daly MJ, Reeve-Daly MP, Lander ES (1996) Parametric and nonparametric linkage analysis: a unified multipoint approach. Am J Hum Genet 58:1347–1363 [PMC free article] [PubMed] [Google Scholar]
- Kumakiri C, Kodama K, Shimizu E, Yamanouchi N, Okada S, Noda S, Okamoto H, Sato T, Shirasawa H (1999) Study of the association between the serotonin transporter gene regulatory region polymorphism and personality traits in a Japanese population. Neurosci Lett 263:205–207 [DOI] [PubMed] [Google Scholar]
- Lake RI, Eaves LJ, Maes HH, Heath AC, Martin NG (2000) Further evidence against the environmental transmission of individual differences in neuroticism from a collaborative study of 45,850 twins and relatives on two continents. Behav Genet 30:223–233 [DOI] [PubMed] [Google Scholar]
- Lesch K-P, Bengel D, Heils A, Sabol SZ, Greenberg BD, Petri S, Benjamin J, Müller CR, Hamer DH, Murphy DL (1996) Association of anxiety-related traits with a polymorphism in the serotonin transporter gene regulatory region. Science 274:1527–1530 [DOI] [PubMed] [Google Scholar]
- Loehlin JC (ed) (1992) Genes and environment in personality development. Sage Publications, London [Google Scholar]
- Loehlin JC, Nichols RC (eds) (1976) Heredity, environment and personality. University of Texas Press, Austin, TX [Google Scholar]
- Martin N, Goodwin G, Fairburn C, Wilson R, Allison D, Cardon LR, Flint J (2000) A population based study of personality in 34,000 sib-pairs. Twin Res 3:310–315 [PubMed] [Google Scholar]
- Martin N, Jardine R (1986) Eysenck's contribution to behavior genetics. In: Modgil S, Modgil C (eds) Hans Eysenck: consensus and controversy. Falmer Press, Lewes, Sussex, pp 13–62 [Google Scholar]
- Mazzanti CM, Lappalainen J, Long JC, Bengel D, Naukkarinen H, Eggert M, Virkkunen M, Linnoila M, Goldman D (1998) Role of the serotonin transporter promoter polymorphism in anxiety-related traits. Arch Gen Psychiatry 55:936–940 [DOI] [PubMed] [Google Scholar]
- Mott R, Talbot CJ, Turri MG, Collins AC, Flint J (2000) A method for fine mapping quantitative trait loci in outbred animal stocks. Proc Natl Acad Sci USA 97:12649–12654 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Munafò MR, Clark TG, Moore LR, Payne E, Walton R, Flint J. Genetic polymorphisms and personality in healthy adults: a systematic review and meta-analysis. Mol Psychiatry (in press) [DOI] [PubMed] [Google Scholar]
- Murakami F, Shimomura T, Kotani K, Ikawa S, Nanba E, Adachi K (1999) Anxiety traits associated with a polymorphism in the serotonin transporter gene regulatory region in the Japanese. J Hum Genet 44:15–17 [DOI] [PubMed] [Google Scholar]
- Nurnberger JI Jr, Foroud T, Flury L, Su J, Meyer ET, Hu K, Crowe R, Edenberg H, Goate A, Bierut L, Reich T, Schuckit M, Reich W (2001) Evidence for a locus on chromosome 1 that influences vulnerability to alcoholism and affective disorder. Am J Psychiatry 158:718–724 [DOI] [PubMed] [Google Scholar]
- O'Connell JR, Weeks DE (1998) PedCheck: a program for identification of genotype incompatibilities in linkage analysis. Am J Hum Genet 63:259–266 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reed PW, Davies JL, Copeman JB, Bennett ST, Palmer SM, Pritchard LE, Gough SCL, Kawaguchi Y, Cordell HJ, Balfour KM, Jenkins SC, Powell EE, Vignal A, Todd JA (1994) Chromosome-specific microsatellite sets for fluorescence-based, semi-automated genome mapping. Nat Genet 7:390–395 [DOI] [PubMed] [Google Scholar]
- Ricketts MH, Hamer RM, Sage JI, Manowitz P, Feng F, Menza MA (1998) Association of a serotonin transporter gene promoter polymorphism with harm avoidance behaviour in an elderly population. Psychiatr Genet 8:41–44 [DOI] [PubMed] [Google Scholar]
- Risch N, Botstein D (1996) A manic depressive history. Nat Genet 12:351–353 [DOI] [PubMed] [Google Scholar]
- Risch N, Zhang H (1995) Extreme discordant sib pairs for mapping quantitative trait loci in humans. Science 268:1584–1589 [DOI] [PubMed] [Google Scholar]
- ——— (1996) Mapping quantitative trait loci with extreme discordant sib pairs: sampling considerations. Am J Hum Genet 58:836–843 [PMC free article] [PubMed] [Google Scholar]
- Rose RJ, Koskenvuo M, Kaprio J, Sarna S, Langinvainio H (1988) Shared genes, shared experiences and similarity of personality: data from 14,288 adult Finnish co-twins. J Pers Soc Psychol 54:161–171 [DOI] [PubMed] [Google Scholar]
- Roy A (1990) Personality variables in depressed patients and normal controls. Neuropsychobiology 23:119–123 [DOI] [PubMed] [Google Scholar]
- Sham PC, Purcell S (2001) Equivalence between Haseman-Elston and variance-components linkage analyses for sib pairs. Am J Hum Genet 68:1527–1532 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sham PC, Purcell S, Cherny SS, Abecasis GR (2002) Powerful regression-based quantitative-trait linkage analysis of general pedigrees. Am J Hum Genet 71:238–253 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sham PC, Sterne A, Purcell S, Cherny S, Webster M, Rijsdijk F, Asherson P, Ball D, Craig I, Eley T, Goldberg D, Gray J, Mann A, Owen M, Plomin R (2000) GENESiS: creating a composite index of the vulnerability to anxiety and depression in a community-based sample of siblings. Twin Res 3:316–322 [DOI] [PubMed] [Google Scholar]
- Smoller JW, Acierno JS Jr, Rosenbaum JF, Biederman J, Pollack MH, Meminger S, Pava JA, Chadwick LH, White C, Bulzacchelli M, Slaugenhaupt SA (2001) Targeted genome screen of panic disorder and anxiety disorder proneness using homology to murine QTL regions. Am J Med Genet 105:195–206 [DOI] [PubMed] [Google Scholar]
- Talbot CJ, Nicod A, Cherny SS, Fulker DW, Collins AC, Flint J (1999) High-resolution mapping of quantitative trait loci in outbred mice. Nat Genet 21:305–308 [DOI] [PubMed] [Google Scholar]
- Terwilliger JD, Speer M, Ott J (1993) Chromosome-based method for rapid computer simulation in human genetic linkage analysis. Genet Epidemiol 10:217–224 [DOI] [PubMed] [Google Scholar]
- Viken RJ, Rose RJ, Kaprio J, Koskenvuo M (1994) A developmental genetic analysis of adult personality: extraversion and neuroticism from 18 to 59 years of age. J Pers Soc Psychol 66:722–730 [DOI] [PubMed] [Google Scholar]
- Visscher PM, Hopper JL (2001) Power of regression and maximum likelihood methods to map QTL from sib-pair and DZ twin data. Ann Hum Genet 65:583–601 [DOI] [PubMed] [Google Scholar]
- Wang J, Guerra R, Cohen J (1998) Statistically robust approaches for sib-pair linkage analysis. Ann Hum Genet 62:349–359 [DOI] [PubMed] [Google Scholar]
- Wright FA (1997) The phenotypic difference discards sib-pair QTL linkage information. Am J Hum Genet 60:740–742 [PMC free article] [PubMed] [Google Scholar]
- Xu X, Weiss S, Wei LJ (2000) A unified Haseman-Elston method for testing linkage with quantitative traits. Am J Hum Genet 67:1025–1028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeiss AM, Lewinsohn PM (1988) Enduring deficits after remissions of depression: a test of the scar hypothesis. Behav Res Ther 26:151–158 [DOI] [PubMed] [Google Scholar]
- Zubenko GS, Hughes HB III, Maher BS, Stiffler JS, Zubenko WN, Marazita ML (2002) Genetic linkage of region containing the CREB1 gene to depressive disorders in women from families with recurrent, early-onset, major depression. Am J Med Genet 114:980–987 [DOI] [PubMed] [Google Scholar]
- Zuckerman M, Kuhlman DM, Camac C (1988) What lies beyond E and N? Factor analysis of scales believed to measure basic dimensions of personality. J Pers Soc Psychol 54:96–107 [Google Scholar]