Abstract
HOXD13, the most 5′ gene of the HOXD cluster, encodes a homeodomain transcription factor with important functions in limb patterning and growth. Heterozygous mutations of human HOXD13, encoding polyalanine expansions or frameshifts, are believed to act by dominant negative or haploinsufficiency mechanisms and are predominantly associated with synpolydactyly phenotypes. Here, we describe two mutations of HOXD13 (923C→G encoding Ser308Cys and 940A→C encoding Ile314Leu) that cause missense substitutions within the homeodomain. Both are associated with distinctive limb phenotypes in which brachydactyly of specific metacarpals, metatarsals, and phalangeal bones is the most constant feature, exhibiting overlap with brachydactyly types D and E. We investigated the binding of synthetic mutant proteins to double-stranded DNA targets in vitro. No consistent differences were found for the Ser308Cys mutation compared with the wild type, but the Ile314Leu mutation (which resides at the 47th position of the homeodomain) exhibited increased affinity for a target containing the core recognition sequence 5′-TTAC-3′ but decreased affinity for a 5′-TTAT-3′ target. Molecular modeling of the Ile314Leu mutation indicates that this mixed gain and loss of affinity may be accounted for by the relative positions of methyl groups in the amino acid side chain and target base.
Introduction
The HOX genes (related to homeobox genes in the Drosophila melanogaster HOM-C cluster) encode a family of highly conserved transcription factors that play key roles in embryonic development. In humans, there are four HOX gene clusters (HOXA, HOXB, HOXC, and HOXD) that are located on different chromosomes and together comprise a total of 39 genes (Scott 1992; Kosaki et al. 2002). The order of genes within each cluster reflects their temporal and spatial expression patterns during development. In general, genes expressed early in development and in anterior and proximal regions are located at the 3′ end of the cluster, whereas genes expressed later and in more posterior and distal regions are located at the 5′ end (reviewed by Krumlauf 1994). HOXD13 is the most 5′ member of the HOXD cluster and, with homologues numbered D9–D12, shows greatest similarity to the D. melanogaster Abd-B gene (Scott 1992). These so-called posterior HOXD genes and their paralogues in the HOXA cluster play critical roles in limb development (Dollé et al. 1993; reviewed by Rijli and Chambon 1997; Zákány et al. 1997).
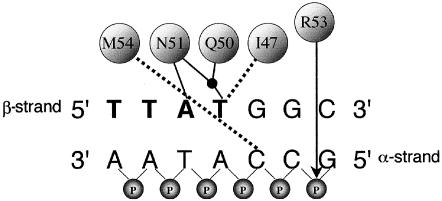
The homeobox region of each HOX gene encodes a domain of 60 amino acids, termed the “homeodomain,” which comprises a flexible N-terminal region followed by three α-helices (I, II, and III) and is highly conserved among eukaryotes (Bürglin 1994; Banerjee-Basu and Baxevanis 2001; Banerjee-Basu et al. 2001). Homeodomains bind DNA, either alone or in heteromeric complexes with partner proteins (Mann and Affolter 1998); their corresponding DNA-binding elements and downstream genetic targets are gradually being elucidated (Banerjee-Basu et al. 2001; Mann and Carroll 2002). In the case of HOXD13, the preferred core-binding sequence is reported to be 5′-TTAC-3′, although 5′-TTAT-3′ is also bound by posterior HOX proteins (Shen et al. 1997b). Crystal structures of several homeodomain-DNA complexes have been obtained (reviewed by Gehring et al. 1994; Billeter 1996). These indicate that helix III is essential for sequence-specific DNA binding by making several contacts in the major groove of DNA; additional minor groove contacts are made by the N-terminal region of the homeodomain. The contacts made by helix III of the D. melanogaster homeodomain protein Ultrabithorax (Ubx) (the orthologue of the seventh vertebrate HOX genes; Scott 1992) to the core DNA consensus sequence 5′-TTAT-3′ are shown in figure 1 (Passner et al. 1999). A similar pattern of contacts is observed for DNA binding by D. melanogaster Antennapedia (Antp) (the orthologue of the sixth vertebrate HOX genes), except that the core consensus sequence is 5′-TAAT-3′ (Fraenkel and Pabo 1998).
Figure 1.
Simplified representation of binding interactions between D. melanogaster Ubx protein and a double-stranded DNA target (Passner et al. 1999). At the top, I47, Q50, N51, and M54 are the four amino acids—Ile, Gln, Asn, and Met, respectively (numbered according to their position in the homeodomain)—that contact specific bases, either through hydrogen bonds (solid lines) or hydrophobic interactions (dashed lines). The black dot represents a water molecule. Designation of the α and β strands follows Billeter (1996). The 5′-TTAT-3′ motif on the β-strand of the DNA target is shown in bold. On the α-strand only, the connecting phosphate (P) groups are included to demonstrate the position (arrow) of the ionic interaction with Arg (R53). Key amino acids in human HOXD13 are identical to those in Ubx, except that M54 is replaced by V54 (Val).
To date, intragenic mutations in only two HOX genes have been confirmed to cause human disorders. Mutations in HOXD13 (located on chromosome 2q31; Ensembl) were first identified in synpolydactyly (SPD [MIM 186000]) (Muragaki et al. 1996), and mutations in HOXA13 cause hand-foot-uterus syndrome (HFUS [MIM 140000]) (Mortlock and Innis 1997). SPD is a rare autosomal dominant limb disorder with incomplete penetrance and variable expressivity (Sayli et al. 1995), typically comprising 3/4 finger syndactyly and 4/5 toe syndactyly with partial or complete digit duplication within the syndactylous web (Muragaki et al. 1996; Goodman et al. 1997; reviewed by Goodman 2002). The causative mutations are heterozygous expansions of a 15-residue polyalanine tract encoded by an imperfect trinucleotide repeat in exon 1 of HOXD13. Longer expansions are associated with greater penetrance and phenotypic severity (Goodman et al. 1997). Analysis of an equivalent mouse model, Hoxd13spdh, suggests a dominant negative mechanism of action by interfering with the function of the remaining wild-type HOXD13 protein and other 5′ HOXD proteins (Bruneau et al. 2001). The mutant protein has pleiotropic actions, which affect patterning, joint formation, and chrondrocyte differentiation during development (Albrecht et al. 2002).
A distinct phenotype, including a novel foot malformation with partial duplication of the base of the second metatarsal in the first web space and a broad hallux, was described in three families with frameshifting deletions of HOXD13 (Goodman et al. 1998; Calabrese et al. 2000). These mutations are predicted to lead to a truncated protein missing part or all of the homeodomain; hence, the phenotype may be caused by haploinsufficiency. A family segregating a missense mutation in the homeodomain of HOXD13, Arg298Trp, also exhibits this phenotype (Debeer et al. 2002). Finally, heterozygous deletions including HOXD13 have been described in split hand–split foot malformation, but the critical region for this phenotype probably lies outside the HOXD cluster (reviewed by Goodman 2002). Although brachydactyly (shortening of the digits) is prominent in both the mouse mutants (Dollé et al. 1993; Bruneau et al. 2001) and in rare human SPD homozygotes (Muragaki et al. 1996), only minor manifestations have previously been associated with heterozygous HOXD13 mutations (fifth-finger clinodactyly, hypoplasia or absence of the middle phalanges of toes 2–5, and occasional shortening of the first metacarpal; reviewed by Goodman 2002).
In view of the diversity of limb malformations associated with HOXD13 mutations, we performed a mutation screen of HOXD13 in 128 consecutive patients with unselected congenital limb abnormalities who required reconstructive surgery. In two subjects, we identified a novel mutation in the homeodomain of HOXD13, Ile314Leu. Detailed characterization of the phenotype associated with these mutations delineated a distinctive polydactyly/brachydactyly pattern, which led us to investigate a family reported elsewhere to have brachydactyly type E (Brailsford 1945; Oude Luttikhuis et al. 1996). A different missense mutation in the homeodomain, Ser308Cys, was found to segregate in the latter family. We have investigated the effect of these mutations on binding to double-stranded (ds) oligonucleotides in vitro and report that the Ile314Leu mutation exhibits mixed gain and loss of affinity to different targets.
Patients and Methods
Patients
Approval for the study was obtained from the Central Oxford Research Ethics Committee. Venous blood samples were obtained at the time of surgery from a consecutive series of 128 patients with congenital limb abnormalities presenting to the Department of Plastic and Reconstructive Surgery, Oxford; from selected relatives; and from an additional family reported elsewhere as being affected with brachydactyly type E (Brailsford 1945; Oude Luttikhuis et al. 1996). Additional buccal samples from relatives were obtained using Cytosoft cytology brushes (Gentra Systems). Genomic DNA was isolated either from venous blood samples by phenol/chloroform extraction or from buccal cells using the Puregene DNA Isolation Kit (Gentra Systems). Metacarpophalangeal profiles (MCPP) were analyzed from direct measurements taken from upper limb radiographs in families A and B using ANTRO software (Garn et al. 1972; Poznanski et al. 1972; Armour et al. 2000).
PCR Amplification and Analysis
Oligonucleotides were designed from HOXD13 genomic and cDNA sequences (GenBank accession numbers AF005219, AF005220, AC009336, and NM_000523). DNA numbering starts from the first nucleotide of the ATG start codon. Primers for genomic PCR were synthesized by Sigma-Genosys. Exon 1 was amplified using two overlapping primer pairs: 1aF 5′-ATGAGCCGCGCCGGGAGCTGGGAC-3′ and 1aR 5′-CGAGGCGTGCGGCGATGACTTGAGCG-3′, 1bF 5′-TACCACTTCGGCAACGGCTACTACAGCTGC-3′ and 1bR 5′-GCACAACTCCCACTCCCAAGTAGGGG-3′, which generated fragments of 474 bp and 501 bp, respectively. Exon 2 was amplified using the primer pair 2F 5′-CTAGGTGCTCCGAATATCCCAGCCT-3′ and 2R 5′-AAGCTGTCTGTGGCCAACCTGGA-3′, which generated a fragment of 333 bp. Reactions took place in a volume of 25 μl that contained 40 ng genomic DNA, 1.5 mM MgCl2, 120 μM dNTPs, 0.4 μM primers, 1 × GeneAmp PCR Buffer II, 0.5 U AmpliTaq Gold DNA polymerase (Applied Biosystems), and 0.05 U Pwo polymerase (Roche). Ten percent DMSO was included in the reaction mix for exon 1 amplifications. Thermocycling was performed on an MJ Research PTC-200 and consisted of 94°C for 10 min, followed by 35 cycles of 94°C for 45 s, 62°C annealing for 45 s, 72°C for 30 s, and a final step of 72°C for 4 min, except that an annealing temperature of 56°C was used for the 1aF-1aR primer pair.
The samples were heteroduplexed by heating to 95°C, followed by slow cooling, and analyzed by denaturing high-performance liquid chromatography (DHPLC) on the WAVE DNA Fragment Analysis System (Transgenomic) at the following temperatures: 1aF-1aR at 64°C, 67°C, and 70°C; 1bF-1bR at 62°C and 64°C; and 2F-2R at 60°C.
Microsatellite markers were purchased from Research Genetics. The Généthon panel of markers used for haplotype analysis was D2S335, D2S326, D2S2307, D2S2188, D2S2257, D2S2314, D2S148, D2S2173, D2S385, and D2S324 (Dib et al. 1996). An additional microsatellite marker, HOXD8 (Sarfarazi et al. 1995), was employed. Microsatellite analysis was performed by blot hybridization of polyacrylamide gels with [α-32P]dCTP-labeled 5′-(CA)10-3′.
DNA Sequencing and Mutation Conformation
PCR products to be sequenced were gel-purified by use of the QIAquick gel extraction kit (Qiagen). Cycle sequencing was performed with primers used for the original PCR by use of the BigDye Terminator Cycle Sequencing Kit (Applied Biosystems) and the ABI 3100 Sequencer.
All mutations were confirmed by an independent method in all available family members. The 923C→G mutation was confirmed by loss of a DdeI restriction site. The 940A→C mutation was confirmed by blot hybridization, using the allele-specific oligonucleotide (ASO) 5′-AGTGACCCTTTGGTTTC-3′ (the position of the mutation is underlined). For each mutation, the entire HOXD13 open reading frame was sequenced in an affected individual (IV-2 from family B; III-2 from family C) and was otherwise normal.
DNA-Protein Binding Studies
The full-length wild-type HOXD13 cDNA open reading frame was obtained by RT-PCR from RNA extracted from a normal fibroblast cell line using the Expand Long Template PCR System (Roche) with primer pair cDNA-F (5′-ATGGACGGGCTGCGGGCAGAC-3′) and cDNA-R (5′-TCAGGAGACAGTATCTTTGAGC-3′) and cloned into pGEM-T easy vector (Promega). This was then subcloned into pRD67:HA vector (Davey et al. 1997), after introducing EcoRI and XhoI restriction sites at either end of the product. Mutant bases were introduced into wild-type HOXD13 clones using the QuikChange site-directed mutagenesis kit (Stratagene), with primers: S308C-F, 5′-GCTACGAACCTATGTGAGAGACAAGTG-3′; I314L-F, 5′-GAGAGACAAGTGACCCTTTGGTTTCAGAACCG-3′; R320A-F, 5′-GTTTCAGAACCGAGCAGTGAAGGACAAG-3′, and their respective complementary primers S308C-R, I314L-R, and R320A-R (mutated residues are underlined). The integrity of the HOXD13 sequence was confirmed in all mutant constructs.
Wild-type and mutated proteins were expressed using the TNT SP6 quick-coupled in vitro transcription/translation system (Promega) in the presence of [35S]-methionine (Redivue Pro-mix L-[35S] in vitro cell labeling mix; Amersham Bioscience). Equalization (to ±20%) of concentrations of wild-type and mutant protein used in binding assays was ensured by counting of 35S-labeled proteins on a PhosphorImager (Molecular Dynamics).
Electrophoretic mobility shift assays (EMSA) were used to detect complex formation between labeled oligonucleotide and HOXD13 proteins. The HPLC-purified oligonucleotides (ThermoHybaid): 5′-GGGATCTGACAGTTTTACGACAGATCT-3′ (β-strand) and 5′-GGAGATCTGTCGTAAAACTGTCAGATC-3′ (α-strand) contain the core recognition sequence (underlined) 5′-TTAC-3′ (Shen et al. 1997a). These oligonucleotides were annealed and radiolabeled by incubation with [α-32P]-dCTP, in the presence of Klenow DNA polymerase (Amersham Bioscience). Other oligonucleotide pairs were synthesized in which the underlined β-strand sequence was varied to 5′-TTAT-3′, 5′-TTAA-3′, 5′-TTAG-3′, and 5′-TTAU-3′, and the complementary sequence was incorporated into the α-strand.
The binding conditions used were similar to those described by Shen et al. (1997a). Binding was carried out in a total volume of 20 μl, using 2 μl of equalized protein and 0.1 pmole 32P-labeled ds probe, incubated on ice for 30 min in buffer (final concentration: 75 mM NaCl, 1 mM EDTA, 1 mM DTT, 10 mM Tris-HCl [pH 7.5], 6% glycerol, 2 μg BSA, 20 ng poly[dI-dC], 0.2 μg single-stranded salmon sperm DNA). Cold competition involved addition of 100-fold excess of unlabeled dsDNA oligonucleotides. Samples were separated on a nondenaturing 6% acrylamide gel in 0.5×TBE at 220 V for 2 h at 4°C. The gels were dried and exposed in a PhosphorImager.
Structural Modeling
The D. melanogaster Ubx/Extradenticle/DNA complex (Protein Data Bank [PDB] ID: 1B8I) (Passner et al. 1999) and Antp/DNA complex (PDB ID: 9ANT) (Fraenkel and Pabo 1998) were used as templates for the modeling experiments. Residues Ile147 in 1B8I and Ile47 in 9ANT are the equivalents of HOXD13 Ile314. The equivalents of the 3′-thymidine (Thy) in 5′-TTAT-3′ are Thy26 and Thy221 in 1B8I and 9ANT, respectively. For each initial template (WT-IT), three “mutants” (Mut-LT, Ile→Leu; Mut-IC, Thy→Cyt [cytosine]; and Mut-LC, Ile→Leu and Thy→Cyt) were generated in silico with CHARMM (Brooks et al. 1983). The backbone conformation was not altered in any case. All calculations were performed in vacuo, with the solvent being approximated using a linear distance-dependent dielectric constant. However, the hydrogen bonding to a water molecule by residues Gln150, Asn151, and Thy26 of 1B8I (Gln50, Asn51, and Thy221 in 9ANT) was kept. Electrostatic interactions beyond 10 Å were not taken into account. The energy of structures was minimized by 12×1,000 steps of conjugate gradient minimization. To study the local influence of mutations, only Ile/Leu147 and Thy/Cyt26 (1B8I numbering), or their equivalents in the 9ANT model, were allowed to move during the minimization. Harmonic constraints were applied to allow smooth minimization. Harmonic forces were decreased every 1,000 steps, from 5,000 to 0 kcal/mol×Å2. Interaction energies between Ile/Leu and Thy/Cyt pairs were calculated with CHARMM. Molecular models were generated with BobScript (Esnouf 1999) and rendered with Raster 3D (Merrit and Bacon 1997).
Results
Identification of Mutations in HOXD13
The mutation screen of HOXD13 in patients with congenital limb abnormalities revealed four probands (of 128 studied) who were heterozygous for pathogenic alterations. One had a novel 21-bp duplication (160_180dup), corresponding to a polyalanine expansion (A54_A60dup) similar to those described elsewhere (Muragaki et al. 1996; Goodman et al. 1997; reviewed by Goodman 2002); another had a novel splice-site mutation (752-2delA), associated with a partial duplication of the second metatarsal within the first web space (Kan et al., in press).
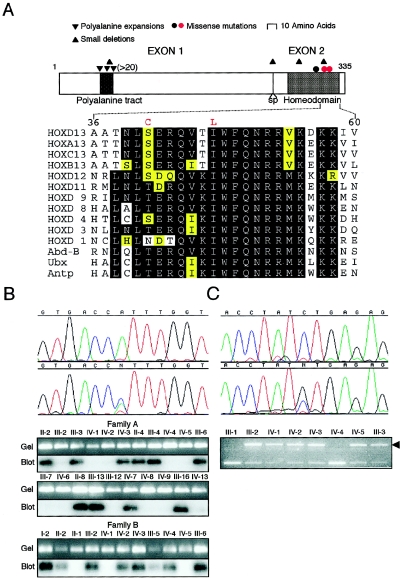
The two remaining probands, both of whom had significant family histories (families A and B; fig. 2), harbored an identical heterozygous transversion in HOXD13 (940A→C), predicting the amino acid substitution Ile314Leu, which locates in helix III at the 47th residue of the conserved homeodomain (fig. 3A). The mutation was confirmed by ASO blot hybridization and identified in all 18 clinically affected family members (10 from family A; 8 from family B) from whom DNA was available but in none of 10 unaffected members at 50% prior risk (fig. 3B). Genotyping, using 11 microsatellite markers spanning HOXD13 (over a distance of 6.2 Mb), revealed sharing of a haplotype of six consecutive markers (D2S2314–D2S324) between affected individuals in the uppermost generations (II-2, -4, and -8 in family A; I-2 in family B; data not shown). Given that these families live 90 km away from each other in southern England, it is likely that the two families are related through a common affected ancestor. A founder effect is also suggested by the identification of the same mutation in a third British family (Caronia et al. 2003).
Figure 2.
Pedigrees of families A, B, and C. Squares represent males; circles represent females; patterns of manifestation (hands only) are categorized as shown in the key at the bottom. The arrows identify the probands, and asterisks indicate individuals who underwent radiographic examination.
Figure 3.
Missense mutations in the homeodomain of HOXD13. A, above, Schematic structure of HOXD13 showing portions encoded by each exon, the position of the splice site (sp), characteristic motifs (polyalanine tract and homeodomain), and the positions of different types of mutation (red dots identify missense mutations reported here). Below, Amino acid sequence conservation in the second half of the homeodomain. The sequence of human HOXD13 is compared with its paralogues, other members of the HOXD group, and the D. melanogaster HOM-C members Abd-B, Ubx, and Antp. The two amino acid substitutions in HOXD13 described in this report are shown above the wild-type sequence in red. Levels of conservation are indicated as follows: black, fully conserved; yellow, moderately conserved; white, poorly conserved. B, Identification of 940A→C mutation in families A and B. The upper panel shows DNA sequence chromatograms from a normal individual (above) and affected individual IV-2 from family B (below). The lower panels show confirmation of the mutation by ASO blot hybridization. C, Identification of 923C→G mutation in family C. The upper panel shows DNA sequence chromatograms from a normal individual (above) and affected individual III-2 (below). The lower panel shows confirmation of the mutation by loss of a DdeI restriction site (arrowhead).
Clinical Phenotype
Clinical examination was performed on 16 affected individuals (8 each from families A and B) as well as 10 unaffected individuals at 50% prior risk (8 from family A; 2 from family B). Clinical photographs of both hands of one additional affected individual in family A were also available. Radiographs of the hands suitable for MCPP (excluding postoperative cases) were available on 15 hands of 8 affected subjects. Apart from moderate generalized brachydactyly, we found four phenotypic patterns associated with the mutation: first, severe middle-finger metacarpal brachydactyly (fig. 4A); second, severe little-finger distal phalanx hypoplasia/aplasia (fig. 4B); third, a combination of the above; and fourth, ring-finger lateral phalangeal duplication accompanied by 3/4 syndactyly and/or additional features (fig. 4C). Five of 17 (29%) of individuals differed in phenotype between their hands (fig. 2). Most affected individuals had little-finger distal phalangeal hypoplasia/aplasia, either alone or in association with other abnormalities, but there were three hands with middle-finger metacarpal brachydactyly but relatively normal 5th digit distal phalanges (figs. 4A and 5A). There was mild ring-finger clinodactyly in 12 hands, and 6 hands showed mild to moderate fifth-finger camptodactyly, usually in association with the little-finger distal phalanx hypoplasia.
Figure 4.
Clinical and radiographic appearance and MCPPs of limbs in individuals heterozygous for the Ile314Leu mutation in HOXD13. A, Individual III-13 from family A. Severe middle-finger metacarpal brachydactyly is present bilaterally and confirmed on the radiograph. Note also the mild clinodactyly of the ring finger. One other hand shows a similar MCPP. B, Individual IV-3 from family B (age 5 years). Absence of the distal phalanx of the little finger is confirmed on the radiograph. Two other hands show similar MCPPs. C, Left and middle, individual II-2 from family B. Note asymmetrical abnormality of the ring fingers: on the right hand there is lateral duplication of the middle phalanx, and on the left hand, there is mild clinodactyly. In addition, the hands show features illustrated in panels A and B. C, Right, The right hand of proband IV-2 in family B (age 1 year) showing lateral duplication of the middle and distal phalanges of the ring finger. D, Foot radiograph of individual III-5 from family B. Note the unilateral shortening of the left fourth metatarsal.
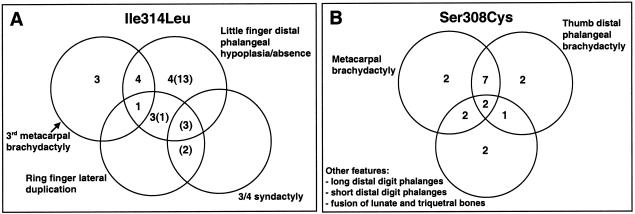
Figure 5.
Combinations of individual abnormalities in hands from subjects with the Ile314Leu mutation (A) and the Ser308Cys mutation (B). In A, figures in parentheses refer to additional cases classified clinically but not radiographically.
Of 20 feet from molecularly confirmed cases that we examined clinically and radiographically, 8 showed mild clinical abnormalities. Seven had external evidence of fifth-toe distal phalangeal hypoplasia/aplasia with absent or rudimentary nail complexes, which was confirmed radiographically. One individual had radiographically confirmed unilateral fourth ray metatarsal brachydactyly with symphalangism of the middle and distal phalanges of the little toe (fig. 4D). Radiographs obtained on the remaining 12 clinically normal feet identified 3 with symphalangism of the middle and distal phalanges of the little toe and one further case of little-toe distal phalanx hypoplasia. Duplication of the second metatarsal within the first web space was not observed in any case. All affected individuals had normal stature.
The presence of brachydactyly of specific metacarpal or metatarsal bones has not been described in association with other HOXD13 mutations, but it is the cardinal feature of brachydactyly type E (BDE [MIM 113300]). Therefore, we sequenced HOXD13 in a family (family C; fig. 2) previously classified with brachydactyly type E (Brailsford 1945; Oude Luttikhuis et al. 1996). We identified a novel heterozygous missense mutation (923C→G) corresponding to the amino acid substitution Ser308Cys, located at the 41st position of the homeodomain (fig. 3A), which segregated in concordance with the phenotype in nine affected individuals (six by direct testing and three intermediate relatives) and one unaffected individual at 50% prior risk (fig. 3C).
The detailed pedigree and phenotype in family C has been reported elsewhere (Brailsford 1945; Oude Luttikhuis et al. 1996). The characteristic features are shortening of one or more of the metacarpals or metatarsals or of both, often occurring asymmetrically, together with either shortening or elongation of specific distal phalanges (notably the first and fifth) and carpal-bone fusion. Published details of the hand phenotype are summarized in figure 5B and document wide intrafamilial variation in these features, which overlap those described in brachydactyly type D as well as type E (see “Discussion” section).
DNA Binding Assays
The distinct phenotype in these three families, notably including metacarpal and metatarsal brachydactyly, suggested that the mutations act by a mechanism distinct from that described in other HOXD13 mutations. Notably, the isoleucine at the 47th position of the homeodomain equivalent to Ile314 in HOXD13 makes hydrophobic contacts with DNA in several available crystal structures, including DNA-bound Ubx/Extradenticle (Passner et al. 1999) (fig. 1), Antennapedia (Fraenkel and Pabo 1998), and HOXB1-PBX1 homeodomain complexes (Piper et al. 1999). We speculated that the HOXD13 missense mutations might affect DNA binding and tested this using EMSA.
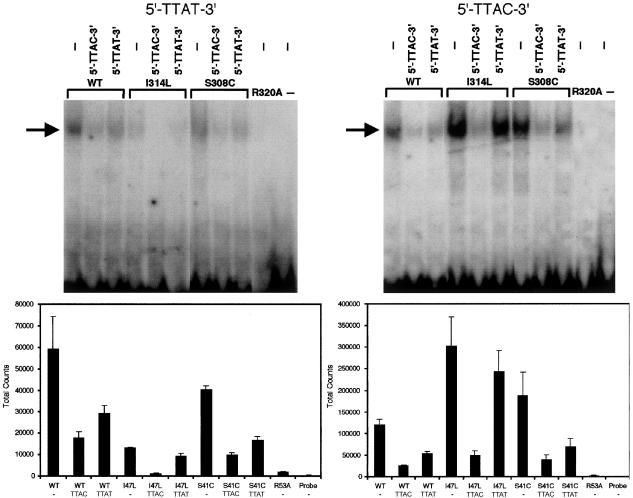
As the DNA contact made by I47 in Ubx is to the last base in the 5′-TTAT-3′ core sequence (fig. 1), we first tested the binding of wild-type and mutant proteins to ds oligonucleotides that contained 5′-TTAT-3′, 5′-TTAC-3′, 5′-TTAA-3′, 5′-TTAG-3′, and 5′-TTAU-3′ (β-strand) sequences. Significant binding was observed only to the 5′-TTAT-3′ and 5′-TTAC-3′ sequences (data not shown). As a negative control, we created an artificial mutation, Arg320Ala. Arg320 lies at the 53rd position of the homeodomain, a highly conserved residue important for contacting with the phosphate backbone of DNA (fig. 1); it is a common site for homeobox gene mutations with demonstrated loss of function (Dattani et al. 1998; Percin et al. 2000; D’Elia et al. 2001). This residue was mutated to alanine, a neutral amino acid with a high propensity to promote α-helix formation. As expected, no DNA binding was obtained using the Arg320Ala mutant protein.
We then compared the affinities of the wild-type and mutant proteins for the 5′-TTAT-3′ and 5′-TTAC-3′ target sequences in more detail. The wild-type protein had similar binding affinities for each target sequence: 50% or more of this binding could be competed off with excess unlabeled competitor oligonucleotide. By contrast, the Ile314Leu mutant protein consistently exhibited greater binding (average 2.4-fold) to 5′-TTAC-3′ but only ∼0.23-fold binding to 5′-TTAT-3′, when compared with wild-type protein. These results were in keeping with experiments involving competition assays with a 100-fold excess of unlabeled ds oligonucleotides (fig. 6). In parallel studies on the mutant Ser308Cys protein, although there was a modest increase in binding to 5′-TTAC-3′ compared with wild-type protein (average of 1.4-fold), this was not consistently replicated in independent experiments (fig. 6).
Figure 6.
In vitro binding of wild-type (WT) and mutant (I314L, S308C, and R320A) HOXD13 proteins to synthetic ds oligonucleotides. Above, representative gel shift assays employing 32P-labeled 5′-TTAT-3′ probe (left) and 5′-TTAC-3′ probe (right) in the absence (−) or presence of unlabeled competitor oligonucleotides. The arrow shows the position of the bound oligonucleotide/protein complexes. Below, quantitation (mean ± SEM) of total counts from 4–5 experiments. Note the difference in scale of absolute counts on the Y-axis.
Molecular Modeling
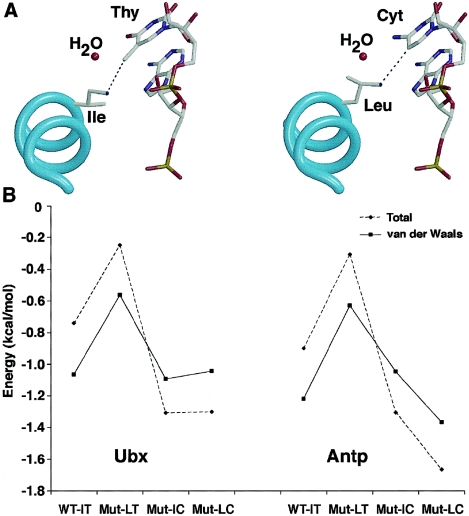
To evaluate the mechanism of the enhanced binding of the Ile314Leu mutant protein to the 5′-TTAC-3′ oligonucleotide, we modeled the interaction on the basis of the crystal structures of D. melanogaster Ubx and Antp homeodomains bound to DNA (fig. 7A). We chose these structures because they are the most closely related proteins for which the three-dimensional structure has been solved. Ubx interacts with a 5′-TTAT-3′ sequence, whereas the Antp homeodomain interacts with 5′-TAAT-3′ in the respective crystal structures. To study the role of residue 314 in HOXD13 (Ile or Leu) and the interacting base (Thy or Cyt), four models were generated, in silico, from each crystal structure to mimic the different possible interactions (WT-IT, Ile/Thy; Mut-LT, Leu/Thy; Mut-IC, Ile/Cyt; and Mut-LC, Leu/Cyt). Visual inspection of the models showed that the orientation of the side chain of residue 314 differs in the different mutants. For instance, in the Ile314Leu model (Mut-LT), the side chain of the leucine residue rotates to avoid steric clashes with the 5-methyl group of Thy (not shown), whereas in the double-mutant model (Mut-LC), the leucine can move toward the cytosine, since the latter lacks the 5-methyl group (fig. 7A). In an attempt to estimate the differences among the models, the interaction energy among each of the four combinations of amino acid and base was computed (fig. 7B). Although the energy differences among the four models are small, they are consistent with the DNA-binding results. The WT-IT and Mut-IC models have comparable interaction energies, in agreement with the similar affinity of HOXD13 for 5′-TTAT-3′ and 5′-TTAC-3′. The higher energy of interaction between Leu and Thy in the Mut-LT model, as compared with Leu and Cyt in the Mut-LC model, is consistent with the preference of the Ile314Leu mutant for 5′-TTAC-3′. The major differences are found in the van der Waals energy term, suggesting that the lower affinity of the Ile314Leu mutant for 5′-TTAT-3′ is mainly due to steric hindrance.
Figure 7.
Interaction between HOXD13 and DNA sequences. A, Models based on the Ubx/DNA complex, highlighting the interaction between Ile and thymine (Thy) in the wild type (left panel) and between Leu and cytosine (Cyt) in the “double mutant” (right panel). The cyan spiral indicates the main chain of homeodomain helix III; atoms (except H) in the position 47 side chain and DNA are color coded as follows: C = gray; O = red; N = dark blue; P = orange. B, Total (dashed line) and van der Waals (solid line) interaction energies (in kcal/mol) between Ile and Thy (WT-IT, wild type), Leu and Thy (Mut-LT, Ile→Leu mutant), Ile and Cyt (Mut-IC, Thy→Cyt mutant), and Leu and Cyt (Mut-LC, double mutant). Models were built using the Ubx (left) and Antp (right) homeodomain/DNA complexes.
Discussion
Pathogenicity of HOXD13 Mutations
Using WAVE DHPLC, we identified mutations of HOXD13 in 4 of 128 (3.1%) consecutive samples tested from patients with a wide range of congenital limb abnormalities that required surgery. This indicates that HOXD13 mutations contribute a small but significant percentage to the overall burden of congenital limb abnormalities. In this report, we focus on two missense mutations of HOXD13: Ile314Leu, detected in two patients in the initial screen, and Ser308Cys, detected in a published family following the insight that HOXD13 mutations may present with metacarpal or metatarsal brachydactyly.
Biochemically, isoleucine and leucine are both nonpolar amino acids and share many similar characteristics (Grantham 1974), so it is important to consider the possibility that the Ile314Leu substitution could be only a silent polymorphism. Indeed, in a tabulation of the relative frequency of pathogenic substitutions in humans, as compared with tolerated substitutions in orthologous proteins between species, Miller and Kumar (2001) found that for the isoleucine-to-leucine change, most substitutions are tolerated. Nevertheless, there is compelling evidence that Ile314Leu in HOXD13 is pathogenic: first, the mutation segregates perfectly with the phenotype in 18 affected and 10 unaffected family members at 50% prior risk (two-point LOD score=7.2, using a penetrance in heterozygotes of 0.99); second, the substitution was not present in 168 chromosomes from unrelated normal individuals; third, the isoleucine residue is highly conserved in HOX proteins (fig. 3A); fourth, structural information from NMR and x-ray crystallographic data support isoleucine 47 as one of four key residues in helix III (I47, Q50, N51, and M54) that are responsible for sequence-specific DNA contacts (fig. 1; reviewed by Billeter 1996); and fifth, functional studies indicate that the substitution affects DNA-binding affinity (fig. 6).
The Ser308Cys substitution occurs at position 41, between helix II and helix III of the homeodomain. Biochemically, this substitution is of intermediate severity (Grantham 1974), but it occurs in a less-conserved region of the protein, represented by a threonine residue in Ubx and Antp (fig. 3A). In both structures, this residue interacts via a water molecule with the oxygen atom of phosphates of A and T (underlined) in 5′-TT/AAT-3′, making the effect of substitution difficult to predict (Fraenkel and Pabo 1998; Passner et al. 1999). No definite functional alteration could be established from the DNA-binding studies (fig. 6), but there are several lines of evidence that Ser308Cys is also pathogenic: first, it segregates concordantly with the phenotype in nine affected and one unaffected individual; second, the nucleotide substitution was not detectable in 170 chromosomes from unrelated normal individuals; third, cysteine has been recorded in only two (of >1,000) homeobox sequences, those of D. melanogaster zerknüllt and mouse Pem (Homeodomain Resource; Banerjee-Basu et al. 2001); and fourth, previous functional analysis of a mutation occurring at the identical position of the homeodomain but involving a different substitution (Thr178Met in NKX2-5, which causes human congenital heart disease) showed reduced DNA binding and transcriptional activation compared with wild type (Zhu et al. 2000).
HOXD13 Mutations Are Associated with Brachydactyly Types D and E
The missense mutation Ile314Leu, identified in two families, exhibits a novel phenotype; as in the case of SPD (Goodman et al. 1997), the variability in expressivity is striking, both between family members and in opposite limbs. The most distinctive manifestations of this mutation, seen in the upper limbs, were isolated middle-finger metacarpal brachydactyly and lateral duplication of the distal elements of the ring finger. However, these features were observed in only a proportion of patients (fig. 5A) and were sometimes asymmetrical (fig. 4C); the lateral duplications did not involve more proximal phalanges or metacarpal elements, in contrast to more severe cases of SPD. Only a minority of individuals exhibited 3/4 syndactyly overlapping the typical appearance of SPD. The occurrence of metacarpal and metatarsal brachydactyly led us to analyze a family published elsewhere (family C) segregating this phenotype through five generations (Brailsford 1945; Oude Luttikhuis et al. 1996). The identification of a heterozygous Ser308Cys mutation in family C confirms that HOXD13 mutations are a cause of metacarpal and metatarsal brachydactyly. The phenotype of this latter mutation was characterized by selective shortening of metacarpals (rays 3, 4, and 5) and metatarsals (rays 1 and 4) as well as shortening of the distal phalanx of the thumb. No family member had syndactyly, but foot radiographs of several affected individuals show an abnormally broad first metatarsal, sometimes with a bony spur somewhat reminiscent of the abnormality described by Goodman et al. (1998), postulated to be associated with haploinsufficiency.
The most commonly adopted classification of the nonsyndromic brachydactylies is that devised by Bell (1951). Type D brachydactyly (BDD [MIM 113200] is defined as a short, broad distal phalanx in the thumb, and BDE is defined as comprising one or more shortened metacarpals and metatarsals. Although both types of brachydactyly may be a feature of several syndromes (Rubinstein-Taybi and Saethre-Chotzen syndromes in type D; Turner syndrome, mutations in the α-subunit of Gs protein, and deletions of 2q37 in type E), no locus has been defined for nonsyndromic BDD or BDE. Although not breeding true for the pure phenotypes, families A–C all exhibit features of BDE; in family C, this is combined in some individuals with BDD. The present study identifies HOXD13 mutations as a cause of both BDD and BDE, and further mutation analysis of patients with these diagnoses may help to refine the clinical classification of these disorders.
Functional Effects of Homeodomain Substitutions
EMSA showed that Ile314Leu confers an increased binding affinity for the target sequence 5′-TTAC-3′ but a decreased affinity for 5′-TTAT-3′ compared with wild-type protein, suggesting that the mutation has mixed gain- and loss-of-affinity effects. Mutations of the homeodomain that increase binding affinity are unusual, but an example reported elsewhere was the Pro148His substitution of MSX2 (located in the N-terminal arm of the homeodomain), which exhibited this effect in a similar assay (Ma et al. 1996). Isoleucine and leucine are isomers; that is, they only differ by the position of a methyl group that is linked to the first (Ile: Cβ) or to the second (Leu: Cγ) carbon atom of the side chain. As a consequence, the Ile314Leu mutation may create steric hindrance between the leucine and the 5-methyl group of the thymine at the 3′ end of the 5′-TTAT-3′ core, explaining the reduced binding affinity of the mutant protein for this sequence. By contrast, in the presence of cytosine, which lacks the 5-methyl group, we propose that the Ile314Leu substitution may complement the lack of methyl in the base, resulting in an increased affinity for 5′-TTAC-3′ (fig. 7). On a cautionary note, in vivo activity is likely to require the binding of partner proteins (such as MEIS1; Shen et al. 1997a) to HOXD13, which might modulate the relative binding affinities of the transcription complex to different DNA targets.
Although EMSA analysis of the Ser308Cys substitution did not identify consistent differences from wild type, our results show that this mutation does not simply abolish DNA binding (in contrast to the Arg320Ala control). The substitution lies between helices II and III and involves relatively little change in molecular volume, compatible with a subtle effect on DNA binding. Further work will be required to determine the pathophysiological mechanism of this mutation.
Three Distinct Classes of Mutation Exist in HOXD13
Three distinct classes of mutations have now been described in HOXD13: polyalanine tract expansions, truncations, and specific amino acid substitutions. Each is likely to exert its effect through different mechanisms, consistent with differences in the phenotypic outcome. Accumulating evidence suggests that polyalanine expansion mutations may exert a dominant negative effect over wild-type protein (Bruneau et al. 2001). The mutations which truncate the HOXD13 protein are likely to cause loss of function (haploinsufficiency), whereas we have presented evidence that the Ile314Leu substitution in the homeodomain exerts both gain and loss of DNA-binding affinity to different targets. This was also speculated to be the pathological mechanism of missense substitutions of two key base-contacting residues (Q50 and N51; see fig. 1) in HOXA13, both of which were associated with severe or variant HFUS phenotypes; however, no functional data were presented in these cases (Goodman et al. 2000; Innis et al. 2002).
Our conclusion is that a leucine at position 47 of the homeodomain causes an altered repertoire of DNA binding rather than simple loss of function, which implies that in a different molecular and developmental context, a homeobox gene encoding Leu47 could act in a physiological manner. It is therefore interesting that several homeodomain-containing proteins from the fungal pathogen Ustilago maydis indeed utilize a leucine at this position (Kronstad and Leong 1990; Schulz et al. 1990; Banerjee-Basu et al. 2001).
Acknowledgments
We thank the families who took part in the study for their help and cooperation. We thank Navaratnam Elanko, for help with DNA extraction; Kevin Clark, for DNA sequencing; Anne Goriely and Christina Tufarelli, for technical advice; and Alasdair Hunter, for access to the ANTRO software. This study was funded by the Wellcome Trust (to D.J. and A.O.M.W.), the Ministry of Education in Taiwan (to S-h.K.), the Overseas Research Students Awards Scheme (to S-h.K.), CNRS France (to P.R.), and MRC-UK (to P.R. and R.M.E.).
Electronic-Database Information
Accession numbers and URLs for data presented herein are as follows:
- Ensembl, http://www.ensembl.org/Homo_sapiens/mapview?chr=2 (for physical map of 2q31 region)
- GenBank, http://www.ncbi.nlm.nih.gov/GenBank/ (for HOXD13 [accession numbers. AF005219, AF005220, AC009336, and NM_000523])
- Homeodomain Resource, http://research.nhgri.nih.gov/homeodomain/ (for homeodomain sequences and DNA binding sites)
- Online Mendelian Inheritance in Man (OMIM), http://www.ncbi.nlm.nih.gov/Omim/(for SPD, HFUS, BDD, and BDE)
- Protein Data Bank, http://www.pdb.org/ (for Antp [ID: 9ANT] and Ubx [ID: 1B8I] homeodomain-DNA structures)
References
- Albrecht AN, Schwabe GC, Stricker S, Böddrich A, Wanker EE, Mundlos S (2002) The synpolydactyly homolog (spdh) mutation in the mouse: a defect in patterning and growth of limb cartilage elements. Mech Dev 112:53–67 [DOI] [PubMed] [Google Scholar]
- Armour CM, Bulman DE, Hunter AGW (2000) Clinical and radiological assessment of a family with mild brachydactyly type A1: the usefulness of metacarpophalangeal profiles. J Med Genet 37:292–296 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Banerjee-Basu S, Baxevanis AD (2001) Molecular evolution of the homeodomain family of transcription factors. Nucleic Acids Res 29:3258–3269 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Banerjee-Basu S, Sink DW, Baxevanis AD (2001) The Homeodomain Resource: sequences, structures, DNA binding sites and genomic information. Nucleic Acids Res 29:291–293 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bell J (1951) On brachydactyly and symphalangism. In: The treasury of human inheritance. Vol 5. Cambridge University Press, Cambridge, pp 1–31 [Google Scholar]
- Billeter M (1996) Homeodomain-type DNA recognition. Prog Biophys Mol Biol 66:211–225 [DOI] [PubMed] [Google Scholar]
- Brailsford JF (1945) Familial brachydactyly. Br J Radiol 18:167–172 [DOI] [PubMed] [Google Scholar]
- Brooks BR, Bruccoleri RE, Olafson BD, States DJ, Swaminathan S, Karplus M (1983) CHARMM: a program for macromolecular energy, minimization, and dynamics calculations. J Comput Chem 4:187–217 [Google Scholar]
- Bruneau S, Johnson KR, Yamamoto M, Kuroiwa A, Duboule D (2001) The mouse Hoxd13spdh mutation, a polyalanine expansion similar to human type II synpolydactyly (SPD), disrupts the function but not the expression of other Hoxd genes. Dev Biol 237:345–353 [DOI] [PubMed] [Google Scholar]
- Bürglin TR (1994) A comprehensive classification of homeobox genes. In: Duboule D (ed) Guidebook to the homeobox genes. Oxford University Press, Oxford, pp 27–71 [Google Scholar]
- Calabrese O, Bigoni S, Gualandi F, Trabanelli C, Camera G, Calzolari E (2000) A new mutation in HOXD13 associated with foot pre-postaxial polydactyly. Eur J Hum Genet 8 S1:140 [Google Scholar]
- Caronia G, Goodman FR, McKeown CME, Scambler PJ, Zappavigna V (2003) An I47L substitution in the HOXD13 homeodomain causes a novel human limb malformation by producing a selective loss of function. Development 130:1701–1712 [DOI] [PubMed] [Google Scholar]
- Dattani MT, Martinez-Barbera J-P, Thomas PQ, Brickman JM, Gupta R, Mårtensson I-L, Toresson H, Fox M, Wales JKH, Hindmarsh PC, Krauss S, Beddington RSP, Robinson ICAF (1998) Mutations in the homeobox gene HESX1/Hesx1 associated with septo-optic dysplasia in human and mouse. Nat Genet 19:125–133 [DOI] [PubMed] [Google Scholar]
- Davey RA, Hamson CA, Healey JJ, Cunningham JM (1997) In vitro binding of purified murine ecotropic retrovirus envelope surface protein to its receptor, MCAT-1. J Virol 71:8096–8102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Debeer P, Bacchelli C, Scambler PJ, De Smet L, Fryns J-P, Goodman FR (2002) Severe digital abnormalities in a patient heterozygous for both a novel missense mutation in HOXD13 and a polyalanine tract expansion in HOXA13. J Med Genet 39:852–856 [DOI] [PMC free article] [PubMed] [Google Scholar]
- D'Elia AV, Tell G, Paron I, Pellizzari L, Lonigro R, Damante G (2001) Missense mutations of human homeoboxes: a review. Hum Mutat 18:361–374 [DOI] [PubMed] [Google Scholar]
- Dib C, Faure S, Fizames C, Samson D, Drouot N, Vignal A, Millaseau P, Marc S, Hazan J, Seboun E, Lathrop M, Gyapay G, Morissette J, Weissenbach J (1996) A comprehensive genetic map of the human genome based on 5,264 microsatellites. Nature 380:152–154 [DOI] [PubMed] [Google Scholar]
- Dollé P, Dierich A, LeMeur M, Schimmang T, Schuhbaur B, Chambon P, Duboule D (1993) Disruption of the Hoxd-13 gene induces localized heterochrony leading to mice with neotenic limbs. Cell 75:431–441 [DOI] [PubMed] [Google Scholar]
- Esnouf RM (1999) Further additions to MolScript version 1.4, including reading and contouring of electron-density maps. Acta Crystallogr D Biol Crystallogr 55:938–940 [DOI] [PubMed] [Google Scholar]
- Fraenkel E, Pabo CO (1998) Comparison of X-ray and NMR structures for the Antennapedia homeodomain-DNA complex. Nat Struct Biol 5:692–697 [DOI] [PubMed] [Google Scholar]
- Garn SM, Hertzog KP, Poznanski AK, Nagy JM (1972) Metacarpophalangeal length in the evaluation of skeletal malformation. Radiology 105:375–381 [DOI] [PubMed] [Google Scholar]
- Gehring WJ, Qian YQ, Billeter M, Furukubo-Tokunaga K, Schier AF, Resendez-Perez D, Affolter M, Otting G, Wüthrick K (1994) Homeodomain-DNA recognition. Cell 78:211–223 [DOI] [PubMed] [Google Scholar]
- Goodman F, Giovannucci-Uzielli ML, Hall C, Reardon W, Winter R, Scambler PJ (1998) Deletions in HOXD13 segregate with an identical, novel foot malformation in two unrelated families. Am J Hum Genet 63:992–1000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goodman FR (2002) Limb malformations and the human HOX genes. Am J Med Genet 112:256–265 [DOI] [PubMed] [Google Scholar]
- Goodman FR, Bacchelli C, Brady AF, Brueton LA, Fryns J-P, Mortlock DP, Innis JW, Holmes LB, Donnenfeld AE, Feingold M, Beemer FA, Hennekam RCM, Scambler PJ (2000) Novel HOXA13 mutations and the phenotypic spectrum of hand-foot-genital syndrome. Am J Hum Genet 67:197–202 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goodman FR, Mundlos S, Muragaki Y, Donnai D, Giovannucci-Uzielli ML, Lapi E, Majewski F, McGaughran J, McKeown C, Reardon W, Upton J, Winter RM, Olsen BR, Scambler PJ (1997) Synpolydactyly phenotypes correlate with size of expansions in HOXD13 polyalanine tract. Proc Natl Acad Sci USA 94:7458–7463 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grantham R (1974) Amino acid difference formula to help explain protein evolution. Science 185:862–864 [DOI] [PubMed] [Google Scholar]
- Innis JW, Goodman FR, Bacchelli C, Williams TM, Mortlock DP, Sateesh P, Scambler PJ, McKinnon W, Guttmacher AE (2002) A HOXA13 allele with a missense mutation in the homeobox and a dinucleotide deletion in the promoter underlies Guttmacher syndrome. Hum Mutat 19:573–574 [DOI] [PubMed] [Google Scholar]
- Kan S-h, Johnson D, Giele H Wilkie AOM. An acceptor splice site mutation in HOXD13 results in variable hand, but consistent foot malformations. Am J Med Genet (in press) [DOI] [PubMed] [Google Scholar]
- Kosaki K, Kosaki R, Suzuki T, Yoshihashi H, Takahashi T, Sasaki K, Tomita M, McGinnis W, Matsuo N (2002) Complete mutation analysis panel of the 39 human HOX genes. Teratology 65:50–62 [DOI] [PubMed] [Google Scholar]
- Kronstad JW, Leong SA (1990) The b-mating type locus of Ustilago maydis contains variable and constant regions. Genes Dev 4:1384–1395 [DOI] [PubMed] [Google Scholar]
- Krumlauf R (1994) HOX genes in vertebrate development. Cell 78:191–201 [DOI] [PubMed] [Google Scholar]
- Ma L, Golden S, Wu L, Maxson R (1996) The molecular basis of Boston-type craniosynostosis: the Pro148→His mutation in the N terminal arm of the MSX2 homeodomain stabilizes DNA binding without altering nucleotide sequence preferences. Hum Mol Genet 5:1915–1920 [DOI] [PubMed] [Google Scholar]
- Mann RS, Affolter M (1998) HOX proteins meet more partners. Curr Opin Genet Dev 8:423–429 [DOI] [PubMed] [Google Scholar]
- Mann RS, Carroll SB (2002) Molecular mechanisms of selector gene function and evolution. Curr Opin Genet Dev 12:592–600 [DOI] [PubMed] [Google Scholar]
- Merritt EA, Bacon DJ (1997) Raster 3D: photorealistic molecular graphics. Methods Enzymol 277:505–524 [DOI] [PubMed] [Google Scholar]
- Miller MP, Kumar S (2001) Understanding human disease mutations through the use of interspecific genetic variation. Hum Mol Genet 10:2319–2328 [DOI] [PubMed] [Google Scholar]
- Mortlock DP, Innis JW (1997) Mutation of HOXA13 in hand-foot-genital syndrome. Nat Genet 15:179–180 [DOI] [PubMed] [Google Scholar]
- Muragaki Y, Mundlos S, Upton J, Olsen BR (1996) Altered growth and branching patterns in synpolydactyly caused by mutations in HOXD13. Science 272:548–551 [DOI] [PubMed] [Google Scholar]
- Oude Luttikhuis MEM, Williams DK, Trembath RC (1996) Isolated autosomal dominant type E brachydactyly: exclusion of linkage to candidate regions 2q37 and 20q13. J Med Genet 33:873–876 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Passner JM, Ryoo HD, Shen L, Mann RS, Aggarwal AK (1999) Structure of a DNA-bound Ultrabithorax-Extradenticle homeodomain complex. Nature 397:714–719 [DOI] [PubMed] [Google Scholar]
- Percin EF, Ploder LA, Yu JJ, Arici K, Horsford DJ, Rutherford A, Bapat B, Cox DW, Duncan AMV, Kalnins VI, Kocak-Altintas A, Sowden JC, Traboulsi E, Sarfarazi M, McInnes RR (2000) Human microphthalmia associated with mutations in the retinal homeobox gene CHX10. Nat Genet 25:397–401 [DOI] [PubMed] [Google Scholar]
- Piper DE, Batchelor AH, Chang C-P, Cleary ML, Wolberger C (1999) Structure of a HoxB1-Pbx1 heterodimer bound to DNA: role of the hexapeptide and a fourth homeodomain helix in complex formation. Cell 96:587–597 [DOI] [PubMed] [Google Scholar]
- Poznanski AK, Garn SM, Nagy JM, Gall JC Jr (1972) Metacarpophalangeal pattern profiles in the evaluation of skeletal malformations. Radiology 104:1–11 [DOI] [PubMed] [Google Scholar]
- Rijli FM, Chambon P (1997) Genetic interactions of Hox genes in limb development: learning from compound mutants. Curr Opin Genet Dev 7:481–487 [DOI] [PubMed] [Google Scholar]
- Sarfarazi M, Akarsu AN, Sayli BS (1995) Localization of the syndactyly type II (synpolydactyly) locus to 2q31 region and identification of tight linkage to HOXD8 intragenic marker. Hum Mol Genet 4:1453–1458 [DOI] [PubMed] [Google Scholar]
- Sayli BS, Akarsu AN, Sayli U, Akhan O, Ceylaner S, Sarfarazi M (1995) A large Turkish kindred with syndactyly type II (synpolydactyly). 1. Field investigation, clinical and pedigree data. J Med Genet 32:421–434 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schulz B, Banuett F, Dahl M, Schlesinger R, Schäfer W, Martin T, Herskowitz I, Kahmann R (1990) The b alleles of U. maydis, whose combinations program pathogenic development, code for polypeptides containing a homeodomain-related motif. Cell 60:295–306 [DOI] [PubMed] [Google Scholar]
- Scott MP (1992) Vertebrate homeobox gene nomenclature. Cell 71:551–553 [DOI] [PubMed] [Google Scholar]
- Shen W-F, Montgomery JC, Rozenfeld S, Moskow JJ, Lawrence HJ, Buchberg AM, Largman C (1997a) AbdB-like Hox proteins stabilize DNA binding by the Meis1 homeodomain proteins. Mol Cell Biol 17:6448–6458 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen W-F, Rozenfeld S, Lawrence HJ, Largman C (1997b) The Abd-B-like Hox homeodomain proteins can be subdivided by the ability to form complexes with Pbx1a on a novel DNA target. J Biol Chem 272:8198–8206 [DOI] [PubMed] [Google Scholar]
- Zákány J, Fromental-Ramain C, Warot X, Duboule D (1997) Regulation of number and size of digits by posterior Hox genes: a dose-dependent mechanism with potential evolutionary implications. Proc Natl Acad Sci USA 94:13695–13700 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu W, Shiojima I, Hiroi Y, Zou Y, Akazawa H, Mizukami M, Toko H, Yazaki Y, Nagai R, Komuro I (2000) Functional analyses of three Csx/Nkx-2.5 mutations that cause human congenital heart disease. J Biol Chem 275:35291–35296 [DOI] [PubMed] [Google Scholar]