Abstract
Juvenile myoclonic epilepsy (JME) is a common form of generalized epilepsy that starts in adolescence. A major JME susceptibility locus (EJM1) was mapped to chromosomal region 6p21 in three independent linkage studies, and association was reported between JME and a microsatellite marker in the 6p21 region. The critical region for EJM1 is delimited by obligate recombinants at HLA-DQ and HLA-DP. In the present study, we found highly significant linkage disequilibrium (LD) between JME and a core haplotype of five single-nucleotide–polymorphism (SNP) and microsatellite markers in this critical region, with LD peaking in the BRD2 (RING3) gene (odds ratio 6.45; 95% confidence interval 2.36–17.58). DNA sequencing revealed two JME-associated SNP variants in the BRD2 (RING3) promoter region but no other potentially causative coding mutations in 20 probands from families with positive LOD scores. BRD2 (RING3) is a putative nuclear transcriptional regulator from a family of genes that are expressed during development. Our findings strongly suggest that BRD2 (RING3) is EJM1, the first gene identified for a common idiopathic epilepsy. These findings also suggest that abnormalities of neural development may be a cause of common idiopathic epilepsy, and the findings have implications for the generalizability of proposed pathogenetic mechanisms, derived from diseases that show Mendelian transmission, to their complex counterparts.
Introduction
The epilepsies comprise an enormous diversity of disorders of heterogeneous etiology, manifestation, and prognosis. Almost half of all epilepsies have some genetic basis (Annegers et al. 1996), but only a small proportion appear to display Mendelian inheritance. Many of our current beliefs about the molecular and cellular mechanisms in epilepsy derive from these examples, which are based on rare, large pedigrees. Such pedigrees received the most attention because they have been the simplest to study genetically. Although mutations in genes for ion channels, neuroreceptors, and neurotransmitters have been demonstrated in such rare, densely affected epilepsy pedigrees (Mulley et al. 2003), the forms of epilepsy commonly seen in the clinic show neither the specific mutations nor any other mutations in genes identified in those pedigrees (Harkin et al. 2002; Kananura et al. 2002; authors' unpublished data). In contrast to the rare Mendelian pedigrees, the common forms of idiopathic generalized epilepsy (IGE) have a complex inheritance, even though the IGEs are thought to have an exclusively genetic basis (Greenberg et al. 1992). Not only do population studies (Tsuboi and Christian 1973; Beck-Mannagetta and Janz 1991) suggest an oligogenic mode of inheritance with interaction between loci, but a genome scan of individuals with adolescent-onset IGE demonstrated strong evidence of linkage to several loci, combinations of which may lead to specific epilepsy syndromes (Durner et al. 2001). It seems unlikely that single gene mutations are sufficient to explain the molecular mechanisms for epilepsies with this model of complex inheritance.
Juvenile myoclonic epilepsy (JME [MIM 254770]) is one of the most easily recognized IGEs of adolescence, diagnosable by the occurrence of bilateral, upper-limb myoclonic jerks on awakening (Janz and Christian 1957). Studies of three separate family collections have reported significant evidence of linkage between JME and the major susceptibility locus EJM1 at chromosome 6p21, designated “EJM1” (Greenberg et al. 1988b; Durner et al. 1991; Weissbecker et al. 1991; Sander et al. 1997; Greenberg et al. 2000). In addition to linkage, there is evidence of allelic association in this region, with a microsatellite allele located in the HLA class II region. This microsatellite is located in an intron of the BRD2 (RING3) gene (Greenberg et al. 2000). Recombination mapping in families with JME has delimited the boundaries of EJM1 to a 1-cM region between the HLA-DQ and HLA-DP (DQ-DP) loci (Sander et al. 1997; Greenberg et al. 2000). In the present study, we aimed to confirm and further refine gene localization of EJM1 and to search for molecular variants that might explain JME susceptibility in a complex genetic model.
Families and Methods
Study Design
Our earlier studies suggested that EJM1 was located between DQ and DP (Sander et al. 1997; Greenberg et al. 2000), so we first sought evidence of association between JME and single SNP marker alleles in this region. Then, to increase informativeness of markers, we reconstructed two-locus haplotypes from data on consecutive SNPs. We performed case-control analysis, using these haplotypes, and, to guard against possible population stratification, we confirmed positive haplotype associations, using intrafamilial controls (untransmitted alleles) in haplotype relative-risk analysis (Falk and Rubenstein 1987). Next, we searched for a common risk haplotype in families with positive LOD scores in the EJM1 region. Finally, we searched for mutations by sequencing exons and promoter sequences in BRD2 and adjacent genes that showed significant linkage disequilibrium (LD) with JME.
Families
We collected probands with JME and their families from physicians’ practices, as described elsewhere (Greenberg et al. 2000). We selected probands with typical forms of JME, in accordance with international classification guidelines (Commission on Classification and Terminology of the International League Against Epilepsy 1989). Twenty parent-offspring trios from families with positive LOD scores (>0.1) at both of two EJM1 microsatellite markers, DQB1 and DRB1, were designated as the “EJM1+ set.” LOD scores for EJM1+ families were 0.15–1.50, with a mean of 0.38. Alleles and haplotypes of probands in the EJM1+ set were used as case data in the case-control analysis to find associations. Transmitted and untransmitted alleles and haplotypes in the EJM1+ set were also used in transmission/disequilibrium testing. Institutional review board approval for this study was obtained from the appropriate institutions. All participating patients and family members gave their informed consent.
Controls
We used three control groups: The first consisted of 53 JME parent-offspring trios with negative LOD scores at EJM1 markers (EJM1−), collected at the same time and from the same population as EJM1+ families. The second comprised laboratory controls, which consisted of 64 carriers of Wilson disease or spinal muscular atrophy from the Columbia Genome Center. We used the epilepsy control group because it represented a population similar to that from which the EJM1+ case set was drawn, thus safeguarding against the possibility of selection bias in the case-control analysis. One theoretical disadvantage of using the epilepsy control group is the possibility of over-matching (i.e., an association cannot be demonstrated because case and control groups with JME might share too many allelic similarities at adjacent markers). We therefore used the laboratory controls as a second control group to check for over-matching in case-control analysis. Finding significant LD using each of the two control groups would therefore demonstrate that associated alleles or haplotypes are neither specific to populations with epilepsy nor a result of selection bias. Third, we used untransmitted alleles in the EJM1+ set as internal controls for EJM1+ transmitted alleles in haplotype relative-risk analysis.
DNA Preparation
DNA was purified from blood or lymphoblastoid cell lines, using the Puregene kit (Gentra Systems) according to the manufacturer’s protocols. PCR amplification for all fragments was performed on either an MJ Research Tetrad thermal cycler or a Hybaid MultiBlock System under the following conditions: 32 cycles at 94°C for 2 min, with denaturation at 94°C for 30 s, annealing at 56°C for 30 s, and an extension at 72°C for 45 s, followed by a final extension at 72°C for 4 min.
SNP Discovery
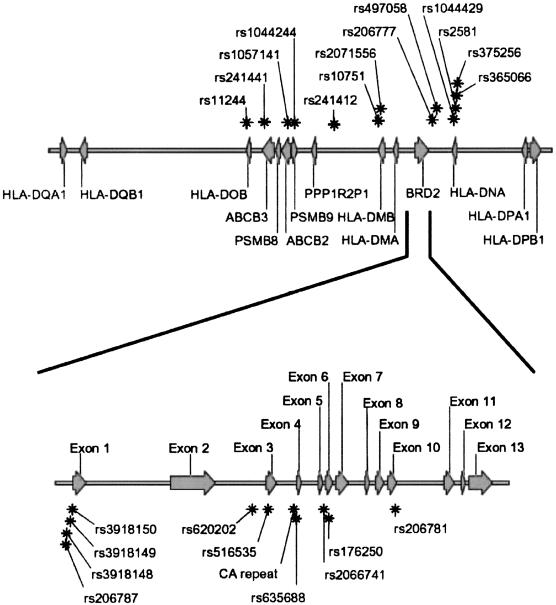
We selected >50 SNPs from the National Center for Biotechnology Information (NCBI) database, but not all of these SNPs were found in our samples. We also identified new SNPs by DNA sequencing of the region, which we entered into the NCBI database. Ultimately, we were able to use 20 SNPs for association analysis, all from the DQ-DP region and with minor allele frequency >10%. The location of these SNPs, in relation to known genes in this region, is shown in figure 1 (top). The genomic structure of BRD2 (RING3) and the CA repeat microsatellite in BRD2 (RING3), with which we had previously demonstrated association, is shown in figure 1 (bottom).
Figure 1.
DQ-DP region on human chromosome 6, SNP markers, and genomic structure of BRD2 (RING3). Top, arrows along the chromosome denote genes, which are labeled, and their direction of transcription; asterisks denote SNP markers. Bottom, BRD2 (RING3) gene exploded to show the relation of the exonic sequences with SNPs, the CA repeat microsatellite, and other DNA variations.
SNP Genotyping
Fluorescence polarization analysis of previously described mutations and polymorphic sites found during this study was done using the AcycloPrime-FP SNP detection kit (Perkin Elmer Life Sciences) according to the manufacturer’s protocol. Fluorescence measurements were performed on fluorescence microplate analyzer (LJL BioSystems).
Sequencing and Mutation Analysis
Sequences of coding and promoter regions of BRD2 (RING3), PPP1R2P1, DMA, DMB, and DNA genes were determined on the ABI 310 automated sequencer using the ABI Taq2 sequencing kit according to the manufacturer’s protocol. Presequencing PCR clean-up was done by enzymatic degradation of primers and dNTPs, using exonuclease I and shrimp alkaline phosphatase cocktail.
Statistical Analysis
We first performed unmatched case-control analyses on single SNPs, generating odds ratios (ORs) with 95% CIs. To increase informativeness of the markers, we then generated consecutive two-locus haplotypes, reconstructing haplotypes from genotypic data, using GENEHUNTER (Kruglyak et al. 1996). We compared allele (for single SNPs) and two-locus haplotype frequencies in the EJM1+ set with frequencies in two separate control groups: EJM1− families and nonepilepsy controls. We used both case-control analysis and the haplotype relative risk (HRR) design (Falk and Rubenstein 1987) to test for LD. The HRR method eliminates potential population stratification, which might occur if underlying allele frequencies differed in the case and control populations from which the families were drawn. However, the HRR design, which is restricted to one control per case, has lower statistical power to detect an association than does the case-control analysis, which uses roughly three controls per case. After finding two-locus associations, we identified a longer common risk haplotype spanning five SNP and microsatellite markers. We calculated ORs of association for the common core haplotype and computed the power of our data set to detect this association, under the assumption of a two-sided type I error rate of 5%. All analyses were performed using Stata for Macintosh (StataCorp 1996).
We used computer simulation (Greenberg et al. 1999) to assess the true significance of two-locus haplotype associations (i.e., type I error). Computer simulation offers a more realistic estimate than does conventional Bonferroni correction. In this genetic context, Bonferroni correction gives an overly conservative estimate of type I error, because it does not take into account the nonindependence of adjacent alleles in the presence of LD. In the simulation, we randomly generated 20 equally spaced biallelic markers that incorporate known LD in the DQ-DP region. We assumed no recombination between the markers and no association with disease. We simulated 10,000 data sets of case and control families, each data set being the same size as the EJM1+ and control sets, and we analyzed the resulting simulated data exactly as we analyzed our actual data. We counted the number of data sets in which one marker, or a consecutive two-locus or longer haplotype, showed random association with disease in these 10,000 data sets, to determine an empirical and more realistic P value for our case-control analyses.
Results
LD with JME
Single-SNP case-control analyses suggested eight allelic associations in the DQ-DP region. These all occurred in or adjacent to the BRD2 (RING3) gene and its promoter region (table 1). Case-control analyses using consecutive two-locus haplotypes confirmed single SNP analysis findings. The results were similar whether the EJM1− set or the nonepilepsy set was used as a control. We found three almost-consecutive JME-associated two-locus haplotypes spanning ∼41 kb of the DQ-DP region. This region included BRD2 (RING3) and HLA-DMB genes (table 2 and fig. 2). Simulations, which incorporated the known LD in the region and the SNP allele frequencies, showed that the association with two consecutive SNP markers was highly significant (P<.0016). Further, the association with three consecutive two-locus haplotypes was very highly significant (P<.0001). In 10 of the 20 EJM1+ individuals, haplotype analysis identified a common core haplotype that was centered between markers rs620202 telomerically and rs2066741 centromerically, a distance of 2,256 bp. A further five individuals had a similar haplotype, differing by only one allele; the remaining five individuals differed by two or more alleles. This core haplotype was located in the BRD2 (RING3) gene itself (table 3) and conferred an increased disease odds of 6.45 (95% CI 2.36–17.58) in case-control analysis with nonepilepsy controls, 9.58 (95% CI 2.97–30.63) with EJM1− controls, and 3.89 (95% CI 1.46–10.37) with HRR analysis. Nine of ten EJM1+ individuals were heterozygous for the risk haplotype, which was consistent with our LOD score maximization under a dominant model at EJM1 (Greenberg et al. 2000). Our association findings therefore confirmed and refined previous localization of EJM1 by linkage.
Table 1.
Case-Control Analysis of Single SNPs
|
Frequency in |
||||||
| Markera | Distanceb(bp) | Location | Allele | EJM1+ | Control | OR (95% CI)c |
| rs11244 | 0 | HLA-DOB | C/t | .82 | .75 | 1.51 (.61–3.68) |
| rs241441 | 5,070 | ABCB3 | C/t | .40 | .37 | 1.15 (.56–2.36) |
| rs1057141 | 37,989 | ABCB2 | G/a | .85 | .84 | 1.09 (.41–2.85) |
| rs1044244 | 44,329 | PSMB9 | T/c | .28 | .26 | 1.08 (.49–2.38) |
| rs241412 | 80,879 | … | T/g | .43 | .34 | 1.44 (.70–2.97) |
| rs10751 | 121,762 | HLA-DMB | T/c | .25 | .13 | 2.14 (.90–5.08) |
| rs2071556 | 123,780 | HLA-DMB | C/a | .55 | .36 | 2.19 (1.07–4.50) |
| rs206787 | 162,081 | BRD2 promoter | T/a | .53 | .33 | 2.21 (1.08–4.52) |
| rs3918149 | 162,216 | BRD2 promoter | T/c | .29 | .13 | 2.80 (1.19–6.64) |
| rs620202 | 161,048 | BRD2 intron 1 | G/t | .85 | .72 | 2.16 (.85–5.46) |
| rs516535 | 161,524 | BRD2 exon 2 | C/t | .48 | .31 | 2.05 (1.00–4.22) |
| rs635688 | 162,370 | BRD2 intron 2 | T/c | .53 | .34 | 2.16 (1.05–4.42) |
| rs2066741 | 163,304 | BRD2 intron 5 | T/c | .68 | .45 | 2.51 (1.20–5.24) |
| rs206781 | 165,351 | BRD2 exon 9 | T/c | .70 | .65 | 1.27 (.59–2.69) |
| rs206777 | 171,733 | … | C/t | .50 | .30 | 2.29 (1.11–4.71) |
| rs497058 | 176,049 | … | T/c | .51 | .34 | 2.08 (1.01–4.28) |
| rs1044429 | 191,853 | … | C/t | .85 | .81 | 1.33 (.51–3.44) |
| rs2581 | 193,612 | HLA-DNA | T/g | .43 | .33 | 1.48 (.72–3.05) |
| rs365066 | 194,349 | HLA-DNA | T/c | .40 | .33 | 1.35 (.65–2.80) |
| rs375256 | 195,031 | HLA-DNA | T/c | .79 | .75 | 1.22 (.50–2.99) |
Markers are listed in order from DQ to DP.
Distances are measured from the first SNP marker, rs11244.
Statistically significant associations are underlined. These associated SNPs span the HLA-DMB gene, the BRD2 (RING3) gene, and promoter region.
Table 2.
Two-Locus and Core Haplotypes: HRR and Case-Control Analyses[Note]
| Markers | Haplotype | T(n = 40) | nT(n = 40) | EJM1−(n = 106) | C(n = 128) | HRR (95% CI)a,bT vs. nT | OR (95% CI)bEJM1+ vs. EJM1− | OR (95% CI)bT vs. C |
| rs241441-rs1057141 | C-G | .28 | .25 | .22 | .30 | 1.14 (.40–3.21) | 1.36 (.60–3.12) | .90 (.41–1.96) |
| rs1044244-rs241412 | T-T | .28 | .26 | .15 | .18 | 1.09 (.38–3.08) | 1.47 (.58–3.72) | 1.42 (.58–3.49) |
| rs10751-rs2071556 | T-C | .20 | .13 | .08 | .09 | 1.69 (.48–5.86) | 2.91 (1.04–8.14) |
2.61 (1.00–6.89) |
| rs206787-rs3918149 | T-T | .25 | .07 | .07 | .07 | 4.83 (1.07–24.02) |
4.62 (1.67–12.81) |
4.37 (1.67–11.45) |
| rs620202-rs516535 | G-C | .48 | .35 | .35 | .28 | 1.65 (.63–4.26) | 1.69 (.81–3.51) | 2.27 (1.10–4.71) |
| CA repeat-rs635688 | 6-T | .35 | .13 | .09 | .13 | 3.63 (1.10–11.85) |
5.44 (2.15–13.75) |
3.45 (1.53–7.82) |
| rs2066741-rs206781 | T-T | .55 | .25 | .51 | .45 | 3.67 (1.35–9.94) |
1.24 (.61–2.56) | 1.52 (.75–3.09) |
| rs206777-rs497058 | C-T | .38 | .16 | .25 | .24 | 3.25 (1.05–9.92) |
1.82 (.84–3.94) | 1.92 (.91–4.08) |
| rs1044429-rs2581 | C-T | .38 | .39 | .37 | .29 | .95 (.37–2.47) | 1.03 (.49–2.17) | 1.45 (.694–3.04) |
| rs365066-rs375256 | T-T | .18 | .13 | .21 | .17 | 1.52 (.42–5.42) | .80 (.32–2.01) | 1.06 (.42–2.68) |
| rs620202-rs516535-brd2-rs635688-rs2066741 | G-C-6-T-T | .28 | .10 | .13 | .06 | 3.89 (1.46–1.37) |
9.58 (2.97–30.63) |
6.45 (2.36–17.58) |
Note.— C = EJM1− controls; Hap = haplotype; nT = nontransmitted chromosomes of EJM1+ trios; T = transmitted. A core haplotype of five markers (last row), centered on these associated haplotypes, confers a 4- to 10-fold increased disease risk over other haplotypes. Case-control analyses for this haplotype had an 87%–89% power to detect this association. A longer haplotype (not shown) of 11 markers (rs2071556 to rs497058) that includes promoter SNPs is found in six probands with EJM but is present only in two laboratory controls and in one EJM1− control.
HRR indicates five consecutive haplotypes in or around BRD2 (RING3) (rs206787 to rs497058) in LD with disease; three of these haplotypes are also significantly associated in case-control analysis using one or both sets of external controls.
Statistically significant associations are underlined.
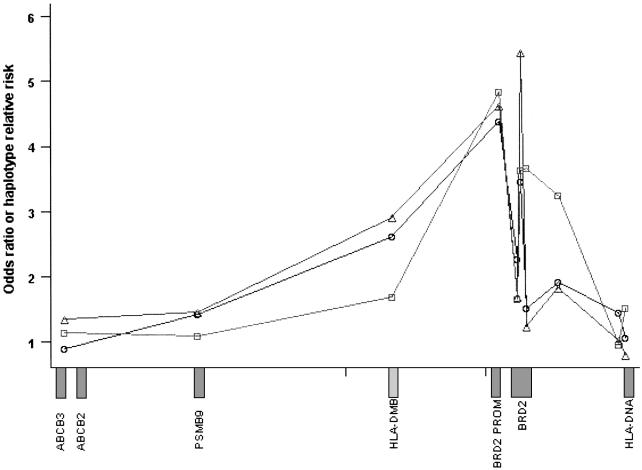
Figure 2.
LD across the EJM1 region. Curves depict the change in two-locus HRR (squares), and ORs against EJM1− controls (triangles) or nonepilepsy controls (circles) at points along the critical region. Each symbol denotes the association at the midpoint between two adjacent SNPs; data points are shown joined by a continuous line. The approximate location of genes is shown along the X-axis. Exact values for relative risk and ORs are given in table 2. Note the sharp increase in LD around the BRD2 (RING3) gene and promoter SNPs.
Table 3.
Rare DNA Variations in the BRD2 (RING3) Gene and Promoter Region[Note]
|
SNP |
No. of Mutations Present/Total Chromosomes |
|||||
| Marker | Mutation Sitea | Name | EJM1+ Tb | EJM1+ nTc | EJM1− Tb | Controls |
| rs3918148 | −175C→G | … | 2/39 | 1/29 | 2/101 | 3/121 |
| rs3918150 | −2C→G | … | 0/39 | 2/32 | 0/96 | 0/125 |
| rs3918144 | c.145_146GC→TG | A49C | 1/40 | 0/32 | 2/104 | 2/126 |
| rs1803864 | c.489C→T | V163A | 0/36 | 0/29 | 0/90 | 0/123 |
| rs176250 | c.712C→T | F238L | 0/40 | 1/33 | 0/103 | 2/127 |
| rs3918141 | c.1339−4C→G | … | 1/40 | 6/38 | 3/104 | 2/128 |
| rs3918143 | c.1421C→T | A474V | 1/38 | 1/30 | 4/104 | 0/118 |
| rs12822 | c.1498C→T | E500X | 0/39 | 0/31 | 0/103 | 0/125 |
| rs3918142d | c.1499_1501delAGG | E500del | 4/40 | 1/32 | 0/104 | 0/128 |
| rs1049369 | c.1640G→A | R547K | 0/40 | 0/32 | 0/104 | 1/128 |
| mutex10 | c.1795G→C | A599P | 1/40 | 1/32 | 3/102 | 0/103 |
Note.— All DNA variants are rare, and no variant is significantly associated with disease.
Marker coordinates are given as distance from the first nucleotide of BRD2 (RING3) mRNA (GenBank accession number NM_005104).
T = transmitted.
nT = nontransmitted.
This deletion has a murine homologue.
Mutation Detection
So far, none of the common SNPs in coding regions were predicted to lead to a change in amino acid sequence. We had noted that two of the strongly associated SNPs occurred in the BRD2 (RING3) promoter region, but with unknown functional significance. In the next step, we sequenced BRD2 (RING3) and neighboring genes in the 20 EJM1+ probands, searching for alterations of DNA sequence leading to known genetic disturbances, such as missense mutations in coding regions, splice variants, or promoter mutations. We assessed 11 rare DNA variations against the EJM1− and control sets (fig. 1 [bottom]): the frequency of variations in EJM1+ sets was not different from control sets (table 3). We then extended mutation screening to adjacent genes that were near enough to BRD2 (RING3) to be within the region implicated by LD mapping and haplotype analysis. We tested for mutations in all exons and splice sites of PPP1R2P1, HLA-DMA, and HLA-DMB genes, which are centromeric to BRD2 (RING3), and HLA-DNA, which is telomeric to BRD2 (RING3), in four families with the highest LOD scores for 6p21 markers. All the DNA variants we found in these genes have been described elsewhere as SNPs and are thought to be neutral variants. In summary, we found no alternative causative mutations or polymorphisms by sequencing of exons and splice sites, but we did find two strongly associated SNPs in the BRD2 (RING3) promoter region, which were of uncertain relevance.
Discussion
This is the first study to precisely localize a gene for a form of idiopathic generalized epilepsy commonly found in the population. The location of a gene for JME at chromosome 6p21 was originally discovered by linkage analysis and confirmed by several independent studies of the common form of JME (Greenberg et al. 1988b; Durner et al. 1991; Weissbecker et al. 1991; Sander et al. 1997). A separate group detected some evidence for linkage of JME in the HLA region (highest LOD score 1.4) at a high recombination fraction, but it was dismissed as not being statistically significant (Whitehouse et al. 1993). These studies, which involve subjects from various geographic regions, also provide evidence that this JME locus is found in several different populations.
The boundaries of the critical region for EJM1 at 6p21 have been delimited, by obligate recombinants in two families, to a 1-cM region between DQ and DP (Sander et al. 1997; Greenberg et al. 2000). We have now demonstrated strong LD between JME and markers in this critical region. LD peaks at markers within the BRD2 (RING3) gene and its promoter region, a gene with which we had previously demonstrated allelic association with a microsatellite marker. We have demonstrated that this strong association is unlikely to have resulted from either chance or population stratification. Our findings therefore strongly suggest that BRD2 (RING3) is EJM1, a major susceptibility gene for a common form of JME.
Sequencing of exons, as well as exon-intron boundaries, failed to reveal any obvious causative mutations in the BRD2 (RING3) gene. However, we found two strongly disease-associated SNP variants in the promoter region, which may lead to altered expression of BRD2 (RING3). Although the significance of these promoter variants is as yet uncertain, several lines of evidence lend support to the suggestion that BRD2 (RING3) is EJM1 and to the likelihood that promoter variants contribute to disease susceptibility. The localizing evidence that ties BRD2 (RING3) to EJM1 is discussed above; below, we discuss the relevance of an oligogenic model of inheritance for JME, and we outline the putative biological role of BRD2 (RING3), showing that it is a credible candidate gene for JME.
JME has an age-dependent, variable phenotype that overlaps with other common forms of adolescent-onset IGEs. Specifically, JME is defined by the occurrence of characteristic myoclonic jerks on awakening, but individuals with JME may also have generalized tonic-clonic or absence seizure types. Our previous linkage findings in the adolescent-onset IGEs (Durner et al. 2001) supported the oligogenic model in which epistatic interactions between loci influenced the expression of these individual seizure types (Greenberg et al. 1988a). Strong evidence of linkage (LOD score 4.4 or 5.2 [multipoint or two-point]) on chromosome 18 suggested that this locus conferred susceptibility to all adolescent-onset IGEs, possibly interacting with a modifying locus (EJM1) on chromosome 6 for myoclonic seizures and with loci on chromosomes 5 and 8 for nonmyoclonic (generalized tonic-clonic and absence) seizures (Durner et al. 2001).
Unlike the rare forms of IGE reported in densely affected pedigrees, the common form of JME is not a monogenic disorder in which single mutations correlate strongly with disease expression. It is apparent from the above oligogenic model that a single critical mutation in BRD2 (RING3), sufficient by itself to cause disease manifestation, would preclude an interactive role for other loci and might not be compatible with the complex pattern of inheritance observed in typical families with JME. A hypothesis more consistent with the known observations is that a disturbance in the transcription of BRD2 (RING3), which might not have severe consequences by itself, could lead to expression of seizures in conjunction with genetic variants at interacting loci. Continuing from this genetic model, one of the interacting genes is likely to be located within the major susceptibility locus that we have studied on chromosome 18 (Durner et al. 2001). To summarize, individual seizure types in IGE may result from the interaction of genetic variants. Separate loci are probably insufficient by themselves to lead to seizure expression, but each contributes to disease susceptibility. This model is consistent with the observed pattern of epilepsy and seizure distribution within families of probands with IGE.
To date, assumptions about the function of genes involved in the pathogenesis of IGEs have largely been drawn from the study of rare, densely affected Mendelian pedigrees (Mulley et al. 2003). Investigators have reported simple gene mutations associated with serious disruption of ion-channel and neuroreceptor function. So far, we have been unable to find mutations in either GABRG2 genes or KCNAB1 genes in probands with common forms of JME or IGE (Evgrafov et al. 2002; authors' unpublished data). These negative findings are in agreement with the original reports of GABRG2 mutations in epilepsy: no additional mutations were found in affected family members from 10 “GEFS-like” families (Baulac et al. 2001), nor were GABRG2 mutations found after screening 200 patients with IGE (Harkin et al. 2002) and 135 patients with idiopathic absence epilepsy (Kananura et al. 2002). These findings in rare pedigrees reinforce our understanding of the role of ion channels and neuroreceptors in normal signal transmission in the CNS. However, the known role of BRD2 (RING3) and related genes suggests the involvement of far more complex and sophisticated mechanisms in the pathogenesis of common forms of IGE than those that are suggested by reports of pedigrees with Mendelian transmission of epilepsy.
BRD2 (RING3) belongs to a highly conserved subfamily of double bromodomain–containing proteins related to the Drosophila female sterile homeotic (fsh) gene, which has an important function in development and appears to interact genetically with the trithorax locus (Gans et al. 1975, 1980; Digan et al. 1986). There are four members of the fsh subfamily in mice and humans, all of which are characterized by the presence of two bromodomains and an extra-terminal (ET) domain. The mouse homologue of BRD2 (RING3), first designated as “Fsrg1,” is expressed ubiquitously but occurs at its highest levels in ovary, testis, placenta, and hormonally modulated epithelia (Rhee et al. 1998). Fsrg1 is also expressed in the mouse embryo, notably in the developing brain and CNS (Rhee et al. 1998; T. Crowley, K. Rhee, M. Brunori, and D. Wolgemuth, personal communication). The Fsrg1 protein participates in nuclear protein complexes that include E2 promoter–binding factor (E2F) proteins, transactivating the promoters of several important cell cycle genes that are dependent on E2F (Denis et al. 2000). The localization of Fsrg1 protein on euchromatin is consistent with its hypothesized function as a transcriptional regulator (Crowley et al. 2002). A nuclear/cytoplasmic translocation of Fsrg1 protein has been observed, both in cultured mouse fibroblasts and in mammary epithelial cells during the reproductive cycle, correlating with both proliferation and apoptosis (Guo et al. 2000; Crowley et al. 2002). The rat homologue of RING3 is also induced during the early stages of programmed neuronal cell death in experimental conditions (Wang et al. 1997), suggesting a role in the modeling of the developing nervous system. BRD2 (RING3) shares at least 95% homology with murine Fsrg1 at the protein level and is expressed in human brain.
Although it has not been extensively studied in humans (Thorpe et al. 1997), a role for BRD2 (RING3) in regulating brain development is likely, and errors in regulation might explain the basis of this form of JME. The likelihood that BRD2 (RING3) plays a role in the brain is especially interesting in light of evidence of abnormal cerebral microanatomy in JME. Neuropathological studies have shown a diffuse increase of single dystopic neurons in the stratum moleculare and in the subcortical white matter in JME and other idiopathic generalized epilepsies (Meencke and Janz 1984). Quantitative magnetic resonance imaging analysis suggests an increase in cortical gray matter in the mesial frontal lobes of living patients with JME, which lends further support for a pathological mechanism resulting in subtle cerebral structural abnormality (Woermann et al. 1999). In the framework of an oligogenic model, we can postulate that BRD2 (RING3) promoter variants may lead to abnormal structural and/or functional interaction with other proteins involved in controlling particular stages of brain development. The function of BRD2 (RING3) as a transcriptional regulator is consonant with an interactive role in a more complex pathway. Abnormalities in a developmental pathway might result in neural cell overgrowth or lack of programmed cell death in specific regions of the brain. These abnormalities may result in disorganized neuronal connectivity and regions of neocortical hyperexcitability, leading to clinical seizures, a mechanism of epileptogenesis already well established in genetic cortical dysplasias (see Flint and Kriegstein [1997] for review). Persisting morphological and functional abnormalities might also explain the poor prognosis for seizure remission in JME.
Taken together, the genetic evidence implicating BRD2 (RING3) as EJM1, the oligogenic model of pathogenesis for common IGE, and the putative role of BRD2 (RING3) in the development of the CNS strongly suggest that BRD2 (RING3) is EJM1 and that variations in the initiation of BRD2 (RING3) transcription may be important in the molecular pathogenesis of JME. Although the SNP variants in the BRD2 (RING3) promoter do not appear to have a dramatic effect, findings in other common complex diseases have suggested that dramatic changes are not the rule. For example, in a recent study of the common forms of migraine, investigators found five associated SNPs in the insulin receptor gene (INSR), which had previously been localized by linkage (McCarthy et al. 2001). None of the INSR SNPs affected transcription, translation, or protein expression. Similarly, SNPs associated with Crohn disease in the 5q cytokine cluster have not been shown to disrupt either amino acid sequence or the regulatory region of a known gene (Rioux et al. 2001). Both these studies, like our own, offer persuasive localizing evidence, but we must await investigation of interacting genes and biological pathways to explain the pathogenetic role of SNP associations.
Acknowledgments
The study was supported in part by National Institutes of Health grants NS27941, MH48858, DK31775 (to D.A.G.), and NS37466 (to M.D.) and by a Royal Society–Fulbright Distinguished Postdoctoral Scholarship, the Dunhill Medical Trust, and the Epilepsy Foundation of America (to D.K.P.). We thank Debra Wolgemuth, Tom Crowley, Rhonda Trousdale, and Enyuan Shang for helpful comments and discussion. Our thanks also to the families participating in the New York Epilepsy Project and to their referring clinicians: Shlomo Shinnar, Stanley Resor, Jeffrey Cohen, Cynthia Harden, Solomon Moshe, David Rosenbaum, Harriet Kang, Karen Ballaban-Gill, Sharon Hertz, Douglas Labar, Daniel Luciano, Sibylle Wallace, and David Yohai.
Electronic-Database Information
URLs for data presented herein are as follows:
- Cooperative Human Linkage Center (CHLC), http://www.chlc.org [DOI] [PubMed]
- GenBank, http://www.ncbi.nlm.nih.gov/Genbank/ (for BRD2 [RING3] accession number NM_005104)
- Online Mendelian Inheritance in Man (OMIM), http://www.ncbi.nlm.nih.gov/Omim/ (for JME) [PubMed]
References
- Annegers JF, Rocca WA, Hauser WA (1996) Causes of epilepsy: contributions of the Rochester Epidemiology Project. Mayo Clin Proc 71:570–575 [DOI] [PubMed] [Google Scholar]
- Baulac S, Huberfeld G, Gourfinkel-An I, Mitropoulou G, Beranger A, Prud’homme JF, Baulac M, Brice A, Bruzzone R, LeGuern E (2001) First genetic evidence of GABA(A) receptor dysfunction in epilepsy: a mutation in the gamma2-subunit gene. Nat Genet 28:46–48 [DOI] [PubMed] [Google Scholar]
- Beck-Mannagetta G, Janz D (1991) Syndrome-related genetics in generalized epilepsy. Epilepsy Res Suppl 4:105–111 [PubMed] [Google Scholar]
- Commission on Classification and Terminology of the International League Against Epilepsy (1989) Proposal for revised clinical and electroencephalographic classification of epileptic seizures. Epilepsia 22:489–501 [DOI] [PubMed] [Google Scholar]
- Crowley TE, Kaine EM, Yoshida M, Nandi A, Wolgemuth DJ (2002) Reproductive cycle regulation of nuclear import, euchromatic localization, and association with components of Pol II mediator of a mammalian double-bromodomain protein. Mol Endocrinol 16:1727–1737 [DOI] [PubMed] [Google Scholar]
- Denis GV, Vaziri C, Guo N, Faller DV (2000) RING3 kinase transactivates promoters of cell cycle regulatory genes through E2F. Cell Growth Differ 11:417–424 [PMC free article] [PubMed] [Google Scholar]
- Digan ME, Haynes SR, Mozer BA, Dawid IB, Forquignon F, Gans M (1986) Genetic and molecular analysis of fs(1)h, a maternal effect homeotic gene in Drosophila. Dev Biol 114:161–169 [DOI] [PubMed] [Google Scholar]
- Durner M, Keddache MA, Tomasini L, Shinnar S, Resor SR, Cohen J, Harden C, Moshe SL, Rosenbaum D, Kang H, Ballaban-Gill K, Hertz S, Labar DR, Luciano D, Wallace S, Yohai D, Klotz I, Dicker D, Greenberg DA (2001) Genome scan of idiopathic generalised epilepsy: evidence for major susceptibility gene and modifying genes influencing the seizure type. Ann Neurol 49:328–335 [PubMed] [Google Scholar]
- Durner M, Sander T, Greenberg DA, Johnson K, Beck-Mannagetta G, Janz D (1991) Localization of idiopathic generalized epilepsy on chromosome 6p in families of juvenile myoclonic epilepsy patients. Neurology 41:1651–1655 [DOI] [PubMed] [Google Scholar]
- Evgrafov OV, Zhang F-L, Tabares P, Durner M, Pal DK, Gilliam TC, Greenberg DA (2002) KCNAB1 is not responsible for predisposition to a subgroup of juvenile myoclonic epilepsy. Am J Hum Genet 71:S481 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Falk CT, Rubenstein P (1987) Haplotype relative risks: an easy reliable way to construct a proper control sample for risk calculations. Ann Hum Genet 51:227–233 [DOI] [PubMed] [Google Scholar]
- Flint AC, Kriegstein AR (1997) Mechanisms underlying neuronal migration disorders and epilepsy. Curr Opin Neurol 10:92–97 [DOI] [PubMed] [Google Scholar]
- Gans M, Audit C, Masson M (1975) Isolation and characterization of sex-linked female-sterile mutants in Drosophila melanogaster. Genetics 81:683–704 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gans M, Forquignon F, Masson M (1980) The role of dosage of the region 7D1-7D5-6 of the X chromosome in the production of homeotic transformations in Drosophila melanogaster. Genetics 96:887–902 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greenberg DA, Delgado-Escueta AV, Maldonado HM, Widelitz H (1988a) Segregation analysis of juvenile myoclonic epilepsy. Genet Epidemiol 5:81–94 [DOI] [PubMed] [Google Scholar]
- Greenberg DA, Delgado-Escueta AV, Widelitz H (1988b) Juvenile myoclonic epilepsy may be linked to the BF and HLA loci on human chromosome 6. Am J Med Genet 31:185–192 [DOI] [PubMed] [Google Scholar]
- Greenberg DA, Durner M, Delgado-Escueta AV (1992) Evidence for multiple gene loci in the expression of the common generalized epilepsies. Neurology 42:56–62 [PubMed] [Google Scholar]
- Greenberg DA, Durner M, Keddache M, Shinnar S, Resor SR, Moshe SL, Rosenbaum D, Cohen J, Harden C, Kang H, Wallace S, Luciano D, Ballaban-Gil K, Tomasini L, Zhou G, Klotz I, Dicker E (2000) Reproducibility and complications in gene searches: linkage on chromosome 6, heterogeneity, association, and maternal inheritance in juvenile myoclonic epilepsy. Am J Hum Genet 66:508–516 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greenberg DA, MacCluer JW, Spence MA, Falk CT, Hodge SE (1999) Simulated data for a complex genetic trait (problem 2 for GAW11): how the model was developed, and why. Genet Epidemiol 17:S449–S459 [DOI] [PubMed] [Google Scholar]
- Guo N, Faller DV, Denis GV (2000) Activation-induced nuclear translocation of RING3. J Cell Sci 113:3085–3091 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harkin LA, Bowser DN, Dibbens LM, Singh R, Phillips F, Wallace RH, Richards MC, Williams DA, Mulley JC, Berkovic SF, Scheffer IE, Petrou S (2002) Truncation of the GABAA-receptor γ2 subunit in a family with generalized epilepsy with febrile seizures plus. Am J Hum Genet 70:530–536 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Janz D, Christian W (1957) Impulsiv-petit mal. Dtsch Z Nervenheilk 176:348–386 [Google Scholar]
- Kananura C, Haug K, Sander T, Runge U, Gu W, Hallmann K, Rebstock J, Heils A, Steinlein OK (2002) A splice-site mutation in GABRG2 associated with childhood absence epilepsy and febrile convulsions. Arch Neurol 59:1137–1141 [DOI] [PubMed] [Google Scholar]
- Kruglyak L, Daly MJ, Reeve-Daly MP, Lander ES (1996) Parametric and nonparametric linkage analysis: a unified multipoint approach. Am J Hum Genet 58:1347–1363 [PMC free article] [PubMed] [Google Scholar]
- McCarthy LC, Hosford DA, Riley JH, Bird MI, White NJ, Hewett DR, Peroutka SJ, et al (2001) Single-nucleotide polymorphism alleles in the insulin receptor gene are associated with typical migraine. Genomics 78:135–149 [DOI] [PubMed] [Google Scholar]
- Meencke H-J, Janz D (1984) Neuropathological findings in primary generalized epilepsy: a study of eight cases. Epilepsia 25:8–21 [DOI] [PubMed] [Google Scholar]
- Mulley JC, Scheffer IE, Petrou S, Berkovic SF (2003) Channelopathies as a genetic cause of epilepsy. Curr Opin Neurol 16:171–176 [DOI] [PubMed] [Google Scholar]
- Rhee K, Brunori M, Besset V, Trousdale R, Wolgemuth DJ (1998) Expression and potential role of Fsrg1, a murine bromodomain-containing homologue of the Drosophila gene female sterile homeotic. J Cell Sci 111:3541–3550 [DOI] [PubMed] [Google Scholar]
- Rioux JD, Daly MJ, Silverberg MS, Lindblad K, Steinhart H, Cohen Z, Delmonte T, et al (2001) Genetic variation in the 5q31 cytokine gene cluster confers susceptibility to Crohn disease. Nat Genet 29:223–228 [DOI] [PubMed] [Google Scholar]
- Sander T, Bockenkamp B, Hildmann T, Blasczyk R, Kretz R, Wienker TF, Volz A, Schmitz B, Beck-Mannagetta G, Riess O, Epplen JT, Janz D, Ziegler A (1997) Refined mapping of the epilepsy susceptibility locus EJM1 on chromosome 6. Neurology 49:842–847 [DOI] [PubMed] [Google Scholar]
- StataCorp (1996) Stata for Macintosh, version 5.0. StataCorp, College Station, TX [Google Scholar]
- Thorpe KL Gorman P, Thomas C, Sheer D, Trowsdale J, Beck S (1997) Chromosomal localization, gene structure and transcription pattern of the ORFX gene, a homologue of the MHC-linked RING3 gene. Gene 200:177–183 [DOI] [PubMed] [Google Scholar]
- Tsuboi T, Christian W (1973) On the genetics of primary generalized epilepsy with sporadic myoclonias of impulsive petit mal: a clinical and electroencephalographic study of 399 probands. Hum Genet 19:155–182 [DOI] [PubMed] [Google Scholar]
- Wang S, Dibenedetto AJ, Pittman RN (1997) Genes induced in programmed cell death of neuronal PC12 cells and developing sympathetic neurons in vivo. Dev Biol 188:322–336 [DOI] [PubMed] [Google Scholar]
- Weissbecker KA, Durner M, Janz D, Scaramelli A, Sparkes RS, Spence MA (1991) Confirmation of linkage between juvenile myoclonic epilepsy locus and the HLA region of chromosome 6. Am J Med Genet 38:32–36 [DOI] [PubMed] [Google Scholar]
- Whitehouse WP, Rees M, Curtis D, Sundqvist A, Parker K, Chung E, Baralle D, Gardiner RM (1993) Linkage analysis of idiopathic generalized epilepsy (IGE) and marker loci on chromosome 6p in families of patients with juvenile myoclonic epilepsy: no evidence for an epilepsy locus in the HLA region. Am J Hum Genet 53:652–662 [PMC free article] [PubMed] [Google Scholar]
- Woermann FG, Free SL, Koepp MJ, Sisodiya SM, Duncan JS (1999) Abnormal cerebral structure in juvenile myoclonic epilepsy demonstrated with voxel-based analysis of MRI. Brain 122:2101–2108 [DOI] [PubMed] [Google Scholar]