Abstract
The prospect of using linkage disequilibrium (LD) for fine-scale mapping in humans has attracted considerable attention, and, during the validation of a set of single-nucleotide polymorphisms (SNPs) for linkage analysis, a set of data for 4,833 SNPs in 538 clusters was produced that provides a rich picture of local attributes of LD across the genome. LD estimates may be biased depending on the means by which SNPs are first identified, and a particular problem of ascertainment bias arises when SNPs identified in small heterogeneous panels are subsequently typed in larger population samples. Understanding and correcting ascertainment bias is essential for a useful quantitative assessment of the landscape of LD across the human genome. Heterogeneity in the population recombination rate, ρ=4Nr, along the genome reflects how variable the density of markers will have to be for optimal coverage. We find that ascertainment-corrected ρ varies along the genome by more than two orders of magnitude, implying great differences in the recombinational history of different portions of our genome. The distribution of 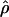 is unimodal, and we show that this is compatible with a wide range of mixtures of hotspots in a background of variable recombination rate. Although
is unimodal, and we show that this is compatible with a wide range of mixtures of hotspots in a background of variable recombination rate. Although 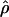 is significantly correlated across the three population samples, some regions of the genome exhibit population-specific spikes or troughs in ρ that are too large to be explained by sampling. This result is consistent with differences in the genealogical depth of local genomic regions, a finding that has direct bearing on the design and utility of LD mapping and on the National Institutes of Health HapMap project.
is significantly correlated across the three population samples, some regions of the genome exhibit population-specific spikes or troughs in ρ that are too large to be explained by sampling. This result is consistent with differences in the genealogical depth of local genomic regions, a finding that has direct bearing on the design and utility of LD mapping and on the National Institutes of Health HapMap project.
Introduction
Before we can assess the likely efficacy of finding genes by genomewide scans for association with a SNP marker, it is essential that the distribution of linkage disequilibrium (LD) across the genome be quantified in more than one target population. Several moderate efforts have been reported that highlight some of the problems and begin to illustrate some of the results of this LD map. Huttley et al. (1999) used data on 5,048 STRs scored in the CEPH families to infer LD within the grandparental generation. They found several regions of the genome in which significant LD spanned >1 Mb and other large regions with very low LD. As forward-thinking as their study was, it suffered from excessive distances separating the STR markers, so it could identify only long-range LD. The magnitude and suddenness of change of LD along a chromosome has been noted by several studies (Eisenbarth et al. 2000; Taillon-Miller et al. 2000), a result implying that judicious choice of markers could greatly improve the ratio of power to cost in LD mapping. Reich et al. (2001) have quantified the decay of LD across 19 clusters of five or six SNPs, with each cluster spanning ∼160 kb. Their study showed not only that were there regions of high and low rates of decay of LD but also that population samples from Nigeria generally had markedly less LD than those from Sweden and Utah. Apart from a bias introduced by the larger sample size of the Nigerians (Weiss and Clark 2002), a key conclusion is that the number of SNPs necessary for a genomewide LD map would not be easy to estimate because of both of these types of heterogeneity. More recently, it has become popular to note that small population samples show a pattern of LD that appears to be locally clustered (Daly et al. 2001; Gabriel et al. 2002), a result widely interpreted as implying that fewer SNPs than originally thought could be used to attain reasonable power in genomewide LD mapping. Assessment of the minimum number of SNPs needed for a whole-genome LD-mapping study with acceptable power requires better knowledge of the landscape of LD across the genome. Examination of chromosome 22 at 15 kb resolution (Dawson et al. 2002) and chromosome 19 at 6 kb resolution (Phillips et al. 2003) identified many regions of high LD, but there remain regions of unusually low LD that will require large numbers of SNPs for coverage by LD mapping.
In addition to the quantification and mapping of genomic regions of high and low LD, it is essential that the degree of heterogeneity in LD among human populations be understood before inferences about the generality of LD associations with diseases can be made. It has been appreciated for many years that the frequency of genetic disorders varies widely across population groups, and it follows that we expect that not all SNP associations will be universal. We can ask whether, as a surrogate for heterogeneity between SNPs and diseases, the pattern of LD among SNPs is consistent across populations. Here, too, there is a growing literature showing heterogeneity among populations, with Africa tending to have low LD and with populations that are more derived or isolated having the highest LD (Dunning et al. 2000; Kidd et al. 2000; Reich et al. 2001; Bonnen et al. 2002).
A number of population genetic inferences can be drawn from surveys of multilocus-SNP genotype frequencies. Because the SNPs used in each of these surveys generally have been discovered in a separate, smaller sample and because these SNPs were then scored in the large sample, of 90, used in the present study, there is a tendency for these SNPs to have a higher population frequency than would SNPs discovered by the sequencing of all 90 individuals. This ascertainment bias has been a target of investigation by analysts concerned that it would distort our view of LD unless a means for correcting the bias could be found (Kuhner et al. 2000; Nielsen 2000; Wakeley et al. 2001). Most of this effort has focused on the frequency spectrum of SNPs, since prior discovery of SNPs in small panels clearly biases against finding rare SNPs. The effect of this ascertainment bias is to underestimate LD, in part because the skew toward SNPs that are more frequent also biases toward older segregating variants that have had more time to recombine. This bias may vary from one population to another, depending on the composition of the panel in which SNPs were discovered (Nielsen and Signorovitch 2003).
Metrics such as D′ and r2 have been widely used to quantify LD, and, although they quantify the statistical dependence between a pair of SNPs, a more appropriate metric for assessing the local landscape of inter-SNP association is the population recombination rate, ρ=4Ner, where Ne is the effective population size and r is the recombination rate between a pair of sites (Pritchard and Przeworski 2001). Linkage studies tell us that the rate of recombination per base pair falls in the range of 2×10-9 to 6×10-8 (Broman et al. 1998). The effective size of the human population has been estimated by several studies to be ∼10,000, so a first guess would be that 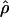 falls in the range of 2×10-5 to 6×10-4. Estimates of
falls in the range of 2×10-5 to 6×10-4. Estimates of  derived in this crude way are closely comparable to the range of
derived in this crude way are closely comparable to the range of 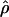 estimated directly from SNP data; however, the empirical data show an excessive rate of LD decay within a window of a few kilobase pairs (Pritchard and Przeworski 2001).
estimated directly from SNP data; however, the empirical data show an excessive rate of LD decay within a window of a few kilobase pairs (Pritchard and Przeworski 2001).
In population genetic models, ρ is a parameter that integrates the effects of mutation, drift, and recombination in giving rise to a particular statistical association across sites. For example, in a population that is in steady state with respect to accumulation of neutral mutations, random drift, and recombination, the expected LD is E(r2)=1/(1+ρ) (Ohta and Kimura 1971; Sved 1971). The value of ρ may vary from one population to another or from one genomic region to another, owing either to differences in local recombination rate or to recent common ancestry of a region. The time of common ancestry may vary widely across genomic regions owing to drift, migration, or natural selection, but, for neutral variation in a panmictic population, the size of the region sharing a most recent common ancestor is 3/2ρ for a pair of chromosomes and 1/ρ for a whole population (Wiuf and Hein 1999).
In the present study, ∼5,500 SNPs were tested in a panel of 30 African Americans, 30 European Americans, and 30 Asians, to validate the SNPs and to obtain information for the purpose of identifying the SNPs to be chosen for a linkage analysis of CEPH families (Matise et al. 2003 [in this issue]). The SNP clusters were defined such that no gap between adjacent SNPs exceeded 50 kb. This produced clusters of 4–11 SNPs (mean 5.7), spanning 16–181 kb (mean 79 kb), with clusters spaced every 2–7 cM on the genetic map. The 4,833 SNPs that were successfully validated were assembled into a data table with map locations and genotypes of all 90 samples, and it was from this table that all subsequent analysis was performed.
Methods
SNP Selection and Typing
The primary data were collected by the performance of TaqMan SNP assays on DNA samples from the SNP Consortium (TSC) diversity panel, consisting of 30 African Americans, 30 European Americans, 20 Japanese, and 10 Chinese (Matise et al. 2003 [in this issue]). Genotype calls were transferred to Cold Spring Harbor Laboratory, where they were placed in the primary database of TSC. A database query of all SNPs in the present study generated a file, which was subsequently filtered to remove those SNPs that departed strongly from Hardy-Weinberg equilibrium (P<10-4) or that did not reveal segregating variation. The final data set consisted of genotypes of 4,833 SNPs (see the SNP Consortium Linkage Map Project Web site). Nearly all SNPs had been placed on both the National Center for Biotechnology Information (NCBI) build 29 physical map and the SNP map of Celera Genomics, and both map locations are in this data file.
Cluster Definition
Clusters were defined for the present analysis by the collection of SNPs into a cluster such that no gap therein exceeds 50 kb. For the analysis of ρ, we also required that the minimum cluster size be 4. This produced a set of 538 clusters when physical-map information from either NCBI or Celera was used. All of the genotype calls, information on physical-map location, and ascertainment methods for each SNP were compiled into a single, easily parsed data file (TSCmap.p1.Celera.txt.gz [freely available at the SNP Consortium Linkage Map Project Web site]). Note that the composition of the clusters described here differs from those in the companion article (Matise et al. 2003 [in this issue]), because the present study examined more SNPs genotyped at Celera Genomics, excluded the Motorola SNPs, and assembled clusters on the basis of physical position only (not on the basis of the level of heterozygosity).
Estimation of Population Recombination Rate
To estimate 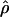 for each cluster of SNPs, we used a modification of Hudson’s (2001) composite-likelihood method, taking the product of the likelihood function calculated for individual pairs of SNPs. The method is modified to take into account the special ascertainment scheme used in the TSC data. For a pair of SNPs, the likelihood function for ρ was calculated as
for each cluster of SNPs, we used a modification of Hudson’s (2001) composite-likelihood method, taking the product of the likelihood function calculated for individual pairs of SNPs. The method is modified to take into account the special ascertainment scheme used in the TSC data. For a pair of SNPs, the likelihood function for ρ was calculated as
where x is the matrix of haplotypic data for the two loci and Ai is the ascertainment condition that variability is observed in the ascertainment sample of locus i. The basic idea is that the sampling probability is calculated conditional on the ascertainment condition. The number of individuals in the ascertainment sample may vary from locus to locus but is known for all loci, and the likelihood function in equation (1) can therefore be calculated using the methods described by Nielsen and Signorovitch (2003). Some modifications were necessary to accommodate genotypic data and variation in the ascertainment condition among loci (these modifications are described in greater detail in appendix A; also see the SNP Consortium Linkage Map Project Web site).
Confidence and Hypothesis Testing
Extensive simulations were performed, to test the hypotheses and to obtain CIs for parameter estimates. In all cases, these simulations were done using a standard coalescent model with recombination (Hudson 1985). One thousand parametric bootstrap samples were collected for each cluster to obtain the variance in the estimates of ρ. These samples were simulated under the estimated parameter values. In some cases, the estimates of ρ were ∞ (i.e., the composite-likelihood functions were nondecreasing functions). In such cases, a bound given by 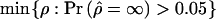 (i.e., by the minimum value of ρ at which the probability of obtaining a composite-likelihood estimate of
(i.e., by the minimum value of ρ at which the probability of obtaining a composite-likelihood estimate of 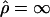 is >0.05) was obtained instead by simulation. Parametric bootstrapping was also used to test for differences in ρ between pairs of clusters. The null hypothesis H0:ρ1=ρ2 was tested using the ratio of the maximum composite likelihood obtained under the null hypothesis and under the alternative hypothesis of ρ1≠ρ. The distribution of this test statistic was then obtained using parametric simulations under the parameter values estimated under the null hypothesis. A Wilcoxon signed-ranks test was used to test for differences in ρ between populations.
is >0.05) was obtained instead by simulation. Parametric bootstrapping was also used to test for differences in ρ between pairs of clusters. The null hypothesis H0:ρ1=ρ2 was tested using the ratio of the maximum composite likelihood obtained under the null hypothesis and under the alternative hypothesis of ρ1≠ρ. The distribution of this test statistic was then obtained using parametric simulations under the parameter values estimated under the null hypothesis. A Wilcoxon signed-ranks test was used to test for differences in ρ between populations.
Estimation of LD
The SNP genotype calls in the present study were not phased, so that doubly heterozygous individuals were ambiguous with respect to haplotypes. For this reason, the LD metrics are all based on the composite LD (Weir 1996), which does not entirely separate departures from multilocus Hardy-Weinberg frequencies of genotypes from true gametic-phase disequilibrium. Nevertheless, studies have shown that, for populations close to Hardy-Weinberg equilibrium, the composite LD metrics are an excellent approximation to the true gametic-phase disequilibrium.
Results
Allele Frequencies and Population Subdivision
The SNPs examined in the present study cannot be construed as a random sample but rather were identified through a variety of different methods and in a variety of discovery panels of different size and composition. Information from TSC indicated that 80% of the SNPs used in the present study were discovered with just one pair of mismatching sequence reads. The consequences of these heterogeneous modes of ascertainment are not easy to assess, but figure 1 shows that the resulting allele-frequency spectrum is rather uniform and that the distribution is similar across populations. Mean frequencies of the minority allele in the African American, Asian, and European American samples were 0.236, 0.247, and 0.253, respectively. The degree of population subdivision can be assessed by the calculation of Wright’s statistic FST, which quantifies the among-population portion of variance. The mean FST for the SNPs in the present study was 0.083, which is consistent with the figure obtained with SNPs that are randomly ascertained by complete sequencing (e.g., see Fullerton et al. 2000). The distribution of FST has a long tail, with 10% of the SNPs having an FST value >0.18 (fig. 2). On the basis of these findings, a principal conclusion is that the SNPs reflect a wide spectrum of among-population variability, including a substantial number with quite strong differentiation. Estimates of FST are also biased by ascertainment of panel SNPs, and a full treatment of bias correction and inferences about past operation of natural selection on the basis of the clustering of elevated, bias-corrected FST will be considered in more detail elsewhere.
Figure 1.
Frequency of the minority SNP allele for all three population samples. Black bars represent African Americans, blue bars represent European Americans, and red bars represent Asians. Apart from the dip in the rarest class, the data are remarkably consistent with a uniform distribution, which one would expect with an ascertainment sample of size 2.
Figure 2.
Distribution of FST for all 4,833 SNPs in the phase 1 set that passed the quality filters. Note the tail of SNPs with exceptionally high FST, possibly indicating local founding effects, random drift, and region-specific differences in natural selection.
Heterogeneity of Rates of Population Recombination, ρ
Before examining classical metrics for LD, we stress that the parameter ρ has key advantages in the context of quantifying the landscape of ancestral recombination in the human genome. Several methods exist for the estimation of ρ, and our approach, outlined in the “Methods” section, is an extension of the pseudolikelihood method of Hudson (2001). This approach explicitly accommodates the ascertainment method in an effort to correct any bias that might be introduced.
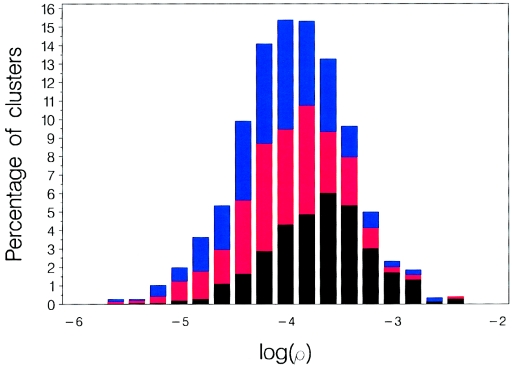
Figure 3 shows that estimates of ρ exhibit striking heterogeneity across the genome, spanning more than two orders of magnitude. Wilcoxon signed-ranks tests reveal that the differences among the three populations are highly significant; however, we stress that the CIs in  are large as a result of the modest size of the samples and the sampling of only 4–10 SNPs per cluster. With a >1,000-fold range in estimates of
are large as a result of the modest size of the samples and the sampling of only 4–10 SNPs per cluster. With a >1,000-fold range in estimates of 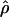 from one genomic region to another, one is tempted to claim that the data identify hotspots and coldspots of recombination. But figure 3 suggests no such bimodality to
from one genomic region to another, one is tempted to claim that the data identify hotspots and coldspots of recombination. But figure 3 suggests no such bimodality to  ; instead, there is a quite smooth log-normal distribution among these clusters in ρ estimates.
; instead, there is a quite smooth log-normal distribution among these clusters in ρ estimates.
Figure 3.
Base 10 logarithm of estimates of the population recombination parameter ρ in the three population samples. Black bars represent African Americans, blue bars represent European Americans, and red bars represent Asians.
The distribution of  is unambiguously unimodal, despite the clear evidence (from other sources) for recombination hotspots (Jeffreys et al. 2000, 2001). The unimodality of
is unambiguously unimodal, despite the clear evidence (from other sources) for recombination hotspots (Jeffreys et al. 2000, 2001). The unimodality of  is not at odds with evidence for hotspots, because of a combination of sampling error and the occurrence of hotspots in locally restricted regions. We demonstrated this by drawing samples from mixtures of normal distributions representing hotspot and nonhotspot rates of recombination, with each component having a variance matching our estimation error. An enormous range of hotspot densities still produced a unimodal distribution of
is not at odds with evidence for hotspots, because of a combination of sampling error and the occurrence of hotspots in locally restricted regions. We demonstrated this by drawing samples from mixtures of normal distributions representing hotspot and nonhotspot rates of recombination, with each component having a variance matching our estimation error. An enormous range of hotspot densities still produced a unimodal distribution of 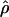 . Departure from a unimodal distribution required a substantial fraction of clusters with a mean
. Departure from a unimodal distribution required a substantial fraction of clusters with a mean 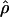 that was 2 SDs greater than background. Given rates of recombination and breadths of recombination hotspots from empirical studies, the clusterwide mean
that was 2 SDs greater than background. Given rates of recombination and breadths of recombination hotspots from empirical studies, the clusterwide mean 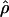 would be perturbed dramatically only if the cluster had dozens of hotspots. Overall, the unimodality shown in figure 3 tells us little about recombination-hotspot density, and substantially greater numbers of markers would be needed to do so. These conclusions are consistent with those of Phillips et al. (2003), who go so far as to say that the heterogeneity in LD across chromosome 19 can be explained without the invocation of recombination hotspots at all.
would be perturbed dramatically only if the cluster had dozens of hotspots. Overall, the unimodality shown in figure 3 tells us little about recombination-hotspot density, and substantially greater numbers of markers would be needed to do so. These conclusions are consistent with those of Phillips et al. (2003), who go so far as to say that the heterogeneity in LD across chromosome 19 can be explained without the invocation of recombination hotspots at all.
Estimates of  vary along the chromosomes in a way that suggests regional differences in recombination rate and/or effective size (fig. 4). Note that some genomic regions exhibit sharp dips or rises in
vary along the chromosomes in a way that suggests regional differences in recombination rate and/or effective size (fig. 4). Note that some genomic regions exhibit sharp dips or rises in 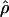 in a single population, other regions show two populations tracking one another, and, elsewhere, all three populations are coherent. This observation is robust over a range of spline widths and is seen with simple moving-average plots as well (not shown). Sharp differences among populations may reflect local perturbations in effective size, perhaps owing to a strong local selection episode. Scanning the whole genome (fig. 5), one can see that no region seems to be immune to this process.
in a single population, other regions show two populations tracking one another, and, elsewhere, all three populations are coherent. This observation is robust over a range of spline widths and is seen with simple moving-average plots as well (not shown). Sharp differences among populations may reflect local perturbations in effective size, perhaps owing to a strong local selection episode. Scanning the whole genome (fig. 5), one can see that no region seems to be immune to this process.
Figure 4.
Base 10 logarithm of ρ in each of the three populations, estimated for each cluster on chromosomes 1 and 2, plotted as a spline fit. Notice how the Asian (red) and European American (blue) samples tend to track together somewhat more than either does with the African American sample (black).
Figure 5.
Base 10 logarithm of ρ, plotted for each cluster (in genome order). Colors alternate for successive chromosomes: odd-numbered chromosomes are denoted by red, and even-numbered chromosomes are denoted by green. The blue line denotes a spline fit.
Standard Metrics for LD
To contrast the above results with the more simply accessible calculations of metrics for LD, we also estimated D, D′, and r2 from the data, without correcting for ascertainment. Because the genotypic data are of unknown linkage phase, all these statistics are based on the composite LD (Weir 1996). Figure 6 shows the relation between r2 and physical distance for 20,894 SNP pairs, and it is clear that many regions have abundant LD, spanning 60 or even 80 kb. But it is crucial to also consider that SNP pairs that are even less than 1 kb apart may have essentially no LD, so that association tests may fail when one of these SNPs is the marker and the other is a determinant of a disease.
Figure 6.
Decay of LD (r2) with physical distance between each respective pair of SNPs. Across this set of comparisons, few cases of strong LD occur for SNPs more distant than ∼80 kb. For every SNP in this data set, there exists, within the same cluster, at least one other SNP that has r2 >0.5.
The estimates of  and of
and of 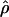 were obtained in radically different ways, yet we expect them to show opposite sides of the same phenomenon. Regions with very high LD would be expected to produce an inference of low population recombination rate. To assess this in an informal way, we examined plots of the estimate of
were obtained in radically different ways, yet we expect them to show opposite sides of the same phenomenon. Regions with very high LD would be expected to produce an inference of low population recombination rate. To assess this in an informal way, we examined plots of the estimate of 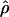 for each cluster versus the mean
for each cluster versus the mean 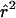 for all pairs in that cluster (fig. 7). The strong negative correlation was observed as expected.
for all pairs in that cluster (fig. 7). The strong negative correlation was observed as expected.
Figure 7.
Clusters with low r2, consistent with higher levels of recombination, generating negative correlation between r2 and ρ. Open circles represent African Americans, blue points represent European Americans, and red points represent Asians.
Heterogeneity in 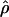 and
and 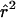 among the Three Population Samples
among the Three Population Samples
One crucial attribute of an LD map is that we have a good understanding of its utility in different human populations. Local differences in LD will likely necessitate selection of some population-specific SNPs for an optimal LD map, but it would be very useful also to have a core set of SNPs that are informative in many populations. For this reason, tests of homogeneity of 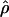 and of LD statistics across populations are vital. A first gross comparison to the Huttley et al. (1999) LD map was performed by examining the 11 regions in the genome that they flagged as having exceptionally high LD. We found that 10 of these 11 regions had a nearby cluster with
and of LD statistics across populations are vital. A first gross comparison to the Huttley et al. (1999) LD map was performed by examining the 11 regions in the genome that they flagged as having exceptionally high LD. We found that 10 of these 11 regions had a nearby cluster with 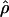 that was <5 × 10−5 in the European American sample, a result that corresponds to unusually low population recombination. It remains an important problem to devise means by which to rigorously integrate and test heterogeneity across LD maps acquired in such distinct ways.
that was <5 × 10−5 in the European American sample, a result that corresponds to unusually low population recombination. It remains an important problem to devise means by which to rigorously integrate and test heterogeneity across LD maps acquired in such distinct ways.
The three population groups considered here were sampled in a uniform way, so that tests of homogeneity have a clear statistical meaning. The homogeneity tests that we applied were performed by the calculation of likelihood ratios. Across the entire map, a Wilcoxon signed-ranks tests showed that the Asian and European American estimates of 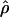 were not different but that the estimates of
were not different but that the estimates of  in the African American sample were significantly elevated (W+ = 80,220 [P<.0001] and 89,142 [P<.0001], for Asian and European American estimates and African American estimates, respectively). Repeating this test in small segments of the genome, one would be hard-pressed to find regions in which
in the African American sample were significantly elevated (W+ = 80,220 [P<.0001] and 89,142 [P<.0001], for Asian and European American estimates and African American estimates, respectively). Repeating this test in small segments of the genome, one would be hard-pressed to find regions in which  is not statistically significantly elevated in the African American sample. Overall, the Pearson correlation coefficient for ρ estimates is 0.608 for African Americans versus Asians, 0.698 for African Americans versus European Americans, and 0.649 for Asians versus European Americans, all significant at P<.0001 (fig. 8). Similarly, log-linear models of genotype counts show significant levels of interpopulation heterogeneity of LD. Despite this heterogeneity, regions of the genome that exhibit very high LD in one population are more likely to exhibit high LD in other populations. This correlation across population groups in rates of decay of LD has been cited previously (e.g., see Abecasis et al. 2001). Nonetheless, even with this interpopulation correlation in LD, there is room for statistically significant difference among the populations in
is not statistically significantly elevated in the African American sample. Overall, the Pearson correlation coefficient for ρ estimates is 0.608 for African Americans versus Asians, 0.698 for African Americans versus European Americans, and 0.649 for Asians versus European Americans, all significant at P<.0001 (fig. 8). Similarly, log-linear models of genotype counts show significant levels of interpopulation heterogeneity of LD. Despite this heterogeneity, regions of the genome that exhibit very high LD in one population are more likely to exhibit high LD in other populations. This correlation across population groups in rates of decay of LD has been cited previously (e.g., see Abecasis et al. 2001). Nonetheless, even with this interpopulation correlation in LD, there is room for statistically significant difference among the populations in 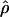 or LD metrics. This heterogeneity arises from two sources: the sampling variation that occurs in drawing the relatively small samples (n=30 per population) and the stochastic variation that occurred during the past ancestry of the populations. The significance tests showed that the heterogeneity exceeds that expected from sampling alone, but whether the heterogeneity exceeds what one expects from neutral drift requires an extensive range of simulations of plausible demographic scenarios and is beyond the scope of the present article.
or LD metrics. This heterogeneity arises from two sources: the sampling variation that occurs in drawing the relatively small samples (n=30 per population) and the stochastic variation that occurred during the past ancestry of the populations. The significance tests showed that the heterogeneity exceeds that expected from sampling alone, but whether the heterogeneity exceeds what one expects from neutral drift requires an extensive range of simulations of plausible demographic scenarios and is beyond the scope of the present article.
Figure 8.
Scatterplot of ρ estimates, showing correlation across populations. The Pearson correlation for Asians versus European Americans is 0.649 (blue) and for Asians versus African Americans is 0.608 (red); not shown is African Americans versus European Americans, for which the correlation is 0.698. Note how the cluster of red points (African Americans) tends to lie above the European American points, consistent with higher population recombination and lower LD in African Americans.
Another approach to the quantification of population subdivision is to consider the estimates of LD among unlinked SNPs. Estimates in the sample pooled across the three population groups give a picture of the spurious LD generated by this known subdivision and can be compared to the LD seen among unlinked SNPs within the same population sample. Mean ± SD  for the pooled sample was 0.0163±0.0259, whereas the respective figures for the Asian, African American, and European American samples were 0.0289±0.0429, 0.0296±0.0445, and 0.0298±0.0434. The greater mean
for the pooled sample was 0.0163±0.0259, whereas the respective figures for the Asian, African American, and European American samples were 0.0289±0.0429, 0.0296±0.0445, and 0.0298±0.0434. The greater mean 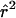 within population samples is almost certainly due to the smaller sample size as compared with the pooled sample. The pooled sample had 4.8% of the pairwise tests of unlinked SNPs, with a significant exact test at the 1% level, indicating some inflation due to population heterogeneity. Within populations, the African American, Asian, and European American samples were much closer to the null expectation, with 1.20%, 1.07%, and 1.09%, respectively, of tests significant at the 1% level. Our overall impression of significance of LD among unlinked SNPs is much less than that of Sinnock and Sing (1972), who found many instances of significant LD among unlinked loci in a sample of 6,756 people from Tecumseh, MI. The large sample size of their study gave it considerable power to detect subtle, but confounded, effects of drift, hidden population stratification, and natural selection.
within population samples is almost certainly due to the smaller sample size as compared with the pooled sample. The pooled sample had 4.8% of the pairwise tests of unlinked SNPs, with a significant exact test at the 1% level, indicating some inflation due to population heterogeneity. Within populations, the African American, Asian, and European American samples were much closer to the null expectation, with 1.20%, 1.07%, and 1.09%, respectively, of tests significant at the 1% level. Our overall impression of significance of LD among unlinked SNPs is much less than that of Sinnock and Sing (1972), who found many instances of significant LD among unlinked loci in a sample of 6,756 people from Tecumseh, MI. The large sample size of their study gave it considerable power to detect subtle, but confounded, effects of drift, hidden population stratification, and natural selection.
Discussion
A SNP sampled from a population can be thought of as possessing an ancestral genealogy joining the members of the sample back to a common ancestor. Samples of two SNPs have two trees of ancestry, and the degree of coupling of these genealogies depends on the rate of recombination between them. To the extent that the trees are correlated, there will be LD between the SNPs. Pairwise LD can thus be thought of through the underlying joint genealogy of the pair of SNPs, and an ancestral recombination graph can be constructed to capture this ancestral history (Wiuf and Hein 1999; Hudson 2001; Nordborg and Tavaré 2002). Processes occurring at the population level that distort the topology of this graph (e.g., migration and population expansion) will likewise have an impact on the pattern of LD. Although it is difficult to tease out the nature of the forces that generate LD in any particular case, examination of a full genome’s breadth of SNPs can provide deeper insights regarding the recombinational history of humans.
Heterogeneity among populations in the LD map has been widely cited in local genomic regions, but the full extent of this heterogeneity becomes quite apparent when a whole-genome scan such as this is done. In fact, among these three major population groups, only 17% of the clusters that span >100 kb produced estimates of ρ that were not statistically heterogeneous across the three population groups. Given an average FST of 0.08 and an FST distribution that includes much higher values, it is perhaps not surprising that some combination of drift and selection would result in such heterogeneity. The role of drift alone in the generation of differences in haplotype frequencies and concomitant variation in LD has been understood for many years (Ohta and Kimura 1971; Sved 1971), but even more striking is the role of local differences in natural selection, driven by pathogens or other local environmental effects (Tishkoff et al. 2001; Hamblin et al. 2002). In addition, one of the major causes of LD across distant sites is admixture, and the African American population appears to have, on average, lower LD than the other population groups, indicating that the past demographic history (larger long-term effective size in Africa) outweighs the admixture effect. On a more local scale, it is also well known that founding events of some population isolates may result in much higher LD over longer distances (Mohlke et al. 2001), but many populations considered as having strong founder effects show LD patterns typical of large populations (Clark et al. 1998; Fullerton et al. 2000).
Many attributes of polymorphisms are considered as means by which to identify interesting departures from the standard neutral theory. If a region of the genome recently had an advantageous mutation sweep to fixation, then it would leave a characteristic signature in local levels of variation and of LD (Kim and Stephan 2002). We are not able to conclude much about local variability, because of the way in which the SNPs were ascertained; however, another attribute that would be distorted by a local selection event is the degree of population subdivision.
The existence of recombination hotspots in the human genome has been known for quite some time, but most of our information comes from the in-depth study of defined regions, rather than from a genomewide scan for hotspot density. The literature on hotspots further leads one to believe that most of the genome has a quite low rate of recombination, and there are relatively sharply defined regions of much higher recombination (Jeffreys et al. 2000, 2001; Daly et al. 2001; Petes 2001). Despite this sharp difference in recombination rate between regions with hotspots versus regions that lack hotspots, the distribution of population recombination rates is smooth and unimodal (fig. 3). Such a unimodal distribution is not unexpected, however, since these estimates of 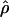 are averaged over regions ⩽180 kb; thus, there is some smoothing that occurs in estimation across regions of this size. In addition, these are estimates of
are averaged over regions ⩽180 kb; thus, there is some smoothing that occurs in estimation across regions of this size. In addition, these are estimates of  , not strictly the recombination rate, so other factors that influence effective population size will also tend to smooth out a bimodal recombination pattern.
, not strictly the recombination rate, so other factors that influence effective population size will also tend to smooth out a bimodal recombination pattern.
Importantly, the rate of recombination within recombination hotspots is known to be highly variable. Jeffreys et al. (2001) showed, both by single-sperm typing and by LD analysis, that a 216-kb segment of chromosome 6 had six recombination hotspots and that each corresponds to well-defined regions of locally lower LD. If the region that Jeffreys et al. (2001) studied reflects the true density of recombination hotspots (one every 36 kb), then we would expect to capture several hotspots in every cluster. Fine-structure analysis of LD from molecularly phased data also provides empirical support for clustering of regions of high and low LD (Olivier et al. 2001). The pattern of LD in the major histocompatibility complex (MHC) region, on chromosome 6, is remarkably consistent across populations, and Kauppi et al. (2003) go so far as to suggest that heterogeneity in LD across this region is dominated by recombination hotspots as opposed to population history. Our data demonstrate that a determination of the role that hotspots play in the shaping of human LD genomewide will require much greater SNP density than was used here. But our data also show clearly that, despite claims of local regions with consistent among-population patterns of LD, the overall picture is that the variability in LD across populations is sufficient to be highly statistically significant. The key point is that, for tests of disease association, the heterogeneity in statistical significance of association with disease is likely to be even greater than the heterogeneity in inter-SNP LD (e.g., because of confounding environmental effects).
We are only beginning to understand the nature of variation in recombination rate across the human genome. In yeast, it is clear that recombination hotspots correspond to regions with elevated frequencies of double-strand DNA breaks (Petes 2001). Deletions and mutations that reduce the incidence of double-strand DNA breaks also reduce the frequency of recombination within hotspots. There also appears to be competition between these sites, in that deletion of a hotspot increases the activity of neighbors and rearrangements that place two hotspots in close proximity reduce the level of activity of both (Fan et al. 1997). In yeast, hotspots are correlated with high GC content, just as there is a correlation between GC content and local recombination rate in humans (Fullerton et al. 2001). Understanding the mechanism behind such correlations may be critical to the success of an LD-mapping approach, because they may provide the clearest way to predict the distance scale over which there are changes in local rates of recombination.
Gene conversion may play a significant role in the decay kinetics of LD in human populations. Ardlie et al. (2001) determined SNP genotypes in 68 STSs and found much less LD among SNPs within very short physical distances than expected on the basis of population genetic theory. They concluded that gene conversion must contribute to the shuffling of gametic phase at this fine scale, another result relevant to fine-scale LD mapping. Frisse et al. (2001) applied Hudson’s (2001) method and found that the inferred ratio of gene conversion to crossing-over is 7.3, with a mean conversion-tract length of 500 bp. Similarly, Wiehe et al. (2000) found that gene conversion appears to play an important role in the decay of LD over short distances within the MHC region, on chromosome 6. Analysis of many genomic regions further supports the idea that local LD is less than expected, and the discrepancy is easily accommodated by inference of gene conversion (Przeworski and Wall 2001). If there is wide variation in the rate of gene conversion across the genome, this could result in distortions in the LD map that may, in turn, adversely affect inferences from a whole-genome association study. Rate of gene conversion could not be estimated from our data, but it is one of potentially many unmeasured factors that may have an impact on the pattern of LD and its utility in the mapping of genes associated with risk of complex disease.
Several approaches have been taken to estimate ρ from a sample of DNA sequences drawn from a steady-state, panmictic population (Hey and Wakeley 1997; Wakeley 1997; Wall 2000; Hudson 2001). All of these estimators are biased downward if the data consist of SNPs ascertained from a small discovery panel, and only recently have methods been devised to accommodate the ascertainment scheme into inferences of  (Nielsen and Signorovitch 2003). To assess the impact of ascertainment bias in this context, we also estimated
(Nielsen and Signorovitch 2003). To assess the impact of ascertainment bias in this context, we also estimated 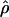 by Hudson’s method, and we found that the squared correlation between the corrected and uncorrected estimates of
by Hudson’s method, and we found that the squared correlation between the corrected and uncorrected estimates of 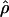 is 0.984, suggesting that the bias correction had very little impact on the
is 0.984, suggesting that the bias correction had very little impact on the 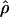 estimates. But correlation is a poor metric for the assessment of performance, because we really need to know how often a false inference would be made using the uncorrected estimates. Of the 527 clusters for which ρ was estimated both with and without correction, we find that 10% of the clusters had an overestimate of ρ >25%. A key observation is that the failure to correct for bias results in estimates of ρ that are greater than the true values. This systematic bias is related to the fact that SNPs ascertained from a small panel will be skewed toward higher allele frequency, which, on average, are older and have had more time to recombine in the population. As pointed out by Nielsen and Signorovitch (2003), the ascertainment biases of D′ and r2 are far greater than is the ascertainment bias of estimates of
estimates. But correlation is a poor metric for the assessment of performance, because we really need to know how often a false inference would be made using the uncorrected estimates. Of the 527 clusters for which ρ was estimated both with and without correction, we find that 10% of the clusters had an overestimate of ρ >25%. A key observation is that the failure to correct for bias results in estimates of ρ that are greater than the true values. This systematic bias is related to the fact that SNPs ascertained from a small panel will be skewed toward higher allele frequency, which, on average, are older and have had more time to recombine in the population. As pointed out by Nielsen and Signorovitch (2003), the ascertainment biases of D′ and r2 are far greater than is the ascertainment bias of estimates of  .
.
An important issue that needs to be elucidated in the near future is the number of SNPs that would be required in order to adequately cover the human genome for an LD map. Great hopes are being placed on the idea that, at a local scale, some genomic regions appear to have relatively few common haplotypes, so that a subset of SNPs might be used to mark these haplotypes (Daly et al. 2001; Johnson et al. 2001; Gabriel et al. 2002); however, these have been sampled regions, not a scan. Patil et al. (2001) covered a whole chromosome, but the focus was on the largest regions, not the long tail of regions with very weak block structure. Zhang et al. (2002) applied dynamic programming to the same data set to obtain a dramatic reduction in numbers of SNPs needed to cover the same regions of locally reduced haplotype diversity. Judson et al. (2002) made an attempt to model the number of SNPs needed for genomewide coverage and found the number to be in the range of 100,000–600,000 SNPs. The very high variability in local recombination rates certainly suggests that the SNP set carefully tailored to this recombination profile could be greatly more efficient than either a uniform or a random distribution. Work is in progress to model this process.
In addition to the caveats about heterogeneity in local LD, all of the above concerns apply even if the disease that is being mapped is fully penetrant and Mendelian in character. Like most other authors writing about dense arrays of SNPs for LD mapping, we have ignored what is probably the biggest impediment to success—namely that the diseased conditions to which we want to apply LD mapping are likely to be genetically heterogeneous and exhibit with low penetrance, low heritability, moderate sibling recurrence risk, and complicated epistatic and genotype-by-environment interactions. These complications may greatly reduce the efficacy of finding genes that underlie such traits through statistical association with a SNP (Weiss and Terwilliger 2000; Weiss and Clark 2002). One limitation that whole-genome LD mapping will face, almost regardless of the landscape of LD, is that the power to detect association drops rapidly with the frequency of the disease-associated alleles (Kaplan and Morris 2001). Only the magnitude of the consequences to health and the fact that many complex disorders have reasonably hopeful sibling recurrence risk motivate us to continue considering LD mapping in this context. As the density of LD maps continues to increase and as we get a clearer picture of the landscape of LD across the human genome, we will get a better idea of the likely efficacy of LD mapping the genes that underlie complex traits.
Acknowledgments
This project was made possible by resources provided by the American Diabetes Association (for the Japanese samples in the diversity panel), the Center for Medical Genetics at the Marshfield Clinic, NCBI, and TSC. Computational resources of the High Performance Computing group of the Cornell Theory Center is gratefully acknowledged. This project was funded by TSC and Motorola Life Sciences. We also thank Applied Biosystems for providing the TaqMan primers and probes. National Institutes of Health grant GM65509 provided additional support to A.G.C., and National Science Foundation grant DEB-0089487 provided additional support to R.N.
Appendix A
Here, we describe how to calculate the likelihood function for a pair of SNPs while taking into account the special ascertainment scheme used in the TSC data. The method is similar to that described by Nielsen and Signorovitch (2003), except that the TSC data lack linkage phase information and there is no consistent ascertainment scheme used for all SNPs.
We distinguish between three samples: the typed sample of diploid individuals, the ascertainment sample of locus 1, and the ascertainment sample of locus 2. Except for loci with missing data, the sample size of typed individuals is x=90. The sample size of the ascertainment samples, called the “depths,” are typically two to five chromosomes and are denoted by d1 and d2. We will assume that the two ascertainment samples are nonoverlapping.
The data in the typed sample can be coded as x=(x00,00,x00,01,x00,11,x01,00,x01,01,x01,11,x11,00,x11,01,x11,11), where x00,00 is the number of individuals of type 00 in the first locus and type 00 in the second locus when two diallelic loci with alleles 0 and 1 are assumed, and so forth. We here assume an arbitrary labeling of alleles, but the method can trivially be extended to the case in which the ancestral state is known. We are interested in calculating
 |
where Ai is the ascertainment condition that variability is observed in the ascertainment sample of locus i, θ=4Neμ, and ρ=2Ner (Ne is the effective population size, μ is the per-site mutation rate, and r is the recombination rate per generation between the two loci). The reason for taking the limit of θ→0 (as in Nielsen 2000 and Hudson 2001) is to eliminate the nuisance parameter θ. The effect of this is very small, since θ is typically on the order of 10−3 and since 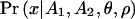 is only weakly dependent on θ. Notice that
is only weakly dependent on θ. Notice that
 |
We will assume that the SNPs have been identified independently in clusters of different reads. Let the joint ascertainment sample for the two loci be na=(na1,na2,na3,na4), where na1, na2, na3, and na4 are the (unknown) numbers of genotypes of types 00, 11, 10, and 01, respectively, in the joint ascertainment sample. Then,
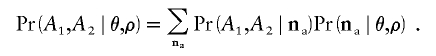 |
Let tij be the length of lineage j of the ascertainment sample of locus i, and let T(i) be the total tree length in locus i. Also, let Ijk be an indicator function that takes on the value 1 if one mutation in lineage j of locus 1 and one mutation in lineage k of locus 2, with no other mutations occurring in the history of the genealogies, generate exactly the data pattern na. Then,
where the expectation is with respect to the joint distribution of genealogies in loci 1 and 2 (Nielsen 2000; Hudson 2001).
Let Pi(0) and Pi(1) be the probabilities of obtaining only alleles of type 0 and only alleles of type 1, respectively, in the ascertainment sample of locus i. Likewise, let Pst be the probability of only obtaining alleles of type s in locus 1 and only obtaining alleles of type t in locus 2 in the ascertainment samples of loci 1 and 2. Then,
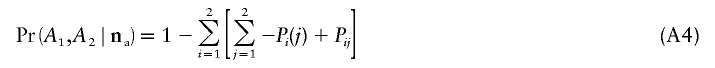 |
and
 |
where I(c) returns 1 if the condition c is true and 0 otherwise. Here, we also use the convention that
 |
if k<0 or k>n.
The numerator in equation (A2) can be calculated by considering the pooled data from typed SNPs and from ascertainment samples, augmented by information regarding haplotypic phase calculated by conditioning on the number of haplotypes of type 11 in the sample of typed SNPs (k). The augmented data may be expressed as n=(n1,n2,n3,n4), where n1, n2, n3, and n4 are the numbers in the augmented sample of genotypes of types 00, 11, 10, and 01, respectively. Then,
 |
Let  be a sample with haplotypic phase known of size x, for which n(x)k=(a,b,c,k), where c=x4+2x7+x8+d, b=x6+x2+2x3+d, a=2x1+x2+x4+x5-d, and d=x5+x6+x8+2x9-k. Since k is fully determined by
be a sample with haplotypic phase known of size x, for which n(x)k=(a,b,c,k), where c=x4+2x7+x8+d, b=x6+x2+2x3+d, a=2x1+x2+x4+x5-d, and d=x5+x6+x8+2x9-k. Since k is fully determined by  and vice versa,
and vice versa,
 |
Here, n=na∪n(x)k. From the study by Hudson (2001),
 |
where xhet is the number of individuals heterozygous in at least one of the loci. Also,
 |
 is given by equation (A3), but with n replaced by na. When considering the limit in equation (A1), we replace
is given by equation (A3), but with n replaced by na. When considering the limit in equation (A1), we replace  with
with 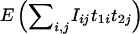 , and we do a similar replacement for
, and we do a similar replacement for  , as in the studies by Nielsen (2000) and Hudson (2001). These expectations can be tabulated for all possible values of n and ρ, assuming Kingman’s (1982) coalescent process, using the computer program by Hudson (2001). After initial tabulation of these expectations, most of the computational time spent calculating the (composite) likelihood function for ρ is devoted to calculating the combinatorial terms in equations (A4)–(A7).
, as in the studies by Nielsen (2000) and Hudson (2001). These expectations can be tabulated for all possible values of n and ρ, assuming Kingman’s (1982) coalescent process, using the computer program by Hudson (2001). After initial tabulation of these expectations, most of the computational time spent calculating the (composite) likelihood function for ρ is devoted to calculating the combinatorial terms in equations (A4)–(A7).
Electronic-Database Information
The URL for data presented herein is as follows:
- SNP Consortium Linkage Map Project, The, http://snp.cshl.org/linkage_maps/
References
- Abecasis GR, Noguchi E, Heinzmann A, Traherne JA, Bhattacharyya S, Leaves NI, Anderson GG, Zhang Y, Lench NJ, Carey A, Cardon LR, Moffatt MF, Cookson WO (2001) Extent and distribution of linkage disequilibrium in three genomic regions. Am J Hum Genet 68:191–197 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ardlie K, Liu-Cordero SN, Eberle MA, Daly M, Barrett J, Winchester E, Lander ES, Kruglyak L (2001) Lower-than-expected linkage disequilibrium between tightly linked markers in humans suggests a role for gene conversion. Am J Hum Genet 69:582–589 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonnen PE, Wang PJ, Kimmel M, Chakraborty R, Nelson DL (2002) Haplotype and linkage disequilibrium architecture for human cancer-associated genes. Genome Res 12:1846–1853 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Broman KW, Murray JC, Sheffield VC, White RL, Weber JL (1998) Comprehensive human genetic maps: individual and sex-specific variation in recombination. Am J Hum Genet 63:861–869 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clark AG, Weiss KM, Nickerson DA, Taylor SL, Buchanan A, Stengård J, Salomaa V, Vartiainen E, Perola M, Boerwinkle E, Sing CF (1998) Haplotype structure and population genetic inferences from nucleotide-sequence variation in human lipoprotein lipase. Am J Hum Genet 63:595–612 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daly MJ, Rioux JD, Schaffner SF, Hudson TJ, Lander ES (2001) High-resolution haplotype structure in the human genome. Nat Genet 29:229–232 [DOI] [PubMed] [Google Scholar]
- Dawson E, Abecasis GR, Bumpstead S, Chen Y, Hunt S, Beare DM, Pabial J, et al (2002) A first-generation linkage disequilibrium map of human chromosome 22. Nature 418:544–548 [DOI] [PubMed] [Google Scholar]
- Dunning AM, Durocher F, Healey CS, Teare MD, McBride SE, Carlomagno F, Xu CF, Dawson E, Rhodes S, Ueda S, Lai E, Luben RN, Van Rensburg EJ, Mannermaa A, Kataja V, Rennart G, Dunham I, Purvis I, Easton D, Ponder BAJ (2000) The extent of linkage disequilibrium in four populations with distinct demographic histories. Am J Hum Genet 67:1544–1554 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eisenbarth I, Vogel G, Krone W, Vogel W, Assum G (2000) An isochore transition in the NF1 gene region coincides with a switch in the extent of linkage disequilibrium. Am J Hum Genet 67:873–880 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fan QQ, Xu F, White MA, Petes TD (1997) Competition between adjacent meiotic recombination hotspots in the yeast Saccharomyces cerevisiae. Genetics 145:661–670 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frisse L, Hudson RR, Bartoszewicz A, Wall JD, Donfack J, Di Rienzo A (2001) Gene conversion and different population histories may explain the contrast between polymorphism and linkage disequilibrium levels. Am J Hum Genet 69:831–843 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fullerton SM, Bernardo Carvalho A, Clark AG (2001) Local rates of recombination are positively correlated with GC content in the human genome. Mol Biol Evol 18:1139–1142 [DOI] [PubMed] [Google Scholar]
- Fullerton, SM, Clark AG, Weiss KM, Nickerson DA, Taylor SL, Stengård JH, Salomaa V, Vartiainen E, Perola M, Boerwinkle E, Sing CF (2000) Apolipoprotein E variation at the sequence haplotype level: implications for the origin and maintenance of a major human polymorphism. Am J Hum Genet 67:881–900 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gabriel SB, Schaffner SF, Nguyen H, Moore JM, Roy J, Blumenstiel B, Higgins J, DeFelice M, Lochner A, Faggart M, Liu-Cordero SN, Rotimi C, Adeyemo A, Cooper R, Ward R, Lander ES, Daly MJ, Altshuler D (2002) The structure of haplotype blocks in the human genome. Science 296:2225–2229 [DOI] [PubMed] [Google Scholar]
- Hamblin MT, Thompson EE, Di Rienzo A (2002) Complex signatures of natural selection at the Duffy blood group locus. Am J Hum Genet 70:369–383 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hey J, Wakeley J (1997) A coalescent estimator of the population recombination rate. Genetics 145:833–846 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hudson RR (1985) The sampling distribution of linkage disequilibrium under an infinite allele model without selection. Genetics 109:611–631 [DOI] [PMC free article] [PubMed] [Google Scholar]
- ——— (2001) Two-locus sampling distributions and their application. Genetics 159:1805–1817 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huttley GA, Smith MW, Carrington M, O’Brien SJ (1999) A scan for linkage disequilibrium across the human genome. Genetics 152:1711–1722 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeffreys AJ, Kauppi L, Neumann R (2001) Intensely punctate meiotic recombination in the class II region of the major histocompatibility complex. Nat Genet 29:217–222 [DOI] [PubMed] [Google Scholar]
- Jeffreys AJ, Ritchie A, Neumann R (2000) High resolution analysis of haplotype diversity and meiotic crossover in the human TAP2 recombination hotspot. Hum Mol Genet 9:725–733 [DOI] [PubMed] [Google Scholar]
- Johnson GC, Esposito L, Barratt BJ, Smith AN, Heward J, Di Genova G, Ueda H, Cordell HJ, Eaves IA, Dudbridge F, Twells RC, Payne F, Hughes W, Nutland S, Stevens H, Carr P, Tuomilehto-Wolf E, Tuomilehto J, Gough SC, Clayton DG, Todd JA (2001) Haplotype tagging for the identification of common disease genes. Nat Genet 29:233–237 [DOI] [PubMed] [Google Scholar]
- Judson R, Salisbury B, Schneider J, Windemuth A, Stephens JC (2002) How many SNPs does a genome-wide haplotype map require? Pharmacogenomics 3:379–391 [DOI] [PubMed] [Google Scholar]
- Kaplan N, Morris R (2001) Prospects for association-based fine mapping of a susceptibility gene for a complex disease. Theor Popul Biol 60:181–191 [DOI] [PubMed] [Google Scholar]
- Kauppi L, Sajantila A, Jeffreys AJ (2003) Recombination hotspots rather than population history dominate linkage disequilibrium in the MHC class II region. Hum Mol Genet 12:33–40 [DOI] [PubMed] [Google Scholar]
- Kidd JR, Pakstis AJ, Zhao H, Lu RB, Okonofua FE, Odunsi A, Grigorenko E, Bonne-Tamir B, Friedlaender J, Schulz LO, Parnas J, Kidd KK (2000) Haplotypes and linkage disequilibrium at the phenylalanine hydroxylase locus, PAH, in a global representation of populations. Am J Hum Genet 66:1882–1899 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim Y, Stephan W (2002) Detecting a local signature of genetic hitchhiking along a recombining chromosome. Genetics 160:765–777 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kingman JFC (1982) On the genealogy of large populations. J Appl Probability 19A:27–43 [Google Scholar]
- Kuhner M, Beerli P, Yamato J, Felsenstein J (2000) Usefulness of single nucleotide polymorphism data for estimating population parameters. Genetics 156:439–447 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matise TC, Sachidanandam R, Clark AG, Kruglyak L, Wijsman E, Kakol J, Buyske S, et al (2003) A 3.9-centimorgan-resolution human single-nucleotide polymorphism linkage map and screening set. Am J Hum Genet 72:271–284 (in this issue) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mohlke KL, Lange EM, Valle TT, Ghosh S, Magnuson VL, Silander K, Watanabe RM, Chines PS, Bergman RN, Tuomilehto J, Collins FS, Boehnke M (2001) Linkage disequilibrium between microsatellite markers extends beyond 1 cM on chromosome 20 in Finns. Genome Res 11:1221–1226 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nielsen R (2000) Estimation of population parameters and recombination rates from single nucleotide polymorphisms. Genetics 154:931–942 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nielsen R, Signorovitch J (2003) Correcting for ascertainment biases when analyzing SNP data: application to the estimation of linkage disequilibrium. Theor Popul Biol 63:245–255 [DOI] [PubMed] [Google Scholar]
- Nordborg M, Tavaré S (2002) Linkage disequilibrium: what history has to tell us. Trends Genet 18:83–90 [DOI] [PubMed] [Google Scholar]
- Ohta T, Kimura M (1971) Linkage disequilibrium between two segregating nucleotide sites under the steady flux of mutations in a finite population. Genetics 68:571–580 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olivier M, Bustos VI, Levy MR, Smick GA, Moreno I, Bushard JM, Almendras AA, Sheppard K, Zierten DL, Aggarwal A, Carlson CS, Foster BD, Vo N, Kelly L, Liu X, Cox DR (2001) Complex high-resolution linkage disequilibrium and haplotype patterns of single-nucleotide polymorphisms in 2.5 Mb of sequence on human chromosome 21. Genomics 78:64–72 [DOI] [PubMed] [Google Scholar]
- Patil N, Berno AJ, Hinds DA, Barrett WA, Doshi JM, Hacker CR, Kautzer CR, Lee DH, Marjoribanks C, McDonough DP, Nguyen BT, Norris MC, Sheehan JB, Shen N, Stern D, Stokowski RP, Thomas DJ, Trulson MO, Vyas KR, Frazer KA, Fodor SP, Cox DR (2001) Blocks of limited haplotype diversity revealed by high-resolution scanning of human chromosome 21. Science 294:1719–1723 [DOI] [PubMed] [Google Scholar]
- Petes TD (2001) Meiotic recombination hot spots and cold spots. Nat Rev Genet 2:360–369 [DOI] [PubMed] [Google Scholar]
- Phillips MS, Lawrence R, Sachidanandam R, Morris AP, Balding DJ, Donaldson MA, Studebaker JF, et al (2003) Chromosome-wide distribution of haplotype blocks and the role of recombination hot spots. Nat Genet 33:382–387 [DOI] [PubMed] [Google Scholar]
- Pritchard JK, Przeworski M (2001) Linkage disequilibrium in humans: models and data. Am J Hum Genet 69:1–14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Przeworski M, Wall J (2001) Why is there so little intragenic linkage disequilibrium in humans? Genet Res 77:143–151 [DOI] [PubMed] [Google Scholar]
- Reich D Cargill M, Bolk S, Ireland J, Sabeti PC, Richter DJ, Lavery T, Kouyoumjian R, Farhadian SF, Ward R, Lander ES (2001) Linkage disequilibrium in the human genome. Nature 411:199–204 [DOI] [PubMed] [Google Scholar]
- Sinnock P, Sing CF (1972) Analysis of multilocus genetic systems in Tecumseh, Michigan. II. Consideration of the correlation between nonalleles in gametes. Am J Hum Genet 24:393–415 [PMC free article] [PubMed] [Google Scholar]
- Sved JA (1971) Linkage disequilibrium and homozygosity of chromosome segments in finite populations. Theor Popul Biol 2:125–141 [DOI] [PubMed] [Google Scholar]
- Taillon-Miller P, Bauer-Sardiña I, Saccone NL, Putzel J, Laitinen T, Cao A, Kere J, Pilia G, Rice JP, Kwok PY (2000) Juxtaposed regions of extensive and minimal linkage disequilibrium in human Xq25 and Xq28. Nat Genet 25:324–328 [DOI] [PubMed] [Google Scholar]
- Tishkoff SA, Varkonyi R, Cahinhinan N, Abbes S, Argyropoulos G, Destro-Bisol G, Drousiotou A, Dangerfield B, Lefranc G, Loiselet J, Piro A, Stoneking M, Tagarelli A, Tagarelli G, Touma EH, Williams SM, Clark AG (2001) Haplotype diversity and linkage disequilibrium at human G6PD: recent origin of alleles that confer malarial resistance. Science 293:455–462 [DOI] [PubMed] [Google Scholar]
- Wakeley JR (1997) Using the variance of pairwise differences to estimate the recombination rate. Genet Res 69:45–48 [DOI] [PubMed] [Google Scholar]
- Wakeley J, Nielsen R, Liu-Cordero SN, Ardlie K (2001) The discovery of single-nucleotide polymorphisms—and inferences about human demographic history. Am J Hum Genet 69:1332–1347 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wall JD (2000) A comparison of estimators of the population recombination rates. Mol Biol Evol 17:156–163 [DOI] [PubMed] [Google Scholar]
- Weir BS (1996) Genetic Data Analysis II. Sinauer Associates, Sunderland, MA [Google Scholar]
- Weiss KM, Clark AG (2002) Linkage disequilibrium and the mapping of complex human traits. Trends Genet 18:19–24 [DOI] [PubMed] [Google Scholar]
- Weiss KM, Terwilliger JD (2000) How many diseases does it take to map a gene with SNPs? Nat Genet 26:151–157 [DOI] [PubMed] [Google Scholar]
- Wiehe T, Mountain J, Parham P, Slatkin M (2000) Distinguishing recombination and intragenic gene conversion by linkage disequilibrium patterns. Genet Res Camb 75:61–73 [DOI] [PubMed] [Google Scholar]
- Wiuf C, Hein J (1999) The ancestry of a sample of sequences subject to recombination. Genetics 151:1217–1228 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang K, Deng M, Chen T, Waterman MS, Sun F (2002) A dynamic programming algorithm for haplotype block partitioning. Proc Natl Acad Sci USA 99:7335–7339 [DOI] [PMC free article] [PubMed] [Google Scholar]