Abstract
Over the past 20 years, the incidence of cutaneous malignant melanoma (CMM) has increased dramatically worldwide. A positive family history of the disease is among the most established risk factors for CMM; it is estimated that 10% of CMM cases result from an inherited predisposition. Although mutations in two genes, CDKN2A and CDK4, have been shown to confer an increased risk of CMM, they account for only 20%–25% of families with multiple cases of CMM. Therefore, to localize additional loci involved in melanoma susceptibility, we have performed a genomewide scan for linkage in 49 Australian pedigrees containing at least three CMM cases, in which CDKN2A and CDK4 involvement has been excluded. The highest two-point parametric LOD score (1.82; recombination fraction [θ] 0.2) was obtained at D1S2726, which maps to the short arm of chromosome 1 (1p22). A parametric LOD score of 4.65 (θ=0) and a nonparametric LOD score of 4.19 were found at D1S2779 in nine families selected for early age at onset. Additional typing yielded seven adjacent markers with LOD scores >3 in this subset, with the highest parametric LOD score, 4.95 (θ=0) (nonparametric LOD score 5.37), at D1S2776. Analysis of 33 additional multiplex families with CMM from several continents provided further evidence for linkage to the 1p22 region, again strongest in families with the earliest mean age at diagnosis. A nonparametric ordered sequential analysis was used, based on the average age at diagnosis in each family. The highest LOD score, 6.43, was obtained at D1S2779 and occurred when the 15 families with the earliest ages at onset were included. These data provide significant evidence of a novel susceptibility gene for CMM located within chromosome band 1p22.
Introduction
Cutaneous malignant melanoma (CMM) represents a significant public health burden in all populations of European origin. In the United States, for example, CMM incidence has increased at a rate greater than that of any other cancer type (477% increase in incidence between 1950 and 1998, based on Surveillance, Epidemiology, and End Results [SEER] data) (Ries et al. 2003). Incidence rates vary by more than an order of magnitude even in susceptible populations, with lifetime risk of CMM ranging from 0.2% in southern Europe to 1% in the United States and Scandinavia and an average of 4% in Australia. According to SEER data, the 5-year survival rate for CMM decreases dramatically (from 96% to 12%) in more advanced stages of disease (Ries et al. 2003). It is hoped that further characterization of the genetic factors contributing to an elevated risk of developing CMM will form a basis for understanding which individuals in susceptible populations are most at risk, how that risk can be modified, and how that knowledge would allow targeted surveillance and recognition of disease at early stages, when treatment strategies are maximally beneficial.
The etiology of CMM is complex, involving both heterogeneous genetic and environmental components. The CDKN2A locus (MIM 600160) accounts for susceptibility in ∼25% of all kindreds with familial CMM (Goldstein et al. 2000), whereas mutations in the CDK4 (MIM 123829) gene have, to date, been documented in only three CMM-prone families (Zuo et al. 1996; Soufir et al. 1998). The average lifetime risk conferred by CDKN2A mutations is ∼70% in multiple-case families but shows significant variation between regions (Bishop et al. 2002). In contrast to these high-penetrance mutations, variants of the melanocortin receptor 1 gene (MC1R) have proven to be low-penetrance or medium-risk susceptibility alleles for CMM (Palmer et al. 2000). These MC1R (MIM 155555) variants also act as modifier alleles, increasing the penetrance of mutations in CDKN2A (Box et al. 2001; van der Velden et al. 2001). Clearly, additional CMM predisposition genes are likely to exist.
Material and Methods
Families
Ascertainment and clinical evaluation of the 49 Australian pedigrees with familial CMM included in our genomewide scan (phase 1) are described elsewhere (Kefford et al. 1991; Nancarrow et al. 1992b). In an effort to maximize the chance of identifying a novel high-risk susceptibility gene, we restricted inclusion in our study to pedigrees that were mutation negative for both CDKN2A and CDK4, by SSCP and/or direct sequencing, and contained at least three CMM cases with DNA from blood available for genotyping. In addition, any pedigree showing evidence of haplotype sharing in the 9p21-p22 region, where CDKN2A is located, was also excluded, to minimize inclusion of families with undetected mutations within the CDKN2A gene or outside the coding region. Several noncoding mutations in CDKN2A have been identified (Liu et al. 1999; Harland et al. 2001). In total, our genomewide scan was conducted in 49 Australian families (phase 1, table 1). Each family was highly loaded with CMM cases (average number of affected individuals 4.3, range 3–8). Although we required that each family have at least three affected individuals, 37% of the families (18/49) had more than five affected individuals. There was no evidence of an excess of pancreatic cancer in these families, as has been previously reported in some CDKN2A mutation–positive families (Goldstein et al. 1995; Borg et al. 2000; Vasen et al. 2000); however, three families contained cases of both cutaneous and ocular melanoma. Pedigrees included in the second phase analyses (phase 2, table 1) were identified and collected in Australia, the United States, the United Kingdom, France, Sweden, and The Netherlands, by members of the Melanoma Genetics Consortium. More details about the families can be found elsewhere (Bergman et al. 1992; Hussussian et al. 1994; Gruis et al. 1995; Platz et al. 1997; Soufir et al. 1998; Hashemi et al. 1999; Newton Bishop et al. 1999; Bishop et al. 2000; Borg et al. 2000; Goldstein et al. 2000; Harland et al. 2000). The goal of the Melanoma Genetics Consortium, established in 1997, is to investigate factors related to the inheritance of an increased risk of melanoma (Kefford et al. 1999; Bishop et al. 2002). Melanoma cases were verified as thoroughly as possible from cancer registries, pathology reports, or clinical records.
Table 1.
Characteristics of Families with Melanoma
| Sample Site | Phase | No. ofFamiliesper Site | Average No.of AffectedIndividualsper Family(Range) | Average No.of AffectedIndividualsTyped perFamily(Range) | Average Ageat Diagnosisin Years perFamily (Range) |
| Queensland Institute of Medical Research, Brisbane, Australia | 1 | 27 | 4.5 (3–8) | 3.9 (3–7) | 45.5 (29.7–76) |
| Westmead Institute for Cancer Research, Sydney, Australia | 1 | 22 | 4.1 (3–7) | 3.6 (3–6) | 43.8 (24–65) |
| Queensland Institute of Medical Research, Brisbane, Australia | 2 | 9 | 4.2 (3–8) | 3.2 (3–4) | 46.6 (30–58.6) |
| Westmead Institute for Cancer Research, Sydney, Australia | 2 | 15 | 3.7(3–8) | 3.4 (3–7) | 44.3 (28.8–62.7) |
| Genetic Epidemiology Branch, National Cancer Institute, Bethesda, U.S.A. | 2 | 2 | 4.5 (3–6) | 3.5 (3–4) | 41.9 (38.8–45) |
| Leiden University Medical Center, Leiden, The Netherlands | 2 | 2 | 5.5 (5–6) | 5.5 (5–6) | 43.9 (39.3–48.4) |
| Cancer Research UK, Leeds, U.K. | 2 | 2 | 3 | 3 | 44.3 (27.3–61.3) |
| Institut Gustave Roussy, Villejuif, France | 2 | 1 | 5 | 4 | 41.2 |
| University Hospital, Lund, Sweden | 2 | 1 | 5 | 4 | 54.6 |
| Karolinska Hospital, Stockholm, Sweden | 2 | 1 | 3 | 3 | 49 |
Genotyping
Genomic DNA was prepared from blood samples through use of standard techniques. DNA samples were genotyped using 414 STR markers with an average intermarker spacing of 10 cM and an average heterozygosity of 80%. PCRs, using fluorescently labeled primers, were set up using a TECAN Genesis 200 robot. All PCRs were performed in a 15-μl volume containing 20 ng of genomic DNA, 0.33 μM each primer, 0.25 mM each dNTP, 2.5 mM MgCl2, 10 mM Tris-HCl, 50 mM KCl, and 0.5 U Taq polymerase. PCR amplification was performed using the GeneAmp 9600 or 9700 thermocyclers (Applied Biosystems). PCR cycling conditions were as follows: 95°C for 12 min, followed by 10 cycles of 94°C for 15 s, 55°C for 15 s, 72°C for 30 s, and 20 cycles of 89°C for 15 s, 55°C for 15 s, and 72°C for 30 s, with a final extension of 72°C for 10 min. Depending on the PCR yield, 5–15 μl of product was combined with those of as many as 20 other markers of appropriate size and fluorescent label. PCR products were separated using the ABI 377/3100 DNA sequencer, allowing multiple fluorescently labeled markers to be run in a single lane. The ROX 400 size standard was run as an internal size standard. Allele sizing was calculated using the local southern algorithm available in the GENESCAN software program (Applied Biosystems). Allele calling and binning were performed using GENOTYPER software (Applied Biosystems). All genotyping was performed with the inclusion of a CEPH control individual (1347–02), for quality control purposes.
Statistical Analyses
Simulations were performed using the SLINK and GENEHUNTER programs. Power analyses were performed under the assumption of a dominant affecteds-only model with a gene frequency of 0.003 and a rate of sporadic disease in nongene carriers that ranged between 0% and 4%, increasing with age. Individuals with ocular or cutaneous melanoma were coded as “affected,” and all other individuals were coded as “unknown.” A linked (i.e., 5 cM from the trait) marker with six equally frequent alleles was simulated using the SLINK program. Two-point parametric LOD scores were calculated using the GENEHUNTER program. The results in table 2 are based on 1,000 replicates for each parameter set.
Table 2.
Power To Detect Linkage In 49 Genomewide Scan Pedigrees: Parametric Analysis (LOD)
|
LOD Score |
% with LOD Score |
||||
| Proportionof FamilieswithLinkage | Mean | Maximum | ⩾1 | ⩾2 | ⩾3 |
| 0% | −21.25 | −6.47 | .0 | .0 | .0 |
| 25% | −15.45 | 4.49 | .1 | .1 | .1 |
| 50% | −7.47 | 9.54 | 12.6 | 8.7 | 6.9 |
| 75% | 2.05 | 17.00 | 65.0 | 61.3 | 55.7 |
| 100% | 13.58 | 20.94 | 100.0 | 99.8 | 99.8 |
The quality of our data was checked in several ways. First, genotyping data were checked for Mendelian inconsistency through use of both the GAS and Pedcheck programs. Any marker violating Mendelian transmission was double checked in the laboratory. Ambiguous marker genotypes were deleted to reduce further problems in statistical analysis. Second, genotype data integrity was further checked using the CRIMAP program. The marker order and distances were estimated from the data and were compared to publicly available genetic and physical maps. Finally, biological kinship was also examined, using the RelCheck computer program. The RelCheck program used the genotypic information of autosomal markers in this genome scan to calculate the most likely relationship of a putative relative pair. Five relationships were considered: MZ twins, parent/offspring, full sibs, half sibs, and unrelated individuals. Allele frequencies were estimated from all genotyped individuals using the Gconvert program (David Duffy's QIMR Homepage).
Parametric analyses were performed using the dominant genetic model derived using data from the Australian cancer registries and age-specific penetrances estimated from pedigree data (Nancarrow et al. 1992b). This model specifies a disease allele frequency of 0.003 and eight liability classes, with penetrances of 0.005, 0.13, 0.31, 0.52, 0.62, 0.7, 0.77, and 0.8 for ages 0–15 years, 16–25 years, 26–35 years, 36–45 years, 46–55 years, 56–65 years, 66–75 years, and >75 years, respectively, for disease gene carriers; and phenocopy rates of 0, 0.001, 0.005, 0.011, 0.018, 0.026, 0.032, and 0.04 for the same age groups for noncarriers. To reduce the impact of incomplete penetrance, all unaffected individuals were coded as “unknown” in our analyses. Two-point parametric analyses were performed using the FASTLINK program (Lathrop et al. 1984).
Nonparametric analyses were also performed using the pseudomarker approach, which approximates a model-free affected relative pair analysis but maintains an important property of LOD score analysis: pedigree correlations between all relatives are considered jointly, and the pedigree is not broken into a set of all possible relative pairs (Göring and Terwilliger 2000). We also assume that the mode of inheritance is dominant in this nonparametric analysis.
The predivided samples test (Morton 1956) was used to test for heterogeneity between the nine families with early onset and the remaining families in the phase 1 data. Ordered sequential analysis was performed on the basis of average age at diagnosis in each family. In this analysis, using the 46 markers spanning the most promising region, we determined the maximum two-point nonparametric LOD score in all families and then removed the family with the highest mean age at diagnosis and repeated the analyses. Multipoint linkage analyses were performed using both parametric and nonparametric methods, implemented by the computer program GENEHUNTER-PLUS (Kruglyak et al. 1996; Kong and Cox 1997). For the parametric analyses, the same autosomal dominant model used in the two-point analyses (indicated above) was assumed. Linkage in the presence of heterogeneity was assessed by use of Smith’s admixture test for heterogeneity.
Results
Simulations were performed to evaluate the empirical type 1 error and the estimated power to detect linkage under the assumption that certain percentages of families (0%, 25%, 50%, 75%, or 100%) show linkage to a given locus (table 2) (Ott 1989; Weeks et al. 1990). When the case in which none of the families was linked is considered, the family set is robust against type 1 errors: none of the simulated parametric results (out of 1,000 replicates) showed a LOD score >1.
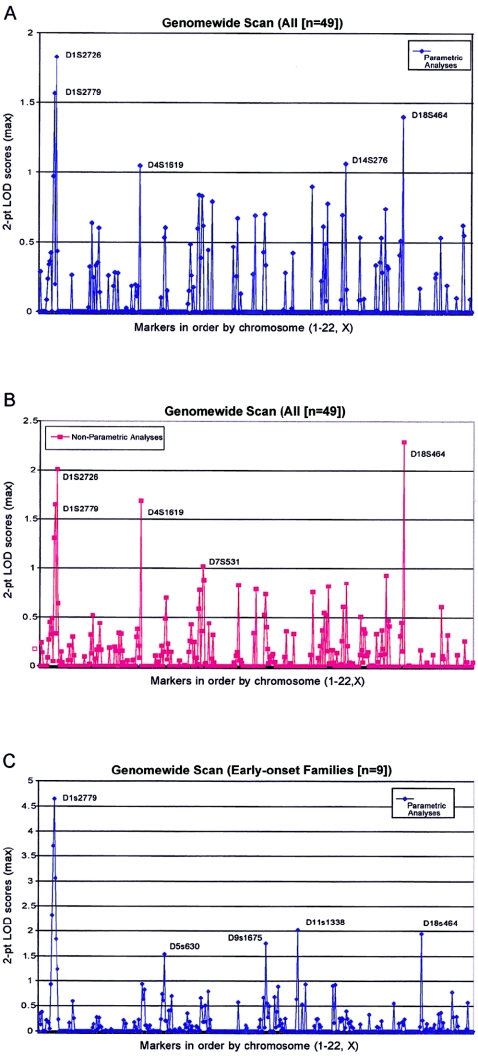
For the parametric analyses of genotype data, we used an affecteds-only model that assumed a dominant mode of transmission for the trait and a gene frequency of 0.003. The use of an affecteds-only model reduces the effect of incomplete penetrance by coding all unaffected individuals as “unknown.” The model also allowed for an age-dependent phenocopy rate, appropriate for Australian populations, that ranged from 0% to 4% (Nancarrow et al. 1992b). Plots of the two-point parametric and nonparametric LOD scores for the genomewide scan are shown in figure 1A and 1B. The highest LOD score observed was 1.82 (recombination fraction [θ] 0.2) at marker D1S2726, which maps to the short arm of chromosome 1 (1p22). The nonparametric LOD score at D1S2726 was 2.06. On the basis of our simulations, the probability of obtaining a parametric LOD score this high in the absence of linkage was <.001 (table 2). In addition to chromosome 1p, several other regions showed LOD scores >1.0 in either parametric or nonparametric analyses, including regions on chromosomes 4, 7, 14, and 18 (fig. 1A and 1B). We have focused our follow-up efforts on chromosome 1, however, since it was the only chromosome for which three closely located markers, D1S207, D1S2779, and D1S2726, all gave LOD scores >1.0.
Figure 1.
A, Parametric two-point LOD scores for the genomewide scan. Affected and unaffected individuals in 49 pedigrees with melanoma were genotyped at 414 loci throughout the genome. Maximum two-point LOD scores were calculated and the results plotted as a function of order along the chromosome. B, Nonparametric two-point LOD scores for the genomewide scan for the same 49 pedigrees with melanoma. C, Maximum two-point parametric LOD scores for the genomewide scan for nine families with the earliest age at onset.
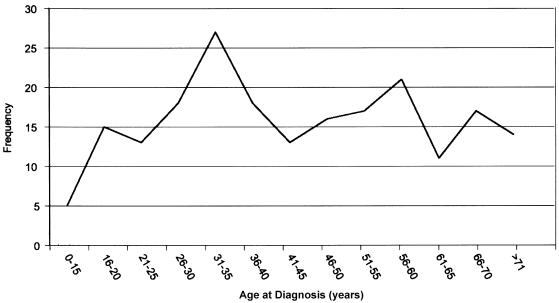
To minimize genetic heterogeneity, we performed subset analyses based on the mean age at diagnosis within each family. To empirically define a cutoff point, we plotted the age at diagnosis from all 212 affected individuals from all 49 families (fig. 2). The distribution of age at diagnosis suggested a mixture of two or three underlying normal distributions with a clear peak at age 35 years. When we assigned this age as a cutoff point, nine families with a mean age at diagnosis ⩽35 years were stratified as “early onset” for the subset analyses. Parametric linkage analysis of our genomewide scan data in these nine families with early-onset disease revealed six consecutive markers on chromosome 1 (1p22) with two-point LOD scores >1.0; the highest LOD score was 4.65 at marker D1S2779 (θ=0) (fig. 1C). The two-point LOD score at D1S2779 (θ=0) in the remaining 40 families without early onset was −13.95. Furthermore, in the same group of 40 families without early onset, the maximum two-point LOD score at this marker was 0.06 (θ=0.4) (table 3). The predivided samples test for heterogeneity between these two classes of families was highly significant (χ2=12.88; 1 df; P<.001). Nonparametric analyses also produced similar results; seven consecutive markers had nonparametric LOD scores >1.0, with the highest nonparametric LOD score, 4.19, at D1S2779.
Figure 2.
Age at onset for 212 affected individuals from the 49 families with melanoma included in genomewide scan for linkage in phase 1.
Table 3.
Chromosome 1 Follow-Up Linkage Results for Susceptibility to Melanoma and Markers on Chromosome 1 in 82 Families
|
Maximum Two-PointParametric LOD Score (θ)d |
Maximum Two-PointNonparametric LOD Scoree |
|||||||||
| Markera | MarshfieldSex-AveragedPositionb(cM) | Distancefrom MarkerAbove(cM) | UCSC 11/02Positionc(bp) | Distance from Marker Above(Mb) | Early Onset(⩽35 years) | Late Onset(>35 years) | All | Early Onset(⩽35 years) | Late Onset(>35 years) | All |
| #D1S468 | 4.22 | … | 3542337 | … | .00 (.50) | .00 (.50) | .00 (.50) | … | … | … |
| #D1S214 | 14.04 | 9.82 | 6770993 | 32.29 | .34 (.20) | .03 (.30) | .29 (.30) | … | … | … |
| #D1S450 | 20.61 | 6.57 | … | … | .13 (.30) | .00 (.50) | .00 (.50) | … | … | … |
| #D1S2667 | 24.68 | 4.07 | 11330682 | … | .38 (.20) | .00 (.50) | .00 (.50) | … | … | … |
| #D1S2697 | 37.05 | 12.37 | 15533516 | 42.03 | .00 (.50) | .00 (.50) | .00 (.50) | … | … | … |
| #D1S199 | 45.33 | 8.28 | 18982096 | 34.49 | .00 (.50) | .00 (.50) | .00 (.50) | … | … | … |
| #D1S234 | 55.1 | 9.77 | 24220507 | 52.38 | .00 (.50) | .00 (.50) | .00 (.50) | … | … | … |
| #D1S255 | 65.47 | 10.37 | 36704662 | 124.84 | .21 (.10) | .01 (.30) | .08 (.30) | … | … | … |
| #D1S2797 | 75.66 | 10.19 | 45890570 | 91.86 | .20 (.30) | .04 (.30) | .24 (.30) | … | … | … |
| #D1S2748 | 75.66 | .00 | … | … | .17 (.30) | .17 (.30) | .34 (.30) | … | … | … |
| #D1S2890 | 85.68 | 10.02 | 56788356 | … | .05 (.30) | .31 (.30) | .36 (.30) | … | … | … |
| #D1S230 | 95.31 | 9.63 | 61525270 | 47.37 | .93 (.10) | .00 (.50) | .42 (.30) | … | … | … |
| #D1S2841 | 106.45 | 11.14 | … | … | 2.32 (.05) | .00 (.50) | .00 (.50) | … | … | … |
| *D1S430 | 109.04 | 2.59 | 80176893 | … | 1.66 (.05) | .09 (.40) | .58 (.30) | 2.40 | .09 | .94 |
| *D1S2862 | 113.69 | 4.65 | 81153641 | .98 | 2.06 (.05) | .03 (.40) | .65 (.30) | 2.27 | .00 | .93 |
| *#D1S207 | 113.69 | .00 | 81455812 | .3 | 3.99 (.00) | .10 (.35) | 1.27 (.25) | 5.46 | .13 | 2.25 |
| D1S1159 | 113.69 | .00 | 81853072 | .4 | 1.78 (.05) | .20 (.30) | 1.11 (.25) | 3.45 | .31 | 2.14 |
| D1S551 | 113.69 | .00 | 81810783 | −.04 | 2.05 (.05) | .00 (.50) | .91 (.20) | 3.14 | .00 | 1.57 |
| D1S226 | 113.69 | .00 | 82049611 | .24 | 2.48 (.05) | .02 (.40) | 1.01 (.25) | 3.46 | .00 | 1.22 |
| *D1S208 | 113.83 | .14 | 82120006 | .07 | 2.73 (.05) | .00 (.50) | .51 (.30) | 3.77 | .00 | .48 |
| D1S2882 | 113.69 | .00 | 81997165 | −.12 | 1.16 (.10) | .11 (.35) | .65 (.25) | 1.80 | .23 | 1.21 |
| *D1S2807 | 114.24 | .41 | 82225024 | .23 | 1.76 (.05) | .29 (.30) | 1.10 (.20) | 1.95 | .24 | 1.22 |
| D1S488 | 114.24 | .00 | 82242659 | .02 | 1.57 (.05) | .00 (.50) | .52 (.25) | 1.54 | .01 | .60 |
| *D1S2889 | 116.72 | 2.48 | 83973823 | 1.73 | .97 (.10) | .00 (.50) | .23 (.35) | 1.41 | .00 | .40 |
| *AFMB331xe9 | NA | … | 84740658 | .77 | .11 (.25) | .09 (.30) | .18 (.30) | 1.24 | .28 | 1.10 |
| *D1S2766 | 118.14 | 1.42 | 85214144 | .47 | 1.91 (.05) | .87 (.20) | 2.23 (.20) | 2.04 | .21 | 1.36 |
| *D1S1618 | NA | … | 85890455 | .68 | 2.46 (.05) | .00 (.50) | 1.22 (.20) | 3.25 | .00 | 1.18 |
| *AL109625.M1 | NA | … | 86992070 | 1.1 | 2.64 (.00) | .10 (.35) | 1.39 (.20) | 4.01 | .23 | 2.26 |
| *AFM263xf9 | NA | … | 87645479 | .65 | .27 (.05) | .31 (.20) | .49 (.20) | .14 | .92 | 1.06 |
| *GATA7b10 | NA | … | 87948697 | .3 | 3.00 (.00) | .00 (.50) | .80 (.25) | 4.84 | .00 | 1.44 |
| D1S2627 | 120.28 | … | 88080460 | .13 | 2.25 (.05) | .16 (.30) | 1.38 (.20) | 1.98 | .00 | .94 |
| *D1S2129 | NA | … | 89285171 | 1.2 | 2.11 (.05) | .00 (.50) | .12 (.35) | 1.82 | .00 | .46 |
| *AFMA205xd5 | NA | … | 89347025 | .06 | 3.07 (.00) | .66 (.20) | 2.29 (.15) | 3.65 | .97 | 2.97 |
| *D1S1572 | NA | … | 89675385 | .33 | 1.82 (.00) | .01 (.35) | .61 (.20) | 4.04 | .00 | 1.46 |
| *D1S435 | 125.51 | 5.23 | 90734206 | 1.06 | 2.46 (.00) | .60 (.25) | 1.81 (.20) | 2.04 | .20 | 1.12 |
| *D1S188 | 126.16 | .65 | 91693598 | .96 | 3.84 (.00) | .00 (.50) | 1.13 (.25) | 3.97 | .00 | 1.14 |
| D1S424 | 126.16 | .00 | 92089831 | .4 | 3.81 (.00) | .34 (.35) | 1.79 (.20) | 3.71 | .30 | 1.77 |
| *D1S2804 | 126.16 | .00 | 92091307 | .00 | 4.61 (.00) | .17 (.35) | 1.68 (.25) | 4.99 | .48 | 2.85 |
| *D1S2776 | 126.16 | .00 | 92416061 | .32 | 4.95 (.00) | .19 (.30) | 2.58 (.15) | 5.37 | .00 | 1.72 |
| D1S2868 | 126.16 | .00 | 92541586 | .13 | 3.19 (.00) | .13 (.40) | .73 (.30) | 3.57 | .30 | 1.51 |
| D1S2849 | 126.16 | .00 | 92725804 | .18 | 3.39 (.00) | .00 (.50) | .86 (.25) | 2.91 | .00 | .91 |
| *#D1S2779 | 126.16 | .00 | 92906395 | .18 | 4.65 (.00) | .06 (.40) | 1.67 (.20) | 6.43 | .05 | 1.98 |
| *D1S2813 | 129.37 | 3.21 | 94469401 | 1.56 | 1.19 (.05) | .19 (.35) | .55 (.30) | 1.52 | .27 | .98 |
| D1S2664 | 129.37 | .00 | 95152266 | .68 | 2.78 (.05) | .00 (.50) | .67 (.30) | 4.32 | .13 | 2.16 |
| *D1S2819 | 129.37 | .00 | 94731975 | −.42 | 2.18 (.05) | .00 (.50) | .37 (.30) | 2.85 | .00 | .57 |
| *D1S2719 | 129.37 | .00 | 95918953 | 1.19 | 1.97 (.05) | .00 (.50) | .18 (.30) | 2.80 | .00 | .53 |
| *D1S2793 | 129.37 | .00 | 96194529 | .28 | 2.90 (.00) | .13 (.40) | 1.04 (.25) | 2.47 | .16 | 1.24 |
| *D1S420 | 128.73 | .00 | 97573027 | 1.38 | 1.76 (.05) | .00 (.50) | .27 (.35) | 1.61 | .10 | .74 |
| D1S2753 | 129.37 | .64 | 97594397 | .02 | 3.48 (.00) | .00 (.50) | .16 (.35) | 3.29 | .00 | .66 |
| *D1S2739 | 130.73 | 1.36 | 97907381 | .31 | 2.33 (.00) | .00 (.50) | .37 (.30) | 2.05 | .19 | 1.03 |
| *D1S2808 | 131.87 | 1.14 | 98382874 | .48 | 2.04 (.00) | .38 (.30) | 1.30 (.20) | 2.21 | .43 | 1.32 |
| D1S2671 | 134.2 | 2.33 | 100447508 | 2.06 | 2.87 (.00) | .14 (.40) | .95 (.25) | 3.07 | .34 | 1.73 |
| *D1S223 | 134.2 | .00 | 100778480 | .33 | 2.05 (.05) | .00 (.50) | .86 (.20) | 2.33 | .00 | .59 |
| *#D1S206 | 134.2 | .00 | 100866397 | .09 | 3.71 (.00) | .00 (.50) | .31 (.30) | 3.44 | .00 | .80 |
| *D1S2896 | 134.2 | .00 | 101153691 | .29 | 3.19 (.05) | .01 (.40) | .83 (.25) | 3.77 | .00 | 1.20 |
| *D1S495 | 136.88 | 2.68 | 101742418 | .59 | 3.42 (.05) | .00 (.50) | .69 (.30) | 3.56 | .11 | 1.58 |
| *D1S2626 | 136.34 | .00 | 102176693 | .43 | 3.08 (.00) | .05 (.40) | .77 (.25) | 1.78 | .00 | .43 |
| *D1S2888 | 136.88 | .54 | 104007624 | 1.83 | 1.75 (.05) | .00 (.50) | .28 (.35) | 1.80 | .09 | .98 |
| *D1S429 | 136.88 | .00 | 104610867 | .6 | 2.24 (.05) | .03 (.40) | .87 (.25) | 1.90 | .00 | .61 |
| #D1S2726 | 144.38 | 7.50 | 110050830 | 5.44 | 1.84 (.05) | .54 (.30) | 1.82 (.20) | … | … | … |
| #D1S252 | 150.27 | 5.89 | 116395341 | 6.34 | 1.23 (.10) | .00 (.50) | .43 (.30) | … | … | … |
| #D1S498 | 155.89 | 5.62 | 147048835 | 30.65 | .23 (.20) | .00 (.50) | .00 (.50) | … | … | … |
| #D1S484 | 169.68 | 13.79 | 156499829 | 9.45 | .00 (.50) | .15 (.30) | .00 (.50) | … | … | … |
| #D1S2705 | 170.84 | 1.16 | 156590216 | .09 | .00 (.50) | .31 (.30) | .00 (.50) | … | … | … |
| #D1S2878 | 177.86 | 7.02 | 161054323 | 4.46 | .00 (.50) | .24 (.30) | .00 (.50) | … | … | … |
| #D1S196 | 181.49 | 3.63 | 163254967 | 2.20 | .00 (.50) | .00 (.50) | .00 (.50) | … | … | … |
| #D1S218 | 191.52 | 10.03 | 169931791 | 6.68 | .00 (.50) | .00 (.50) | .00 (.50) | … | … | … |
| #D1S238 | 202.73 | 11.21 | 183574128 | 13.64 | .00 (.50) | .00 (.50) | .00 (.50) | … | … | … |
| #D1S413 | 212.44 | 9.71 | 194078844 | 10.50 | .00 (.50) | .02 (.30) | .00 (.50) | … | … | … |
| #D1S249 | 220.65 | 8.21 | 201176606 | 7.10 | .00 (.50) | .00 (.50) | .00 (.50) | … | … | … |
| #D1S425 | 231.11 | 10.46 | 207926397 | 6.75 | .00 (.50) | .00 (.50) | .00 (.50) | … | … | … |
| #D1S213 | 242.34 | 11.23 | … | … | .16 (.30) | .00 (.50) | .00 (.50) | … | … | … |
| #D1S2800 | 252.12 | 9.78 | 230030530 | … | .00 (.50) | .00 (.50) | .00 (.50) | … | … | … |
| #D1S2785 | 266.27 | 14.15 | 236330149 | 6.30 | .00 (.50) | .00 (.50) | .00 (.50) | … | … | … |
| #D1S2842 | 273.46 | 7.19 | 238119266 | 1.79 | .60 (.20) | .00 (.50) | .26 (.30) | … | … | … |
| #D1S2836 | 285.75 | 12.29 | … | … | .26 (.30) | .00 (.50) | .00 (.50) | … | … | … |
Asterisks (*) indicate markers used in multipoint analyses; number signs (#) indicate GWS markers.
Positions are based on the Genetic Map Index, Center for Medical Genetics Web site.
Positions are based on the human genome map available from the UCSC Genome Bioinformatics Web site (11/02).
Results are for phase 1.
Results are totals for phase 1 and phase 2.
To further validate our results and narrow the region of interest, 43 additional markers covering ∼30 cM (flanking marker D1S2779) in the 1p22 region were typed (table 3). These markers provided further evidence for linkage. The overall maximum two-point parametric LOD score increased to 2.58 (θ=0.15) at marker D1S2776. In the families with early onset, there were 3 markers with two-point parametric LOD scores ⩾4 and 12 markers with LOD scores >3. The highest two-point parametric LOD score increased to 4.95 (θ=0) at D1S2776. The results from nonparametric analyses were similar: one marker with a LOD score >5.0, four markers with LOD scores >4, and 13 markers with LOD scores >3, with the highest two-point nonparametric LOD score, 5.37, at D1S2776.
In the second phase of evaluating the evidence for linkage in this region, we genotyped 63 additional families, through use of our dense panel of 46 markers from 1p22. Of these families, 33 met our original criteria for inclusion into the study—that is, they had DNA available from blood for at least three individuals affected with CMM, they were both CDKN2A- and CDK4-mutation negative, and, furthermore, they showed no common haplotype in the 9p21-p22 region among affected individuals (i.e., they showed no evidence of linkage to 9p). The remaining 30 families did not meet our original criteria for inclusion into the study, because the three affected individuals showed evidence of haplotype sharing in the 9p21-p22 region (i.e., they showed evidence of linkage to 9p). Since these families might have undetected CDKN2A mutations, their evidence for linkage was examined independently. All families came from the Melanoma Genetics Consortium and represented eight different sites in Australia, Europe, and the United States (table 1). Our analyses focused on the 82 families that met our original criteria (phase 1 [n=49] and phase 2 [n=33]) (table 1). Given that these families were ascertained from countries reporting varying melanoma incidences and penetrances (Bishop et al. 2002), our subsequent analyses were performed using nonparametric methods only.
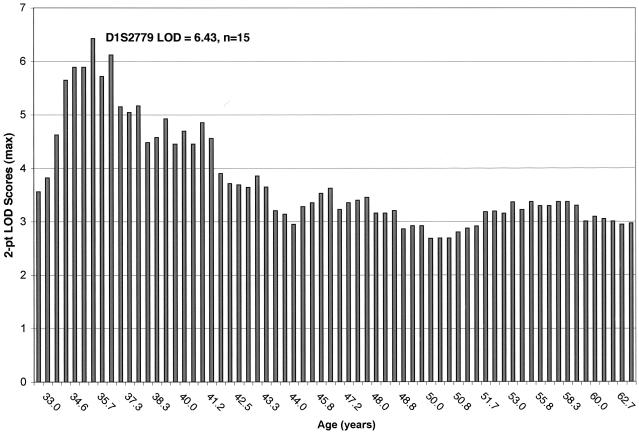
Inclusion of the additional 33 phase 2 families increased our evidence for linkage in the 1p22 region. Without stratification by average age at diagnosis, the maximum two-point nonparametric LOD score increased to 2.97 at marker AA205XD5 (table 3). Furthermore, six markers in the region showed overall two-point nonparametric LOD scores >2.0, and eight families showed individual family LOD scores >1.0. Again, we were interested in looking at the evidence for linkage in the families with early onset but were reluctant to use a single age cutoff, given the differing age-dependent risks of CMM in mutation carriers in the various countries from which our families originated. Ordered sequential analysis was therefore performed, on the basis of the average age at diagnosis in each family. In this analysis, we looked at the evidence for linkage in all families, then removed the family with the highest mean age at diagnosis and repeated the analyses. Figure 3 shows the evidence for linkage in the families with the lowest mean age at diagnosis to the highest (left to right). In these nonparametric analyses, the highest two-point LOD score, 6.43, was obtained at D1S2779 and occurred when the 15 families with earliest onset were included for analysis (fig. 3). The oldest average age at diagnosis for this empirically defined subset of families was 35.7 years, very similar to the cutoff used in the 49 phase 1 Australian kindreds (⩽35 years). When defined in this way, the proportion of families with linkage to 1p22 is 18% of the initial 49 families (n=9; all from Australia), as well as 18% of the total cohort of 82 families (n=15; 14/15 from Australia). We attempted to evaluate the empirical significance of these results in two ways. First, we evaluated the empirical significance of this subset of 15 families by resampling 15 families randomly from the set of 82 families and recalculating the total LOD score on this subset. This process was repeated 1,000,000 times. Only five replicates gave LOD scores greater than or equal to our observed value of 6.43, resulting in an empirical significance level of .000005. This empirical P value is equal to an empirical LOD score of 4.2. Second, we performed 100,000 replicates, simulating an unlinked marker in these 15 families with early onset. No replicate gave a LOD score that was equal to the observed value of 6.43. In fact, only 10 replicates out of 100,000 gave LOD scores >2, of which only 2 were >3 (maximum LOD score 3.04).
Figure 3.
Ordered sequential analysis, based on the mean age at diagnosis in each family. Overall maximum two-point nonparametric LOD scores for chromosome 1 markers for families with the lowest mean age at onset to the highest (left to right).
Results from parametric and nonparametric multipoint analyses were consistent with the two-point results. Given inconsistencies in the order of markers between the draft human genome sequence and published genetic maps of this region, we performed multipoint analyses that included only the 33 (of 46) markers in this region that could be unambiguously ordered. (A YAC-based physical map has been developed that spans the entire candidate region with one small gap, through use of Whitehead Institute’s YAC database; STS content mapping of these YACs with primers for the genetic markers has provided us with a precise order of 33 markers.) In the complete family set (n=82), the highest parametric multipoint LOD score allowing for heterogeneity was 2.36, between loci D1S430 and D1S2862. The estimated proportion of families with linkage was 17%, which is consistent with our estimate using the ordered sequential analysis (18%). HLOD scores were >1.0 across an ∼16-cM region. In the 15 families with early onset defined by the ordered sequential analysis, the maximum multipoint LOD score allowing for heterogeneity was 4.83, between markers D1S435 and D1S188. In this analysis, the estimated proportion of families linked was 65%.
To define the region of interest more precisely, we looked for potential recombinants in the families with linkage—that is, the 15 families with early onset contributing to the ordered sequential analysis peak. Several informative recombination events were noted, and they delimited the region to D1S430 on the telomeric side and D1S2664 on the centromeric side, an interval of ∼20 cM. There are several good candidate genes within this region—namely, the TGFBR3 and CDC7 genes. We are currently working to determine whether either gene is involved in melanoma susceptibility.
Discussion
We report the results from the first complete genomewide scan for linkage for melanoma susceptibility. Our goal was to localize novel genes involved in melanoma susceptibility. We restricted inclusion into our study to highly loaded melanoma pedigrees, which were mutation negative for both CDKN2A and CDK4. Furthermore, we excluded any family with evidence of haplotype sharing in the 9p21-22 region where CDKN2A is located. This strategy may prove conservative, but it helped to minimize the possibility of including families with undetected CDKN2A involvement. Genetic heterogeneity is a significant factor contributing to the difficulty of mapping genes involved in complex traits; therefore, our strict inclusion criteria, as well as subset analyses, may have improved the power of our study.
In the first phase of our study, we performed both parametric and nonparametric analyses. Both methods provided evidence for linkage between a melanoma susceptibility locus and markers on chromosome 1p. The highest parametric LOD score observed was 1.82 (θ=0.2), at marker D1S2726, which maps to the short arm of chromosome 1 (1p22). The highest nonparametric LOD score at D1S2726 was 2.06. On the basis of our simulations, the probability of obtaining a parametric LOD score this high by chance alone was small (<.001).
In our parametric analyses, we employed two strategies, which were also aimed at increasing the power of our study. First, using an affecteds-only model reduces the effect of incomplete penetrance by coding all unaffected individuals as “unknown.” Our model still allowed for phenocopies in non–gene carriers. The allowance for sporadic cases of melanoma in the model is critical, since sun exposure is known to be a significant environmental risk factor for CMM. Ideally, we would have reliable sun-exposure data for each individual in the study, thus allowing its incorporation into the model as a covariate. However, since these data are currently unavailable, we attempted to allow for the effect of this unmeasured environmental risk factor by including nongenetic cases in our model. Second, the utilization of liability classes can maximize power in the investigation of complex traits, which often exhibit reduced penetrance and phenocopies. Specifically, in our analyses, liability classes allowed us to incorporate the observed increase in sporadic rate with increasing age into our model. Thus, with the incorporation of liability classes defined by age, we maximized linkage information from younger affected individuals who are more likely to be gene carriers.
Since parametric analyses are model dependent and it is well known that misspecification of the disease model (especially misspecification of a trait’s degree of dominance) can lead to a decrease in power as well as a biased estimate of θ (Clerget-Darpoux et al. 1986), we also used nonparametric analyses to further examine linkage data in this region. Because the families in the present study were identified and collected from populations that report varying melanoma incidences and penetrances (Bishop et al. 2002), the second phase of analyses was performed using nonparametric methods only. In contrast to parametric methods, in which information regarding the genetic model of the trait is essential, the nonparametric methods do not require that details regarding the underlying genetic model of the trait be specified. Without stratification by average age at diagnosis, the maximum two-point nonparametric LOD score was 2.97 at marker AA205XD5. It should be noted that this region on chromosome 1 (1p22) is genetically unlinked (>50 cM away) to the 1p36 region where linkage to CMM has previously been reported (MIM 155600) (Bale et al. 1989; Goldstein et al. 1993), though not confirmed, in other cohorts (van Haeringen et al. 1989; Cannon-Albright et al. 1990; Kefford et al. 1991; Nancarrow et al. 1992a).
The previous identification of two genes, CDKN2A and CDK4, that independently confer susceptibility to CMM suggests that genetic heterogeneity is likely to be a complicating variable in the gene mapping of CMM risk alleles (despite the analysis method). We therefore explored subset analysis, using age at diagnosis as a criterion. This strategy has been critical to the successful mapping of other cancer genes (Hall et al. 1990; Smith et al. 1996). Furthermore, examining the evidence of linkage in the families with early onset is consistent with epidemiological data, which suggests that, like other hereditary cancers, familial melanomas develop at an earlier age than nonfamilial melanomas. By using an ordered sequential analysis, we avoided using a single age at diagnosis to define families with “early onset.” Figure 3 clearly shows that the evidence for linkage to this 1p locus is strongest in the families with earlier onset. The highest nonparametric LOD score, 6.43, was obtained at D1S2779 and occurred when the 15 families with earliest onset were included. On the basis of extensive simulations performed to evaluate the empirical significance of these results, it is highly unlikely that these results would have occurred by chance. Future analyses will include stratification based on other phenotypic criteria, such as segregation of both ocular and cutaneous melanoma.
Although the significant evidence of linkage to 1p obtained in this study was identified in families showing no evidence of linkage to 9p, we were interested in looking at the evidence for linkage in the 30 families with linkage to 9p. Since these families might have undetected CDKN2A mutations, their evidence for linkage was examined independently. As expected, examining the family-by-family two-point LOD scores suggests that this 1p locus is not significant in families with evidence of linkage to 9p. Of the 46 1p22 markers typed, 45 had negative LOD scores out to θ values of .4 (data not shown).
Our current candidate interval spans a region of ∼15 Mb across chromosomal bands 1p31.1 to 1p21.3 and contains 60 known genes, several partial transcripts, and several predicted transcripts. Identification of the susceptibility gene by the positional candidate cloning approach will most likely require further narrowing of the candidate region. Availability of the draft human genome sequence will provide a major resource to identify new markers for this purpose. However, identification of new kindreds with early age at diagnosis for melanoma and subsequent genotyping for markers from the candidate region may be more helpful in pinpointing the precise localization of the susceptibility gene within the candidate interval. To this end, phase 3 of our study is focused on identification of new families with melanoma that again have had CDKN2A and CDK4 involvement excluded by both mutation detection and linkage analysis.
Within the 15 families with early onset contributing to the ordered sequential analysis peak, haplotypes were constructed using the 46 markers typed on 1p. Segregation of the haplotypes was determined and recombinants were considered useful only in those families in which all affected individuals carried the same haplotype for at least part of the region typed on 1p. Recombinants were detected in seven individuals. Pooling this information across families allowed us to narrow our region of interest to a 20-cM region between markers D1S430 and D1S2664. However, the potential complexity of the underlying genetic model of melanoma should generate caution. Specifically, an affected individual does not necessarily share the susceptibility allele and might instead represent a sporadic case of melanoma. However, in our families, the recombinant individuals ranged in age from 24 to 36 years and are thus likely to be gene carriers.
Given that both CDKN2A and CDK4 are known to function in cell-cycle regulation, two genes within our candidate region—transforming growth factor β receptor 3 (TGFBR3) and cell division cycle 7–related kinase (CDC7)—could be considered as candidates for contributing to melanoma susceptibility. Both genes map close to the markers with the peak LOD scores (D1S2776 and D1S2779) and play important biological roles in cell differentiation and cell cycle progression. TGFBR3 plays an essential role in the TGF-β signaling pathway (Brown et al. 1999), which, if disrupted, can lead to rapid cell growth (Derynck et al. 2001). CDC7 protein kinase is involved in regulation of DNA replication (Sato et al. 1997) and is, therefore, essential in maintaining the cell cycle.
In summary, we have localized a novel CMM susceptibility gene to 1p22, through use of a cohort of 82 familial melanoma kindreds in which CDKN2A and CDK4 involvement has been excluded. Evidence for linkage is strongest in the ∼20% of families with the earliest age at diagnosis. Ordered sequential nonparametric analyses indicated that the highest two-point LOD score was 6.43, at marker D1S2779; results from multipoint analyses were consistent with the two-point results. These data provide significant evidence of a novel CMM susceptibility gene located within chromosome band 1p22. By analogy to other hereditary cancers, the identification of genes that confer an inherited genetic predisposition to CMM may provide insights into the etiology of all melanomas. Furthermore, characterization of the molecular basis of these genetic effects will enhance melanoma risk assessment and may ultimately lead to interventions that can reduce melanoma-associated morbidity and mortality.
Acknowledgments
We are very grateful to all the family members who willingly participated in this study. The authors wish to acknowledge the contributions to this work that were made by Mary C. Fraser, M.S.N.; Laura Fontaine, B.S.N.; and Deborah Zametkin, M.S.N. We are grateful to the Sydney Melanoma Unit—in particular, to Bill McCarthy and John Thompson—and to John Kelly (Victorian Melanoma Service) for referring families, and we acknowledge the contributions of Barbara Peters, Judy Salmon, and Helen Shaw to their characterization. The work in Sydney was supported by grants from the Australian National Health and Medical Research Council, the Cancer Council NSW, the Melanoma and Skin Cancer Research Institute, and the University of Sydney. Work in Queensland was supported by National Institutes of Health grant CA88363 and by the National Health and Medical Research Council of Australia. The Department of Oncology-Pathology, Radiumhemmet, Karolinska Hospital was supported by grants from The Cancer Society of Stockholm and The King Gustav V Jubilee Fund, Stockholm. We would also like to thank Dr. Izabela Makalowska and Michal Galdzicki for their contributions in the analysis of the candidate interval through use of bioinformatics tools. The following groups from the Melanoma Genetics Consortium participated in this research: Queensland Cancer Fund Research Unit, Joint Experimental Oncology Programme of the Queensland University and Queensland Institute of Medical Research, Brisbane, Australia: Nicholas Hayward, Derek Nancarrow, Jane Palmer, Nicholas Martin, Adele Green, Joanne Aitken, David Whiteman, Marilyn Walters, and Megan Campbell; Westmead Institute for Cancer Research, University of Sydney at Westmead Millennium Institute, Sydney, Australia: Richard F. Kefford, Graham J. Mann, Helen Schmid, Elizabeth A. Holland; Department of Dermatology and Center for Human and Clinical Genetics, Leiden University Medical Center, Leiden, The Netherlands: Wilma Bergman, Rune Frants, Nelleke Gruis, Pieter van der Velden, and Femke de Snoo; Genetic Epidemiology Division, Cancer Research UK Clinical Centre, Leeds, United Kingdom: Jenny Barrett, Julia Newton Bishop, Timothy Bishop, and Mark Harland; Genetic Epidemiology Branch, Division of Cancer Epidemiology and Genetics, National Cancer Institute, National Institutes of Health, United States: Alisa M. Goldstein and Margaret A. Tucker; Institut Gustave Rousy, Villejuif, France: Marie-Françoise Avril, Brigitte Bressac de Paillerets, and Agnès Chompret; Department of Oncology-Pathology, Radiumhemmet, Karolinska Hospital, Stockholm, Sweden: Johan Hansson, Anton Platz, and Jamileh Hashemi; and Lund Cancer Center, Department of Oncology, University Hospital, Lund, Sweden: Christian Ingvar, Åke Borg, Johan Westerdahl, Anna Måsbäck, and Håkan Olsson.
Electronic-Database Information
The URLs for data presented herein are as follows:
- David Duffy's QIMR Homepage, http://www2.qimr.edu.au/davidD (for the Gconvert program)
- Genetic Map Index, Center for Medical Genetics, http://research.marshfieldclinic.org/genetics/Map_Markers/maps/IndexMapFrames.html
- Online Mendelian Inheritance in Man (OMIM), http://www.ncbi.nlm.nih.gov/Omim/ (for CDKN2A, CDK4, MC1R, and the CMM locus on 1p36)
- UCSC Genome Bioinformatics, http://genome.ucsc.edu/
References
- Bale SJ, Dracopoli NC, Tucker MA, Clark WH Jr, Fraser MC, Stanger BZ, Green P, Donis-Keller H, Housman DE, Greene MH (1989) Mapping the gene for hereditary cutaneous malignant melanoma-dysplastic nevus to chromosome 1p. N Engl J Med 320:1367–1372 [DOI] [PubMed] [Google Scholar]
- Bergman W, Gruis NA, Frants RR (1992) The Dutch FAMMM family material: clinical and genetic data. Cytogenet Cell Genet 59:161–164 [DOI] [PubMed] [Google Scholar]
- Bishop DT, Demenais F, Goldstein AM, Bergman W, Bishop JN, Bressac-de Paillerets B, Chompret A, Ghiorzo P, Gruis N, Hansson J, Harland M, Hayward N, Holland EA, Mann GJ, Mantelli M, Nancarrow D, Platz A, Tucker MA (2002) Geographical variation in the penetrance of CDKN2A mutations for melanoma. J Natl Cancer Inst 94:894–903 [DOI] [PubMed] [Google Scholar]
- Bishop JA, Wachsmuth RC, Harland M, Bataille V, Pinney E, Mac KP, Baglietto L, Cuzick J, Bishop DT (2000) Genotype/phenotype and penetrance studies in melanoma families with germline CDKN2A mutations. J Invest Dermatol 114:28–33 [DOI] [PubMed] [Google Scholar]
- Borg A, Sandberg T, Nilsson K, Johannsson O, Klinker M, Masback A, Westerdahl J, Olsson H, Ingvar C (2000) High frequency of multiple melanomas and breast and pancreas carcinomas in CDKN2A mutation-positive melanoma families. J Natl Cancer Inst 92:1260–1266 [DOI] [PubMed] [Google Scholar]
- Box NF, Duffy DL, Chen W, Stark M, Martin NG, Sturm RA, Hayward NK (2001) MC1R genotype modifies risk of melanoma in families segregating CDKN2A mutations. Am J Hum Genet 69:765–773 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown CB, Boyer AS, Runyan RB, Barnett JV (1999) Requirement of type III TGF-β receptor for endocardial cell transformation in the heart. Science 283:2080–2082 [DOI] [PubMed] [Google Scholar]
- Cannon-Albright LA, Goldgar DE, Wright EC, Turco A, Jost M, Meyer LJ, Piepkorn M, Zone JJ, Skolnick MH (1990) Evidence against the reported linkage of the cutaneous melanoma-dysplastic nevus syndrome locus to chromosome 1p36. Am J Hum Genet 46:912–918 [PMC free article] [PubMed] [Google Scholar]
- Clerget-Darpoux F, Bonaiti-Pellie C, Hochez J (1986) Effects of misspecifying genetic parameters in lod score analysis. Biometrics 42:393–399 [PubMed] [Google Scholar]
- Derynck R, Akhurst RJ, Balmain A (2001) TGF-β signaling in tumor suppression and cancer progression. Nat Genet 29:117–129 [DOI] [PubMed] [Google Scholar]
- Goldstein AM, Dracopoli NC, Ho EC, Fraser MC, Kearns KS, Bale SJ, McBride OW, Clark WH Jr, Tucker MA (1993) Further evidence for a locus for cutaneous malignant melanoma- dysplastic nevus (CMM/DN) on chromosome 1p, and evidence for genetic heterogeneity. Am J Hum Genet 52:537–550 [PMC free article] [PubMed] [Google Scholar]
- Goldstein AM, Fraser MC, Struewing JP, Hussussian CJ, Ranade K, Zametkin DP, Fontaine LS, Organic SM, Dracopoli NC, Clark WH Jr, Tucker MA (1995) Increased risk of pancreatic cancer in melanoma-prone kindreds with p16INK4 mutations. N Engl J Med 333:970–974 [DOI] [PubMed] [Google Scholar]
- Goldstein AM, Struewing JP, Chidambaram A, Fraser MC, Tucker MA (2000) Genotype-phenotype relationships in U.S. melanoma-prone families with CDKN2A and CDK4 mutations. J Natl Cancer Inst 92:1006–1010 [DOI] [PubMed] [Google Scholar]
- Göring HH, Terwilliger JD (2000) Linkage analysis in the presence of errors IV: joint pseudomarker analysis of linkage and/or linkage disequilibrium on a mixture of pedigrees and singletons when the mode of inheritance cannot be accurately specified. Am J Hum Genet 66:1310–1327 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gruis NA, van der Velden PA, Sandkuijl LA, Prins DE, Weaver-Feldhaus J, Kamb A, Bergman W, Frants RR (1995) Homozygotes for CDKN2 (p16) germline mutation in Dutch familial melanoma kindreds. Nat Genet 10:351–353 [DOI] [PubMed] [Google Scholar]
- Hall JM, Lee MK, Newman B, Morrow JE, Anderson LA, Huey B, King MC (1990) Linkage of early-onset familial breast cancer to chromosome 17q21. Science 250:1684–1689 [DOI] [PubMed] [Google Scholar]
- Harland M, Holland EA, Ghiorzo P, Mantelli M, Bianchi-Scarra G, Goldstein AM, Tucker MA, Ponder BA, Mann GJ, Bishop DT, Newton Bishop J (2000) Mutation screening of the CDKN2A promoter in melanoma families. Genes Chromosomes Cancer 28:45–57 [PubMed] [Google Scholar]
- Harland M, Mistry S, Bishop DT, Bishop JA (2001) A deep intronic mutation in CDKN2A is associated with disease in a subset of melanoma pedigrees. Hum Mol Genet 10:2679–2686 [DOI] [PubMed] [Google Scholar]
- Hashemi J, Linder S, Platz A, Hansson J (1999) Melanoma development in relation to non-functional p16/INK4A protein and dysplastic naevus syndrome in Swedish melanoma kindreds. Melanoma Res 9:21–30 [DOI] [PubMed] [Google Scholar]
- Hussussian CJ, Struewing JP, Goldstein AM, Higgins PA, Ally DS, Sheahan MD, Clark WH Jr, Tucker MA, Dracopoli NC (1994) Germline p16 mutations in familial melanoma. Nat Genet 8:15–21 [DOI] [PubMed] [Google Scholar]
- Kefford RF, Newton Bishop JA, Bergman W, Tucker MA (1999) Counseling and DNA testing for individuals perceived to be genetically predisposed to melanoma: a consensus statement of the Melanoma Genetics Consortium. J Clin Oncol 17:3245–3251 [DOI] [PubMed] [Google Scholar]
- Kefford RF, Salmon J, Shaw HM, Donald JA, McCarthy WH (1991) Hereditary melanoma in Australia: variable association with dysplastic nevi and absence of genetic linkage to chromosome 1p. Cancer Genet Cytogenet 51:45–55 [DOI] [PubMed] [Google Scholar]
- Kong A, Cox NJ (1997) Allele-sharing models: LOD scores and accurate linkage tests. Am J Hum Genet 61:1179–1188 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kruglyak L, Daly MJ, Reeve-Daly MP, Lander ES (1996) Parametric and nonparametric linkage analysis: a unified multipoint approach. Am J Hum Genet 58:1347–1363 [PMC free article] [PubMed] [Google Scholar]
- Lathrop GM, Lalouel JM, Julier C, Ott J (1984) Strategies for multilocus linkage analysis in humans. Proc Natl Acad Sci USA 81:3443–3446 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu L, Dilworth D, Gao L, Monzon J, Summers A, Lassam N, Hogg D (1999) Mutation of the CDKN2A 5′ UTR creates an aberrant initiation codon and predisposes to melanoma. Nat Genet 21:128–132 [DOI] [PubMed] [Google Scholar]
- Morton NE (1956) The detection and estimation of linkage between the genes for elliptocytosis and the Rh blood type. Am J Hum Genet 8:80–96 [PMC free article] [PubMed] [Google Scholar]
- Nancarrow DJ, Palmer JM, Walters MK, Kerr BM, Hafner GJ, Garske L, McLeod GR, Hayward NK (1992a) Exclusion of the familial melanoma locus (MLM) from the PND/D1S47 and MYCL1 regions of chromosome arm 1p in 7 Australian pedigrees. Genomics 12:18–25 [DOI] [PubMed] [Google Scholar]
- Nancarrow DJ, Walker GJ, Weber JL, Walters MK, Palmer JM, Hayward NK (1992b) Linkage mapping of melanoma (MLM) using 172 microsatellite markers. Genomics 14:939–947 [DOI] [PubMed] [Google Scholar]
- Newton Bishop JA, Harland M, Bennett DC, Bataille V, Goldstein AM, Tucker MA, Ponder BA, Cuzick J, Selby P, Bishop DT (1999) Mutation testing in melanoma families: INK4A, CDK4 and INK4D. Br J Cancer 80:295–300 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ott J (1989) Computer-simulation methods in human linkage analysis. Proc Natl Acad Sci USA 86:4175–4178 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palmer JS, Duffy DL, Box NF, Aitken JF, O'Gorman LE, Green AC, Hayward NK, Martin NG, Sturm RA (2000) Melanocortin-1 receptor polymorphisms and risk of melanoma: is the association explained solely by pigmentation phenotype? Am J Hum Genet 66:176–186 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Platz A, Hansson J, Mansson-Brahme E, Lagerlof B, Linder S, Lundqvist E, Sevigny P, Inganas M, Ringborg U (1997) Screening of germline mutations in the CDKN2A and CDKN2B genes in Swedish families with hereditary cutaneous melanoma. J Natl Cancer Inst 89:697–702 [DOI] [PubMed] [Google Scholar]
- Ries LAG, Eisner MP, Kosary CL, Hankey BF, Miller BA, Clegg L, Mariotto A, Fay MP, Feuer EJ, Edwards BK (eds) (2003) SEER Cancer Statistics Review, 1975–2000, National Cancer Institute. Bethesda, MD, http://seer.cancer.gov/csr/1975_2000/ (accessed July 3, 2003)
- Sato N, Arai K, Masai H (1997) Human and Xenopus cDNAs encoding budding yeast Cdc7-related kinases: in vitro phosphorylation of MCM subunits by a putative human homologue of Cdc7. Embo J 16:4340–4351 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith JR, Freije D, Carpten JD, Gronberg H, Xu J, Isaacs SD, Brownstein MJ, Bova GS, Guo H, Bujnovszky P, Nusskern DR, Damber JE, Bergh A, Emanuelsson M, Kallioniemi OP, Walker-Daniels J, Bailey-Wilson JE, Beaty TH, Meyers DA, Walsh PC, Collins FS, Trent JM, Isaacs WB (1996) Major susceptibility locus for prostate cancer on chromosome 1 suggested by a genome-wide search. Science 274:1371–1374 [DOI] [PubMed] [Google Scholar]
- Soufir N, Avril MF, Chompret A, Demenais F, Bombled J, Spatz A, Stoppa-Lyonnet D, the French Familial Melanoma Study Group, Bénard J, Bressac-de Paillerets B (1998) Prevalence of p16 and CDK4 germline mutations in 48 melanoma-prone families in France. Hum Mol Genet 7:209–216 [DOI] [PubMed] [Google Scholar]
- van der Velden PA, Sandkuijl LA, Bergman W, Pavel S, van Mourik L, Frants RR, Gruis NA (2001) Melanocortin-1 receptor variant R151C modifies melanoma risk in Dutch families with melanoma. Am J Hum Genet 69:774–779 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Haeringen A, Bergman W, Nelen MR, van der Kooij-Meijs E, Hendrikse I, Wijnen JT, Khan PM, Klasen EC, Frants RR (1989) Exclusion of the dysplastic nevus syndrome (DNS) locus from the short arm of chromosome 1 by linkage studies in Dutch families. Genomics 5:61–64 [DOI] [PubMed] [Google Scholar]
- Vasen HF, Gruis NA, Frants RR, van Der Velden PA, Hille ET, Bergman W (2000) Risk of developing pancreatic cancer in families with familial atypical multiple mole melanoma associated with a specific 19 deletion of p16 (p16-Leiden). Int J Cancer 87:809–811 [PubMed] [Google Scholar]
- Weeks DE, Ott J, Lathrop GM (1990) SLINK: a general simulation program for linkage analysis. Am J Hum Genet Suppl 47:A204 [Google Scholar]
- Zuo L, Weger J, Yang Q, Goldstein AM, Tucker MA, Walker GJ, Hayward N, Dracopoli NC (1996) Germline mutations in the p16INK4a binding domain of CDK4 in familial melanoma. Nat Genet 12:97–99 [DOI] [PubMed] [Google Scholar]