Abstract
Recessive mutations in two of the three collagen VI genes, COL6A2 and COL6A3, have recently been shown to cause Ullrich congenital muscular dystrophy (UCMD), a frequently severe disorder characterized by congenital muscle weakness with joint contractures and coexisting distal joint hyperlaxity. Dominant mutations in all three collagen VI genes had previously been associated with the considerably milder Bethlem myopathy. Here we report that a de novo heterozygous deletion of the COL6A1 gene can also result in a severe phenotype of classical UCMD precluding ambulation. The internal gene deletion occurs near a minisatellite DNA sequence in intron 8 that removes 1.1 kb of genomic DNA encompassing exons 9 and 10. The resulting mutant chain contains a 33–amino acid deletion near the amino-terminus of the triple-helical domain but preserves a unique cysteine in the triple-helical domain important for dimer formation prior to secretion. Thus, dimer formation and secretion of abnormal tetramers can occur and exert a strong dominant negative effect on microfibrillar assembly, leading to a loss of normal localization of collagen VI in the basement membrane surrounding muscle fibers. Consistent with this mechanism was our analysis of a patient with a much milder phenotype, in whom we identified a previously described Bethlem myopathy heterozygous in-frame deletion of 18 amino acids somewhat downstream in the triple-helical domain, a result of exon 14 skipping in the COL6A1 gene. This deletion removes the crucial cysteine, so that dimer formation cannot occur and the abnormal molecule is not secreted, preventing the strong dominant negative effect. Our studies provide a biochemical insight into genotype-phenotype correlations in this group of disorders and establish that UCMD can be caused by dominantly acting mutations.
Introduction
Congenital muscular dystrophy (CMD) comprises a heterogeneous group of disorders characterized by muscle weakness at birth or in early infancy (Voit 1998; Tubridy et al. 2001; Muntoni et al. 2002a). Mutations in the extracellular matrix protein laminin α2 chain, a subunit of laminin 2 in the muscle basement membrane, account for a substantial proportion of CMD cases without mental retardation. An emerging group of CMD affects O-mannose–linked glycosylation of α-dystroglycan, an important laminin-2 receptor in muscle, causing a variable secondary deficiency in laminin α2 (Muntoni et al. 2002b). Recently, another extracellular matrix protein, collagen type VI, has been shown to cause a distinct type of laminin α2–positive CMD (Bertini and Pepe 2002), initially described as “atonic-sclerotic muscular dystrophy” by Otto Ullrich (Ullrich 1930) and now known as “CMD type Ullrich” (MIM 254090). The signature clinical feature of the Ullrich congenital muscular dystrophy (UCMD) phenotype is congenital muscle weakness with proximal joint contractures and coexisting marked distal joint hyperlaxity (Nonaka et al. 1981; De Paillette et al. 1989). Cases of variable severity have been described; however, in the classical most severe presentation, independent walking is not achieved or is achieved briefly and then lost, owing to progression of weakness and contractures (Bertini and Pepe 2002).
Collagen type VI is a ubiquitous connective tissue component, present primarily in the stroma and also close to the basement membrane of most tissues (Timpl and Engel 1987; Timpl and Chu 1994). It consists of three separate chains, the α1(VI), α2(VI) and α3(VI) collagen chains, encoded by the COL6A1, COL6A2, and COL6A3 genes, respectively. The α1(VI) and α2(VI) collagen chains are similar in size (140 kDa), and their genes are linked in a head-to-tail orientation on chromosome 21q22.3 (Weil et al. 1988; Heiskanen et al. 1995). The α3(VI) collagen chain is much larger (260–330 kDa) and is encoded by the gene located on chromosome 2q37 (Weil et al. 1988). The three chains share the same core structure: a relatively short triple-helical domain of 335–336 amino acids with repeating Gly-Xaa-Yaa amino acid sequences flanked by globular domains made up of motifs of 200 amino acids each that are homologous to von Willebrand factor type A (vWF A) domains (Chu et al. 1988, 1989, 1990). The three chains fold into triple-helical monomers, which are then assembled in an antiparallel manner into dimers with the N-terminal globular domains protruding (Furthmayr et al. 1983; Chu et al. 1988). The dimers associate laterally into tetramers, which are subsequently secreted into the extracellular space and associate, end-to-end, into double-beaded collagen VI microfibrils.
Collagen VI mutations in UCMD reported to date are exclusively recessive mutations in either the COL6A2 or COL6A3 genes (Camacho Vanegas et al. 2001; Higuchi et al. 2001; Demir et al. 2002; Ishikawa et al. 2002) (table 1), whereas dominant mutations in all three collagen genes have been associated with Bethlem myopathy (MIM 158810) (Jöbsis et al. 1996; Pan et al. 1998), a milder childhood-onset disorder characterized by muscle weakness and later manifesting multiple joint contractures (Bethlem and Wijngaarden 1976). In three families, a form of autosomal dominant limb girdle muscular dystrophy has also been attributed to heterozygous mutations in the COL6A1 and COL6A2 genes (Scacheri et al. 2002). All of the COL6A2 mutations described in UCMD so far lead to translational frameshifts and subsequent premature stop codons (Camacho Vanegas et al. 2001; Higuchi et al. 2001; Ishikawa et al. 2002). In two of these patients with UCMD, the COL6A2 mRNA has been shown to be nearly absent because of nonsense-mediated mRNA decay (Zhang et al. 2002), resulting in a severe deficiency of collagen VI protein in the muscle. Of the three COL6A3 homozygous mutations in UCMD reported more recently, two are nonsense mutations—one in the N-terminal globular domain and one in the triple-helical domain—and the third one leads to skipping of an exon encoding the triple-helical domain (Demir et al. 2002). The patients with the COL6A3 gene mutations exhibited complete or partial deficiency of collagen VI protein in the muscle, but the biochemical mechanisms underlying the gene mutations have not yet been investigated.
Table 1.
Summary of COL6 Mutations in UCMD[Note]
| Patient and Locationof Mutation | Mutation | Consequence | Status | Reference |
| 1: | ||||
| COL6A2 exon 13 | Ins C | Frameshift | Homozygous | Camacho Vanegas et al. 2001 |
| 2: | ||||
| COL6A2 intron 17 | −2a→g | Del 28 bp | Compound heterozygous | Camacho Vanegas et al. 2001 |
| COL6A2 intron 23 | −1g→a | Exon 24 (46 bp) skipping | Compound heterozygous | Camacho Vanegas et al. 2001 |
| 3a: | ||||
| COL6A2 intron 17 | −2a→g | Del 28 bp | Compound heterozygous | Camacho Vanegas et al. 2001 |
| 4: | ||||
| COL6A2 exon 18 | Del 26 bp | Frameshift | Homozygous | Higuchi et al. 2001 |
| 5: | ||||
| COL6A2 intron 14 | −1g→c | Ins 53 bp or 247 bp from intron sequence | Compound heterozygous | Ishikawa et al. 2002 |
| COL6A2 intron 23 | −3c→g | Exon 24 (46 bp) skipping | Compound heterozygous | Ishikawa et al. 2002 |
| 6: | ||||
| COL6A3 intron 29 | +5a→g | In-frame del of 17 amino acids in the triple-helical domain | Homozygous | Demir et al. 2002 |
| 7: | ||||
| COL6A3 exon 5 | Nonsense | Alternatively spliced variant | Homozygous | Demir et al. 2002 |
| 8: | ||||
| COL6A3 exon 31 | Nonsense | Premature termination | Homozygous | Demir et al. 2002 |
Note.— Ins = insertion; del = deletion.
Only one mutation has been found in this presumably compound heterozygous patient.
Here we characterized two patients affected with CMD. One patient displays a classical, severe phenotype of UCMD, whereas the other patient exhibits a relatively milder CMD phenotype with distal joint hyperlaxity in the absence of contractures. We show that both patients carry de novo heterozygous in-frame deletions of different sizes and locations in the triple-helical domain of the COL6A1 mRNA. We provide biochemical and immunohistochemical evidence for a genotype/phenotype correlation and show that the severe UCMD phenotype can arise from a heterozygous mutation in collagen VI through a dominant negative mechanism.
Subjects and Methods
Patients and Their Families
Two patients (UC-1 and UC-4) affected with CMD associated with distal joint hyperlaxity were studied, as were their unaffected family members. Both families were from Brazil. Dermal fibroblast cultures were established from skin biopsies of the patients, and genomic DNA was extracted from peripheral blood samples from their families. Control fibroblasts and DNA samples were obtained from unaffected volunteers. The genetic and clinical studies were performed in accordance with institutional review board–approved protocols and informed consent.
Cell Culture
Dermal fibroblasts were grown in Dulbecco’s modified Eagle medium with 10% fetal bovine serum (Life Technologies) in 5% CO2 at 37°C. For immunoblotting, immunoprecipitation, and immunofluorescence staining, cells were cultured in the presence of 50 μg/ml L-ascorbic acid phosphate (Wako).
Immunostaining of Fibroblasts
Fibroblasts were grown in the presence of L-ascorbic acid phosphate until 4 d postconfluency. Cells were fixed in 3.7% formaldehyde and were incubated at 25°C with a 1:1,000 dilution of a polyclonal antibody specific for the α1(VI) collagen chain (antibody 1026) (Tillet et al. 1994) for 1 h. The antibody labeling reaction was detected with Cy3-conjugated goat anti-rabbit IgG (1:1,000 dilution; Jackson ImmunoResearch Laboratories), and nuclei were counterstained with DAPI (4′,6′-diamidino-2-phenylindole hydrochloride). Images were obtained using a Zeiss Axioskop epifluorescence microscope with a Toshiba 3CCD camera.
RNA Extraction and Northern Blot Analysis
Total RNA was isolated from confluent fibroblasts with the RNeasy Mini Kit (Qiagen). The total RNA (5 μg) was separated on a 1% agarose gel containing formaldehyde, was transferred to Duralon-UV membrane (Stratagene), and was hybridized with cDNA probes labeled with [32P]dCTP by the Random Prime Labeling system (Amersham Pharmacia) in Ultrasensitive Hybridization Buffer (Ambion), following protocols suggested by the suppliers. The probes were a mixture of cDNA clones encoding the α1(VI), α2(VI), and α3(VI) collagen chains (clones F157, F225, and FO19) (Chu et al. 1989, 1990).
Mutational Analysis by RT-PCR and DNA Sequencing
Total RNA isolated from fibroblasts was reverse transcribed using Superscript II reverse transcriptase (Life Technologies) and oligo(dT) primer for 1 h at 42°C. The resulting cDNA was used as a template for PCR amplification of the entire coding regions of the COL6A1, COL6A2, and COL6A3 mRNA transcripts (GenBank accession numbers NM_0018489, NM_001849, and X52022, respectively) with AmpliTaq Gold polymerase (Applied Biosystems). The COL6A1, COL6A2, and COL6A3 transcripts were amplified with 5, 4, and 14 primer pairs, respectively, yielding overlapping cDNA fragments 400–1,200 bp long. The PCR conditions were 10 min at 94°C; 35 cycles of 94°C for 30 s, 58–60°C for 30 s, and 72°C for 1 min; followed by a final extension of 72°C for 7 min. PCR products were subjected to DNA sequencing with an ABI377 DNA Sequencer (Applied Biosystems). The primer pairs revealing the COL6A1 deletions were as follows: forward, 5′-AACGTTGAGCAAGTGTGCTG-3′; and reverse, 5′-TTCACCAGGGTCTCCTCTTG-3′.
Characterization of the COL6A1 Gene Mutations
Genomic DNA isolated from patient UC-1 and his family was PCR amplified with a forward primer in intron 6 (5′-GACTCGTCTCCATGCTTTCC-3′) and a reverse primer in intron 11 (5′-CATGGGGACTGACAGTGATG-3′), through use of the Expand Long Template PCR system (Roche). The PCR conditions were 94°C for 2 min; 30 cycles at 94°C for 10 s, 58°C for 30 s, and 68°C for 8 min; followed by a final extension at 68°C for 7 min. The PCR products were analyzed on 1% agarose gels. PCR products yielded from patient UC-1 were cloned into the pCR-TOPO vector (TOPO TA Cloning Kit; Invitrogen) and were subjected to DNA sequencing.
Genomic DNA isolated from patient UC-4 and his family was PCR amplified with a forward primer located in the boundary of exon 12/intron 12 (5′-AGGGCTACAAGGTGAGCGTG-3′) and a reverse primer in intron 15 (5′-CAGGACTTGCTTCCAGGAGA-3′), through use of AmpliTaq Gold polymerase (Applied Biosystems) and the cycling conditions described above for RT-PCR. The PCR products from UC-4 and his family were subjected to DNA sequencing.
Protein Analysis by Immunoblotting and Immunoprecipitation
Immunoblot analysis of culture medium and cell layers from confluent fibroblasts were performed using 1026 antibody specific for the α1(VI) collagen chain (Tillet et al. 1994), as described elsewhere (Pepe et al. 1999; Camacho Vanegas et al. 2002). For immunoprecipitation of collagen VI protein, confluent fibroblasts were metabolically labeled with [35S]cysteine overnight and were precipitated with 1014 antibody specific for the α3(VI) collagen chain (Specks et al. 1992), as described elsewhere (Camacho Vanegas et al. 2001). Samples were electrophoresed on 3%–8% SDS-polyacrylamide gels under reduced conditions (25 mM dithiothreitol [DTT]) or on composite 0.5% agarose, 2.4% polyacrylamide gels without reduction with DTT (Lamandé et al. 1998). Reduced (230 and 400 kDa) and unreduced (900 kDa) forms of laminin were used as the molecular mass standard on the composite gel.
Immunostaining of Muscle Biopsies
Biopsies were cut in 10-μm sections through use of a Microm HM505E cryostat and were mounted onto microscope slides before being stored at −80°C until further use. After allowing the slides to acclimate to room temperature, they were fixed with methanol at room temperature for 20 s and were briefly washed with ice-cold 1× PBS (0.01 M KH2PO4, 0.1 M Na2HPO4, 1.37 M NaCl, 0.027 M KCl, pH 7.4). The sections were preincubated in vehicle (10% fetal bovine serum, 1× PBS, 0.1% Triton X-100) at room temperature for 1 h. Primary antibodies at the appropriate dilution in vehicle were applied to the sections and allowed to incubate overnight at 4°C. A mouse monoclonal antibody recognizing collagen type VI (MAB3303; 1:2,500) was obtained from Chemicon International. The rabbit polyclonal antibody against perlecan domain III (1030; 1:2,000) was graciously provided by R. Timpl of the Max Planck Institute for Biochemistry, Germany. Slides were washed in 1× PBS (3 × 20 min) prior to applying the secondary antibodies in vehicle at room temperature for 1 h in the dark. Alexa Fluor 488–conjugated goat anti-mouse IgG and Alexa Fluor 568–conjugated goat anti-rabbit IgG (Molecular Probes) were used as secondary antibodies at 1:500 dilution. The slides were washed in 1× PBS and were post-fixed at room temperature for 8 min in the dark, using cold methanol (−25°C). The slides were briefly washed with 1× PBS before being coverslipped with either Vectashield or Biomedia Gel/Mount media mount.
Images were obtained on a Leica Inverted DM IRE2 HC fluo TCS 1-B-UV microscope coupled to a TCS SP2 spectral confocal system/UV with a scan head containing an AOBS acousto-optical tunable filter. The green HeNe (543 nm/1.2 mW) and blue argon (488 nm/20 mW) laser lines were used to visualize the fluorophores. The images were captured at 1,024×1,024 resolution under a 40× oil lens.
Results
Patients UC-1 and UC-4 Display Severe and Mild Symptoms of CMD, Respectively
Patient UC-1, age 12 years, had congenital-onset muscular hypotonia and weakness, followed by severely delayed motor development. He never stood or walked without support, and his maximal motor ability was sitting without support. He was first evaluated by us at age 6 years, when he presented with severe muscular hypotonia and weakness. The muscle weakness was graded as 3/5 MRC (Medical Research Council) proximally and distally, with minimal bilateral facial weakness. There was marked generalized muscle atrophy. The tendon reflexes were absent or depressed. Severe joint contractures were prevalent, affecting preferentially proximal but also distal joints in the finger flexors of the hands while maintaining small joint hyperlaxity at the same time. He had protruding calcanei. His cognitive development had been completely normal. Since age 11 years, he had suffered from respiratory insufficiency, necessitating ventilatory assistance. His creatine kinase (CK) level was normal, and electromyography (EMG) results were interpreted as showing a myopathic pattern. Results of a CT scan of the brain, cardiac evaluation, and ophthalmological evaluation were all normal. A muscle biopsy, which had been obtained at age 9 years 5 mo, showed moderate proliferation of endomysial and perimysial connective tissue and variation of muscle fiber diameter, as well as scattered degenerating fibers. Patient UC-1 thus displays a classical severe UCMD phenotype.
Patient UC-4, age 6 years, also presented with hypotonia since birth. However, he was able to walk since age 1 year and able to climb stairs, but he got up from the floor with a Gowers' maneuver and suffered frequent falls. Muscle weakness was moderate, at ∼4+/5 MRC, with some proximal predominance. The patient had generalized hyperextensibility of his joints in the upper and lower extremities, in particular in the small joints. There were slight asymmetric contractures of the Achilles tendons, but no other contractures were found. CK level was normal. EMG demonstrated a myopathic pattern, and muscle biopsy exhibited a mildly dystrophic pattern, with variation in muscle fiber diameter, moderate increase in endomysial connective tissue, and occasional degenerating fibers.
Collagen VI Microfibrils Are Absent in Fibroblasts from UC-1 and Markedly Reduced in Fibroblasts from UC-4
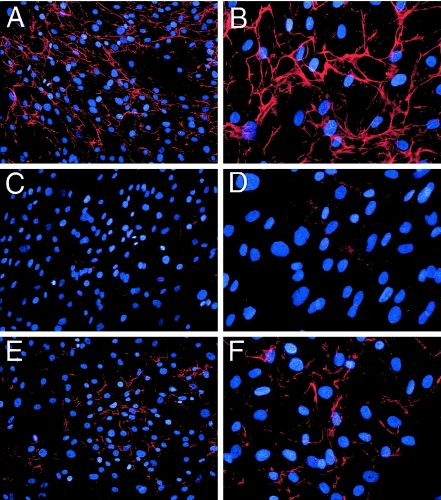
Fibroblasts from patients UC-1 and UC-4 were immunostained with an antibody specific to the α1(VI) collagen chain. Whereas the control fibroblasts exhibited an extensive collagen VI microfibrillar network in the extracellular matrix, fibroblasts from UC-1 were found to deposit few, if any, collagen VI microfibrils (fig. 1). In contrast, fibroblasts from UC-4 deposited collagen VI microfibrils in markedly reduced amounts as compared with the control fibroblasts. Similar immunostaining results were observed when the α3(VI) collagen chain–specific 1014 antibody was used (data not shown). The fibroblasts from both UC-1 and UC-4 deposited amounts of type I collagen fibrils similar to that deposited by the control fibroblasts (data not shown). The results suggested that both patients might carry collagen VI mutations.
Figure 1.
Deposition of collagen VI microfibrils in the extracellular matrix of dermal fibroblasts. Fibroblasts from an unaffected control individual (A and B), patient UC-1 (C and D), and patient UC-4 (E and F) were grown in the presence of 50 μg/ml L-ascorbic acid phosphate for 4 d postconfluency and were stained with the α1(VI) collagen-specific antibody. The antibody reaction was detected with Cy3-conjugated goat anti-rabbit IgG (red), and cells were counterstained with DAPI (blue) to visualize nuclei. Original magnifications ×20 (A, C, and E) and ×40 (B, D, and F).
The COL6A1 Transcripts in UC-1 and UC-4 Each Contain a Heterozygous In-Frame Deletion within the Triple-Helical Domain
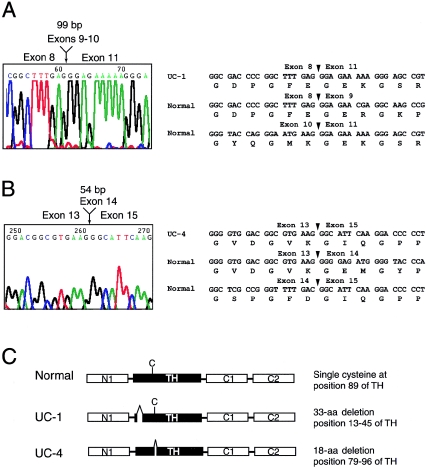
To search for possible collagen VI mutations in UC-1 and UC-4, the coding regions of the COL6A1, COL6A2, and COL6A3 mRNA transcripts were amplified from fibroblast RNA by RT-PCR. A primer pair flanking the COL6A1 triple-helical coding region amplified two products of equal intensity in both patients, one of which was the expected size, whereas the other one was slightly smaller (data not shown). DNA sequencing revealed that the shorter product from UC-1 contained a deletion of 99 bp, extending from nucleotide 805 to 903, encoded by exons 9 (54 bp) and 10 (45 bp) of the COL6A1 gene (fig. 2). The shorter product from UC-4 harbored a smaller deletion, of 54 bp, that was located farther downstream and corresponded to nucleotides 1003–1056, encoded by exon 14. DNA sequences of the larger PCR products from both patients showed no deviations from the reference COL6A1 cDNA sequence. The results indicated that UC-1 and UC-4 carry heterozygous COL6A1 in-frame deletions of different sizes located at different positions within the triple-helical domain.
Figure 2.
In-frame deletions in the triple-helical domain of the COL6A1 mRNA in patients UC-1 and UC-4. A and B, DNA sequence chromatograms showing a deletion of 99 bp encoded by exons 9 and 10 in UC-1 (A) and a deletion of 54 bp encoded by exon 14 in UC-4 (B). DNA and amino acid sequences of the normal and mutant COL6A1 mRNA surrounding the mutations are shown. Arrowheads mark exon borders. C, Schematic diagram of normal and mutant α1(VI) collagen chains. The normal α1(VI) collagen chain consists of a triple-helical domain (TH) of 336 amino acids flanked by N- and C-terminal globular domains made up of vWF-A–like motifs (N1, C1, and C2). The TH domain contains a single cysteine residue (depicted as “C”) at position 89, which is important for dimer assembly. Patient UC-1 contains a 33–amino acid deletion near the N-terminus of the triple-helical domain, whereas patient UC-4 contains an 18–amino acid deletion somewhat downstream, which removes the cysteine residue.
Because all previously reported patients with UCMD carry homozygous or double heterozygous mutations, it was possible that a second mutation in one of the collagen VI genes could be present in UC-1 and UC-4 in a compound heterozygous state with the identified mutations. Therefore, the entire coding regions of the COL6A1, COL6A2, and COL6A3 transcripts were analyzed by DNA sequencing. The analyses yielded a number of silent nucleotide polymorphisms and single-nucleotide changes that resulted in amino acid substitutions (table 2). All of the amino acid substitutions were found in genomic DNA from control individuals and thus were not pathological mutations.
Table 2.
Polymorphisms in Collagen VI Genes That Result in Amino Acid Changes
| Patient, Gene,and Exon | NucleotideChange | Amino AcidChange | Status |
| UC-1: | |||
| COL6A1: | |||
| 35 | 2669C→T | S889L | Heterozygous |
| COL6A2: | |||
| 26 | 2039A→G | H680R | Heterozygous |
| 28 | 2803G→A | G935R | Heterozygous |
| COL6A3: | |||
| 25 | 6653C→T | P2218L | Heterozygous |
| 28 | 6860C→G | P2287R | Homozygous |
| 41 | 9203C→T | T3068I | Homozygous |
| UC-4: | |||
| COL6A2: | |||
| 18 | 1466G→A | R489Q | Heterozygous |
| 26 | 2039A→G | H680R | Homozygous |
| COL6A3: | |||
| 4 | 872G→A | R291K | Heterozygous |
| 36 | 7292T→A | V2431D | Homozygous |
| 39 | 8491G→C | D2831T | Homozygous |
| 40 | 8780C→T | T2927M | Heterozygous |
| 40 | 8870-8871insTGC | 2957insA | Homozygous |
| 40 | 8959G→A | V2987M | Homozygous |
| 41 | 9203C→T | T3068I | Heterozygous |
An Internal COL6A1 Gene Deletion in UC-1 Removes Exons 9 and 10, Whereas a Splice Donor Site Mutation in UC-4 Results in Skipping of Exon 14
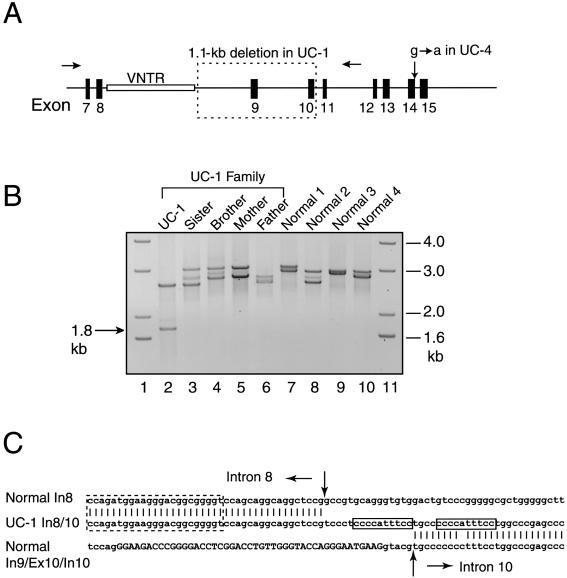
To delineate the mRNA mutations at the genomic level and determine whether the mutations were inherited, genomic DNA from the patients and their families was PCR amplified and DNA sequenced. Primers in introns 6 and 11 were used to amplify genomic DNA from UC-1 and his family (parents, elder brother, and sister) (fig. 3A). The genomic region was found to be highly variable in length, since two major PCR products, ranging in size from 2.6 to 3.2 kb, were amplified from DNA of the family members and unaffected controls (fig. 3B). The size variation was due to the presence of a minisatellite near the 5′ end of intron 8. In the reference sequence (GenBank accession number NT_011515), the minisatellite consists of 32 tandem repeats of the sequence ccagatggagggga(t/c)ggcggg(a)gt. In UC-1, a 2.6-kb PCR product and a substantially smaller 1.8-kb fragment were obtained. DNA sequencing indicated that the 1.8-kb DNA fragment contained a deletion of 1,095 bp that removed the genomic region encompassing exons 9 and 10 and the intervening intron (fig. 3A). The proximal deletion breakpoint was located in intron 8 at 17 bp downstream of the last repeat in the minisatellite sequence (fig. 3C). The distal breakpoint was in intron 10 at 5 bp downstream of exon 10. At the distal breakpoint, a 15-bp inserted sequence (tccctccccatttcc) was found, of which the sequence tccccatttcc was a direct duplication of adjacent intron 10 sequence. The DNA sequence of the 2.6-kb fragment corresponded well with the reference COL6A1 genomic sequence. Comparison of the sizes of the PCR products from UC-1 and his family indicated that the genomic deletion in UC-1 was not present in his parents and two siblings (fig. 3B) and, hence, had arisen de novo. It also indicated that the normal 2.6-kb COL6A1 allele in UC-1 was inherited from the father; therefore, the novel genomic deletion in UC-1 had occurred on his maternal COL6A1 allele.
Figure 3.
Mutational analysis of the COL6A1 gene. A, Schematic diagram of the COL6A1 genomic region including exons 7–15 (blackened boxes). Exon 8 encodes the beginning of the triple-helical domain, and exons 9–15 each encode discrete numbers of Gly-Xaa-Yaa repeats in the triple-helical domain. A minisatellite (unblackened box labeled “VNTR”) is present at the 5′ end of intron 8. In patient UC-1, one of the COL6A1 alleles contains a 1.1-kb gene deletion (dotted box) extending from intron 8 to intron 10. Patient UC-4 carries a heterozygous G→A transition at the +1 position of intron 14. B, PCR amplification of genomic DNA from patient UC-1 and his family (lanes 2–6) and from four unaffected control individuals (lanes 7–10) with primers in introns 6 and 12 (arrows in A). Lanes 1 and 11 contain DNA size markers. C, DNA sequence of the 1.8-kb PCR product from UC-1, showing the breakpoint of the internal gene deletion (middle line) and its alignment with the normal COL6A1 genomic sequence of intron 8 (top line) and of intron 9–exon 10–intron 10 (bottom line). Exon sequences are in capital letters. Between the arrows are 15-bp inserted sequences in the deletion junction, which contains an 11-bp direct duplication of the sequence in intron 10 (boxed). The dotted box depicts the last repeat of the minisatellite sequence in intron 8. In = intron; Ex = exon.
Genomic DNA from UC-4 and his family (parents, elder brother and sister) was PCR amplified with a primer pair flanking exon 14, which yielded a single PCR product of 854 bp in all family members. DNA sequencing of the fragments showed a heterozygous G→A transition at +1 position of the splice donor site in intron 14 in UC-4 (fig. 3A). The mutation was not found in his parents and siblings and therefore also had arisen de novo.
Mutant α1(VI) Collagen Chain Is Secreted Extracellularly in UC-1 but Not in UC-4
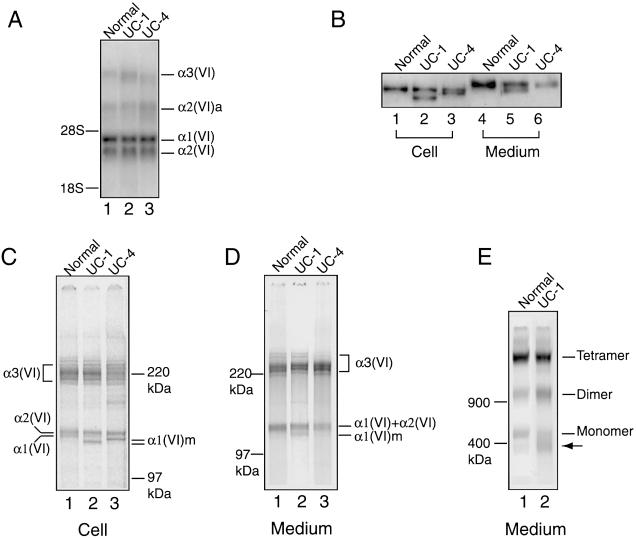
The effects of the COL6A1 gene mutations on collagen VI mRNA and protein synthesis were studied. Analysis of total RNA from UC-1 and UC-4 fibroblasts by northern blot hybridization showed that the steady-state levels of collagen VI mRNA for all three chains in UC-1 and UC-4 were comparable to those in the control (fig. 4A). The results suggest that the mutations do not significantly affect transcription and stability of the COL6A1 mRNA, unlike previous findings of other patients with UCMD (Zhang et al. 2002).
Figure 4.
Analysis of collagen VI mRNA and protein in fibroblasts from UC-1 and UC-4. A, Northern blot analysis of total RNA (5 μg) from normal fibroblasts and those from UC-1 and UC-4, with a mixture of [32P]dCTP-labeled cDNA probes for the three collagen VI mRNA. The probes hybridized with the α1(VI), α2(VI), and α3(VI) collagen mRNA of 4.2, 3.5, and 9–10 kb, respectively, plus an alternatively spliced α2(VI) collagen mRNA, α2(VI)a, of 6.0 kb. B, Immunoblot analysis of cell layer extracts (lanes 1–3) and culture media (lanes 4–6) from normal fibroblasts (lanes 1 and 4) and those from UC-1 (lanes 2 and 5) and UC-4 (lanes 3 and 6), with the antibody specific for the α1(VI) collagen chain. Lanes 1–3 contain 10 μg of total protein from the cell layers, and lanes 4–6 contain 100 μl of culture medium precipitated with 0.9 ml of 100% EtOH. C–E, Immunoprecipitation of cell layers (C) and culture media (D, E) from normal fibroblasts (lane 1) and those from UC-1 (lane 2) and UC-4 (lane 3), with the antibody specific for the α3(VI) collagen chain. Fibroblasts were labeled with [35S]cysteine overnight. Samples in B, C, and D were reduced with 25 mM DTT and separated on 3%–8% polyacrylamide gels. Samples in E were separated on a composite agarose polyacrylamide gel in the absence of 25 mM DTT. The mutant α1(VI) collagen chains are depicted as α1(VI)m. The arrow indicates a protein present in higher amounts in fibroblasts from UC-1 compared with the normal control fibroblasts.
To assess the synthesis and secretion of the mutant α1(VI) collagen chains in fibroblasts from UC-1 and UC-4, cell layers and culture media were analyzed by immunoblotting using the antibody specific for the α1(VI) collagen chain. In the cell layers of UC-1 and UC-4, approximately equal amounts of normal and shorter α1(VI) collagen chains were observed (fig. 4B, lanes 1–3), indicating that the mutant chains in both patients were synthesized efficiently and were stable. Because of posttranslational modifications, the α1(VI) collagen chains that were secreted extracellularly migrated more slowly than those found in the cell layers. Both normal and shorter chains could be detected in the medium of UC-1, whereas only the normal chain was seen in the medium of UC-4. Quantification of three separate immunoblots with a densitometer revealed that the amount of mutant chain secreted by the fibroblasts from UC-1 was 53% of the normal counterpart.
Fibroblasts from UC-1 and UC-4 were metabolically labeled with [35S]cysteine and were immunoprecipitated with the antibody against the α3(VI) collagen chain. The antibody would precipitate free α3(VI) collagen chain as well as α1(VI) and α2(VI) collagen chains that are folded together with the α3(VI) collagen chain in a triple helix. In the cell layers of fibroblasts from both UC-1 and UC-4, the antibody precipitated both the normal and shorter α1(VI) collagen chains (fig. 4C), indicating that the mutant chains from both patients were able to incorporate into triple helices. As predicted from the sizes of the deletions, the mutant chain in UC-1 migrated faster than that in UC-4. In contrast, the mutant α1(VI) collagen chains could be detected only in the medium of UC-1 but not of UC-4 (fig. 4D), consistent with the results of the immunoblot analysis above.
To determine whether the shorter α1(VI) collagen chains secreted by fibroblasts from UC-1 were present in dimers and tetramers, the immunoprecipitated material was analyzed on a composite agarose/polyacrylamide gel without reduction using DTT. The majority of the secreted collagen VI chains in UC-1 were found in tetramers similar to the normal control fibroblasts (fig. 4E). This implied that the mutant α1(VI) collagen chain secreted extracellularly was most likely incorporated into the tetramers. It was noted that there were fewer monomers in UC-1 than in the control, whereas a protein complex that migrated below the monomers was present in higher amounts in UC-1 as compared with the control. This was a consistent finding in two separate experiments. The nature of this faster-migrating protein band is unclear; its apparent molecular mass appears larger than that of a mutant monomer containing a shorter α1(VI) collagen chain.
Collagen VI Protein Is Aberrantly Localized in the UC-1 Muscle but Reduced in the UC-4 Muscle
Muscle biopsies from UC-1 and UC-4 were immunostained with antibodies against collagen VI and perlecan simultaneously (fig. 5). In normal muscle, collagen VI colocalized with perlecan in the basement membrane surrounding muscle fibers. In the UC-1 muscle, collagen VI protein was not seen in the basement membrane, even though the basement membrane appeared intact, as shown by the uniform perlecan staining. Abundant collagen VI protein, however, was found in the interstitial and perivascular regions between muscle fibers. In contrast, some collagen VI protein was present in the basement membrane of the UC-4 muscle, but at a markedly reduced level relative to the staining with the perlecan antibody.
Figure 5.
Immunostaining of muscle biopsies from UC-1, UC-4, and an unaffected control individual. Sections were stained with the monoclonal antibody against collagen VI (green in A, D, and G) and the polyclonal antibody against perlecan (red in B, E, and H). C, F, and I show a merge of green and red fluorescence. In normal muscle (A, B, and C), collagen VI and perlecan colocalize in the basement membrane. In the muscle from UC-1 (D, E, and F), collagen VI is absent in the basement membrane surrounding muscle fibers; instead, it is accumulated in the interstitial and perivascular space between muscle fibers. In the muscle from UC-4 (G, H, and I), collagen VI is present in the basement membrane but appears patchy and significantly reduced relative to the perlecan staining. Collagen VI staining is increased in the interstitial and perivascular space.
Discussion
Here we demonstrate that a patient affected with the severe phenotype of UCMD (UC-1) carries a heterozygous in-frame deletion in the COL6A1 gene but no additional mutations in the COL6A1, COL6A2, and COL6A3 genes. The gene deletion is not present in the unaffected family members and has arisen de novo. The findings indicate that the heterozygous COL6A1 gene deletion is the underlying cause of the severe UCMD phenotype in this patient.
The deletion in UC-1 is located very close to the minisatellite in intron 8 of the COL6A1 gene, suggesting a role for the minisatellite sequence in the gene deletion. PCR analysis of the COL6A1 minisatellite alleles in the UC-1 family and 15 unaffected control individuals indicates that the length of this minisatellite varies greatly (fig. 3B and data not shown). This result supports previous findings that minisatellites can be found in the vicinity of recurrent translocation breakpoints and constitute chromosomal fragile sites (Vergnaud and Denoeud 2000). Minisatellites have, in addition, been shown to affect transcription and intron splicing (Kennedy et al. 1995; Turri et al. 1995). For instance, an expansion of an intronic minisatellite in the USH1C gene has been correlated with the Usher syndrome type 1C in the Acadian population (Verpy et al. 2000). It remains to be tested whether the variation in the size of the minisatellite in intron 8 of the COL6A1 gene interferes with intron splicing and/or affects the steady-state mRNA level.
The milder CMD phenotype of patient UC-4 is caused by a heterozygous skipping of exon 14 in the COL6A1 transcript. This is a recurring mutation in the COL6A1 gene, since skipping of exon 14 has previously been found in two unrelated patients with Bethlem myopathy resulting from mutations in either the first or second nucleotide of the splice donor site in intron 14 (Lamandé et al. 1999; Pepe et al. 1999). Patient UC-4 does not display the significant joint contractures characteristic of patients with Bethlem myopathy at this time. However, he is still young and may develop contractures in the future; therefore, his phenotype at this age could well be compatible with sporadic Bethlem myopathy. It has been pointed out that Bethlem myopathy may present as a mild congenital muscular dystrophy (Jöbsis et al. 1999).
The deletion in UC-1 removes 11 contiguous Gly-Xaa-Yaa repeats in the α1(VI) collagen chain very close to the N-terminus of the triple-helical domain (amino acids 13–45 of the triple-helical domain). On the other hand, UC-4 contains a smaller deletion of six Gly-Xaa-Yaa repeats at amino acids 79–96, which is also located at the N-terminal portion of the triple-helical domain, but somewhat downstream. For fibrillar collagens, the triple-helical folding starts from the C-terminal noncollagenous domains and propagates in a zipperlike fashion toward the N-terminus (Bachinger et al. 1981; Engel and Prockop 1991). If we assume that this is also the case for collagen VI, deletions in the N-terminal portion of the triple-helical domain such as those found in UC-1 and UC-4 would not prevent the mutant chains from folding into a triple helix. Indeed, immunoprecipitation of the cell layers from the fibroblasts of UC-1 and UC-4 indicates that the shorter α1(VI) collagen chains in both patients are incorporated into triple-helical monomers (fig. 4C). The mutant monomers presumably would contain shorter triple-helical domains, because the deletions in the mutant chain are likely to be accommodated by looping out the corresponding regions of the normal chains in a manner similar to a triple-helical deletion in type II collagen described elsewhere (Weis et al. 1998).
The two different deletions, however, have distinct effects on the subsequent step of collagen VI supramolecular assembly. Unlike most collagens, the triple-helical collagen VI monomers are assembled into dimers and tetramers prior to secretion into the extracellular space (fig. 6A) (Timpl and Engel 1987; Timpl and Chu 1994). Dimers are formed by the antiparallel, staggered assembly of two monomers, and the single cysteine located at amino acid 89 of the triple-helical domains in the α1(VI) or α2(VI) collagen chains is responsible for the assembly/stability of the dimers (Furthmayr et al. 1983; Chu et al. 1988). Tetramers are assembled by the lateral association of two dimers, whereby the single cysteine residue at amino acid 50 in the triple-helical domain of the α3(VI) collagen chain is involved in tetramer formation/stability. The critical regions involved in dimer and tetramer formation are all retained in the mutant chain of UC-1; thus, the mutant chain should be able to assemble into dimers and tetramers and be secreted. By contrast, cysteine 89 is deleted in the mutant chain of UC-4, which would prevent the abnormal monomer from assembling into dimers. Consequently, only normal dimers and tetramers will be formed.
Figure 6.
Schematic diagram showing the effects of two different internal deletions on collagen VI assembly. A, Intracellular events. Normally (left), the α1(VI), α2(VI), and α3(VI) collagen chains presumably fold from the C-terminal end into a triple-helical monomer. Two key cysteine residues (vertical lines) in the N-terminal portion of the triple helix are critical for subsequent supramolecular assembly. Dimers are formed by the staggered antiparallel association of monomers with the N-terminal globular domains protruding at both ends. The outer cysteines contributed by either the α1(VI) or α2(VI) collagen chains are important for the dimer assembly. The dimers associate laterally into tetramers, which are stabilized by the outer cysteines contributed by the α3(VI) collagen chain. In UC-1 (middle) and UC-4 (right), equal amounts of normal and mutant (gray) α1(VI) collagen chains assemble into equal amounts of normal and mutant monomers. Because the cysteines critical for dimer and tetramer formation are preserved in UC-1, both normal and mutant monomers can assemble into dimers and tetramers in any combination. Thus, only 1/4 of the dimers and 1/16 of the tetramers are entirely composed of normal chains, whereas 15/16 of the tetramers contain a mixture of normal and mutant chains (gray striped lines). The deletion in UC-4 removes the cysteine in the triple-helical domain of the α1(VI) collagen chain, which prevents the abnormal monomers from assembling into dimers. Therefore, only normal tetramers are formed and secreted. The mutant chain in UC-4 may interfere with the assembly of the normal dimers, resulting in the formation and secretion of normal tetramers at a level ⩽1/2 that of the control. B, Extracellular event. Normally, tetramers are secreted and associated end-to-end into double-beaded microfibrils (top). The microfibrillar assembly involves interactions of the N- and C-globular domains and triple-helical domain. The deletion in UC-1 is located very close to the N-terminal end of the triple-helical domain (broken thick lines). Presumably, the triple helix in the mutant monomer is shortened because the normal α2(VI) and α3(VI) collagen chains are looped out at the region corresponding to the deletion (bottom). As a result, the mutant tetramers cannot properly align with normal tetramers to form the double-beaded structure. Because the majority of the secreted tetramers are abnormal, long microfibrils cannot be assembled.
The immunoprecipitation and immunoblot analyses indeed confirm that the shorter α1(VI) collagen chain in UC-1 is incorporated into tetramers and secreted (fig. 4B, 4D, and 4E). Theoretically, equal amounts of the normal and mutant chains synthesized by UC-1 would result in assembly of equal amounts of the normal and mutant monomers. However, because the mutant chain can be incorporated in the subsequent assembly steps, the effect of the mutation would be greatly amplified. It is expected that only 1/4 of the dimers and only 1/16 of the tetramers would be entirely composed of normal chains, whereas 15/16 of the tetramers would be abnormal, with 1–4 mutant chains (fig. 6A). Experimentally, however, it appears that certain types of abnormal tetramers may not be formed and/or secreted, because immunoblot analysis shows that only 53% of the shorter α1(VI) collagen chains are secreted (fig. 4B). The effect of the deletion likely is further amplified in the microfibrillar assembly step after the mutant tetramers are secreted. Since microfibrils are assembled by end-to-end association of the tetramers via the N-terminus, mutant tetramers with structurally abnormal N-termini cannot correctly align with their neighboring tetramers and thus would drastically impair microfibrillar assembly (fig. 6B).
In contrast, the shorter α1(VI) collagen chain in UC-4 cannot be detected in the culture medium under different electrophoretic conditions (fig. 4 and data not shown). This observation is consistent with previous findings in the two patients with Bethlem myopathy who had the same deletion (Lamandé et al. 1999; Pepe et al. 1999). However, it should be noted that, owing to posttranslational modifications of the secreted chains and relatively small size of the deletion, the possibility remains that the mutant chain may be secreted but cannot be distinguished from the normal chain experimentally. The mutant chain may, in addition, form a partially assembled dimer with its normal counterpart intracellularly (fig. 6). In either case, the mutant chain might exert a slight dominant negative effect. Therefore, the deletion in UC-4 would lead to assembly of collagen VI microfibrils at a level ⩽50% that of the control.
It is interesting to consider another in-frame deletion in the triple helical domain reported in a patient with UCMD (table 1). This patient carried a homozygous splice mutation in the COL6A3 gene, causing skipping of exon 29 and an in-frame deletion of 51 amino acids in the C-terminus of the triple-helical domain (Demir et al. 2002). However, the heterozygous parents were not affected, suggesting that the mutation does not act in a dominant negative fashion. Studies using introduced triple-helical deletions as well as glycine substitutions in the α3(VI) collagen chain have shown that the biosynthetic consequences of the mutations depend on their positions in the triple-helical domain (Lamandé et al. 1999, 2002). Whether the mutant α3(VI) collagen chain with the C-terminal triple-helical deletion is incorporated into the triple helix to potentially act in a dominant negative way remains an interesting question.
The distinct effects of these two triple-helical deletions on collagen VI supramolecular assembly are supported by immunofluorescent labeling of fibroblasts, showing an almost complete absence of collagen VI microfibrils in the fibroblasts of UC-1 and a significant reduction in the fibroblasts of UC-4 (fig. 1). The differential effects are also reflected in their muscle biopsies, which display an absence and substantial reduction of collagen VI immunostaining in the muscle basement membrane of UC-1 and UC-4, respectively (fig. 5). This correlates well with the clinical severity of these two patients. The substantial reduction in collagen VI immunofluorescence observed in fibroblasts and muscle biopsy from UC-4 compared with the control suggests that the mutant chain in UC-4 may exert a slight dominant negative effect, either intracellularly or extracellularly.
Of particular interest is the observation that the mutation in UC-1 leads to loss or mislocalization of collagen VI protein in the muscle basement membrane (fig. 5). By immunoelectron microscopy, collagen VI filaments have previously been shown to localize in close juxtaposition to the muscle basement membrane (Kuo et al. 1997). The results from the muscle of UC-1 suggests that only the assembled collagen VI microfibrils can be properly anchored to the basement membrane. It also suggests that the substantial amounts of mutant collagen VI tetramers secreted into the extracellular space are not degraded but instead are aberrantly accumulated in the interstitial and perivascular space. It is not clear what molecule anchors the collagen VI network to the basement membrane. Possible candidates include collagen type IV, perlecan, and fibronectin, all of which are present in the basement membrane and interact with collagen VI in vitro (Tillet et al. 1994; Kuo et al. 1997). The results highlight the critical role of collagen VI in the muscle basement membrane and suggest that mislocalization of collagen VI is a potential pathological mechanism for the severe UCMD phenotype.
Mutations in collagen type VI have now been associated with seemingly distinct types of muscular dystrophy, which vary widely in clinical severity and connective tissue involvement. Previously, heterozygous mutations are found in the milder phenotypes of Bethlem myopathy and a form of dominant limb girdle muscular dystrophy, whereas homozygous or double heterozygous mutations are observed in the severe phenotype of UCMD. The identification of a heterozygous mutation in a severe phenotype of UCMD thus is a significant departure from previous observations. However, this finding should not be entirely unexpected, because heterozygous mutations in the triple-helical domain of collagen genes are known to cause severe or even lethal phenotypes owing to dominant negative effects (procollagen suicide) exerted by structurally defective chains (Prockop 1990; Myllyharju and Kivirikko 2001).
In conclusion, we have established the COL6A1 gene as the third locus for UCMD and, furthermore, have identified the first example of a multiexon deletion in collagen VI genes. We have also provided evidence that a heterozygous mutation in collagen VI can cause a severe UCMD phenotype through a dominant negative mechanism and suggested a correlation between mutation location and clinical severity. This finding also has considerable relevance for genetic counseling in UCMD.
Acknowledgments
We are grateful to the patients and their families for their participation in this study. We thank Dr. Rupert Timpl for providing polyclonal antibodies to collagen VI and perlecan. The work was supported by National Institutes of Health grant AR38912 (to M.-L.C.). This publication was made possible with funding to C.G.B. from the Florence R. C. Murray Fellowship Program at The Children’s Hospital of Philadelphia. Its contents are solely the responsibility of the authors and do not necessarily represent the official views of the Hospital. C.G.B. is a Pew Scholar in the Biomedical Sciences.
Electronic-Database Information
Accession numbers and URLs for data presented herein are as follows:
- GenBank, http://www.ncbi.nlm.nih.gov/Genbank/ (for COL6A1 [accession numbers NM_0018489 and NT_011515], COL6A2 [accession numbers NM_001849 and NT_011515], and COL6A3 [accession numbers X52022 and NT_005120])
- Online Mendelian Inheritance in Man (OMIM), http://www.ncbi.nlm.nih.gov/Omim/ (for UCMD and Bethlem myopathy)
References
- Bachinger HP, Fessler LI, Timpl R, Fessler JH (1981) Chain assembly intermediate in the biosynthesis of type III procollagen in chick embryo blood vessels. J Biol Chem 256:13193–13199 [PubMed] [Google Scholar]
- Bertini E, Pepe G (2002) Collagen type VI and related disorders: Bethlem myopathy and Ullrich scleroatonic muscular dystrophy. Eur J Paediatr Neurol 6:193–198 [DOI] [PubMed] [Google Scholar]
- Bethlem J, Wijngaarden GK (1976) Benign myopathy, with autosomal dominant inheritance: a report on three pedigrees. Brain 99:91–100 [DOI] [PubMed] [Google Scholar]
- Camacho Vanegas O, Bertini E, Zhang RZ, Petrini S, Minosse C, Sabatelli P, Giusti B, Chu ML, Pepe G (2001) Ullrich scleroatonic muscular dystrophy is caused by recessive mutations in collagen type VI. Proc Natl Acad Sci USA 98:7516–7521 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Camacho Vanegas O, Zhang RZ, Sabatelli P, Lattanzi G, Bencivenga P, Giusti B, Columbaro M, Chu ML, Merlini L, Pepe G (2002) Novel COL6A1 splicing mutation in a family affected by mild Bethlem myopathy. Muscle Nerve 25:513–519 [DOI] [PubMed] [Google Scholar]
- Chu ML, Conway D, Pan TC, Baldwin C, Mann K, Deutzmann, Timpl R (1988) Amino acid sequence of the triple-helical domain of human collagen type VI. J Biol Chem 263:18601–18606 [PubMed] [Google Scholar]
- Chu ML, Pan TC, Conway D, Kuo HJ, Glanville RW, Timpl R, Mann K, Deutzmann R (1989) Sequence analysis of alpha 1(VI) and alpha 2(VI) chains of human type VI collagen reveals internal triplication of globular domains similar to the A domains of von Willebrand factor and two alpha 2(VI) chain variants that differ in the carboxy terminus. EMBO J 8:1939–1946 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chu ML, Zhang RZ, Pan TC, Stokes D, Conway D, Kuo HJ, Glanville R, Mayer U, Mann K, Deutzmann R (1990) Mosaic structure of globular domains in the human type VI collagen alpha 3 chain: similarity to von Willebrand factor, fibronectin, actin, salivary proteins and aprotinin type protease inhibitors. EMBO J 9:385–393 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Demir E, Sabatelli P, Allamand V, Ferreiro A, Moghadaszadeh B, Makrelouf M, Topaloglu H, Echenne B, Merlini L, Guicheney P (2002) Mutations in COL6A3 cause severe and mild phenotypes of Ullrich congenital muscular dystrophy. Am J Hum Genet 70:1446–1458 [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Paillette L, Aicardi J, Goutieres F (1989) Ullrich’s congenital atonic sclerotic muscular dystrophy: a case report. J Neurol 236:108–110 [DOI] [PubMed] [Google Scholar]
- Engel J, Prockop DJ (1991) The zipper-like folding of collagen triple helices and the effects of mutations that disrupt the zipper. Annu Rev Biophys Biophys Chem 20:137–152 [DOI] [PubMed] [Google Scholar]
- Furthmayr H, Wiedemann H, Timpl R, Odermatt E, Engel J (1983) Electron-microscopical approach to a structural model of intima collagen. Biochem J 211:303–311 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heiskanen M, Saitta B, Palotie A, Chu ML (1995) Head to tail organization of the human COL6A1 and COL6A2 genes by fiber-FISH. Genomics 29:801–803 [DOI] [PubMed] [Google Scholar]
- Higuchi I, Shiraishi T, Hashiguchi T, Suehara M, Niiyama T, Nakagawa M, Arimura K, Maruyama I, Osame M (2001) Frameshift mutation in the collagen VI gene causes Ullrich’s disease. Ann Neurol 50:261–265 [DOI] [PubMed] [Google Scholar]
- Ishikawa H, Sugie K, Murayama K, Ito M, Minami N, Nishino I, Nonaka I (2002) Ullrich disease: collagen VI deficiency: EM suggests a new basis for muscular weakness. Neurology 59:920–923 [DOI] [PubMed] [Google Scholar]
- Jöbsis GJ, Boers JM, Barth PG, de Visser M (1999) Bethlem myopathy: a slowly progressive congenital muscular dystrophy with contractures. Brain 122:649–655 [DOI] [PubMed] [Google Scholar]
- Jöbsis GJ, Keizers H, Vreijling JP, de Visser M, Speer MC, Wolterman RA, Baas F, Bolhuis PA (1996) Type VI collagen mutations in Bethlem myopathy, an autosomal dominant myopathy with contractures. Nat Genet 14:113–115 [DOI] [PubMed] [Google Scholar]
- Kennedy GC, German MS, Rutter WJ (1995) The minisatellite in the diabetes susceptibility locus IDDM2 regulates insulin transcription. Nat Genet 9:293–298 [DOI] [PubMed] [Google Scholar]
- Kuo HJ, Maslen CL, Keene DR, Glanville RW (1997) Type VI collagen anchors endothelial basement membranes by interacting with type IV collagen. J Biol Chem 272:26522–26529 [DOI] [PubMed] [Google Scholar]
- Lamandé SR, Morgelin M, Selan C, Jöbsis GJ, Baas F, Bateman JF (2002) Kinked collagen VI tetramers and reduced microfibril formation as a result of Bethlem myopathy and introduced triple helical glycine mutations. J Biol Chem 277:1949–1956 [DOI] [PubMed] [Google Scholar]
- Lamandé SR, Shields KA, Kornberg AJ, Shield LK, Bateman JF (1999) Bethlem myopathy and engineered collagen VI triple helical deletions prevent intracellular multimer assembly and protein secretion. J Biol Chem 274:21817–21822 [DOI] [PubMed] [Google Scholar]
- Lamandé SR, Sigalas E, Pan TC, Chu ML, Dziadek M, Timpl R, Bateman JF (1998) The role of the α3(VI) chain in collagen VI assembly: expression of an α3(VI) chain lacking N-terminal modules N10-N7 restores collagen VI assembly, secretion, and matrix deposition in an α3(VI)-deficient cell line. J Biol Chem 273:7423–7430 [DOI] [PubMed] [Google Scholar]
- Muntoni F, Bertini E, Bonnemann C, Brockington M, Brown S, Bushby K, Fiszman M, Korner C, Mercuri E, Merlini L, Hewitt J, Quijano-Roy S, Romero N, Squarzoni S, Sewry CA, Straub V, Topaloglu H, Haliloglu G, Voit T, Wewer U, Guicheney P (2002a) 98th ENMC International Workshop on Congenital Muscular Dystrophy (CMD), 7th Workshop of the International Consortium on CMD, 2nd Workshop of the MYO CLUSTER project GENRE. 26–28th October, 2001, Naarden, The Netherlands. Neuromuscul Disord 12:889–896 [DOI] [PubMed] [Google Scholar]
- Muntoni F, Brockington M, Blake DJ, Torelli S, Brown SC (2002b) Defective glycosylation in muscular dystrophy. Lancet 360:1419–1421 [DOI] [PubMed] [Google Scholar]
- Myllyharju J, Kivirikko KI (2001) Collagens and collagen-related diseases. Ann Med 33:7–21 [DOI] [PubMed] [Google Scholar]
- Nonaka I, Une Y, Ishihara T, Miyoshino S, Nakashima T, Sugita H (1981) A clinical and histological study of Ullrich’s disease (congenital atonic-sclerotic muscular dystrophy). Neuropediatrics 12:197–208 [DOI] [PubMed] [Google Scholar]
- Pan TC, Zhang RZ, Pericak-Vance MA, Tandan R, Fries T, Stajich JM, Viles K, Vance JM, Chu ML, Speer MC (1998) Missense mutation in a von Willebrand factor type A domain of the α3(VI) collagen gene (COL6A3) in a family with Bethlem myopathy. Hum Mol Genet 7:807–812 [DOI] [PubMed] [Google Scholar]
- Pepe G, Giusti B, Bertini E, Brunelli T, Saitta B, Comeglio P, Bolognese A, Merlini L, Federici G, Abbate R, Chu ML (1999) A heterozygous splice site mutation in COL6A1 leading to an in-frame deletion of the α1(VI) collagen chain in an Italian family affected by Bethlem myopathy. Biochem Biophys Res Commun 258:802–807 [DOI] [PubMed] [Google Scholar]
- Prockop DJ (1990) Mutations that alter the primary structure of type I collagen: the perils of a system for generating large structures by the principle of nucleated growth. J Biol Chem 265:15349–15352 [PubMed] [Google Scholar]
- Scacheri PC, Gillanders EM, Subramony SH, Vedanarayanan V, Crowe CA, Thakore N, Bingler M, Hoffman EP (2002) Novel mutations in collagen VI genes: expansion of the Bethlem myopathy phenotype. Neurology 58:593–602 [DOI] [PubMed] [Google Scholar]
- Specks U, Mayer U, Nischt R, Spissinger T, Mann K, Timpl R, Engel J, Chu ML (1992) Structure of recombinant N-terminal globule of type VI collagen alpha 3 chain and its binding to heparin and hyaluronan. EMBO J 11:4281–4290 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tillet E, Wiedemann H, Golbik R, Pan TC, Zhang RZ, Mann K, Chu ML, Timpl R (1994) Recombinant expression and structural and binding properties of alpha 1(VI) and alpha 2(VI) chains of human collagen type VI. Eur J Biochem 221:177–185 [DOI] [PubMed] [Google Scholar]
- Timpl R, Chu ML (1994) Microfibrillar collagen type VI. In: Yurchenco PD, Birk DE, Mecham RP (eds) Extracellular matrix assembly and structure. Academic Press, San Diego, pp 207–242 [Google Scholar]
- Timpl R, Engel J (1987) Type VI collagen. In: Mayne R, Burgeson R (eds) Structure and function of collagen types. Academic Press, Orlando, FL, pp 105–143 [Google Scholar]
- Tubridy N, Fontaine B, Eymard B (2001) Congenital myopathies and congenital muscular dystrophies. Curr Opin Neurol 14:575–582 [DOI] [PubMed] [Google Scholar]
- Turri MG, Cuin KA, Porter AC (1995) Characterisation of a novel minisatellite that provides multiple splice donor sites in an interferon-induced transcript. Nucleic Acids Res 23:1854–1861 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ullrich O (1930) Kongenitale, atonish-sklerotische Muskeldystrophie. Monatsschr Kinderheikd 47:502–510 [Google Scholar]
- Vergnaud G, Denoeud F (2000) Minisatellites: mutability and genome architecture. Genome Res 10:899–907 [DOI] [PubMed] [Google Scholar]
- Verpy E, Leibovici M, Zwaenepoel I, Liu XZ, Gal A, Salem N, Mansour A, Blanchard S, Kobayashi I, Keats BJ, Slim R, Petit C (2000) A defect in harmonin, a PDZ domain-containing protein expressed in the inner ear sensory hair cells, underlies Usher syndrome type 1C. Nat Genet 26:51–55 [DOI] [PubMed] [Google Scholar]
- Voit T (1998) Congenital muscular dystrophies: 1997 update. Brain Dev 20:65–74 [DOI] [PubMed] [Google Scholar]
- Weil D, Mattei MG, Passage E, N’Guyen VC, Pribula-Conway D, Mann K, Deutzmann R, Timpl R, Chu ML (1988) Cloning and chromosomal localization of human genes encoding the three chains of type VI collagen. Am J Hum Genet 42:435–445 [PMC free article] [PubMed] [Google Scholar]
- Weis MA, Wilkin DJ, Kim HJ, Wilcox WR, Lachman RS, Rimoin DL, Cohn DH, Eyre DR (1998) Structurally abnormal type II collagen in a severe form of Kniest dysplasia caused by an exon 24 skipping mutation. J Biol Chem 273:4761–4768 [DOI] [PubMed] [Google Scholar]
- Zhang RZ, Sabatelli P, Pan TC, Squarzoni S, Mattioli E, Bertini E, Pepe G, Chu ML (2002) Effects on collagen VI mRNA stability and microfibrillar assembly of three COL6A2 mutations in two families with Ullrich congenital muscular dystrophy. J Biol Chem 277:43557–43564 [DOI] [PubMed] [Google Scholar]