Abstract
Amyotrophic lateral sclerosis (ALS) is a fatal adult-onset disease in which motor neurons in the brain and spinal cord degenerate by largely unknown mechanisms. ALS is familial (FALS) in 10% of cases, and the inheritance is usually dominant, with variable penetrance. Mutations in copper/zinc super oxide dismutase (SOD1) are found in 20% of familial and 3% of sporadic ALS cases. Five families with ALS and frontotemporal dementia (ALS-FTD) are linked to 9q21, whereas one family with pure ALS is linked to 18q21. We identified two large European families with ALS without SOD1 mutations or linkage to known FALS loci and conducted a genomewide linkage screen using 400 microsatellite markers. In both families, two-point LOD scores >1 and a haplotype segregating with disease were demonstrated only across regions of chromosome 16. Subsequent fine mapping in family 1 gave a maximum two-point LOD score of 3.62 at D16S3137 and a three-point LOD score of 3.85 for markers D16S415 and D16S3137. Haplotype analysis revealed no recombination >∼30 cM, (flanking markers at D16S3075 and D16S3112). The maximum two-point LOD score for family 2 was 1.84 at D16S415, and the three-point LOD score was 2.10 for markers D16S419 and D16S415. Definite recombination occurred in several individuals, which narrowed the shared haplotype in affected individuals to a 10.1-cM region (flanking markers: D16S3396 and D16S3112). The region shared by both families on chromosome 16q12 corresponds to ∼4.5 Mb on the Marshfield map. Bioinformatic analysis of the region has identified 18 known genes and 70 predicted genes in this region, and sequencing of candidate genes has now begun.
Amyotrophic lateral sclerosis (ALS) is a greatly disabling and ultimately fatal neurodegenerative disease. Muscles become progressively atrophic and weak because of degeneration of motor neurons in the brain and spinal cord. Death due to respiratory failure occurs, on average, 3.5 years after symptom onset (Haverkamp et al. 1995; Shaw et al. 2001). The incidence of ALS is 1–2 per 100,000, but, because survival is relatively short, the prevalence is only ∼4–7 per 100,000 (Kondo 1995). In familial ALS (FALS), the vast majority of affected individuals have an autosomal dominant pattern of inheritance. FALS accounts for 5%–10% of cases, and they are clinically and pathologically indistinguishable from sporadic disease (Rouleau et al. 1996; Shaw et al. 1997).
Linkage to chromosome 21q22.11 was demonstrated in a subset of kindreds with FALS (Siddique et al. 1991), and mutations in SOD1 (ALS1 [MIM 105400]) were subsequently found in 20% of FALS and 3% of sporadic ALS cases (Rosen et al. 1993; Shaw et al. 1998). Over 100 SOD1 mutations have now been reported, and the vast majority are missense, which result in the substitution of a single amino acid (Shaw et al. 2001). Mice transgenic for mutant SOD1 develop motor neuron degeneration by a toxic gain of function, but whether this is due to altered metal ion handling, catalytic function, or protein misfolding and aggregation is still debated (Cleveland et al. 2001).
A recessive form of juvenile-onset primary lateral sclerosis (ALS2 [MIM 205100]) was linked to chromosome 2q33 (Hentati et al. 1994), and truncating mutations have been detected in ALS2/alsin (Hadano et al. 2001; Yang et al. 2001). No genes have yet been identified at the other FALS loci: 18q 21 in one family with dominant classical ALS (ALS3 [MIM 606640] (Hand et al. 2002), 9q34 in one family with a dominant nonfatal juvenile-onset ALS-like disorder (ALS4 [MIM 602433]) (Chance et al. 1998), 15q15-21 in kindreds with recessive juvenile onset (Hentati et al. 1998), and 9q21 in five families with ALS with frontotemporal dementia (ALS-FTD [MIM 105550]) (Hosler et al. 2000). Mutations in tau have been detected in families with frontotemporal dementia (FTD) and Parkinsonism (FTDP) linked to 17q21 (MIM 157140) (Hutton et al. 1998), but only rarely are family members affected by an ALS-like syndrome. The vast majority (80%) of kindreds with dominant FALS are unlinked.
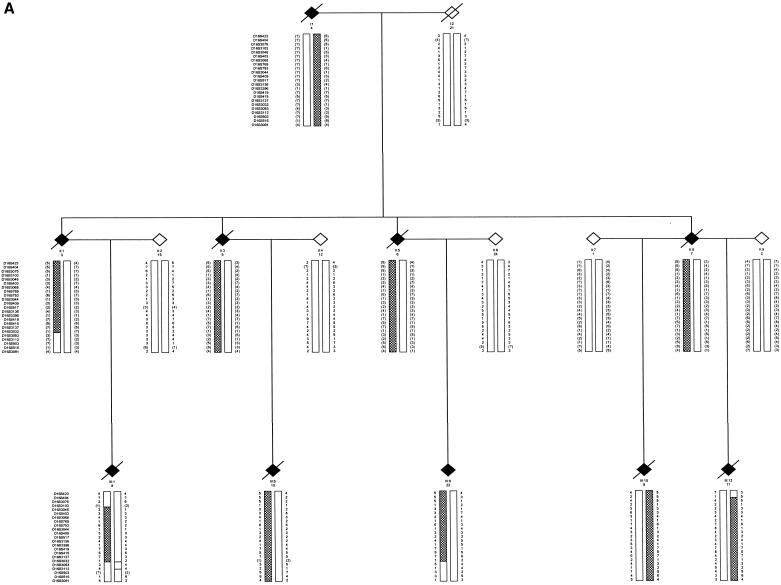
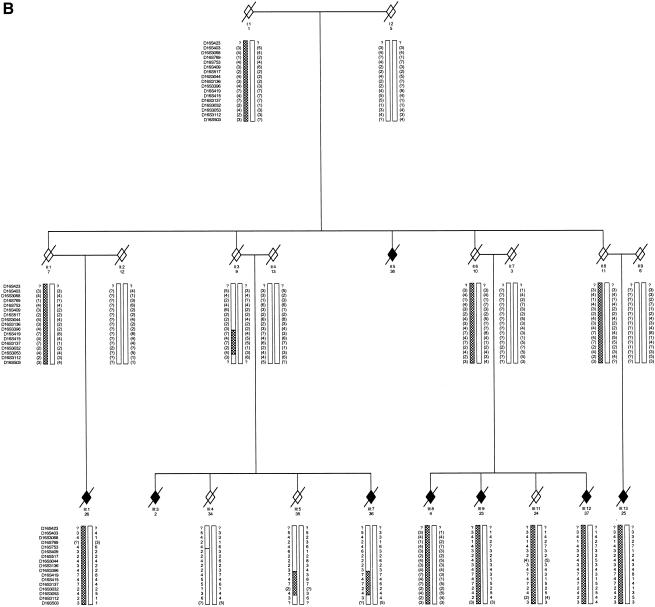
We identified two large unrelated European kindreds with ALS that were suitable for linkage. A consultant neurologist examined all individuals (F1: C.E.S. and P.N.L.; F2: V.D.J. and E.P.M.). ALS was confirmed, on detailed pathological assessment at post mortem examination, in two affected individuals from F1 and three from F2. Family F1 had 15 affected individuals, autosomal dominant inheritance, and a disease penetrance of 83% by age 70 years. All individuals had a limb onset of disease, mean age at onset was 38 years (range 29–51 years), and mean survival was 13 mo (range 7–27 mo). Family F2 had eight affected individuals but several obligate gene carriers, and disease penetrance was low (39%) by age 70 years. The F2 kindred was bigenerational, without consanguinity, and fitted best with an autosomal dominant pattern of inheritance. Four individuals had a bulbar onset of disease, and two developed hallucinations and cognitive impairment during their illness, suggestive of FTD. The mean age at onset was 61 years (range 40–72 years), and mean survival was 35.5 mo (range 12–90 mo).
Ethical committees in both clinical centers approved genetic research on ALS. Following receipt of informed consent, blood was drawn and DNA extracted from 48 members of F1 (5 affected) and 146 members of F2 (5 affected). By genotyping the spouses and offspring, we were able to recreate the genotypes and haplotypes of deceased affected individuals (5 in F1 and 1 in F2) (see fig. 1). No mutations in SOD1 were detected by sequencing two affected members from each family (as described in Shaw et al. [1998]). Tau mutations were excluded by direct sequencing in two members of F2 (as described in Rizzu et al. [1999]). In both families, linkage to reported ALS loci was confidently excluded for 18q21 (maximal two-point LOD scores of 0.33 for D18S846 [F1] and of 0.31 at D18S35 [F2]) and 9q21 (LOD scores of 0.32 at D9S1122 [F1] and of −0.8 at D9S922 [F2]).
Figure 1.
Pedigree drawing of families 1 and 2. Affected individuals are indicated by blackened diamonds. The informative marker alleles are shown. All inferred haplotypes reconstructed (in brackets) were done on the basis of minimal recombination. A hatched bar indicates the disease haplotype. Information is shown only for relevant individuals. The sex of all individuals has been disguised, and unaffected at-risk individuals have been omitted to preserve confidentiality.
Power calculations were made, by use of SLINK, for a single marker with six alleles (Ott 1989; UK Human Genome Mapping Project Resource Centre). The maximum achievable LOD score, for 19 members of F1, was 3.06 and, for 35 members of F2, was 2.04 (by use of 100 replications and identical assumptions as for FASTLINK; see below). A fluorescent dye-labeled medium-density microsatellite marker set (∼10-cM map, PRISM) was used for genotyping, and the PCR products were run out on ABI 377 and 3100 sequencers and analyzed by use of GENOTYPER software (ABI Biosystems). A consistent genotype was obtained in 95% of alleles, and inheritance was checked, by use of PEDCHECK (O’Connell and Weeks 1998; UK Human Genome Mapping Project Resource Centre), prior to linkage. FASTLINK (Cottingham et al. 1993; UK Human Genome Mapping Project Resource Centre) was used with the following assumptions: autosomal dominant inheritance with age-dependent penetrance, no phenocopies, a disease gene frequency of 0.00001, and equal marker allele frequencies. The age cutoff for liability classes was based on reports published elsewhere (Hand et al. 2002) and modified with penetrance calculated in each family: (1) age <46 years, F1=50%, F2=20%; (2) age >46 years, F1=85%, F2=40%; and (3) spouses were given the background population risk.
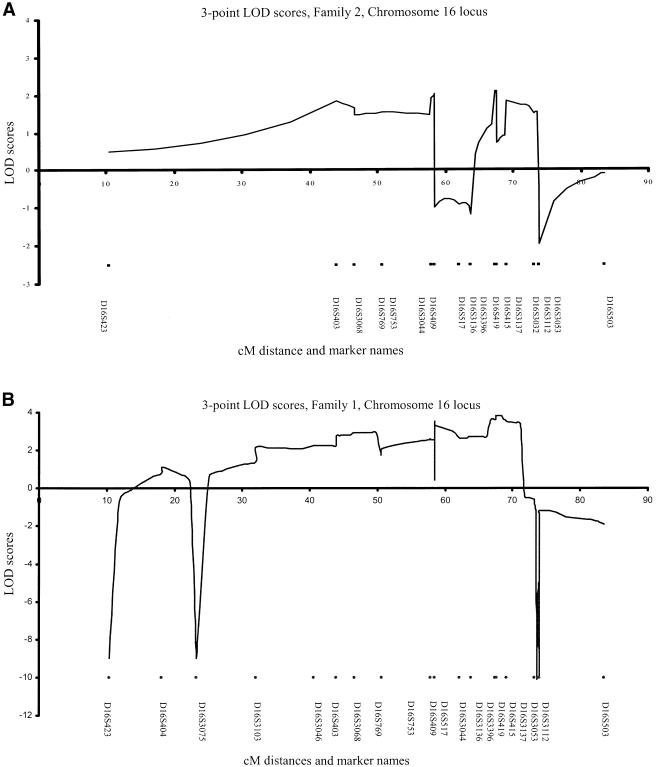
Given the very large number of individuals in both pedigrees, we chose an “affecteds-only” analysis. Genotyping of 385 markers was initially performed on 5 affected individuals from F1 (with an additional 12 unaffected spouses and offspring to recreate 5 further affected individuals) and 5 affected individuals from F2 (with an additional 3 unaffected offspring and 1 spouse and to recreate another affected individual). When alleles were not shared between affected individuals, markers were excluded from further analysis. When alleles were shared in three or more affected individuals in either family, markers were regenotyped (with additional neighboring markers) in the extended kindred (as for SLINK; see above) and tested for linkage by use of FASTLINK (data submitted for review but not shown). In F1, only two regions generated LOD scores >1.0: chromosome 16 (D16S415, LOD=3.5, and D16S3136, LOD=2.27) and chromosome 9 (D9S1677 LOD=1.68). The maximum three-point LOD score across chromosome 9 was only 0.251, and SIMWALK 2.82 (Weeks et al. 1995) produced no shared haplotype in the affected individuals. In F2, there were three regions with LOD scores ∼1.0: chromosome 16 (D16S415, LOD=1.84), chromosome 6 (D6S460, LOD=1.01), and chromosome 11 (D11S904, LOD=0.97), but the maximum three-point LOD scores were 0.18 and 0.23 for chromosomes 6 and 11, respectively; again, haplotype analysis excluded these regions.
The region on chromosome 16 was fine mapped with 24 additional markers in both families. Table 1 details the two-point LOD scores and shows six markers to be above 2 in F1, with a maximal two-point LOD score of 3.62 (θ=0) at D16S3137. Sine the pedigrees were too large for GENEHUNTER, FASTLINK was used to calculate three-point LOD scores, and a maximum of 3.85 was reached for markers D16S415 and D16S3137 (fig. 2). Haplotype analysis in F1 demonstrated recombinant events, with flanking markers D16S3075 and D16S3112 narrowing the region to ∼30 cM (fig. 1; table 1). In F2, the scores were more modest, but seven markers had LOD scores >1.0, with a maximum of 1.84 at D16S415 and 1.80 at D16S3137 (θ=0) (table 1). The maximum three-point LOD score for F2 was 2.10 for markers D16S419 and D16S415 (fig. 2). Empirical P values for the maximum two-point LOD scores at chromosome 16, for F1, are P<.0001 and, for F2, are P<.01, by use of SLINK (Ott 1989). Although positive LOD scores were recorded between D16S403 and D16S517 in F2, a consistent haplotype could not be created from these genotypes, thus excluding linkage to this region. A shared haplotype was observed in the affected individuals of F2, and recombinant events in the affected individual III:7 narrowed the conserved haplotype to a 10.1-cM region (flanking markers: D16S3396 and D16S3112) (fig. 1). This haplotype was also seen in one unaffected sibling and one offspring of III:7 (reviewed but not depicted, to maintain confidentiality).
Table 1.
Two-Point LOD Score Analysis[Note]
| LOD Score at θ= |
||||||
| Family andMarker | Marker Position | .0 | .1 | .2 | .3 | .4 |
| 1: | ||||||
| D16S423 | 10.36 | −∞ | −1.31 | −.55 | −.2 | −.05 |
| D16S404 | 18.07 | 1.06 | .78 | .49 | .24 | .06 |
| D16S3075 | 23.28 | −∞ | .87 | .91 | .71 | .39 |
| D16S3103 | 32.07 | 1.75 | 1.44 | 1.10 | .73 | .36 |
| D16S3046 | 40.65 | 2.48 | 2.15 | 1.73 | 1.22 | .65 |
| D16S403 | 43.89 | 1.12 | .98 | .76 | .50 | .23 |
| D16S3068 | 48.53 | 1.49 | 1.19 | .89 | .58 | .28 |
| D16S769 | 50.6 | 1.21 | 1.06 | .83 | .55 | .26 |
| D16S753 | 57.79 | 2.08 | 1.81 | 1.43 | .98 | .49 |
| D16S409 | 58.46 | .18 | .1 | .04 | .01 | −.01 |
| D16S517 | 58.46 | .41 | .3 | .19 | .10 | .03 |
| D16S3044 | 58.46 | 3.34 | 2.77 | 2.15 | 1.48 | .76 |
| D16S3136 | 62.11 | 2.27 | 1.95 | 1.55 | 1.09 | .57 |
| D16S3396 | 63.78 | 1.15 | .89 | .63 | .38 | .17 |
| D16S419 | 67.4 | 1.42 | 1.14 | .85 | .55 | .26 |
| D16S415 | 67.62 | 3.50 | 2.9 | 2.25 | 1.54 | .79 |
| D16S3137 | 69.05 | 3.62 | 3.01 | 2.34 | 1.61 | .83 |
| D16S3032 | 73.2 | .26 | .22 | .17 | .10 | .04 |
| D16S3053 | 73.91 | −.99 | 1.01 | .88 | .61 | .29 |
| D16S3112 | 73.92 | −2.32 | .11 | .20 | .16 | .08 |
| D16S503 | 83.55 | −1.75 | .35 | .36 | .25 | .11 |
| 2: | ||||||
| D16S423 | 10.36 | .09 | .05 | .02 | .00 | .00 |
| D16S403 | 43.89 | 1.81 | 1.44 | 1.03 | .60 | .20 |
| D16S3068 | 48.53 | 1.18 | .92 | .65 | .36 | .11 |
| D16S769 | 50.6 | .92 | .68 | .44 | .22 | .06 |
| D16S753 | 57.79 | .77 | .55 | .32 | .13 | .02 |
| D16S409 | 58.46 | 1.12 | .85 | .56 | .29 | .08 |
| D16S517 | 58.46 | .62 | .43 | .25 | .12 | .03 |
| D16S3044 | 58.46 | −2.12 | −.06 | .07 | .06 | .03 |
| D16S3136 | 62.11 | −1.14 | .44 | .41 | .25 | .08 |
| D16S3396 | 63.78 | −1.79 | .10 | .13 | .07 | .02 |
| D16S419 | 67.4 | 1.65 | 1.29 | .90 | .51 | .16 |
| D16S415 | 67.62 | 1.84 | 1.45 | 1.03 | .59 | .20 |
| D16S3137 | 69.05 | 1.80 | 1.37 | .93 | .50 | .15 |
| D16S3032 | 73.2 | .14 | .09 | .05 | .02 | .00 |
| D16S3053 | 73.91 | 1.16 | .88 | .57 | .29 | .07 |
| D16S3112 | 73.92 | −2.07 | −.31 | −.09 | −.02 | .00 |
| D16S503 | 83.55 | .27 | .16 | .08 | .03 | .01 |
Note.— Markers within the disease haplotype are in italics.
Figure 2.
Graphical representation of the three-point LOD scores for F1 and F2
The overlapping region shared between the two families corresponds to 10.14 cM on the Marshfield map and ∼4.5 Mb of sequenced genome. A systematic search of publicly available databases (Ensembl, Genome Database, NCBI, and UCSC) has identified 18 known and 70 putative genes in this region. Candidate genes, screened but, to date, without a mutation identified that segregates with disease, include: CAGF9, containing a polyglutamine tract of nine repeats; FTS, an ubiquitin-conjugating enzyme; CAPNS2, a cysteine protease subunit; and SALL1, a transcriptional regulator of higher chromatin order. We have begun screening the remaining genes for mutations, prioritized on the basis of their expression in the nervous system and potential role in the pathogenesis of ALS.
ALS is a rapidly fatal late-onset neurodegenerative disorder for which families suitable for linkage are very rare. Here, we report two unrelated kindreds that are linked to an overlapping region on chromosome 16q12-13. We are aware of two other unrelated families with ALS linked to regions of chromosome 16 that overlap this locus (R. H. Brown and J. de Belleroche, personal communication). This suggests that mutation of a gene in this region is a significant cause of FALS. The identification of another FALS gene will enable more families to undergo predictive and prenatal testing. It will also allow the development of transgenic and cellular models of ALS that will advance our understanding of disease mechanisms and help develop more effective therapies.
Acknowledgments
First and foremost, the authors thank the families who contributed to this project. This work was supported by clinical training fellowships from the Medical Research Council U.K. (to D.R.) and Wellcome Trust (to M.J.P.), Joint Research Committee of King’s College Hospital London (to B.N.S), Psychiatry Research Trust of the Institute of Psychiatry (to B.N.S. and C.V.), and the Motor Neurone Disease Association U.K.
Electronic-Database Information
Accession numbers and URLs for data presented herein are as follows:
- Center for Medical Genetics, Marshfield Medical Research Foundation, http://research.marshfieldclinic.org/genetics/ (for marker position)
- Ensembl, http://www.ensembl.org/ (for sequence and protein function information)
- Genome Database, http://www.gdb.org/ (for marker and gene position)
- National Center for Biotechnology Information (NCBI), http://www.ncbi.nlm.nih.gov/mapview/ (for marker and gene position, as well as sequence and protein function information)
- Online Mendelian Inheritance in Man (OMIM), http://www.ncbi.nlm.nih.gov/Omim/ (for ALS1, ALS2, ALS3, ALS4, ALS-FTD, FTDP, and background information on loci)
- UCSC Genome Bioinformatics, http://genome.ucsc.edu/ (for marker and gene position and sequence information)
- U.K. Human Genome Mapping Project Resource Centre, http://www.hgmp.mrc.ac.uk/Bioinformatics/ (for statistical analysis)
References
- Chance PF, Rabin BA, Ryan SG, Ding Y, Scavina M, Crain B, Griffin JW, Cornblath DR (1998) Linkage of the gene for an autosomal dominant form of juvenile amyotrophic lateral sclerosis to chromosome 9q34. Am J Hum Genet 62:633–640 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cleveland DW, Rothstein JD (2001) From Charcot to Lou Gehrig: deciphering selective motor neuron death in ALS. Nat Rev Neurosci 2:806–819 [DOI] [PubMed] [Google Scholar]
- Cottingham RW Jr, Idury RM, Schaffer AA (1993) Faster sequential genetic linkage computations. Am J Hum Genet 53:252–263 [PMC free article] [PubMed] [Google Scholar]
- Hadano S, Hand CK, Osuga H, Yanagisawa Y, Otomo A, Devon RS, Miyamoto N, Showguchi-Miyata J, Okada Y, Singaraja R, Figlewicz DA, Kwiatkowski T, Hosler BA, Sagie T, Skaug J, Nasir J, Brown RH Jr, Scherer SW, Rouleau GA, Hayden MR, Ikeda JE (2001) A gene encoding a putative GTPase regulator is mutated in familial amyotrophic lateral sclerosis 2. Nat Genet 29:166–173 [DOI] [PubMed] [Google Scholar]
- Hand CK, Khoris J, Salachas F, Gros-Louis F, Lopes AAS, Mayeux-Portas V, Brown RH Jr, Meininger V, Camu W, Rouleau GA (2002) A novel locus for familial amyotrophic lateral sclerosis, on chromosome 18q. Am J Hum Genet 70:251–256 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haverkamp LJ, Appel V, Appel SH (1995) Natural history of amyotrophic lateral sclerosis in a database population: validation of a scoring system and a model for survival prediction. Brain 118:707–719 [DOI] [PubMed] [Google Scholar]
- Hentati A, Bejaoui K, Pericak Vance MA, Hentati F, Speer MC, Hung WY, Figlewicz DA, Haines J, Rimmler J, Ben Hamida C, Ben Hamida M, Brown RH Jr, Siddique T (1994) Linkage of recessive familial amyotrophic lateral sclerosis to chromosome 2q33-q35. Nat Genet 7:425–428 [DOI] [PubMed] [Google Scholar]
- Hentati A, Ouahchi K, Pericak-Vance MA, Nijhawan D, Ahmad A, Yang Y, Rimmler J, Hung W, Schlotter B, Ahmed A, Ben Hamida M, Hentati F, Siddique T (1998) Linkage of a commoner form of recessive amyotrophic lateral sclerosis to chromosome 15q15-q22 markers. Neurogenetics 2:55–60 [DOI] [PubMed] [Google Scholar]
- Hosler BA, Siddique T, Sapp PC, Sailor W, Huang MC, Hossain A, Daube JR, Nance M, Fan C, Kaplan J, Hung WY, McKenna-Yasek D, Haines JL, Pericak-Vance MA, Horvitz HR, Brown RH Jr (2000) Linkage of familial amyotrophic lateral sclerosis with frontotemporal dementia to chromosome 9q21-q22. JAMA 284:1664–1669 [DOI] [PubMed] [Google Scholar]
- Hutton M, Lendon CL, Rizzu P, Baker M, Froelich S, Houlden H, Pickering-Brown S, et al (1998) Association of missense and 5′-splice-site mutations in tau with the inherited dementia FTDP-17. Nature 393:702–705 [DOI] [PubMed] [Google Scholar]
- Kondo K (1995) Epidemiology of motor neuron disease. In: Leigh PN, Swash M (eds) Motor neuron disease: biology and management, Springer Verlag, London, pp 19–33 [Google Scholar]
- O’Connell JR, Weeks DE (1998) PedCheck: a program for identification of genotype incompatibilities in linkage analysis. Am J Hum Genet 63:259–266 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ott J (1989) Computer-simulation methods in human linkage analysis. Proc Natl Acad Sci USA 86:4175–4178 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rizzu P, van Swieten JC, Joosse M, Hasegawa M, Stevens M, Tibben A, Niermeijer MF, Hillebrand M, Ravid R, Oostra BA, Goedert M, van Duijn CM, Heutink P (1999) High prevalence of mutations in the microtubule-associated protein tau in a population study of frontotemporal dementia in the Netherlands. Am J Hum Genet 64:414–421 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosen DR, Siddique T, Patterson D, Figlewicz DA, Sapp P, Hentati A, Donaldson D, et al (1993) Mutations in Cu/Zn superoxide dismutase gene are associated with familial amyotrophic lateral sclerosis. Nature 362:59–62 [DOI] [PubMed] [Google Scholar]
- Rouleau GA, Clark AW, Rooke K, Pramatarova A, Krizus A, Suchowersky O, Julien JP, Figlewicz D (1996) SOD1 mutation is associated with accumulation of neurofilaments in amyotrophic lateral sclerosis. Ann Neurol 39:128–131 [DOI] [PubMed] [Google Scholar]
- Shaw CE, Al Chalabi A, Leigh N (2001) Progress in the pathogenesis of amyotrophic lateral sclerosis. Curr Neurol Neurosci Rep 1:69–76 [DOI] [PubMed] [Google Scholar]
- Shaw CE, Enayat ZE, Chioza BA, Al-Chalabi A, Radunovic A, Powell JF, Leigh PN (1998) Mutations in all five exons of SOD-1 may cause ALS. Ann Neurol 43:390–394 [DOI] [PubMed] [Google Scholar]
- Shaw CE, Enayat ZE, Powell JF, Anderson VE, Radunovic A, al-Sarraj S, Leigh PN (1997) Familial amyotrophic lateral sclerosis: molecular pathology of a patient with a SOD1 mutation. Neurology 49:1612–1616 [DOI] [PubMed] [Google Scholar]
- Siddique T, Figlewicz DA, Pericak Vance MA, Haines JL, Rouleau G, Jeffers AJ, Sapp P, et al (1991) Linkage of a gene causing familial amyotrophic lateral sclerosis to chromosome 21 and evidence of genetic-locus heterogeneity. N Engl J Med 324:1381–1384 [DOI] [PubMed] [Google Scholar]
- Weeks DE, Sobel E, O’Connell JR, Lange K (1995) Computer programs for multilocus haplotyping of general pedigrees. Am J Hum Genet 56:1506–1507 [PMC free article] [PubMed] [Google Scholar]
- Yang Y, Hentati A, Deng HX, Dabbagh O, Sasaki T, Hirano M, Hung WY, Ouahchi K, Yan J, Azim AC, Cole N, Gascon G, Yagmour A, Ben Hamida M, Pericak-Vance M, Hentati F, Siddique T (2001) The gene encoding alsin, a protein with three guanine-nucleotide exchange factor domains, is mutated in a form of recessive amyotrophic lateral sclerosis. Nat Genet 29:160–165 [DOI] [PubMed] [Google Scholar]