Abstract
Amyotrophic lateral sclerosis (ALS) is a rapidly progressive, adult-onset motor neuron disease that arises as a dominantly inherited trait in ∼10% of ALS cases. Mutations in one gene, cytosolic Cu/Zn superoxide dismutase (SOD1), account for ∼25% of familial ALS (FALS) cases. We have performed a genetic linkage screen in 16 pedigrees with FALS with no evidence for mutations in the SOD1 gene and have identified novel ALS loci on chromosomes 16 and 20. The analysis of these genes will delineate pathways implicated as determinants of motor-neuron viability and provide insights into possible therapies for ALS.
Much effort has been directed toward identification of gene defects that underlie the 10% of ALS cases that are inherited as dominant traits. To date, only one ALS gene has been identified (Rosen et al. 1993). It is striking that forced expression in mice (Gurney et al. 1994; Ripps et al. 1995; Wong et al. 1995) and rats (Nagai et al. 2001; Howland et al. 2002) of ALS-associated mutant forms of this gene, “SOD1” (MIM 147450), produces a rodent motor-neuron disorder that recapitulates most features of ALS. Two additional motor neuron–disease genes have been described recently, although their mutations are not associated with the phenotype of typical ALS (Hadano et al. 2001; Yang et al. 2001 [MIM 205100]; Puls et al. 2003 [MIM 607376]). Additional ALS loci have been identified on chromosomes 18 (Hand et al. 2002 [MIM 606640]) and X (Siddique et al. 1998), and pedigrees with ALS–frontotemporal dementia (MIM 105550) have been genetically linked to chromosome 9 (Hosler et al. 2000).
In this study, we analyzed genetic linkage in 16 families in our Boston data set with multigenerational, autosomal dominant transmission of ALS and no mutations in the SOD1 gene. The families range in size from 11 to 67 individuals (table 1). All patients with ALS met El Escorial criteria for diagnosis (Brooks 1994). Informed consent and internal review board approval were received for this study. Genotype data were generated, by use of genomic DNA, for 43 patients with familial ALS (FALS) and 265 relatives, which allowed reconstruction of genotypes in 450 individuals. Fifty-one spouses provided allele frequency control subjects. Families were genotyped with 423 simple sequence-repeat polymorphisms, with an average spacing of 8 cM (maximum 14 cM). Fifty-five additional markers defined regions around neurological disease loci and refined linked haplotypes. Families not displaying male-to-male transmission of ALS were screened with 12 markers on chromosome X. Markers were selected from the Weber set (8, 8A, 10; Center for Medical Genetics) or the Genome Database. Mapping information was from National Center for Biotechnology Information (NCBI) and Whitehead Institute databases.
Table 1.
Composition of Family with FALS
| Family | TotalAffectedIndividuals | CollectedAffectedIndividuals | TotalCollectedIndividuals | SIMLINK Max (θ=0) | Mean Ageat Onset(years) | Mean DiseaseDuration(years) | Male-to-Male Transmission |
| 4 | 5 | 3 | 16 | 1.54 | 59.7 ± 2.3 | 3.8 ± 2.0 | Y |
| 13 | 4 | 2 | 18 | 1.50 | 54.5 ± 11.1 | 4.3 ± 2.5 | N |
| 20 | 10 | 4 | 19 | 2.56 | 54.3 ± 9.9 | 5.3 ± 3.4 | Y |
| 35 | 3 | 1 | 21 | 1.38 | 50.7 ± 2.3 | 4.7 ± 1.5 | N |
| 55 | 13 | 5 | 32 | 4.06 | 38.6 ± 13.6 | 2.0 ± 0.6 | Y |
| 64 | 4 | 1 | 8 | .66 | 55.0 ± 8.1 | 2.9 ± 1.3 | Y |
| 81 | 2 | 1 | 9 | 1.20 | 45.6 ± 16.3 | 7.5 ± 7.8 | N |
| 86 | 6 | 3 | 7 | .82 | 43.8 ± 5.0 | 3.0 ± .8 | Y |
| 101 | 2 | 1 | 10 | .43 | 36.0 ± 1.4 | 5.5 ± 3.5 | N |
| 124 | 4 | 2 | 17 | 1.42 | 71.0 ± 9.4 | 1.9 ± .6 | N |
| 159 | 9 | 5 | 25 | 2.26 | 45.7 ± 6.3 | 3.0 ± 1.9 | Y |
| 164 | 3 | 3 | 10 | .73 | 69.3 ± 4.0 | 3.2 ± 2.5 | N |
| 166 | 6 | 4 | 26 | 2.04 | 62.7 ± 7.4 | 2.3 ± .9 | Y |
| 171 | 4 | 4 | 11 | 1.01 | 48.8 ± 5.0 | 1.9 ± .6 | N |
| 388 | 4 | 2 | 25 | 1.88 | 56.5 ± 1.6 | 2.9 ± 1.6 | N |
| 391 | 9 |
2 |
23 |
.80 |
57.0 ± 6.1 | 15.0 ± 4.2 | Y |
| Total | 88 | 43 | 277 | 24.29 | 53.1 ± 9.6 | 4.2 ± 3.2 |
Forward primers contained a 19-bp 5′ extension of M13F sequence, which allowed incorporation of dye during PCR amplification. Products were separated on 6% acrylamide gels by use of LiCor 4200 (LiCor) automated sequencers, and images were analyzed with RFLPscan (Scanalytics). Genotype data were used to calculate two-point and multipoint logarithm-of-odds (LOD) scores by use of FASTLINK routines MLINK, ILINK, and LINKMAP (NCBI, version 4.1P) (Lathrop et al. 1984, 1986). These calculations assume 90% penetrance by age 70 years. Haplotypes were displayed in Cyrillic (Cherwell Scientific), arranged by hand, and compared with SIMWALK2 output (Sobel and Lange 1996).
SIMLINK analysis (Ploughman and Boehnke 1989) demonstrated that these 16 pedigrees should be appropriately powered to identify one or more ALS loci (table 1). Thus, for a marker directly on the locus (θ=0 cM), the total maximal LOD score was 24.28.
Initial analysis of genotype data from 423 markers in the 16 families identified several regions with two-point LOD scores >1.5. Of particular interest were regions in which contiguous markers produced positive scores within the same family. Two such regions were evident in a pattern that suggested genetic linkage to FALS. The first, identified in pedigree 55, spans 51.2 cM (37.8 Mb) from chromosome 16p12.1 to 16q21. A peak two-point LOD score of 2.65 at marker D16S753 was observed. We ran 14 additional markers in this region in all families, at an average density of 2 cM. These confirmed the LOD scores and defined a haplotype that comigrates with the disease in all affected members of pedigree 55 (table 2; fig. 1A). Analysis of these markers and the haplotype indicated that the putative ALS locus is flanked by crossovers at D16S764 and D16S3053 (fig. 1A). A multipoint LOD score was calculated for this family, with a peak value of 3.29 at D16S403 (table 3; fig. 1B). Further analysis revealed that the markers that define the minimum candidate region in family 55 also define haplotypes in six other pedigrees that segregate consistently with ALS; lower LOD scores and additional possible linked regions across the genome preclude the assignment of this locus as ALS-associated in these families.
Table 2.
Marker Order and Two-Point LOD Scores in Family 55 in Chromosome 16 Haplotype
| Marker | Location onChromosome 16 (cM) | Peak LOD | Distance of Peak LOD from Marker(cM) |
| D16S764 | 30 | .58 | 13.8 |
| D16S403 | 44 | 2.28 | 3.0 |
| D16S769 | 51 | 1.35 | 10.7 |
| D16S411 | 60 | 2.87 | 2.1 |
| D16S3253 | 72 | .80 | 11.9 |
| D16S3053 | 74 | .18 | 19.8 |
| D16S2620 | 81 | 2.03 | .1 |
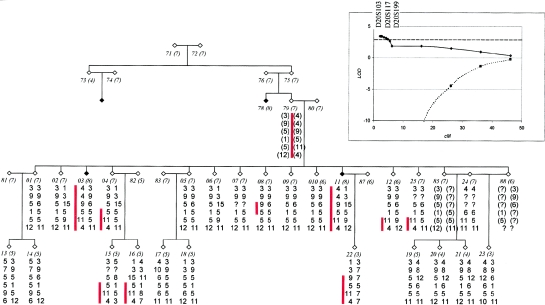
Figure 1.
Pedigree 55. Vertical bars designate the region affected individuals share between markers D16S764 and D16S3053. Alleles are displayed for the following markers (centromere to q-telomere): D16S764, D16S403, D16S769, D16S411, D16S3253, D16S3053, and D16S2620. Penetrance is displayed as class and is based on age. There are six age classes at 60 years or younger. Class 7 corresponds to age 61 or older. Class 8 denotes individuals with ALS. The numbers in italics denote individual number and (in parentheses) the class number. Inset, Multipoint LOD scores for family 55 (solid lines) and for all 16 families studied (dotted lines). Dashed line, LOD 3.0. Calculations were at five intervals between markers D16S403, D16S753, and D16S415, by use of the LINKMAP utility in FASTLINK. In figure 1 and figure 2, affected individuals are denoted by blackened symbols; unaffected individuals are denoted by unblackened symbols. Males and females are both represented by diamonds to preserve anonymity. Genotypes in parentheses are inferred.
Table 3.
Multipoint LOD Scores in Family 55 on Chromosome 16
|
Multipoint LOD for |
||
| Locus (cM) [Marker] | Family 55 | All Families |
| 34.0 | 3.12 | −10.34 |
| 44.0 [D16S403] | 3.29 | −59.44 |
| 52.3 | 3.09 | −25.74 |
| 57.9 [D16S753] | 2.53 | −43.25 |
| 64.9 | 2.90 | −20.66 |
| 69.6 [D16S415] | 2.96 | −42.24 |
| 79.0 | 2.89 | −6.42 |
After identifying this linkage to chromosome 16 with these data, we subsequently investigated unlinked families from an independent FALS data set (Northwestern University [NWU]) for linkage to chromosome 16. We identified a single family (NWU 9968) that generated a peak multipoint LOD score of 1.47 at D16S3396 (64 cM). This family had a mean age at onset of 67 years and a mean disease duration of 2.1 years. There were eight affected family members, five of whom were sampled. DNA was available on 24 family members. The maximal expected LOD score for NWU 9968 was 2.25.
In the Boston family set, a second potential ALS-linked region was identified in family 388. In this pedigree, two affected individuals of 15 siblings share a unique 5-Mb segment at chromosome 20ptel (fig. 2A and 2B). Two contiguous markers (D20S103 and D20S117) produce two-point LOD scores >3.0, whereas two additional contiguous markers yield scores >1.5. These were the highest LOD scores produced in this family in the entire genome scan of 423 markers. Haplotype analysis revealed a pattern of alleles that is coinherited with the disease in this family. A sibling who was unaffected at age 70 years shared alleles below D20S199 with the two affected individuals, thus potentially implicating the region above this marker as ALS-associated. Given these initial positive scores, we analyzed nine additional markers in this region in all families, at an average marker density of 1.5 cM. The additional markers further elucidate the haplotype associated with the disease in the two affected individuals (table 4; fig. 2A). The ALS-associated haplotype spans the region from the telomere of the p short arm to D20S199, a distance of ∼6.25 cM (1 Mb). No other families are genetically linked to this locus by LOD scores or a shared haplotype. A multipoint LOD score was calculated for this family, with a peak value of 3.46 at D20S103 (table 5; fig. 2B).
Figure 2.
Pedigree 388. Vertical bars designate a region shared by the affected individuals, from the telomere of chromosome 20p to marker D20S199; one unaffected individual shares the distal-most markers. Alleles are displayed for the following markers (from p-telomere to centromere): D20S103, D20S117, D20S199, D20S473, D20S889, and D20S95. Numbers in italics are defined in figure 1. Inset, Multipoint LOD scores for family 388 (solid lines) and for all 16 families studied (dotted lines). Dashed line, LOD=3.0. Calculations were at 5 intervals between markers D20S103, D20S117, and D20S199, by use of the LINKMAP utility in FASTLINK.
Table 4.
Marker Order and Two-Point LOD Scores in Family 388 in Chromosome 20
| Marker | Location onChromosome 20(cM) | Peak LOD | Distance of Peak LOD from Marker(cM) |
| D20S103 | 2 | 3.39 | .1 |
| D20S117 | 3 | 3.17 | .1 |
| D20S199 | 6 | 1.70 | 4.6 |
| D20S473 | 10 | 1.22 | 11.8 |
| D20S889 | 11 | .64 | 19.6 |
| D20S95 | 17 | 1.12 | 12.2 |
Table 5.
Multipoint LOD Scores in Family 388 on Chromosome 20
|
Multipoint LOD for |
||
| Locus (cM) [Marker] | Family 388 | All Families |
| 2.1 [D20S103] | 3.46 | −21.33 |
| 2.5 | 3.46 | −13.41 |
| 2.8 [D20S117] | 3.46 | −9.12 |
| 4.8 | 2.95 | −39.51 |
| 6.2 [D20S199] | 1.87 | −29.51 |
| 16.2 | 1.86 | −40.39 |
From a genome scan of 16 familial pedigrees with ALS without evidence of SOD1 gene mutations, we have identified two novel ALS gene loci on chromosomes 16 and 20. In our pedigree 55, the linkage to chromosome 16 seems valid for three reasons. First, the markers producing the highest LOD scores in this genomewide scan of 423 polymorphic markers were all located within the linked region; no similar evidence of linkage was demonstrated for markers at other loci. Second, the linked markers define a clear haplotype, delineating a minimal region spanning 51.2 cM (37.8 Mb) of chromosome 16 that is coinherited with the disease in all affected individuals. Third, we are now aware that two other investigators each have identified two families linked to this region (C. Shaw and J. de Belleroche, personal communication). In our pedigree 388, the linkage to chromosome 20 seems probable though somewhat less secure. It is true that, for this family, the best LOD scores were produced by the markers on the distal short arm of chromosome 20; no analogous scores were seen among the 415 other markers tested across the genome. However, we do not have evidence of similar linkage in any other family. Moreover, the linkage rests on genotypes of only two affected individuals, both in the same generation. Efforts are under way to collect additional members of this pedigree.
Our understanding of the pathogenesis of ALS will be enhanced by knowledge of the molecular biological properties of new ALS genes. In the 10 years since its identification as the first ALS gene, studies of SOD1 have provided valuable insights into aspects of the biology of cell death in ALS, although numerous fundamental points of controversy remain in understanding the neurotoxicity of mutant SOD1 protein.
For this reason, the identity of the putative ALS genes on chromosomes 16 and 20 will be of considerable interest. The haplotype we have defined for chromosome 16 spans 51 cM (38 Mb). At present, the number of genes in this region ranges between 322 and 380, according to Ensembl database and NCBI genome browsers. The haplotype on chromosome 20 spans 6 cM (1 Mb), a region that encompasses 20–24 genes, as defined by the Ensembl and NCBI databases. That we identified ALS loci in only 2 of 16 families with ALS indicates that there is heterogeneity in the genetic causes of ALS. It seems likely that many genes will be implicated in the pathogenesis of this disease.
Acknowledgments
We are indebted to the patients and their families for their participation in this research. This research was supported by the Angel Fund for ALS Research, Project ALS, the Muscular Dystrophy Association (to D.M.Y., R.H.B., and J.L.H.), the Amyotrophic Lateral Sclerosis Association (to R.H.B.), the Pierre de Bourgknecht ALS Research Foundation (to R.H.B.), National Institutes of Health grants 1PO1NS31248-02 (to R.H.B.) and RO1NS37912 (to R.H.B.), and the Howard Hughes Medical Institute (to H.R.H.). H.R.H. is an Investigator of the Howard Hughes Medical Institute.
Electronic-Database Information
Accession numbers and URLs for data presented herein are as follows:
- Center for Medical Genetics, Marshfield, http://research.marshfieldclinic.org/genetics/
- Ensembl, http://www.ensembl.org/Homo_sapiens/
- FASTLINK version 4.1P, http://www.ncbi.nlm.nih.gov/CBBresearch/Schaffer/fastlink.html
- Genome Database, http://www.gdb.org/
- National Center for Biotechnology Information (NCBI), http://www.ncbi.nlm.nih.gov/mapview/ (for NCBI map viewer)
- National Center for Biotechnology Information (NCBI), http://www.ncbi.nlm.nih.gov/genome/guide/human/ (for NCBI genes)
- Online Mendelian Inheritance in Man (OMIM), http://www.ncbi.nlm.nih.gov/Omim/ (for ALS, ALS–frontotemporal dementia, and SOD1)
- SIMLINK version 4.11, http://www.sph.umich.edu/statgen/boehnke/simlink.html
- SIMWALK version 2.82, http://watson.hgen.pitt.edu/register/
- Whitehead Institute, http://www-genome.wi.mit.edu/resources.html
References
- Brooks B (1994) El Escorial world federation of neurology criteria for the diagnosis of amyotrophic lateral sclerosis. J Neurol Sci 124:96–107 [DOI] [PubMed] [Google Scholar]
- Gurney ME, Pu H, Chiu AY, Dal Canto MC, Polchow CY, Alexander DD, Caliendo J, Hentati A, Kwon YW, Deng H-X, Chen W, Zhai P, Sufit RL, Siddique T (1994) Motor neuron degeneration in mice that express a human Cu,Zn superoxide dismutase mutation. Science 264:1772–1775 [DOI] [PubMed] [Google Scholar]
- Hadano S, Hand CK, Osuga H, Yanagisawa Y, Otomo A, Devon RS, Miyamoto N, Showguchi-Miyata J, Okada Y, Singaraja R, Figlewicz DA, Kwiatkowski T, Hosler BA, Sagie T, Skaug J, Nasir J, Brown RH Jr, Scherer SW, Rouleau GA, Hayden MR, Ikeda JE (2001) A gene encoding a putative GTPase regulator is mutated in familial amyotrophic lateral sclerosis 2. Nat Genet 29:166–173 [DOI] [PubMed] [Google Scholar]
- Hand CK, Khoris J, Salachas F, Gros-Louis F, Lopes AAS, Mayeux-Portas V, Brown RH Jr, Meininger V, Camu W, Rouleau GA (2002) A novel locus for familial amyotrophic lateral sclerosis, on chromosome 18q. Am J Hum Genet 70:251–256 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hosler BA, Siddique T, Sapp PC, Sailor W, Huang MC, Hossain A, Daube JR, Nance M, Fan C, Kaplan J, Hung WY, McKenna-Yasek D, Haines JL, Pericak-Vance MA, Horvitz HR, Brown RH Jr (2000) Linkage of familial amyotrophic lateral sclerosis with frontotemporal dementia to chromosome 9q21-q22. JAMA 284:1664–1669 [DOI] [PubMed] [Google Scholar]
- Howland DS, Liu J, She Y, Goad B, Maragakis NJ, Kim B, Erickson J, Kulik J, DeVito L, Psaltis G, DeGennaro LJ, Cleveland DW, Rothstein JD (2002) Focal loss of the glutamate transporter EAAT2 in a transgenic rat model of SOD1 mutant-mediated amyotrophic lateral sclerosis (ALS). Proc Natl Acad Sci USA 99:1604–1609 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lathrop GM, Lalouel JM, Julier C, Ott J (1984) Strategies for multilocus linkage analysis in humans. Proc Natl Acad Sci USA 81:3443–3446 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lathrop GM, Lalouel JM, White RL (1986) Construction of human linkage maps: likelihood calculations for multilocus linkage analysis. Genet Epidemiol 3:39–52 [DOI] [PubMed] [Google Scholar]
- Nagai M, Aoki M, Miyoshi I, Kato M, Pasinelli P, Kasai N, Brown RH Jr, Itoyama Y (2001) Rats expressing human cytosolic copper-zinc superoxide dismutase transgenes with amyotrophic lateral sclerosis: associated mutations develop motor neuron disease. J Neurosci 21:9246–9254 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ploughman LM, Boehnke M (1989) Estimating the power of a proposed linkage study for a complex genetic trait. Am J Hum Genet 44:543–551 [PMC free article] [PubMed] [Google Scholar]
- Puls I, Jonnakuty C, LaMonte BH, Holzbaur EL, Tokito M, Mann E, Floeter MK, Bidus K, Drayna D, Oh SJ, Brown RH Jr, Ludlow CL, Fischbeck KH (2003) Mutant dynactin in motor neuron disease. Nat Genet 33:455–456 [DOI] [PubMed] [Google Scholar]
- Ripps ME, Huntley GW, Hof PR, Morrison JH, Gordon JW (1995) Transgenic mice expressing an altered murine superoxide dismutase gene provide an animal model of amyotrophic lateral sclerosis. Proc Natl Acad Sci USA 92:689–693 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosen DR, Siddique T, Patterson D, Figlewicz DA, Sapp P, Hentati A, Donaldson D, et al (1993) Mutations in Cu/Zn superoxide dismutase gene are associated with familial amyotrophic lateral sclerosis. Nature 362:59–62 [DOI] [PubMed] [Google Scholar]
- Siddique T, Hong S-T, Brooks BR, Hung WY, Siddique NA, Rimmler J, Kaplan JP, Haines JL, Brown RH Jr, Pericak-Vance MA (1998) X-linked dominant locus for late-onset familial amyotrophic lateral sclerosis. Am J Hum Genet A308 [Google Scholar]
- Sobel E, Lange K (1996) Descent graphs in pedigree analysis: applications to haplotyping, location scores, and marker-sharing statistics. Am J Hum Genet 58:1323–1337 [PMC free article] [PubMed] [Google Scholar]
- Wong P, Pardo C, Borchelt D, Lee M, Copeland N, Jenkins N, Sisodia S, Cleveland D, Price D (1995) An adverse property of a familial ALS-linked SOD1 mutation causes motor neuron disease characterized by vacuolar degeneration of mitochondria. Neuron 14:1105–1116 [DOI] [PubMed] [Google Scholar]
- Yang Y, Hentati A, Deng HX, Dabbagh O, Sasaki T, Hirano M, Hung WY, Ouahchi K, Yan J, Azim AC, Cole N, Gascon G, Yagmour A, Ben-Hamida M, Pericak-Vance M, Hentati F, Siddique T (2001) The gene encoding alsin, a protein with three guanine-nucleotide exchange factor domains, is mutated in a form of recessive amyotrophic lateral sclerosis. Nat Genet 29:160–165 [DOI] [PubMed] [Google Scholar]