Abstract
Hypertensive nephropathy (HN) and focal segmental glomerulosclerosis (FSGS) are significant causes of end-stage renal disease (ESRD), but no genes or loci have been associated with this phenotype among African Americans, a group at high risk. We performed a genomewide linkage scan with ∼400 microsatellite markers on 23 individuals of a large four-generation African American family with 18 affected individuals (7 with ESRD), in which the 13-year-old proband (also with ESRD) presented with hypertension and proteinuria (2–4 g/day) and underwent a kidney biopsy that revealed FSGS-like lesions with arteriolar thickening. A genomewide scan revealed LOD scores of >2.5 for markers on chromosomes 3 and 9, and fine mapping was performed on 5 additional members (total 28 members) that showed a maximum multipoint LOD score of 5.4 in the 9q31-q32 region, under an autosomal dominant model with 99% penetrance. This 8-cM (6-Mb) region is flanked by markers D9S172 and D9S105, and further candidate gene sequencing studies excluded the coding regions of three genes (ACTL7A, ACTL7B, and CTNNAL1). To our knowledge, this is the first report of a locus, denoted as “HNP1,” for the HN/FSGS phenotype in a large African American family with dominantly inherited nephropathy characterized by ESRD, hypertension, and some features of FSGS.
Hypertension is a major public health problem in the Western world and may result from a variety of renal disorders (Caetano et al. 2001; Adamczak et al. 2002; Torpy et al. 2002; Hayden et al. 2003). Although the number of hypertensive patients in the United States has doubled since 1980, the frequency of end-stage renal disease (ESRD) has been increasing geometrically (Caetano et al. 2001). Previous epidemiological studies found that only a few hypertensive patients develop renal injury; however, more recent clinical observations have revealed that the relative risk of developing ESRD is increased by as much as 20-fold in hypertensive patients (Klag et al. 1996; Moore et al. 1999; Agodoa et al. 2001). A genetic predisposition to develop both hypertension and hypertensive ESRD is suggested by several observations. Family studies have shown that hypertension is more frequent in first-degree relatives of patients with primary glomerulopathies or diabetic nephropathy than in the general population (Adamczak et al. 2002). Also, several studies have shown that African Americans are more likely than their white counterparts to develop hypertension-related ESRD (Marcantoni et al. 2002; Hayden et al. 2003; Kaperonis and Bakris 2003). Hypertension-related ESRD, or hypertensive nephropathy (HN), is a distinct histologic entity; however, in clinical practice, a kidney biopsy is not always performed when an affected individual presents with long-standing hypertension and renal failure. Clinically diagnosed HN can thus be a heterogeneous entity and may include several distinct disorders, including focal segmental glomerulosclerosis (FSGS).
Etiological factors responsible for HN and FSGS are largely unknown, but there appears to be a strong genetic risk for both conditions. Recently, several genomic loci (i.e., 2p24-p25, 15q, 17q21, 12p13, 4p16, and 3q21) have been reported to be linked to essential hypertension, but they are not typically associated with renal failure or ESRD (Xu et al. 1999a, 1999b; Lifton et al. 2001; Angius et al. 2002). Indeed, evidence in rodent models suggests that the predisposition to nephropathy and hypertension may be under separate genetic control (Brown et al. 1996; Churchill et al. 1997). Recent molecular genetic studies of FSGS, a progressive and potentially inherited nephropathy, have identified several genes on chromosomes 19q13 (i.e., nephrin [MIM 602716], α actinin 4 [MIM 60438]), and 1q25 (podocin [MIM 604766]), as well as a locus on 11q21 (FSGS2 [MIM 603965]) (Lenkkeri et al. 1999; Winn et al 1999; Boute et al. 2000; Kaplan et al. 2000; Winn 2002). Hypertension is generally a key feature in these genetic causes of renal failure, which are characterized by either significant proteinuria or full-blown nephrotic syndrome. Although no definitive genomic loci have been described for HN, a locus for a progressive nephropathy with hypertension was recently reported on 1q21 in a large multigenerational Israeli family of Iraqi Jewish origin (RFH1 [MIM 161900]) (Cohn et al. 2000). It is quite likely that several genes are responsible for the phenotypes that resemble HN or FSGS.
Here, we describe a large, four-generation African American family (CHP 109), residing in western Pennsylvania (fig. 1), with an autosomal dominant nephropathy. The disorder was clinically diagnosed as HN in multiple family members, and one member demonstrated features of FSGS on renal biopsy. We performed a genomewide linkage scan on 23 members, to identify the putative genetic locus associated with HN in this family. To our knowledge, this is the first locus to be identified for the HN/FSGS phenotype in African Americans, a group that carries a high risk of HN.
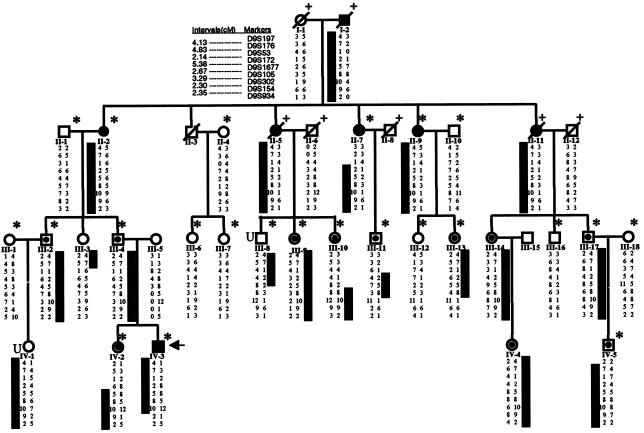
Figure 1.
Pedigree of family CHP 109. Squares indicate males, and circles indicate females. Slashes indicate deceased individuals. Blackened symbols indicate affected individuals; unblackened symbols indicate unaffected individuals. Symbols that contain small blackened circles indicate that the individual had hypertension alone (i.e., urinalysis revealed no abnormal results). Asterisk (*) indicates individual whose DNA was used for genomewide scan. Plus symbol (+) indicates that alleles for the individual were inferred. “U” indicates that disease status of individual was unknown.
Evaluation of family members included a complete history, urinalyses, and hematological and biochemical investigations; in addition, renal histological studies were performed in the proband. Since the majority of the family members in this pedigree presented with hypertension in the 4th or 5th decade of life, they were classified as affected on the basis of the following criteria: (a) if, at any age, they had ESRD requiring dialysis or renal transplantation or if they had elevated blood urea nitrogen (BUN) and serum creatinine levels (more than twice as high as normal for their age); (b) if they had hematuria (>10 red blood cells [RBC] per high-power field [HPF]) and/or >2+ protein in urine, as shown by qualitative urinalysis, in the absence of other systemic diseases that are likely to lead to proteinuria or hematuria; or (c) if, by the age of 40 years, they had documented hypertension, with or without urinary or renal functional abnormalities, that required antihypertensive therapy. They were classified as unaffected if they were >40 years of age and were normotensive and had no detectable hematuria or proteinuria on qualitative urinalysis or if they were unrelated married-in spouses. Disease status was categorized as unknown if no clinical information was available or if the individual was ⩽40 years of age and had no evidence of high blood pressure or anomalies of urinary or renal function.
The proband (individual IV-3 [indicated by an arrow in fig. 1]), a 13-year-old boy, presented to the pediatric renal clinics at the Children’s Hospital of Pittsburgh, with hypertension (systemic blood pressure range 150–170/80–100 mmHg) and mild renal insufficiency (serum creatinine 1.6 mg/dl; BUN 22 mg/dl). Investigations revealed 3–4+ nonorthostatic proteinuria and hematuria (20–40 RBC/HPF), hemoglobin 10 g/dl, white blood cell count 7,800/mm3, and normal serum complements and lupus serology. Kidney biopsy showed a range of glomerular changes (fig. 2). Of the 17 glomeruli seen, 2 were morphologically normal, and the rest had focal and segmental sclerosing features or were globally sclerotic. Collapsed lobules in some of the segmentally affected glomeruli showed a hyaline appearance that did not extend to afferent arterioles and imparted a slightly nodular-sclerotic appearance. Amyloid stains in the hyalinized segments were negative. Residual capillary membranes in the unaffected segments were of normal thickness. Mesangial cellularity and matrix were not increased. There was patchy tubular atrophy and interstitial fibrosis without an inflammatory component. Immunostains revealed moderate deposits of C3 in most glomeruli in a diffuse but coarsely granular pattern that also extended to afferent arterioles, and the deposits were graded as 2–4+. The muscular walls of the small arteries and arterioles were prominent, and immunostaining for smooth muscle actin showed them to be thicker than arterioles seen in kidney biopsy specimens of other individuals with nephrotic syndrome or FSGS. Deposits of IgM were present only in the collapsed segments and in the mesangium of sclerosing lobules, which correlated with moderately dense deposits within the basement membranes seen on electron microscopy (EM). The electron dense deposits are generally not seen in typical FSGS. The tissue examined by EM, however, had only sclerosing and collapsed glomeruli, and the ultrastructural details of the more preserved glomeruli could not be evaluated. After the initial presentation, this patient rapidly progressed to ESRD within 1 year and was normoglycemic throughout the period prior to cadaveric renal transplantation at 14 years of age. The posttransplant course was also complicated by severe hypertension, and the patient had an abrupt loss of allograft function ∼2 years post transplantation.
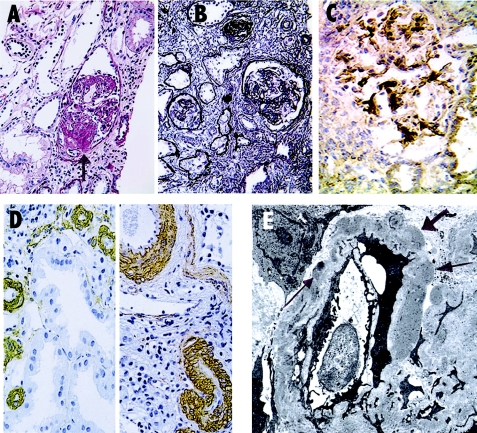
Figure 2.
Kidney biopsy findings in CHP family 109. A, PAS stain shows a glomerulus with sclerosing lesions in a number of lobules, imparting a “nodular” appearance in collapsed segment (arrow). B, PAS-M stain (Jones silver methenamine) shows the range of glomerular lesions from globally sclerotic to partial glomerular preservation. Interstitial fibrosis is present with entrapped atrophic tubules. C, Immunoperoxidase-DAB stain for the presence of IgM reveals mesangial/perimesangial deposition. D, Two views of smooth muscle actin stains showing prominent and thickened muscular walls of small arteries and arterioles. E, Tissue for electron microscopy consisted of only partly or fully sclerosed glomeruli. A partially preserved segment of a sclerosing glomerulus is shown with moderately dense deposits (arrows) in basement membrane in a distribution similar to that of the IgM in panel C.
There were 18 affected individuals in the family, as defined by the presence of hypertension, renal failure, and abnormal urinalysis results. Besides the proband, six additional family members (I-2, II-2, II-5, II-7, II-9, and II-11) had ESRD that typically developed in the 5th or 6th decade of life after 20–30 years of hypertension. Hypertensive renal failure was diagnosed in individual I-2 when he was in his 50s, 10 years prior to his death while receiving hemodialysis (HD). II-2 was diagnosed clinically to have hypertensive nephropathy and first received HD in her early 70s, after several decades of being hypertensive. II-5 was also diagnosed with HN and died while on HD at 72 years of age. II-11 died in her 80s, after undergoing dialysis for almost 4 years, with a diagnosis of HN. II-7 is currently receiving HD, and II-9 is hypertensive with renal failure. Other affected individuals in generations III and IV (except GPID IV-2) were found to be hypertensive prior to age of 40 years and generally required multiple agents to control blood pressure. No abnormalities were detected in the urine of these individuals (except IV-4 and IV-5, for whom results of urinalysis were not available). GPID IV-2 had 2+ protein in urine and 1–2+ hematuria, and urinalysis showed white blood cell and mixed cellular casts. In general, none of the affected individuals (except III-17) were obese, and, except in the proband, the diagnosis of HN was made on clinical grounds rather than kidney biopsy.
The maximum attainable LOD score for this pedigree was estimated by the SLINK program (Weeks et al. 1990), under the assumption of a four-allele system with equal frequencies, a 5% recombination with the disease locus, and a disease allele frequency of 0.0001. A set of 1,000 iterations was generated, and maximum attainable LOD score was calculated using the MSIM program for various θ. These studies showed that family CHP 109 was capable of generating a maximum two-point LOD score of 4.8, under an autosomal dominant model with a penetrance of 99%. We then obtained genomic DNA from EDTA-anticoagulated whole blood, by use of the salting-out procedure, for the genomewide scan (Miller et al. 1988; Vats et al. 2000). These studies were performed after obtaining informed consent and were approved by the human rights committee of the Children’s Hospital of Pittsburgh. A genomewide linkage scan was performed on 23 individuals, and 5 additional family members (total of 28 individuals) underwent fine mapping. Fluorescent genotyping was performed by mammalian genotyping service facilities at Marshfield Clinics, using ∼400 microsatellite markers of screening set 10, which provided an average spacing of 9 cM across the genome. The PCR for fine mapping and candidate gene sequencing was performed by standard methods, and PCR products were analyzed on an ABI 3700 capillary analyzer (Applied Biosystems) at the Center for Human Genetics and Integrative Biology of the University of Pittsburgh. The genotyping data were analyzed using GENOTYPER software (version 3.7; Applera) and were entered in a PROGENY2000 database management system (Progeny Software). The sequencing reactions for candidate genes were performed using the BigDye technique, and data were analyzed using Sequencher software (Gene Codes).
The genotyping data were assessed by use of the PedCheck program, to identify and correct Mendelian errors (O’Connell and Weeks 1998). The linkage was calculated under a dominant model with a penetrance of 0.99, without phenocopy, and under the assumption of a disease allele frequency of 0.0001, using the GENEHUNTER and VITESSE statistical programs for each marker in the genomewide scan (O’Connell and Weeks 1995; Kruglyak et al. 1996). Marker allele frequencies were estimated from the unrelated spouses in the family. Two-point and multipoint LOD scores (Z scores), and Z-1 CIs were calculated for fine mapping data, using the VITESSE algorithm as described elsewhere (O’Connell and Weeks 1995). Haplotype analysis was performed by use of SIMWALK software and also via visual inspection (Weeks et al. 1995).
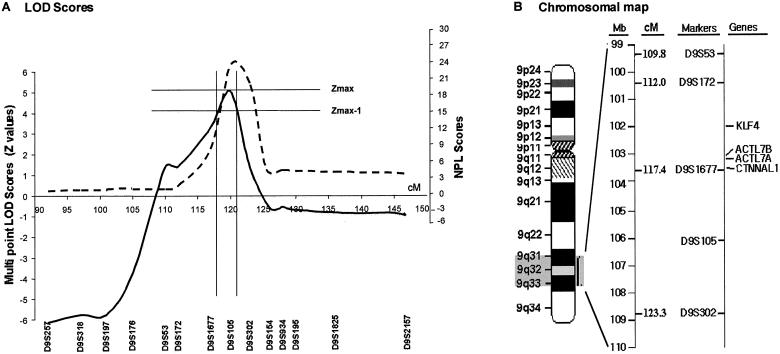
Analyses of genotyping data revealed negative LOD scores for chromosomal regions 1q25, 19q13, and 11q21-22, which are known to contain loci for familial FSGS genes, and also for 1q21 markers associated with the RFH1 locus (table 1). Analysis of genomewide data by GENEHUNTER identified two chromosomal regions with LOD scores >2. These regions, chromosomes 9q31-32 and 3p24-pter, were further investigated by fine mapping, using markers spaced 1–5 cM apart and using the following fluorescent dye–labeled markers obtained from Research Genetics/Invitrogen: D9S257, D9S318, D9S197, D9S176, D9S53, D9S172, D9S1677, D9S105, D9S302, D9S154, D9S934, D9S195, D9S1825, and D9S2157 for chromosome 9q31-32; and D3S1307, D3S1297, D3S3050, D3S1304, and D3S1286 for chromosome 3p24-pter. A peak two-point LOD score of 3.93 for marker D9S105 was obtained through VITESSE analysis, with 99% penetrance and a disease allele frequency of 0.0001 at θ=0 (table 2). A Z score of 5.42 was obtained for the trait locus, denoted “HNP1,” in the 9q32 region between markers D9S1677 and D9S105. These LOD score calculations were robust to changes in presumed marker allele and disease allele frequencies. Similar results were generated when computation of LOD scores was performed with disease allele frequencies of 0.1–0.00001, as well as when marker alleles were assumed to be equally frequent. Figure 3 shows the multipoint LOD score and nonparametric linkage (NPL) score plots as well as an ideogram of the linked region. Linkage to chromosome 3 was excluded upon fine mapping (LOD scores ranging from −1.2 to −10.0 for the all the analyzed markers). By using the “maximum LOD −1” method, we constructed a support interval with an asymptotic CI >95% (Winn et al. 1999). The HNP1 support interval was flanked by marker D91677 proximally and by D9S302 distally. The haplotypes for nine markers—in the order cen-D9S197, D9S176, D9S53, D9S172, D9S1677, D9S105, D9S302, D9S154, D9S934-tel—are shown below each individual genotyped (fig. 1). Recombinant events in individuals III-10, III-11, III-13, and IV-2 also localize the linked region to an ∼8-cM (6-Mb) interval bounded by markers D9S172 and D9S105.
Table 1.
LOD Scores of Several Candidate Regions for HN/FSGS
| Location andLinked Marker | Two-PointLOD Scoreat θ=0 | Gene |
| 1q21: | ||
| D1S534 | −16. 51 | RFH1 |
| D1S1653 | −4. 98 | |
| D1S1679 | −3. 27 | |
| 1q25: | ||
| D1S1677 | −. 01 | NPHS2 |
| D1S1589 | −7. 18 | |
| D1S518 | −5. 81 | |
| 11q21-q22: | ||
| D11S1998 | −14. 45 | FSGS2 |
| D11S4464 | −7. 78 | |
| D11S1304 | −9. 19 | |
| 19q13: | ||
| D19S245 | −6. 39 | ACTN4 |
| D19S559 | −6. 31 | NPHS1 |
| D19S246 | −6. 73 |
Table 2.
Two-Point LOD Scores for Analyzed Markers in Chromosome 9q31-q32 Region
|
LOD Score at θ = |
||||||||||
| Marker | .00 | .05 | .10 | .15 | .20 | .25 | .30 | .35 | .40 | .45 |
| D9S922 | −6.09 | −2.18 | −1.24 | −.73 | −.42 | −.22 | −.10 | −.04 | −.01 | .00 |
| D9S1120 | −6.41 | −2.22 | −1.21 | −.68 | −.37 | −.18 | −.07 | −.01 | .00 | .00 |
| D9S257 | −8.68 | −1.00 | −.28 | .04 | .19 | .22 | .19 | .12 | .04 | −.01 |
| D9S318 | −6.16 | −1.98 | −.96 | −.47 | −.19 | −.05 | .02 | .04 | .03 | .01 |
| D9S197 | −8.33 | −1.42 | −.44 | .01 | .22 | .29 | .27 | .19 | .08 | .00 |
| D9S176 | −6.11 | −.42 | .25 | .51 | .59 | .57 | .47 | .34 | .19 | .07 |
| D9S53 | 1.07 | 2.45 | 2.38 | 2.18 | 1.91 | 1.60 | 1.26 | .90 | .53 | .19 |
| D9S172 | 2.28 | 3.54 | 3.43 | 3.17 | 2.83 | 2.44 | 2.00 | 1.52 | 1.01 | .48 |
| D9S1677 | 2.67 | 3.91 | 3.76 | 3.47 | 3.10 | 2.67 | 2.20 | 1.68 | 1.12 | .56 |
| D9S105 | 3.93 | 3.64 | 3.32 | 3.00 | 2.61 | 2.28 | 1.89 | 1.47 | 1.02 | .53 |
| D9S302 | −.28 | 2.64 | 2.81 | 2.72 | 2.50 | 2.20 | 1.83 | 1.41 | .95 | .47 |
| D9S154 | −3.03 | 1.53 | 2.01 | 2.10 | 2.02 | 1.82 | 1.54 | 1.20 | .80 | .39 |
| D9S934 | .20 | 1.58 | 1.58 | 1.46 | 1.27 | 1.05 | .81 | .56 | .32 | .12 |
| D9S195 | −4.05 | .55 | 1.07 | 1.20 | 1.17 | 1.04 | .85 | .62 | .37 | .14 |
| D9S1825 | −.12 | −.07 | −.03 | −.01 | .01 | .02 | .02 | .02 | .02 | .01 |
| D9S2157 | −2.50 | −.20 | .22 | .38 | .42 | .40 | .35 | .27 | .18 | .09 |
| D9S1838 | −1.54 | .40 | .78 | .90 | .90 | .82 | .69 | .52 | .33 | .14 |
Figure 3.
Multipoint linkage analysis of chromosome 9q31-q32. A, Multipoint LOD score (Z score) plot based on VITESSE analyses (solid line) and NPL score plot generated by GENEHUNTER analyses (dashed line). B, Chromosomal map of 9q31-q32. Several candidate genes are located around the D9S1677 marker.
An analysis of the region of interest on chromosome 9q31-32 showed several strong candidate genes, including Kruppel-like factor 4 (KLF4 [MIM 602253]), actin-like 7A (ACTL7A [MIM 604303]), actin-like 7B (ACTL7B [MIM 604304]), and catenin α–like 1 (CTNNAL1 [MIM 604785]) (fig. 3). KLF4 is a transcription factor, whose mRNA is expressed, at a high level, in the metanephric kidney during early embryonic development (Garrett-Sinha et al. 1996). ACTL7A and ACTL7B are intronless genes situated next to each other, which encode for actin-like proteins (Chadwick et al. 1999). CTNNAL1 encodes a cadherin-associated protein that is involved in cell-cell interactions (Zhang et al. 1998). We have thus far sequenced ACTL7A, ACTL7B, and CTNNAL1 genes, using the primers shown in table 3. The results of gene sequencing experiments showed several polymorphisms that did not segregate with affected status (also shown in table 3).
Table 3.
Mutation Screen for Candidate Genes in 9q31-q32
| Gene and Region | PCRProduct(bp) | Forward and Reverse Primers | Variations |
| ACTL7A: | |||
| Promoter | 527 | 3′-TGAGGCCTACCATGAGCTGCTTGC-5′ | SNP at −739 from start codon (A/G) |
| 5′-ATCCCTTATTCTAGTCGGTGGCGG-3′ | |||
| 5′ UTR, N-term | 512 | 3′-CCACCCTGCTTTTGCTCTTAACG-5′ | |
| 5′-GGCCATCCCTTAAAGAGGCAGTC-3′ | |||
| Coding | 498 | 3′-ACCAAGAAGGTGGGCAACCAG-5′ | Missense mutation (Ala481Pro) |
| 5′-GTTGGTGTGTGGGCTCAGTGG-3′ | |||
| Coding | 540 | 3′-TCCGACAAGAGATGAAGATCGCC-5′ | |
| 5′-CCAGCTGCATGGACTTGATGAGA-3′ | |||
| Coding | 527 | 3′-TGGGAAGGAGATTCAGCTGTGCC-5′ | Missense mutation (Val1018Met) |
| 5′-CACACTGTGGAGGGGCATATGAG-3′ | |||
| C terminal, 3′ UTR | 446 | 3′-GTACGAGGAACACGGGCCTTTCT-5′ | |
| 5′-TGCCCCAACAGCATTTACCCTCT-3′ | |||
| ACTL7B: | |||
| Promoter | 480 | 3′-GCA TCACCTCTCTAGAGCACC-5′ | |
| 5′-GCATTTCCATGCCTAAGGTGTTCA-3′ | |||
| 5′ UTR, N terminal | 496 | 3′-TGCCTTACTGGAGGGGCTCAAAT-5′ | |
| 5′-GGGCTTCATCTTGAGCTGAGTGG-3′ | |||
| Coding | 498 | 3′-GAGGCAGATGGCGACAAGGAACA-5′ | |
| 5′-GCGTACTTCTCCCGGTTGCTGCT-3′ | |||
| Coding | 496 | 3′-TGGGAGTACATCTTCCGCACCGCC-5′ | |
| 5′-GCTCCTGGCCAATAGTGATGAGTT-3′ | |||
| Coding | 524 | 3′-GGACGACCACCTGCACATCATAG-5′ | |
| 5′-ATGCCGAGGCTCAGCACTTGCTG-3′ | |||
| C terminal, 3′ UTR | 511 | 3′-CAGCTCTGGGTCAGCAAGGAAGA-5′ | |
| 5′-GAAACAGGAGACTGGAGTCGGAC-3′ | |||
| CTNNAL1 Exon: | |||
| 1 | 477 | 3′-AGAAGAGATAAGGGCGGGGCCAT-5′ | |
| 5′-GGCGGGAAGGAGAAGAGGGGC-3′ | |||
| 2 | 429 | 3′-TGCCAGGCTTCATTTTCTTTGAC-5′ | |
| 5′-GTATTCGCGGAAGCTAATCCAGG-3′ | |||
| 3 | 380 | 3′-GCTGAGTTTCTCCTCATTGTGCC-5′ | |
| 5′-ACAAGCCAAGCAATCCACCACTA-3′ | |||
| 4 | 354 | 3′-CCCCTTGTGTGTCTTCTATCTGGC-5′ | SNP at intron 3 (−84 from exon 4): C/G |
| 5′-CCAGTCCAAACAATCCTAACGATC-3′ | |||
| 5 | 367 | 3′-TCACATGGTCTTAGACTCCTCCGT-5′ | |
| 5′-ACACCATGTTCTGCTGAGCTGTTC-3′ | |||
| 6 | 421 | 3′-GTGAGCCAGGGACCCAGGAGATT-5′ | |
| 5′-GAGCCGAGATCACACCACTGCAC-3′ | |||
| 7 | 442 | 3′-TCCCCTCAGCTTGGGAAAAGGAA-5′ | |
| 5′-CCTGTCCCATTCTGGAACAAATTC-3′ | |||
| 8 | 289 | 3′-GAGATGCGGTAACATTACATGTAT-5′ | |
| 5′-GAATTACGGGCATGAGCCACCAT-3′ | |||
| 9 | 494 | 3′-AGTGGAACCCATGGAACTGCTTT-5′ | CA repeats at intron 8 (−8 from exon 9): (CA)5/(CA)4 |
| 5′-TTTAGAGGCCAGACTGTTGCAGG-3′ | |||
| 10 | 500 | 3′-CACCTTGCCATGATTGGATAGGA-5′ | SNP at intron 10 (94 from exon 10): C/T |
| 5′-GCCATGTTCTGGCACACTACCAA-3′ | |||
| 11 | 388 | 3′-ATTGCCCAACATGGAGCTTGTGT-5′ | |
| 5′-TCATTTGCAGTTGAGGTTTGGCA-3′ | |||
| 12 | 363 | 3′-GGAGAGGTGGAGATTGACCAGCA-5′ | |
| 5′-TTGTGGTAGAGGGGAGCAGCAAC-3′ | |||
| 13, 14 | 609 | 3′-GGCACTGACTTGCTTTCTCTGGC-5′ | Deletion (G/D) at intron 12 (−139) |
| 5′-ACCGACTTTAACCTGTCAATGATT-3′ | SNP (T/G) at intron 12 (−122) | ||
| 15 | 554 | 3′-TTCAGTGATATGCCTCATCAAGGTA-5′ | |
| 5′-CCCACTTTTAGCCCGAAATTAAA-3′ | |||
| 16, 17 | 476 | 3′-ACCAACTATCCATAGACATCAGTC-5′ | |
| 5′-AGGAAATAAATTGCCCTTCTGTGG-3′ | |||
| 18, 19 | 610 | 3′-AATGGTAAGGTCTTTTAAGTACTA-5′ | |
| 5′-GACAGTTGGGATATTTGGTTTGA-3′ |
Our results provide strong evidence, in a large African American family, of an HN susceptibility locus (HNP1) mapping to chromosome 9q31-q32 that was identified on a genomewide scan and confirmed by fine mapping. We ruled out loci on 1q21, 1q25, 11p21, and 19q13 regions, which have been previously linked with familial FSGS or RFH1, because they were consistently associated with negative or low LOD scores (Cohn et al. 2000; Winn et al 2002). An examination of the UCSC genome browser showed 33 genes, of which 15 are known and the rest are computer predicted, in the 6-Mb HNP1 locus flanked by D9S172 and D9S105. We further analyzed several candidate genes from this region. ACTL7A and ACTL7B appeared to be strong candidates, in view of the involvement of ACTN4, located on 19q13, with familial FSGS (Kaplan et al. 2000). CTNNAL1, an adhesion molecule, was also studied, because several related molecules, including catenin, are involved in the maintenance of podocyte architecture. However, we have excluded the coding regions of these genes as being responsible for this phenotype. Studies, including sequencing of KLF4 gene, are ongoing to narrow the interval and to identify the responsible gene.
Previous linkage studies have suggested significant genetic heterogeneity for FSGS and HN phenotypes. We had reported linkage between chromosome 19q13 and an autosomal dominant FSGS with late-onset hypertension in another large African American family (CHP 101) that also resides in western Pennsylvania (Vats et al. 2000). Genomic DNA sequencing excluded the two main candidate genes (NPHS1 and ACTN4) and several other genes (authors' unpublished data). We also recently found a linkage between 11q24 and a nephropathy characterized by hypertension, thin basement membranes, and deafness in an Asian Indian family (CHP 177) (Prakash et al. 2003). Although several genes for the FSGS phenotype have been identified, they are mostly associated with the disease in white populations. In contrast to the FSGS loci, the locus described by Cohn et al. (2000) is the closest description of another possible HN locus, but it is reported in a different ethnic group. Cohn et al. ascertained a large Israeli family of Iraqi Jewish origin with an autosomal dominant form of adult-onset nephropathy. Hypertension was the main presenting sign in all patients and was associated with progressive renal failure. The report of Cohn et al. (2000), however, did not describe any specific histological entity. One individual with a creatinine level similar to that of our proband, had 3 sclerotic and 7 normal glomeruli (of the 10 examined), along with interstitial fibrosis and mild tubular atrophy. Another hypertensive individual in that report had a serum creatinine level of 1.7 mg/dl and showed mild thinning of the glomerular basement membranes on electron microscopy, but no other significant alterations were revealed by microscopy or immunostaining. By performing a genomewide linkage search, Cohn et al. localized the disease locus to an 11.6-cM region on 1q21, which is flanked by markers D1S2696 and D1S2635 (maximum LOD score 4.71, with D1S305 at θ=0). They considered atrial natriuretic peptide receptor-1 (NPR1 [MIM 108969]), which maps to 1q21, as a candidate disease gene, since a polymorphic allele of this gene is associated with essential hypertension (Nakayama et al. 2000); however, they concluded that the disorder was distinct from a form of medullary cystic kidney disease (MCKD1 [MIM 174000]) that also maps to 1q21. These findings, along with those of the family described here, underscore the genetic heterogeneity of the HN/FSGS phenotype.
Although HN and FSGS are two distinct disorders and HN is typically associated with histologic lesions characterized by ischemic changes and an absence of immunofluorescence findings, HN is usually diagnosed, in clinical practice, without renal biopsy or any evidence that hypertension actually preceded the renal disorder (Torpy et al. 2002; Kaperonis and Bakris 2003). Thus, HN can clinically resemble many other nephropathies including FSGS (Freedman et al. 1995; Preston et al. 1996; Bohle et al. 1998; Martinez-Maldonado 1998; Ruilope and Segura 2002). In contrast to HN, FSGS is characterized by segmental glomerular scars involving some (focal) glomeruli (Habib 1973), and, although severe proteinuria is a hallmark of FSGS, low grades of proteinuria are not uncommon in HN. Recent studies show a trend toward an increase in the incidence of FSGS and HN, which is especially evident in African Americans. The reason for this trend is unclear, but genetic factors have been implicated (Haas et al. 1997; Marcantoni et al. 2002; Tucker 2002). Also, FSGS is frequently associated with hypertension and tends to progress to ESRD (Schwartz et al. 1995). The overlapping nature of the HN and FSGS phenotypes was also evident in the current study, in which several members presented with nonsyndromic HN and progressed to ESRD. Many of the members were clinically diagnosed with HN, whereas the index case was found to have features of FSGS. Indeed, in one study, “true” HN was seen in only 48% of 56 white patients in whom HN was clinically diagnosed (Zucchelli and Zuccala 1996), and another study showed that 19% of a cohort of patients with clinical HN actually had FSGS lesions (Caetano et al. 2001).
In summary, we report linkage of HN to a locus on chromosome 9q31-q32 in a large African American family with dominantly inherited ESRD and hypertension and with features of FSGS. With the identification of the first such locus, further studies can be performed to identify the causative genes and to evaluate the importance of this locus in other African American families with this phenotype. The detection of HN genes involved in this high-risk population will allow dissection of molecular mechanisms involved in this important public health problem.
Acknowledgments
This study was supported by a Young Investigator award from the National Kidney Foundation (to A.V.) and by NIH grants DK02854 and DK064933 (to A.V.) and HL54526 (to R.E.F.). We thank the mammalian genotyping service at Marshfield Clinics for performing the genomewide scans on this family as a part of a larger project. We thank the members of the reported family for their help and participation. This article was partially presented in abstract form at the annual meeting of the Society of Nephrology, held in Philadelphia in November 2002.
Electronic-Database Information
URLs for data presented herein are as follows:
- Center for Medical Genetics, Marshfield Medical Research Foundation http://research.marshfieldclinic.org/genetics/ (for Marshfield Clinics screening set 10)
- NCBI Entrez, http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?db=Nucleotide (for human genome maps of 1q21-25, 9q31-32, 11q21-22 and 19q13).
- Online Mendelian Inheritance in Man (OMIM), http://www.ncbi.nlm.nih.gov/Omim/
- UCSC Genome Bioinformatics, http://genome.ucsc.edu/ (for human genome working draft)
References
- Adamczak M, Zeier M, Dikow R, Ritz E (2002) Kidney and hypertension. Kidney Int Suppl 62–67 [DOI] [PubMed] [Google Scholar]
- Agodoa LY, Appel L, Bakris GL, Beck G, Bourgoignie J, Briggs JP, Charleston J, et al (2001) Effect of ramipril vs amlodipine on renal outcomes in hypertensive nephrosclerosis: a randomized controlled trial. JAMA 285:2719–2728 [DOI] [PubMed] [Google Scholar]
- Angius A, Petretto E, Maestrale GB, Forabosco P, Casu G, Piras D, Fanciulli M, Falchi M, Melis PM, Palermo M, Pirastu M (2002) A new essential hypertension susceptibility locus on chromosome 2p24- p25, detected by genomewide search. Am J Hum Genet 71:893–905 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bohle A, Wehrmann M, Greschniok A, Junghans R (1998) Renal morphology in essential hypertension: analysis of 1177 unselected cases. Kidney Int Suppl 67:S205–206 [DOI] [PubMed] [Google Scholar]
- Boute N, Gribouval O, Roselli S, Benessy F, Lee H, Fuchshuber A, Dahan K, Gubler MC, Niaudet P, Antignac C (2000) NPHS2, encoding the glomerular protein podocin, is mutated in autosomal recessive steroid-resistant nephrotic syndrome. Nat Genet 24:349–354 [DOI] [PubMed] [Google Scholar]
- Brown DM, Provoost AP, Daly MJ, Lander ES, Jacob HJ (1996) Renal disease susceptibility and hypertension are under independent genetic control in the fawn-hooded rat. Nat Genet 12:44–51. [DOI] [PubMed] [Google Scholar]
- Caetano ER, Zatz R, Saldanha LB, Praxedes JN (2001) Hypertensive nephrosclerosis as a relevant cause of chronic renal failure. Hypertension 38:171–176 [DOI] [PubMed] [Google Scholar]
- Chadwick BP, Mull J, Helbling LA, Gill S, Leyne M, Robbins CM, Pinkett HW, Makalowska I, Maayan C, Blumenfeld A, Axelrod FB, Brownstein M, Gusella JF, Slaugenhaupt SA (1999) Cloning, mapping, and expression of two novel actin genes, actin-like-7A (ACTL7A) and actin-like-7B (ACTL7B), from the familial dysautonomia candidate region on 9q31. Genomics 58:302–309 [DOI] [PubMed] [Google Scholar]
- Churchill PC, Churchill MC, Bidani AK, Griffin KA, Picken M, Pravenec M, Kren V, St Lezin E, Wang JM, Wang N, Kurtz TW (1997) Genetic susceptibility to hypertension-induced renal damage in the rat: evidence based on kidney-specific genome transfer. J Clin Invest 100:1373–1382 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cohn DH, Shohat T, Yahav M, Ilan T, Rechavi G, King L, Shohat M (2000) A locus for an autosomal dominant form of progressive renal failure and hypertension at chromosome 1q21. Am J Hum Genet 67:647–651 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Freedman BI, Iskandar SS, Appel RG (1995) The link between hypertension and nephrosclerosis. Am J Kidney Dis 25:207–221 [DOI] [PubMed] [Google Scholar]
- Garrett-Sinha LA, Eberspaecher H, Seldin MF, de Crombrugghe B (1996) A gene for a novel zinc-finger protein expressed in differentiated epithelial cells and transiently in certain mesenchymal cells. J Biol Chem 271:31384–31390 [DOI] [PubMed] [Google Scholar]
- Haas M, Meehan SM, Karrison TG, Spargo BH (1997) Changing etiologies of unexplained adult nephrotic syndrome: a comparison of renal biopsy findings from 1976–1979 and 1995–1997. Am J Kidney Dis 30:621–631 [DOI] [PubMed] [Google Scholar]
- Habib R (1973) Focal glomerular sclerosis. Kidney Int 4:355–361 [DOI] [PubMed] [Google Scholar]
- Hayden PS, Iyengar SK, Schelling JR, Sedor JR (2003) Kidney disease, genotype and the pathogenesis of vasculopathy. Curr Opin Nephrol Hypertens 12:71–78 [DOI] [PubMed] [Google Scholar]
- Kaperonis N, Bakris G (2003) Blood pressure, antihypertensive therapy and risk for renal injury in African-Americans. Curr Opin Nephrol Hypertens 12:79–84 [DOI] [PubMed] [Google Scholar]
- Kaplan JM, Kim SH, North KN, Rennke H, Correia LA, Tong HQ, Mathis BJ, Rodriguez-Perez JC, Allen PG, Beggs AH, Pollak MR (2000) Mutations in ACTN4, encoding alpha-actinin-4, cause familial focal segmental glomerulosclerosis. Nat Genet 24:251–256 [DOI] [PubMed] [Google Scholar]
- Klag MJ, Whelton PK, Randall BL, Neaton JD, Brancati FL, Ford CE, Shulman NB, Stamler J (1996) Blood pressure and end-stage renal disease in men. N Engl J Med 334:13–18 [DOI] [PubMed] [Google Scholar]
- Kruglyak L, Daly MJ, Reeve-Daly MP, Lander ES (1996) Parametric and nonparametric linkage analysis: a unified multipoint approach. Am J Hum Genet 58:1347–1363 [PMC free article] [PubMed] [Google Scholar]
- Lenkkeri U, Mannikko M, McCready P, Lamerdin J, Gribouval O, Niaudet PM, Antignac CK, Kashtan CE, Homberg C, Olsen A, Kestila M, Tryggvason K (1999) Structure of the gene for congenital nephrotic syndrome of the Finnish type (NPHS1) and characterization of mutations. Am J Hum Genet 64:51–61 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lifton RP, Gharavi AG, Geller DS (2001) Molecular mechanisms of human hypertension. Cell 104:545–556 [DOI] [PubMed] [Google Scholar]
- Marcantoni C, Ma LJ, Federspiel C, Fogo AB (2002) Hypertensive nephrosclerosis in African Americans versus Caucasians. Kidney Int 62:172–180 [DOI] [PubMed] [Google Scholar]
- Martinez-Maldonado M (1998) Hypertension in end-stage renal disease. Kidney Int Suppl 68:S67–S72 [DOI] [PubMed] [Google Scholar]
- Miller SA, Dykes DD, Polesky HF (1988) A simple salting out procedure for extracting DNA from human nucleated cells. Nucleic Acids Res 16:1215 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moore MA, Epstein M, Agodoa L, Dworkin LD (1999) Current strategies for management of hypertensive renal disease. Arch Intern Med 159:23–28 [DOI] [PubMed] [Google Scholar]
- Nakayama T, Soma M, Takahashi Y, Rehemudula D, Kanmatsuse K, Furuya K (2000) Functional deletion mutation of the 5′-flanking region of type A human natriuretic peptide receptor gene and its association with essential hypertension and left ventricular hypertrophy in the Japanese. Circ Res 86:841–845 [DOI] [PubMed] [Google Scholar]
- O’Connell JR, Weeks DE (1995) The VITESSE algorithm for rapid exact multilocus linkage analysis via genotype set-recoding and fuzzy inheritance. Nat Genet 11:402–408 [DOI] [PubMed] [Google Scholar]
- ——— (1998) PedCheck: a program for identification of genotype incompatibilities in linkage analysis. Am J Hum Genet 63:259–266 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prakash S, Chung K, Sinha S, Barmada M, Ellis D, Finegold D, Ferrell E, Randhawa PS, Dinda A, Vats AN (2003) Autosomal dominant progressive nephropathy with deafness: linkage to a new locus on chromosome 11q24. J Am Soc Nephrol 14:1794–1803 [DOI] [PubMed] [Google Scholar]
- Preston RA, Singer I, Epstein M (1996) Renal parenchymal hypertension: current concepts of pathogenesis and management. Arch Intern Med 156:602–611 [DOI] [PubMed] [Google Scholar]
- Ruilope LM, Segura J (2002) Renal protection by antihypertensive therapy. Curr Hypertens Rep 4:324–328 [DOI] [PubMed] [Google Scholar]
- Schwartz MM, Korbet SM, Rydell J, Borok R, Genchi R (1995) Primary focal segmental glomerular sclerosis in adults: prognostic value of histologic variants. Am J Kidney Dis 25:845–852 [DOI] [PubMed] [Google Scholar]
- Torpy J, Lynm C, Glass RM (2002) JAMA patient page: hypertensive kidney disease. JAMA 288:2498 [DOI] [PubMed] [Google Scholar]
- Tucker JK (2002) Focal segmental glomerulosclerosis in African Americans. Am J Med Sci 323:90–93 [DOI] [PubMed] [Google Scholar]
- Vats A, Nayak A, Ellis D, Randhawa PS, Finegold DN, Levinson KL, Ferrell RE (2000) Familial nephrotic syndrome: clinical spectrum and linkage to chromosome 19q13. Kidney Int 57:875–881 [DOI] [PubMed] [Google Scholar]
- Weeks DE, Ott J, Lathrop GM (1990) SLINK: a general simulation program for linkage analysis. Am J Hum Genet Suppl 47:A204 [Google Scholar]
- Weeks DE, Sobel E, O’Connell JR, Lange K (1995) Computer programs for multilocus haplotyping of general pedigrees. Am J Hum Genet 56:1506–1507 [PMC free article] [PubMed] [Google Scholar]
- Winn MP (2002) Not all in the family: mutations of podocin in sporadic steroid-resistant nephrotic syndrome. J Am Soc Nephrol 13:577–579 [DOI] [PubMed] [Google Scholar]
- Winn MP, Conlon PJ, Howell DN, Slotterbeck BD, Smith AH, Graham FL, Bembe M, Quarles LD, Pericak-Vance MA, Vance JM (1999) Linkage of a gene causing familial focal segmental glomerulosclerosis to chromosome 11 and further evidence of genetic heterogeneity. Genomics 58:113–120 [DOI] [PubMed] [Google Scholar]
- Xu X, Rogus JJ, Terwedow HA, Yang J, Wang Z, Chen C, Niu T, Wang B, Xu H, Weiss S, Schork NJ, Fang Z (1999a) An extreme-sib-pair genome scan for genes regulating blood pressure. Am J Hum Genet 64:1694–1701 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu X, Yang J, Rogus J, Chen C, Schork N (1999b) Mapping of a blood pressure quantitative trait locus to chromosome 15q in a Chinese population. Hum Mol Genet 8:2551–2555 [DOI] [PubMed] [Google Scholar]
- Zhang JS, Nelson M, Wang L, Liu W, Qian CP, Shridhar V, Urrutia R, Smith DI (1998) Identification and chromosomal localization of CTNNAL1, a novel protein homologous to alpha-catenin. Genomics 54:149–154 [DOI] [PubMed] [Google Scholar]
- Zucchelli P, Zuccala A (1996) Recent data on hypertension and progressive renal disease. J Hum Hypertens 10:679–682 [PubMed] [Google Scholar]