Abstract
Microdeletions and microduplications, not visible by routine chromosome analysis, are a major cause of human malformation and mental retardation. Novel high-resolution, whole-genome technologies can improve the diagnostic detection rate of these small chromosomal abnormalities. Array-based comparative genomic hybridization allows such a high-resolution screening by hybridizing differentially labeled test and reference DNAs to arrays consisting of thousands of genomic clones. In this study, we tested the diagnostic capacity of this technology using ∼3,500 flourescent in situ hybridization–verified clones selected to cover the genome with an average of 1 clone per megabase (Mb). The sensitivity and specificity of the technology were tested in normal-versus-normal control experiments and through the screening of patients with known microdeletion syndromes. Subsequently, a series of 20 cytogenetically normal patients with mental retardation and dysmorphisms suggestive of a chromosomal abnormality were analyzed. In this series, three microdeletions and two microduplications were identified and validated. Two of these genomic changes were identified also in one of the parents, indicating that these are large-scale genomic polymorphisms. Deletions and duplications as small as 1 Mb could be reliably detected by our approach. The percentage of false-positive results was reduced to a minimum by use of a dye-swap-replicate analysis, all but eliminating the need for laborious validation experiments and facilitating implementation in a routine diagnostic setting. This high-resolution assay will facilitate the identification of novel genes involved in human mental retardation and/or malformation syndromes and will provide insight into the flexibility and plasticity of the human genome.
Introduction
Mental retardation, with or without additional malformations, occurs in 2%–3% of the general population. Although a considerable number of cases can be explained by the presence of gross chromosomal abnormalities or other factors, such as metabolic and/or neurological anomalies, the etiology of mental retardation remains unexplained for ∼50% of patients (Anderson et al. 1996; de Vries et al. 1997). Submicroscopic, subtelomeric chromosome rearrangements contribute significantly to mental retardation and malformation, comprising up to 5% of the previously unexplained cases (Flint et al. 1995; Knight et al. 1999; Biesecker 2002; de Vries et al. 2003). These findings underscore the potential importance of submicroscopic chromosomal anomalies as a major cause of human mental retardation and malformation. To routinely detect these changes in a diagnostic setting, an efficient and robust technology is needed that screens the entire genome for copy-number abnormalities with a resolution beyond the level of a light microscope (5–10 Mb). Array-based comparative genomic hybridization (arrayCGH) technology measures submicroscopic DNA copy-number changes and allows the simultaneous high-resolution mapping of these changes onto the genome sequence (Solinas-Toldo et al. 1997; Pinkel et al. 1998; Snijders et al. 2001). We previously developed an array-based subtelomeric assay that screens all human subtelomeric regions in a single hybridization reaction (Veltman et al. 2002). Here we report the construction and application of a genomewide microarray for the identification of known and novel microdeletions and duplications in patients with mental retardation and malformations.
Patients and Methods
Patients
Genomic DNAs, isolated from blood lymphocytes of four cytogenetically normal, healthy individuals (two males and two females), were used for array validation and as normal reference DNAs. Additional genomic DNAs were isolated from three patients with FISH-verified known microdeletion syndromes (Prader-Willi syndrome [PWS; MIM 176270] on chromosome 15q11-15q12, Smith-Magenis syndrome [SMS; MIM 182290] on chromosome 17p11.2, and trichorhinophalangeal syndrome [TRPS; MIM 190350] on chromosome 8q23.1–q24.11), as well as from 20 patients with mental retardation and additional dysmorphisms of unknown etiology. The latter patients were all seen by a clinical geneticist and had undergone extensive diagnostic work-up, including routine chromosome analysis without a diagnosis. They all had a phenotype suggestive of a chromosomal abnormality, and all scored three points or higher on the checklist developed by de Vries et al. (2001). Genomic DNAs from patients and controls were isolated and purified using a QIAamp kit (Qiagen), according to the instructions of the manufacturer.
Array-Based Comparative Genomic Hybridization
Clone Set
A total of 3,569 well-characterized, colony-purified, and FISH-verified BAC clones were used for array construction. Most of the BACs were derived from the RPCI-11 BAC library used as the main intermediate substrate for the sequencing and mapping of the human genome (Osoegawa et al. 2001). The set includes ∼3,200 clones selected through a collaboration of the Children’s Hospital Oakland Research Institute, BACPAC Resources Center, and several other groups to cover the genome with a 1-Mb resolution (Cheung et al. 2001). Information on this clone set and its availability can be obtained at the BACPAC Resources Center Web site. Additional clones were added to the array, resulting in an even higher-resolution coverage of genomic regions known to be involved in human malformation and mental retardation, including the subtelomeric regions of all human chromosomes (77 clones) (Knight et al. 2000) and regions associated with known microdeletion syndromes (30 clones). Finally, chromosome 12 and chromosome 18 were covered with a higher density through the addition of clones used in previous studies (Veltman et al. 2003b; Zafarana et al. 2003).
Array Preparation
Genomic target DNAs were isolated from 12-ml bacterial cultures using Qiagen R.E.A.L. Prep 96 BioRobot kits on a Qiagen BioRobot 9600 (Qiagen), following the instructions of the manufacturer. Degenerate oligonucleotide-primed (DOP) PCR was performed on isolated DNA from all clones, essentially as described elsewhere (Telenius et al. 1992), with minor modifications (Veltman et al. 2002). Taq2000 (Stratagene) was used as a thermostable polymerase. DOP-PCR products were dissolved at a concentration of 1μg/μl in a 50% DMSO solution and robotically spotted in triplicate onto CMT-GAPS coated glass slides (Corning, UltraGaps) using an OmniGrid 100 arrayer (Genomic Solutions). The array consisted of 48 subgrids, and replicates were printed in different subgrids across the array.
Labeling and Hybridization
Labeling and hybridization were performed essentially as described elsewhere (Veltman et al. 2002). In brief, genomic DNA was labeled by random priming with Cy3-dUTP or Cy5-dUTP (Amersham Biosciences). Test and reference samples were mixed with 120 μg Cot-1 DNA (Roche), coprecipitated, and resuspended in 130 μl of a hybridization solution containing 50% formamide, 10% dextran sulfate, 2 × SSC, 4% SDS, and 10 μg/μl yeast tRNA (Invitrogen). After denaturation of probe and target DNA, hybridization and posthybridization washing procedures were performed using a GeneTAC Hybridization Station (Genomic Solutions), according to the manufacturer's instructions. In brief, an 18-h hybridization with active circulation of the probe was performed, followed by five posthybridization wash cycles in 50% formamide/2 × SSC at 45°C and five wash cycles in phosphate-buffered saline at 20°C. Slides were dried by centrifugation after a brief wash in water.
Image Analysis and Processing
Slides were scanned and imaged on an Affymetrix 428 scanner (Affymetrix) using the Affymetrix 428 scanner software package (version 1.0). The acquired microarray images were analyzed using GenePix Pro 4.0 (Axon Instruments), as described elsewhere (Veltman et al. 2002). For all further analyses, the median of the pixel intensities minus the median local background was used for every spot on the array (Cy3 and Cy5, calculated separately). Data normalization was performed in the software package SAS version 8.0 (SAS Institute) for each array subgrid, by applying Lowess curve fitting with a smoothing factor of 0.1 to predict the log2-transformed test-over-reference (T/R) value on the basis of the average logarithmic fluorescent intensities (Cleveland 1979). This smoothing factor was shown to result in the lowest percentage of false-positive results while not increasing the amount of false-negative results in the validation experiments. A consequence of this smoothing procedure is that the ratios of the clones with a copy-number gain or loss are closer to the normal range of log2 ratios than in normalization procedures without this smoothing.
Quality Control
Clones with an SD of the triplicates >0.3 were excluded in individual experiments, as well as clones with fewer than two replicates remaining after this analysis. Excluded from all experiments were 63 clones that did not show reliable hybridization results in at least four of the five normal-versus-normal control experiments. Clones that mapped to the sex chromosomes (n=163) were not analyzed in detail. The array contains a final set of 3,343 autosomal clones with a coverage of at least 1 clone per Mb. From this final set, 3% of the clones, on average, were excluded per experiment on the basis of the quality criteria.
Thresholds for copy-number gain and loss were determined by examining the results of the control experiments and of previously published work and were set at log2 T/R values of 0.3 and −0.3, respectively. Experiments were excluded when >5% of the clones showed intensity ratios outside of these regions. Of the 40 experiments performed in this study, 5 experiments did not meet these quality criteria. These experiments were successfully repeated. The final data set is available as a downloadable electronic supplement in three file formats, Excel (*.xls), comma-delimited text (*.csv), and tab-delimited text (*.txt).
Analysis of Replicate Experiments
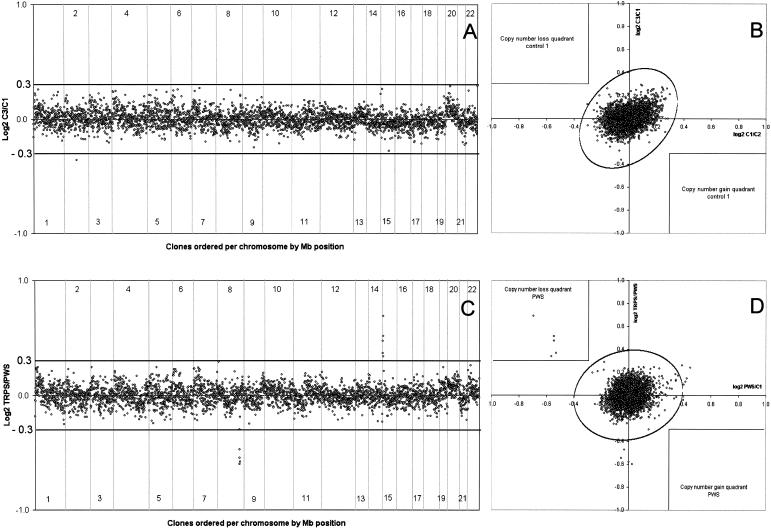
In this study, we performed a dye-swap experiment for each case (patient or control). For statistical analysis of these two experiments, we developed a two-dimensional assay in the software package SAS version 8.0 (SAS Institute) in which reference regions were calculated containing 99.999% of the data points (P=.99999), assuming that the pairs of normalized ratios follow a bivariate normal distribution (fig. 1B and 1D). Under the assumption of no deleted or duplicated regions, the number of data points outside the resulting ellipse is expected to be 1/100,000 × the number of clones on the array—in our case, 1/100,000×3,343=0.03. Clones represented by data points outside this reference region in the scatterplot are candidates for a microduplication or deletion event. However, since a dye-swap experiment was performed for each case, the data points also have to be located in the correct quadrant of the scatterplot (i.e., a positive sign for experiment 1 [patient 1 vs. control 1] and a negative sign for experiment 2 [control 1 vs. patient 1] indicates a potentially duplicated clone, whereas a deleted clone shows opposite signs in both experiments). The a priori thresholds for copy-number gain (log2 T/R value 0.3) or loss (log2 T/R value −0.3) are therefore integrated into the scatterplot to indicate the candidate clones for microdeletion or duplication events.
Figure 1 .
ArrayCGH genomic profiles of validation experiments. Arrays contained 3,343 human autosomal clones (indicated by small circles representing the mean log2-transformed and Lowess-normalized T/R intensity ratios), ordered in A and C from 1pter to 22qter on the basis of the physical mapping positions obtained from the November 2002 freeze of the UCSC genome browser. In panels A and C, chromosome boundaries are indicated by vertical lines. Panel A shows the result of a normal-versus-normal hybridization (control 3 vs. control 1). Nearly all clones fall within the a priori–set thresholds for copy-number gain (log2T/R value 0.3) and copy-number loss (log2 T/R value −0.3) indicated by the horizontal lines. One clone on chromosome 2 shows an intensity ratio outside these thresholds and might represent a false-positive result. Panel B shows the result of the combined analysis of the two hybridizations performed with control 1 (X-axis: control 1 vs. control 2; Y-axis: control 3 vs. control 1). The ellipse represents the border of the reference regions containing 99.999% of the data points; the thresholds for copy-number gain and loss are also integrated into this figure (see the “Patients and Methods” section for details). As can be seen, there is only one clone outside the reference region; however, this clone does not pass the thresholds for copy-number loss in both experiments and can therefore be discarded from further analyses. The clone on chromosome 2 that fell outside the threshold for copy-number loss in panel A is clearly within the normal reference region and can therefore also be discarded for further analyses. Panel C shows the result of the hybridization of DNA from a patient with TRPS against DNA from a patient with PWS. A total of four clones, spanning 2.7 Mb of genomic sequence on 8q23.3-q24.11, showed log2 TRPS-over-PWS intensity ratios below the threshold for copy-number loss, confirming the presence of a deletion of this genomic region in the TRPS patient. In addition, five clones, spanning 2.9 Mb of sequence on 15q11.2, show log2 intensity ratios above the (reverse) threshold for copy-number gain, indicating a deletion of this genomic region in the PWS patient. No clones outside these target genomic regions show potential false-positive results. The combined results of two experiments involving the PWS patient are shown in panel D. The five target clones on 15q11.2 are reproducibly deleted in both experiments and fall outside the bivariate normal distribution reference region (P=.99999) and within the copy-number loss quadrant indicated in the upper left quadrant.
FISH Validation Experiments
FISH validation experiments were performed on metaphase spreads prepared from patient-derived lymphoblast cell lines using routine procedures. Probe labeling, slide preparation, and hybridization were carried out essentially as described elsewhere (de Bruijn et al. 2001). A Zeiss epifluorescence microscope, equipped with appropriate filters, was used for visual examination of the slides. Digital images were captured using a high-performance cooled CCD camera (Photometrics) coupled to a Macintosh Quadra 950 computer. The Image FISH software package (Intergen) was used for analysis of the FISH images. Inverted images of DAPI-stained slides were used for chromosome identification.
Results
Validation Experiments
To test the specificity and sensitivity of the whole genome BAC array, we performed a series of five normal-versus-normal control hybridizations using four normal healthy blood donors (including a dye-swap experiment for each control). Figure 1A shows a representative genomic profile resulting from such an experiment. Nearly all clones show log2 intensity ratios in between the a priori thresholds for copy-number gain (0.3) or loss (−0.3). In the five normal-versus-normal experiments, an average of six clones (0.18%) passed these thresholds. Although very low, this level of background noise would still require a substantial number of FISH experiments to distinguish true microdeletions and microduplications from false-positive results. Therefore, the combination of two experiments (with dye swap) was analyzed for each control case, using stringent criteria for the presence of copy-number gain or loss (see the “Patients and Methods” section and fig. 1B). The added value of combining data of two separate (T/R) experiments was clearly shown by the fact that the number of false positives was reduced to zero in all four cases tested.
Next, we tested the sensitivity of the technology by hybridizing DNA from three patients with known, FISH-confirmed microdeletion syndromes (i.e., one patient with PWS, one patient with SMS, and one patient with TRPS) to the genomewide array. Similar to the normal-versus-normal experiments, DNA samples were hybridized not only against each other but also against one of the normal controls. Figure 1C shows the result of a hybridization of DNA from the patient with TRPS against DNA from the patient with PWS. A total of four clones, representing 2.7 Mb of consensus genomic sequence on 8q23.3-q24.11, showed TRPS-over-PWS intensity ratios below the threshold for copy-number loss, thus confirming the presence of a deletion of this genomic region in the TRPS patient. In addition, five clones, spanning 2.9 Mb of sequence on 15q11.2, showed log2 intensity ratios above the (reverse) threshold for copy-number gain, indicating a deletion of this genomic interval encompassing the genes SNRPN and UBE3A in the patient with PWS. The combined results of two experiments involving the patient with PWS are shown in figure 1D. The five target clones on 15q11.2 are reproducibly deleted in both experiments and fall outside the bivariate normal distribution reference region (P=.99999) and within the upper left quadrant, indicating copy-number loss. Detailed analysis of the DNA from these two patients and the patient with SMS (containing a deletion of two genomic clones spanning a 1.5-Mb region on 17p11.2) showed that the target microdeletion region could be readily identified in each individual experiment with the target clones present on the array. Individual hybridizations showed an average of seven clones (0.21%) with log2 intensity ratios outside the thresholds for copy-number gain or loss, very similar to the percentage of false positives obtained in the normal-versus-normal control experiments described above. Statistical analysis of the duplicate experiments for each case failed to reproduce any of these aberrant ratios, strongly indicating that these were indeed false-positive results.
In conclusion, the sensitivity to detect submicroscopic (1.5–2.9 Mb) deletions was reproducibly validated, and the specificity of the technology was assured by performing two hybridizations for each case and applying a stringent statistical analysis.
Detection of Novel Deletions and Duplications in Patients with Unexplained Mental Retardation and Dysmorphisms
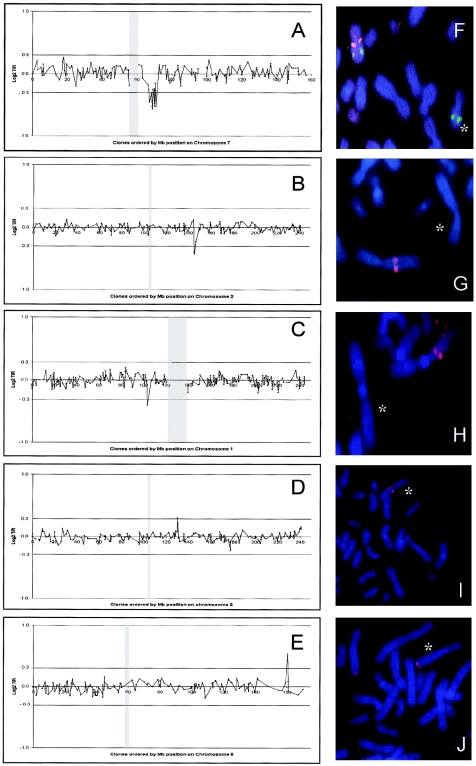
We selected a test series of 20 patients with mental retardation of unknown etiology to investigate whether the high-resolution, genomewide, microarray-based, copy-number screening would allow the identification of small genomic changes not detectable by routine karyotyping. In concordance with the validation experiments, each case was hybridized once against a normal control sample and once against another patient with a different clinical phenotype. This procedure was chosen to minimize the costs and, at the same time, the risks of hybridizing cases with identical genetic abnormalities against each other, which would result in masking of the abnormality. In 7 of the 20 patients, copy-number alterations were reproducibly detected by arrayCGH on our genomewide microarray. In five of these cases, the results could be confirmed by FISH on metaphase spreads of the patients. These included three microdeletions and two microduplications (table 1; fig. 2). De novo occurrence was checked by investigating DNA samples from the parents. As expected, the microduplications proved difficult to validate by FISH. We therefore repeated the arrayCGH procedure in the two cases with microduplications on a small, high-density array containing only the regions of interest and confirmed the presence of a microduplication in both cases (data not shown).
Table 1.
Phenotypes and Detected Microdeletions/-duplications
| Patient | Sex | Age (years) | Degree of MRa | Phenotypeb | ArrayCGH Result |
| 1 | Male | 2 | Severe | Microcephaly, facial dysmorphism, pulmonary arterial and valve stenosis | del(7)(q11.21q11.23), 17 clones, 8 Mb, de novo |
| 2 | Female | 12 | Severe | Microcephaly, facial dysmorphism, short stature | del(2)(q22.3q23.2), 3 clones, 2 Mb, de novo |
| 3 | Female | 23 | Moderate | Facial dysmorphism, short stature, ventricular septum defect | del(1)(p21p21), 1 clone, <1 Mb, also detected in father |
| 4 | Male | 2 | Severe | Microcephaly, facial dysmorphism | dup(2)(q21.2q21.2), 1 clone, <1 Mb, also detected in father |
| 5 | Male | 19 | Mild | Facial dysmorphism, polydactyly, polycystic kidney (familial) | dup(6)(q25.3q26), 1 clone, <1 Mb, parents not available |
MR = mental retardation.
Detailed clinical information is given in the text.
Figure 2 .
Detailed genomic profiles and FISH validation of copy-number abnormalities identified in five cases with unexplained mental retardation. Panels A–E represent individual profiles of the affected chromosomes for each case, with clones ordered for each chromosome from pter to qter on the basis of the physical mapping positions obtained from the November 2002 freeze of the UCSC genome browser. The centromeric region is indicated by a vertical gray dash, the thresholds for copy-number gain (log2 T/R value 0.3) and copy-number loss (log2 T/R value −0.3) are indicated by horizontal lines. Panels F–J represent the FISH validation using (one of) the target clone(s) identified by arrayCGH. Affected chromosomes are indicated by an asterisk (*). Panel A shows the deletion on 7q11 in patient 1, with 14 clones in this region showing an average ratio of −0.5. FISH validation of this case is shown in the adjacent panel F, in which one of the deleted clones on 7q11 is shown in red and a not-deleted control probe is shown in green. Panel B shows the microdeletion on 2q22 in patient 2 with a total of three clones crossing the threshold for copy-number loss, with FISH validation in the adjacent panel G. Deletion of a single clone on 1p21 is shown in panel C for patient 3; this clone was confirmed by FISH to be deleted not only in the patient (panel H) but also in the father of the patient. Copy-number gain detected in a single clone is shown in panels D and E for patients 4 and 5, with FISH confirmation in panels I and J.
The largest deletion identified in patient 1 was verified by FISH (fig. 2F) and targeted 17 clones on the array, spanning a total region of 8.6 Mb on 7q11.21-q11.23. The karyotype of this case was re-examined because of the relatively large size of this deletion, but no abnormalities were identified. It is interesting that this genomic region contains the complete common deletion segment for the Williams-Beuren syndrome (Bayes et al. 2003).
The other deletions and duplications were considerably smaller. In patient 2, the deletion on 2q22.3-q23.2 was encompassed by three clones mapping in a 2-Mb genomic interval, whereas, in the other three cases, only one clone was involved, indicating the presence of abnormalities <1 Mb in size. In one patient (patient 3) with a deletion on 1p21, the deletion was identified by FISH in the father of the patient as well, indicating that this might be a novel genomic polymorphism. Similarly, the duplication of a single clone on 2q21.2 in patient 4 was identified by FISH in the father. Unfortunately, the parents of patient 5, whose DNA contained a duplication of a single clone on 6q25.3-q26, were not available for checking de novo occurrence.
Discussion
ArrayCGH provides a high spatial genomic resolution and allows a fully automated evaluation of thousands of genomic loci. Previous applications have been mainly directed at genomic abnormalities in cancer (Snijders et al. 2001; Veltman et al. 2003a; Wessendorf et al. 2003). In this study, we demonstrate the application of arrayCGH in detecting known and novel submicroscopic abnormalities. The specificity and sensitivity of this approach was tested and validated in cytogenetically normal and healthy individuals, as well as in patients with known microdeletion syndromes. Deletions and duplications were detected reliably in a single overnight hybridization experiment without a priori knowledge of the genomic region involved. Individual experiments were performed with a low level of false-positive results that was reduced further by performing a replicate dye-swap experiment followed by a thorough statistical analysis. From our data, we conclude that such replicate experiments are essential for implementation in a diagnostic setting, since they considerably reduce the need for laborious confirmation experiments while greatly improving the validity of the results.
Application of this approach in a pilot study of 20 patients with unexplained mental retardation and additional malformations resulted in the detection of five copy-number abnormalities, three deletions, and two duplications, all beyond the microscopic resolution (∼10 Mb). The patient with the largest deletion (8.6 Mb on 7q11.21-q11.23) was a 2-year-old boy (patient 1) with severe mental retardation, plagio- and microcephaly, postnatal growth retardation, facial dysmorphism (downward-slanting palpebral fissures, periorbital fullness, epicanthus and telecanthus, broad mouth with full lips, and sagging cheeks), short neck, unilateral simian crease, and a peripheral and valvular pulmonary stenosis. The deletion completely overlaps with the common 1.6-Mb region deleted in patients with Williams syndrome (Bayes et al. 2003). Although this boy had some facial features fitting the diagnosis of Williams syndrome, his clinical presentation was more severe than is commonly observed in patients with Williams syndrome, most notably the severe retardation. Moreover, a pulmonary valve stenosis is less often observed in this syndrome (Eronen et al. 2003). It seems likely that the severity of the phenotype is related to the large size of the deletion, a finding that has also been observed in four cases with even larger deletions, three of which also showed characteristics typical of the Williams syndrome (Valentine and Sergovich 1977; Frydman et al. 1986; Mizugishi et al. 1998; Wu et al. 1999).
A 2-Mb deletion on 2q22.3-q23.2 was identified in a 12-year-old girl (patient 2) who had severe mental retardation, short stature (height 3 SDs below normal), microcephaly (head circumference 2.5 SDs below normal), obesitas, facial dysmorphism (coarse facies, upward-slanting palpebral fissures, hypotelorism, abnormally shaped ears, high nasal bridge, small, carp-shaped mouth with downward-turned corners, narrow, flat palate, and broad chin), and long, narrow hands with short digiti V. The deletion was just distal to a more common deletion on 2q22 and did not include the SIP1 gene, which is associated with Mowat-Wilson syndrome (Mowat et al. 2003). The clinical presentation of this girl differs also from the phenotype seen in this relatively new syndrome. The deletion is de novo, so haploinsufficiency of one or more genes within the deleted region could be causative for the phenotype. So far, no obvious candidate gene appears to be present in this genomic region.
In a 23-year-old female (patient 3) with moderate mental retardation, autism, short stature, minor facial dysmorphism (upward-slanting palpebral fissures, deep-set eyes, short philtrum), short broad feet, a small ventricular septum defect, and childhood absences, we detected a deletion of a single clone on 1p21. This deletion was present in her healthy father, as well. Although familial occurrence of this small deletion (<1 Mb) does not rule out a causative role, it may very well be a novel genomic polymorphism. It is known that similar polymorphisms (either deletions or duplications) without any clinical significance are present in the genome, but, at present, they are underrecognized. Examples of known normal genomic variations include a 2.5-Mb duplication of 8p23.1 (Barber et al. 1998; Engelen et al. 2000) and a number of subtelomeric polymorphisms (Ballif et al. 2000; Linardopoulou et al. 2001; Der-Sarkissian et al. 2002).
It is interesting that two microduplications were also detected using this comparative microarray technology. One duplication on 2q21.2 (<1 Mb) occurred in a 2-year-old boy (patient 4) with severe mental retardation, microcephaly, and facial dysmorphism (metopic ridge, synophrys, arched eyebrows, long eyelashes, upward-slanting palpebral fissures, low-set, posteriorly rotated malformed ears, and high nasal bridge). This duplication was detected also in his father, indicating the presence of another genomic polymorphism. The other duplication was found to be present on 6q25.3-q26 (<1 Mb) in a 19-year-old male (patient 5) with mild mental retardation, postaxial polydactyly of hands and feet, facial dysmorphism (upward-slanting palpebral fissures, high, narrow nasal bridge, short philtrum, retrognathia, and irregular teething), and medullary polycystic kidneys. The kidney abnormality was also present in his two mentally normal sisters. It is interesting to note that, in a series of 36 patients with a larger duplication, including the 6q25.3-q26 region, four cases have been reported with a polydactyly (Schinzel 2001). Unfortunately, the parents of this patient were unavailable for checking de novo occurrence. However, a common copy-number variation affecting a single BAC at 6q26 was recently reported by Albertson and Pinkel (2003). In this publication, the observed copy-number differences between individuals were explained by variation in the length of the apolipoprotein (a) gene, which is highly polymorphic in the human genome because of variation in the number of copies of a 5.5-kb sequence encoding kringle repeats (Kamboh et al. 1991; Lackner et al. 1993). Indeed, this gene is located within the BAC affected in this patient (RP11-43B19), and therefore it is highly likely that this copy-number change also represents a genomic polymorphism.
The detection of five microdeletions/-duplications in a series of 20 patients with mental retardation of unknown etiology in this pilot study underscores the strength of the arrayCGH technique. It should be mentioned that the patients in this study were selected on the basis of a phenotype suggestive of a chromosomal abnormality (de Vries et al. 2001). Therefore, this cohort may not be representative of the population of individuals with mental retardation as a whole. Also, two of the genomic abnormalities were identified in one of the parents as well, reducing the chance that these abnormalities are underlying the disorder. On the other hand, the resolution of the current microarray is, on average, 1 Mb, and, therefore, cases with more subtle anomalies may have been missed. To this end, we and other groups are in the process of constructing microarrays completely covering the genome with an average resolution of ∼50 kb (for more information, see the BACPAC Resources Center's Human BAC Minimal Tiling Set Web site).
There are three main applications for using arrayCGH in patients with mental retardation and malformations. First, it is to be expected that the number of interstitial microdeletions/-duplications will be comparable or may even exceed the ∼5% submicroscopic, subtelomeric rearrangements currently reported among individuals with mental retardation (de Vries et al. 2003). For this reason, this 1-Mb–resolution array is currently being evaluated in a diagnostic setting in our department. Ultimately, this array-based copy-number screening may partially replace karyotyping in this patient group. Second, these studies will facilitate the detection of genes involved in physical and mental development. Several such genes have already been identified as a result of systematic deletion mapping using microscopic chromosomal abnormalities (Romeo et al. 1994; Belloni et al. 1996; Robinson et al. 2003). The high resolution of the arrayCGH method allows for rapid and precise mapping of candidate genes for specific malformations. Third, a systematic analysis of genomic polymorphisms will give more insight into the flexibility and plasticity of the human genome. This latter may prove to be a fruitful field of study, given the fact that such large rearrangements will often involve multiple genes and may therefore serve as predisposing factors for multifactorial disorders.
Supplementary Material
Acknowledgments
We thank Yvonne Jonkers, Martin Elferink, and Monique Kersten-Niessen, for expert technical assistance; Ben Hamel, for referring cases; and Rolph Pfundt, for useful discussions. B.B.A.d.V. was supported by a grant from the Netherlands Organization for Health Research and Development (Zon-MW).
Electronic-Database Information
Accession numbers and URLs for data presented herein are as follows:
- BACPAC Resources Center, http://www.chori.org/bacpac/
- BACPAC Resources Center's Human BAC Minimal Tiling Set Web site, http://bacpac.chori.org/pHumanMinSet.htm
- Online Mendelian Inheritance in Man (OMIM), http://www.ncbi.nlm.nih.gov/Omim/ (for PWS, SMS, and TRPS)
References
- Albertson DG, Pinkel D (2003) Genomic microarrays in human genetic disease and cancer. Hum Mol Genet: 12:R145–152 [DOI] [PubMed] [Google Scholar]
- Anderson G, Schroer RJ, Stevenson RE (1996) Mental retardation in South Carolina. II. Causation. In: Saul RA, Phelan MC (eds) Proceedings of the Greenwood Genetic Center. Greenwood Genetic Center, Greenwood, SC, pp 32–44 [Google Scholar]
- Ballif BC, Kashork CD, Shaffer LG (2000) The promise and pitfalls of telomere region-specific probes. Am J Hum Genet 67:1356–1359 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barber JC, Joyce CA, Collinson MN, Nicholson JC, Willatt LR, Dyson HM, Bateman MS, Green AJ, Yates JR, Dennis NR (1998) Duplication of 8p23.1: a cytogenetic anomaly with no established clinical significance. J Med Genet 35:491–496 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bayes M, Magano LF, Rivera N, Flores R, Perez Jurado LA (2003) Mutational mechanisms of Williams-Beuren syndrome deletions. Am J Hum Genet 73: 131–151 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belloni E, Muenke M, Roessler E, Traverso G, Siegel-Bartelt J, Frumkin A, Mitchell HF, Donis-Keller H, Helms C, Hing AV, Heng HH, Koop B, Martindale D, Rommens JM, Tsui LC, Scherer SW (1996) Identification of sonic hedgehog as a candidate gene responsible for holoprosencephaly. Nat Genet 14:353–356 [DOI] [PubMed] [Google Scholar]
- Biesecker LG (2002) The end of the beginning of chromosome ends. Am J Med Genet 107:263–266 10.1002/ajmg.10160.abs [DOI] [PubMed] [Google Scholar]
- Cheung VG, Nowak N, Jang W, Kirsch IR, Zhao S, Chen XN, Furey TS, et al. (2001) Integration of cytogenetic landmarks into the draft sequence of the human genome. Nature 409:953–958 10.1038/35057192 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cleveland WS (1979) Robust locally weighted regression and smoothing scatterplots. J Amer Stat Assoc 74: 829–836 [Google Scholar]
- de Bruijn DR, Kater-Baats E, Eleveld M, Merkx G, Geurts Van Kessel A (2001) Mapping and characterization of the mouse and human SS18 genes, two human SS18-like genes and a mouse Ss18 pseudogene. Cytogenet Cell Genet 92:310–319 [DOI] [PubMed] [Google Scholar]
- Der-Sarkissian H, Vergnaud G, Borde YM, Thomas G, Londono-Vallejo JA (2002) Segmental polymorphisms in the proterminal regions of a subset of human chromosomes. Genome Res 12:1673–1678 10.1101/gr.322802 [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Vries BB, van den Ouweland AM, Mohkamsing S, Duivenvoorden HJ, Mol E, Gelsema K, van Rijn M, Halley DJ, Sandkuijl LA, Oostra BA, Tibben A, Niermeijer MF (1997) Screening and diagnosis for the fragile X syndrome among the mentally retarded: an epidemiological and psychological survey. Am J Hum Genet 61:660–667 [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Vries BB, White SM, Knight SJ, Regan R, Homfray T, Young ID, Super M, McKeown C, Splitt M, Quarrell OW, Trainer AH, Niermeijer MF, Malcolm S, Flint J, Hurst JA, Winter RM (2001) Clinical studies on submicroscopic subtelomeric rearrangements: a checklist. J Med Genet 38:145–150 10.1136/jmg.38.3.145 [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Vries BBA, Winter R, Schinzel A, Van Ravenswaaij-Arts C (2003) Telomeres: a diagnosis at the end of the chromosomes. J Med Genet 40:385–398 10.1136/jmg.40.6.385 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Engelen JJ, Moog U, Evers JL, Dassen H, Albrechts JC, Hamers AJ (2000) Duplication of chromosome region 8p23.1→p23.3: a benign variant? Am J Med Genet 91:18–21 [DOI] [PubMed] [Google Scholar]
- Eronen M, Peippo M, Raatikka M, Arvio M, Johansson R, Kahkonen M (2002) Cardiovascular manifestations in 75 patients with Williams syndrome. J Med Genet 39:554–558 10.1136/jmg.39.8.554 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flint J, Wilkie AO, Buckle VJ, Winter RM, Holland AJ, McDermid HE (1995) The detection of subtelomeric chromosomal rearrangements in idiopathic mental retardation. Nat Genet 9:132–140 [DOI] [PubMed] [Google Scholar]
- Frydman M, Steinberger J, Shabtai F, Steinherz R (1986) Interstitial 7q deletion [46,XY,del(7)(pter→cen::q112→qter)] in a retarded quadriplegic boy with normal beta glucuronidase. Am J Med Genet 25:245–249 [DOI] [PubMed] [Google Scholar]
- Kamboh MI, Ferrell RE, Kottke BA (1991) Expressed hypervariable polymorphism of apolipoprotein (a). Am J Hum Genet 49: 1063–1074 [PMC free article] [PubMed] [Google Scholar]
- Knight SJL, Regan R, Nicod A, Horsley SW, Kearney L, Homfray T, Winter RM, Bolton P, Flint J (1999) Subtle chromosomal rearrangements in children with unexplained mental retardation. Lancet 354:1676–1681 10.1016/S0140-6736(99)03070-6 [DOI] [PubMed] [Google Scholar]
- Knight SJ, Lese CM, Precht KS, Kuc J, Ning Y, Lucas S, Regan R, Brenan M, Nicod A, Lawrie NM, Cardy DL, Nguyen H, Hudson TJ, Riethman HC, Ledbetter DH, Flint J (2000) An optimized set of human telomere clones for studying telomere integrity and architecture. Am J Hum Genet 67:320–332 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lackner C, Cohen JC, Hobbs HH (1993) Molecular definition of the extreme size polymorphism in apolipoprotein (a). Hum Mol Genet 2: 933–940 [DOI] [PubMed] [Google Scholar]
- Linardopoulou E, Mefford HC, Nguyen O, Friedman C, van den Engh G, Farwell DG, Coltrera M, Trask BJ (2001) Transcriptional activity of multiple copies of a subtelomerically located olfactory receptor gene that is polymorphic in number and location. Hum Mol Genet 10:2373–2383 10.1093/hmg/10.21.2373 [DOI] [PubMed] [Google Scholar]
- Mizugishi K, Yamanaka K, Kuwajima K, Kondo I (1998) Interstitial deletion of chromosome 7q in a patient with Williams syndrome and infantile spasms. J Hum Genet 43:178–181 10.1007/s100380050064 [DOI] [PubMed] [Google Scholar]
- Mowat DR, Wilson MJ, Goossens M (2003) Mowat-Wilson syndrome. J Med Genet 40:305–310 10.1136/jmg.40.5.305 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osoegawa K, Mammoser AG, Wu C, Frengen E, Zeng C, Catanese JJ, de Jong PJ (2001) A bacterial artificial chromosome library for sequencing the complete human genome. Genome Res 11:483–496 10.1101/gr.169601 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pinkel D, Segraves R, Sudar D, Clark S, Poole I, Kowbel D, Collins C, Kuo WL, Chen C, Zhai Y, Dairkee SH, Ljung BM, Gray JW, Albertson DG (1998) High resolution analysis of DNA copy number variation using comparative genomic hybridization to microarrays. Nat Genet 20:207–211 10.1038/2524 [DOI] [PubMed] [Google Scholar]
- Robinson SW, Morris CD, Goldmuntz E, Reller MD, Jones MA, Steiner RD, Maslen CL (2003) Missense mutations in CRELD1 are associated with cardiac atrioventricular septal defects. Am J Hum Genet 72:1047–1052 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Romeo G, Ronchetto P, Luo Y, Barone V, Seri M, Ceccherini I, Pasini B, Bocciardi R, Lerone M, Kaariainen H, et al. (1994) Point mutations affecting the tyrosine kinase domain of the RET proto-oncogene in Hirschsprung's disease. Nature 367:377–378 10.1038/367377a0 [DOI] [PubMed] [Google Scholar]
- Schinzel A (2001) Catalogue of unbalanced chromosome aberrations in man, 2nd ed. Walter de Gruyter, Berlin [Google Scholar]
- Snijders AM, Nowak N, Segraves R, Blackwood S, Brown N, Conroy J, Hamilton G, Hindle AK, Huey B, Kimura K, Law S, Myambo K, Palmer J, Ylstra B, Yue JP, Gray JW, Jain AN, Pinkel D and Albertson DG (2001) Assembly of microarrays for genome-wide measurement of DNA copy number by CGH. Nat Genet 29:263–264 10.1038/ng754 [DOI] [PubMed] [Google Scholar]
- Solinas-Toldo S, Lampel S, Stilgenbauer S, Nickolenko J, Benner A, Dohner H, Cremer T, Lichter P (1997) Matrix-based comparative genomic hybridization: biochips to screen for genomic imbalances. Genes Chromosomes Cancer 20:399–407 [DOI] [PubMed] [Google Scholar]
- Telenius H, Carter NP, Bebb CE, Nordenskjold M, Ponder BA, Tunnacliffe A (1992) Degenerate oligonucleotide-primed PCR: general amplification of target DNA by a single degenerate primer. Genomics 13:718–725 [DOI] [PubMed] [Google Scholar]
- Valentine H, Sergovich F (1977) A syndrome associated with interstitial deletion of chromosome 7q. Birth defects, OAS (3b):261–262 [Google Scholar]
- Veltman JA, Fridlyand J, Pejavar S, Olshen AB, Korkola JE, DeVries S, Carroll P, Kuo WL, Pinkel D, Albertson D, Cordon-Cardo C, Jain AN, Waldman FM (2003a) Array-based comparative genomic hybridization for genome-wide screening of DNA copy number in bladder tumors. Cancer Res 63:2872–2880 [PubMed] [Google Scholar]
- Veltman JA, Jonkers Y, Nuijten I, Janssen I, Van Der Vliet W, Huys E, Vermeesch J, Van Buggenhout G, Fryns JP, Admiraal R, Terhal P, Lacombe D, Geurts van Kessel A, Smeets D, Schoenmakers EFPM, Van Ravenswaaij-Arts CM (2003b) Definition of a critical region on chromosome 18 for congenital aural atresia by arrayCGH. Am J Hum Genet 72:1578–1584 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Veltman JA, Schoenmakers EFPM, Eussen BH, Janssen I, Merkx G, van Cleef B, van Ravenswaaij CM, Brunner HG, Smeets D, Geurts van Kessel A (2002) High-throughput analysis of subtelomeric chromosome rearrangements by use of array-based comparative genomic hybridization. Am J Hum Genet 70:1269–1276 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wessendorf S, Schwaenen C, Kohlhammer H, Kienle D, Wrobel G, Barth TF, Nessling M, Moller P, Dohner H, Lichter P, Bentz M (2003) Hidden gene amplifications in aggressive B-cell non-Hodgkin lymphomas detected by microarray-based comparative genomic hybridization. Oncogene 22:1425–1429 10.1038/sj.onc.1206297 [DOI] [PubMed] [Google Scholar]
- Wu YQ, Nickerson E, Shaffer LG, Keppler-Noreuil K, Muilenburg A (1999) A case of Williams syndrome with a large, visible cytogenetic deletion. J Med Genet 36:928–932 [PMC free article] [PubMed] [Google Scholar]
- Zafarana G, Grygalewicz B, Gillis AJM, Vissers LE, van der Vliet W, van Gurp RJ, Debiec-Rychter M, Oosterhuis JW, Geurts van Kessel A, Schoenmakers EF, Looijenga LH, Veltman JA (2003) 12p-amplicon structure analysis in testicular germ cell tumors of adolescents and adults by arrayCGH. Oncogene 22:7695–7701 10.1038/sj.onc.1207011 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.