Abstract
Nonsyndromic X-linked mental retardation (MRX) is defined by an X-linked inheritance pattern of low IQ, problems with adaptive behavior, and the absence of additional specific clinical features. The 13 MRX genes identified to date account for less than one-fifth of all MRX, suggesting that numerous gene defects cause the disorder in other families. In a female patient with severe nonsyndromic mental retardation and a de novo balanced translocation t(X;7)(p11.3;q11.21), we have cloned the DNA fragment that contains the X-chromosomal and the autosomal breakpoint. In silico sequence analysis provided no indication of a causative role for the chromosome 7 breakpoint in mental retardation (MR), whereas, on the X chromosome, a zinc-finger gene, ZNF41, was found to be disrupted. Expression studies indicated that ZNF41 transcripts are absent in the patient cell line, suggesting that the mental disorder in this patient results from loss of functional ZNF41. Moreover, screening of a panel of patients with MRX led to the identification of two other ZNF41 mutations that were not found in healthy control individuals. A proline-to-leucine amino acid exchange is present in affected members of one family with MRX. A second family carries an intronic splice-site mutation that results in loss of specific ZNF41 splice variants. Wild-type ZNF41 contains a highly conserved transcriptional repressor domain that is linked to mechanisms of chromatin remodeling, a process that is defective in various other forms of MR. Our results suggest that ZNF41 is critical for cognitive development; further studies aim to elucidate the specific mechanisms by which ZNF41 alterations lead to MR.
Introduction
Developmental delay, also referred to as “mental retardation” (MR), affects an estimated 2%–3% of the population (Chelly and Mandel 2001). Although the etiology of MR is complex and poorly understood, recent investigations have highlighted the importance of genetic factors in cognitive development. In particular, studies of the X chromosome have confirmed that there are numerous specific monogenic forms of MR. Of significant historical importance is the recognition of fragile X syndrome (FRAXA) and the identification of the FMR1 gene (MIM 309550). FRAXA is caused by a CGG repeat expansion in the FMR1 5′ UTR, which is then abnormally methylated. Accounting for 2%–2.5% of the established X-linked forms of MR (XLMR), this syndrome is the most common cause of XLMR known at present (for review, see Jin and Warren [2003]). XLMR is now divided into two subgroups: syndromic XLMR (MRXS), which includes FRAXA and other MR-associated disorders that can be defined by a set of specific clinical features, and MRX, which includes all X-linked forms of MR for which the only consistent clinical feature is MR. To date, 30 genes responsible for MRXS and 13 genes responsible for MRX have been cloned (Frints et al. 2002; Hahn et al. 2002; Vervoort et al. 2002). The recent discovery that mutations in ARX (MIM 300382)—the human homologue of the Drosophila gene Aristaless—are responsible for syndromic MRX with infantile spasms, Partington syndrome (MIM 309510), and MRX (Bienvenu et al. 2002; Stromme et al. 2002) clearly illustrates that mutations in a single disease gene may result in a relatively broad spectrum of clinical features. This phenomenon has been observed for an increasing number of genes implicated in both MRXS and MRX, including MECP2 (MIM 300005) (Amir et al. 1999; Couvert et al. 2001; Yntema et al. 2002), AGTR2 (MIM 300034) (Vervoort et al. 2002), FGD1 (MIM 305400) (Pasteris et al. 1994; Lebel et al. 2002), RSK2 (MIM 300075) (Trivier et al. 1996; Hanauer and Young 2002), ATRX (MIM 300032) (Gibbons et al. 1995; Villard et al. 1996b), and L1CAM (MIM 308840) (Jouet et al. 1994; Brewer et al. 1996). Studies of L1CAM mutations in a family with a variety of neurologic abnormalities (Fryns et al. 1991; Ruiz et al. 1995) highlighted the fact that a specific mutation can result in widely variable phenotypes even within a family.
Given the low mutation frequency in currently known MRX genes, together with the number of families for which linkage data implicate unidentified X-chromosomal genes, it can be assumed that there are as many as 30–100 MRX genes that remain to be discovered (Ropers et al. 2003). In the search for candidate MRX genes, molecular characterization of de novo chromosome abnormalities has proved to be a promising starting point. Given the large percentage of noncoding DNA in the human genome, it is not surprising that balanced translocations, which occur at a frequency of ∼1 in 2,000 live births (Warburton 1991), are often present in healthy individuals. However, among carriers of balanced translocations, the frequency of congenital abnormalities is double that among individuals with normal karyotypes (Warburton 1991), suggesting that, in approximately half of disease-associated balanced translocations, there is a causal relationship between the translocation and the disorder. Since many balanced translocations do not disrupt genes, it is reasonable to expect that, among those that do disrupt genes or control elements, a causal relationship is much more frequent. X-chromosomal genes that are disrupted by balanced translocations in individuals with developmental anomalies are especially good candidate-disease genes because X-chromosomal genes typically have only one functional copy. This applies in the case of females as it does in the case of males because female balanced-translocation carriers exhibit skewed X inactivation, was 118limiting transcription of X-chromosomal genes to those on the translocated chromosome (Schmidt and DuSart 1992).
In the current study, we investigated the chromosome breakpoints in a female patient with severe MRX who carries a de novo balanced X;7 translocation. Cloning of the breakpoint-spanning DNA fragment, gene expression studies, and mutation screening in unrelated families have enabled us to identify a novel candidate gene for nonsyndromic XLMR.
Subjects and Methods
Patient with Translocation
The proband is a 7-year-old girl who presents with severe developmental delay and carries a de novo balanced chromosome rearrangement t(X;7)(p11.3;q11.21) (depicted in fig. 1A). She is the daughter of healthy unrelated parents, and there is no family history of mental disability or related illness. She has two healthy siblings. After normal pregnancy and birth, she had an Apgar score of 10/10, birth weight was 3,130 g (25th percentile, on the basis of the National Center for Disease Control and Prevention), length was 51 cm (50th percentile), and head circumference was 33.5 cm (25th percentile). At age 1 wk, she had metatarsus adductus. At age 3 mo, an umbilical hernia was observed.
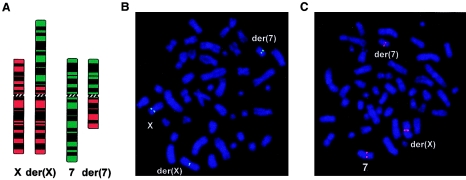
Figure 1.
A, Ideograms depicting normal chromosomes X and 7 and their derivatives der(X) and der(7). Patient metaphase chromosomes were used for FISH of X-chromosomal breakpoint-spanning cosmid clone LAII14H20 with signals on the normal X and split signals on der(X) and der(7) as indicated (B), and chromosome 7 breakpoint-spanning BAC clone RP11-196D18 (GenBank accession number AC069285) with signals on the normal 7 and split signals on der(7) and der(X) as indicated (C).
At age 2 years, the patient did not yet walk or speak, and she exhibited clearly delayed psychomotor development. At age 3 years 9 mo, occasional convulsions, predominantly localized to the shoulders and upper back, were reported, which led to the diagnosis of myoclonic epilepsy. At age 5 years 6 mo, her physical development was in the normal range. She had no dysmorphic features, and her skin was normal; that is, there was no evidence of genetic mosaicism. She could speak, but she was unable to create sentences of more than four words.
On clinical examination at age 6 years 4 mo, her height was 118 cm (50th percentile), her weight was 25 kg (90th percentile), and she was friendly and cooperative. She had some difficulty standing on one leg and walking along a line, which suggested minor ataxia. Routine laboratory tests, including thyroid hormone and ammonium levels, had normal results. Screening for inborn errors of metabolism resulted in no indication of amino acidopathies or organic acidurias. MRI results were normal; the electroencephalogram (EEG) exhibited a spike-wave pattern. The patient continued to suffer from myoclonic seizures and was treated with valproate and ethosuximide. EEG was normal 1 year later. Samples from patient and parents were obtained after receiving informed consent.
Families with XLMR and Control Individuals
DNA from 200 families collected by the European XLMR Consortium, including 14 families for which linkage data implicates the Xp11 region, was available for mutation screening. DNA from an additional 10 Australian families with MRX linked to the Xp11 region was also included in the panel for screening. The control panel consisted of 275 normal males and 65 normal females. All samples were obtained after receiving informed consent.
FISH
Chromosome analysis was performed using standard high-resolution techniques. For breakpoint mapping, we used CEPH and ICRFy900 YAC probes—selected from the Whitehead Institute and the Integrated X Chromosome Database contigs—and BACs and PACs from the Sanger Institute and the University of California–Santa Cruz (UCSC) “Golden Path.” Cosmid clones were selected from the chromosome X–specific cosmid library (LANL), available from the RZPD Deutsches Ressourcenzentrum für Genomforschung (RZPD). Clones were prepared by standard techniques, were labeled with appropriately coupled dUTPs by nick translation or were directly labeled by degenerate oligonucleotide primer–PCR, and were used as probes in FISH, as described elsewhere (Wirth et al. 1999).
Southern Blotting and Breakpoint Cloning
DNA isolation and Southern blotting were performed according to standard protocols. Following overnight restriction endonuclease digestion, genomic DNA was separated by agarose gel electrophoresis, was transferred in 20×SSC to Nylon membranes (Roth), and was crosslinked by UV. Probes of PCR-amplified genomic DNA were labeled with 32[P]dCTP, by Klenow reaction with random hexamer primers. After preannealing to sonicated human placental DNA (HT Biotechnology), probes were hybridized to membranes in polyethylene glycol hybridization buffer (10% PEG 6000; 0.125 M NaPO4; 0.25 M NaCl; 0.001 M EDTA; 7% SDS) supplemented with salmon sperm DNA. Blots were washed in 0.2×SSC/0.2% SDS before exposure to radiographic film. Primer pairs for amplification of probes were based on the genomic sequence of clone RP13-83I24 (GenBank accession number AL590283) and are as follows: 13 forward 5′-GTG TGT TGA AAT GTG ATT TCT GG-3′ and 1,055 reverse 5′-AAT TCC TGG AGG AAT TTT CTT CC-3′, 5,705 forward 5′-AAA AGA CTG AAC AGG AGA AAA GG-3′ and 6,553 reverse 5′-TCT CCA TAT CCA TTC TTT ATG CC-3′, 9,087 forward 5′-ATG CAC TAC AGA GTT TAT GTG AG-3′ and 10,112 reverse 5′-TTC TCT TGG GCT TTA CTC TAT TG-3′, 15,123 forward 5′-AAC TCG CTT CAG TTA TAC CAT TC-3′ and 16,128 reverse 5′-TTT TCA TTA AAC ATC AGG GAG CG-3′, 26,828 forward 5′-GAG TGA AAA AAG AAG CTA TTG CC-3′ and 27,466 reverse 5′-GGT CAA AGG CAA TTT TCC TTA TG-3′. Breakpoint cloning was performed essentially as described elsewhere (Siebert et al. 1995). Patient genomic DNA was subject to restriction digest with EcoRI, was ligated to adaptors, and was amplified with adaptor-specific primers (AP1 and AP2) and nested X chromosome sequence-specific primers 5′-CAC ACA GAT AGA GAC GAT ACA-3′ (primer 1) and 5′-TCT TAC AGA CCA TCT CTA AAC C-3′ (primer 2). Following nested PCR, DNA of the expected size was gel purified with the QIAquick gel extraction kit (Qiagen) and was cloned into pGEMTeasy (Promega), and the inserted breakpoint fragment was sequenced with the BigDye Terminator Chemistry (PE Biosystems) and was separated on an ABI 377 DNA sequencer.
RNA Isolation and Expression Studies
RNA was isolated from lymphoblastoid cells by use of Trizol (Invitrogen), according to the manufacturer’s recommendations. For reverse transcription (RT) with Superscript II (Invitrogen), 5 μg of RNA was used. RT reactions were performed essentially according to the manufacturer’s protocol but in the presence of RNAguard (Amersham Pharmacia), by use of either oligo dT or random hexamers for priming. Amplification of hypoxanthine phosphoribosyltransferase (HPRT) with intron-spanning primers (TGG CGT CGT GAT TAG TGA TG and TAT CCA ACA CTT CGT GGG GT) served as a cDNA loading control. Breakpoint-spanning primers (ATG GCA GCT AAT GGG GAC TC and CCT GAA CAG CTC TGA TGT GG) or primers 3′ to the breakpoint (AGT CAG AGG CTG CCT TCA AG and AGT AGA GGA AGG GCT ATG GG) were used to amplify translocation patient and control ZNF41 transcripts. Splice variants in patients and control individuals were amplified with one of the two forward primers—F1 (CTT CGG AGC TGA CAC TAA GC) or F2 (ATG GCA GCT AAT GGG GAC TC)—and the reverse primer, CTG TTC AAG GCA GCA TGG TC, that is specific for known ZNF41 variants 41.3 and 41.6. Human multiple-tissue northern blots (Clontech) were probed with a gel-purified 3′ ZNF41 fragment that had been amplified by RT-PCR as described above and had been labeled as for Southern blotting. Blots were hybridized in QuikHyb (Stratagene), according to manufacturer’s protocol. To control for RNA loading, blots were hybridized either to the β-actin probe provided or to a G3PDH fragment amplified with the following primers: ACC CCT TCA TTG ACC TCA ACT AC and TGC TTC ACC ACC TTC TTG ATG TC.
Mutation Screening by Denaturing High-Performance Liquid Chromatography (DHPLC)
PCR products with a range of 100–300 nt in length and corresponding to the complete known human ZNF41 coding sequence plus ⩽40 nt of neighboring intronic sequences were amplified from genomic DNA and were analyzed by DHPLC. The screened coding sequence corresponds to ZNF41 mRNA transcript variant 1 (also referred to as “ZNF41.1” [GenBank accession number NM_007130]) and to the 3′ exon 2 extension indicated by the sequence ZNF41.6 (GenBank accession number AJ010021) and the 5′ exon 3 extension indicated by the sequences ZNF41.6 and ZNF41.3 (GenBank accession number AJ010018). Exon 5 was amplified in eight overlapping fragments (exons 5a–5h). PCR reactions were carried out in 50-μl reaction volumes containing 100 ng of genomic DNA, 300 pmol of each primer, 0.4 mM dNTPs, 1 U Amplitaq DNA polymerase (Perkin Elmer), and 2–4 mM MgCl2, with the exception of exon 5d, which was amplified using 1.5 U Taq DNA polymerase and Q-Solution (Qiagen). An initial 5-min denaturation at 95°C was followed by 35 cycles of 1 min at 95°C, 1 min of annealing, a 1.5-min extension at 72°C, and a final extension step of 10 min at 72°C. Sequences of primer pairs and specific amplification conditions are indicated in table 1. Products were checked by 1.5% agarose gel electrophoresis before analysis by WAVE nucleic acid fragment analysis system (Transgenomic). Paired products were denatured at 95°C for 5 min and were slowly brought to room temperature. Reannealed DNA duplexes were injected and eluted with a linear acetonitrile gradient at a flow rate of 0.9 ml/min, with a mobile phase consisting of a mixture of buffer A (0.1 M triethylammonium acetate or TEAA with 0.1% acetonitrile) and buffer B (0.1 M TEAA with 25% acetonitrile).
Table 1.
Primers for Mutation Detection and PCR Amplification Conditions
| Exon | Forward Primer (5′→3′) | Reverse Primer (5′→3′) | Annealing Temperature | MgCl2 Concentration |
| 2 | GTTGTCAGCAGGAGAGCCTG | TGTCTCCAGGGTGGCAGGTC | 64°C | 3 mM |
| 3 | GCATTAAAAACAACTTGCTATC | ATGGAAATACCCTCATTAGCAG | 56°C | 2 mM |
| 4 | CCAATCAGCTGGACCCAATC | GTGATACTGACTTCCATGTCC | 62°C | 4 mM |
| 5a | CTGCTGATTGTTCTTTGTGTCG | ATTGTGTCACAGTTATGGAGTC | 61°C | 4 mM |
| 5b | ATAATTCATGTGACTACCAAGC | CCATTACACATTCAGTACACAC | 61°C | 4 mM |
| 5c | CCACCCACCATCAGAAAATTC | TCAGATGTCTAAAGAGGTCTG | 59°C | 3 mM |
| 5d | AGAAACCCTACAAATGCAGTG | CTCCAGTATGAATTCTCTGATG | 59°C | 2.25 mM |
| 5e | GGCCTTCATCCAGAAATCAC | CCACAGCCATTGCACTTATAG | 61°C | 4 mM |
| 5f | AAAGGCTTTTACTGACCAGTC | TGCATTCATAAGGCTTCTCTC | 59°C | 4 mM |
| 5g | AGAAATCGCACTTCATTGCGC | TCTGGATGAAAGCTTTCCCAC | 61°C | 4 mM |
| 5h | AGTCTCATACTGGAGAAAGAC | CTGCTATTAGATACCTGATGC | 59°C | 3 mM |
Melting profiles and resolution temperatures for each fragment were predicted by the Transgenomic WAVEMAKER software, version 4.1. For each pair of patient or control samples exhibiting exceptional elution profiles, DNA was reamplified and gel purified using the QIAquick gel extraction kit (Qiagen), was sequenced using the BigDye Terminator Chemistry (PE Biosystems), and was separated on an ABI 377 DNA sequencer.
Results
X Chromosome Breakpoint Mapping by FISH
The translocation patient in this study had been screened for abnormalities in the FMR1 and FMR2 (MIM 309548) genes; disease-causing triplet repeats were ruled out. The de novo translocation t(X;7)(p11.3;q11.21), depicted in fig. 1A, indicates a disruption in a region of the X chromosome to which several families with MRX have been mapped (Chelly and Mandel 2001; Ropers et al. 2003). Successive FISHs of genomic YAC clones to patient metaphase chromosomes localized the breakpoint to a region of <1 cM (table 2). Clone ICRFy900C1228, harboring marker DXS426, was breakpoint spanning (data not shown). Hybridization of BAC and PAC clones within the region of the overlapping YAC (table 3) led to the identification of two clones, RP13-83I24 (GenBank accession number AL590283) and RP13-479F17 (GenBank accession number AL590223)—with partially overlapping sequence—that gave proximal and distal hybridization signals, respectively (data not shown), suggesting that the breakpoint might lie in the region of sequence overlap. PCR products from within this region (nts 1–20585 of clone RP13-83I24) were used for screening of a human chromosome X-specific cosmid library. Positive clones were FISH mapped (table 3), and cosmid clone LAII14H20 gave split signals (fig. 1B), indicating that it spans the breakpoint.
Table 2.
FISH Results for CEPH or ICRF YAC Clones Mapping to Chromosome X
| YAC Clonea | Location on Xb | FISH Result |
| iA1034 | 42 cM | Distal |
| 749G05 | 68 cM | Distal |
| iF02014 | NA | Distal |
| iB01003 | NA | Distal |
| iC04013 | NA | Distal |
| 896E11 | 73 cM | Distal |
| iB01128 | NA | Distal |
| iB07021 | NA | Distal |
| iB01026 | 77 cM | Distal |
| iB02007 | 77 cM | Distal |
| iD01151 | 77 cM | Distal |
| 964C07 | 77 cM | Distal |
| iC1228 | 77 cM | Overlapping |
| iA0120 | NA | Proximal |
| iA1220 | 77 cM | Proximal |
| iC1022 | 77 cM | Proximal |
| iC0874 | NA | Proximal |
i = ICRF.
NA = not available.
Table 3.
FISH Results for BAC, PAC, and Cosmid Clones
| Clone | GenBank Accession Numbera | FISH Result |
| RP11-30G10 | AQ003212 | Distal |
| RP4-733D15 | AL023623 | Distal |
| RP11-75A9 | AL139811 | Distal |
| RP1-71L16 | AL022165 | Distal |
| RP1-250J21 | AL162583 | Distal |
| RP3-484G21 | AL163597 (STS) | Distal |
| RP11-553I6 | AL359698 | Distal |
| RP1-306D1 | Z83822 | Distal |
| RP1-659F15 | AL096791 | Distal |
| RP11-571E6 | AL591503 | Distal |
| RP11-479F17 | AL590223 | Distal |
| LAII14H20 | NA | Overlapping |
| LAII33P05 | NA | Proximal |
| LAII59H11 | NA | Proximal |
| LAII61J11 | NA | Proximal |
| RP11-83I24 | AL590283 | Proximal |
| RP1-1147B6 | NA | Proximal |
| RP1-230G1 | AQ489515 | Proximal |
| RP1-212G6 | AL009172 | Proximal |
| RP3-473A14 | AL031967 | Proximal |
| RP3-393P12 | AL022578 | Proximal |
NA = not available.
Localization of the X chromosome Breakpoint by Southern Hybridization and Subsequent Breakpoint Cloning
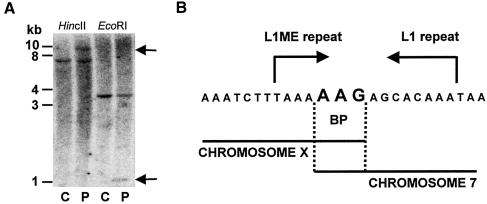
Further breakpoint localization within the region of the overlapping cosmid was done by Southern blot hybridizations. Aberrant restriction fragments in patient DNA were observed in several independent digests, by use of a probe corresponding to nts 9529–10112 of clone RP13-83I24 (fig. 2A). In particular, the aberrant fragment of ∼1 kb in the patient EcoRI digest enabled us to locate precisely the breakpoint. Subsequent cloning and sequencing of this hybrid fragment (fig. 2B) indicated that the chromosome 7 breakpoint lies within BAC clone RP11-196D18 (GenBank accession number AC069285). This result was confirmed by FISH (fig. 1C).
Figure 2.
A, Southern blot of patient (P) and control (C) genomic DNA probed with a PCR product corresponding to nucleotides 9529–10112 of clone RP11-83I24. Aberrant fragments specific to patient DNA (black arrows) in HincII (left) and EcoRI (right) digests correspond to sizes of ∼10 kb and ∼1 kb, respectively. B, Sequence of the breakpoint region indicating (in large type) the three nucleotides common to both chromosome X and chromosome 7.
The Rearrangement Disrupts the X-Chromosomal ZNF41 Gene, and Full-Length Transcripts Are Absent in the Patient Cell Line
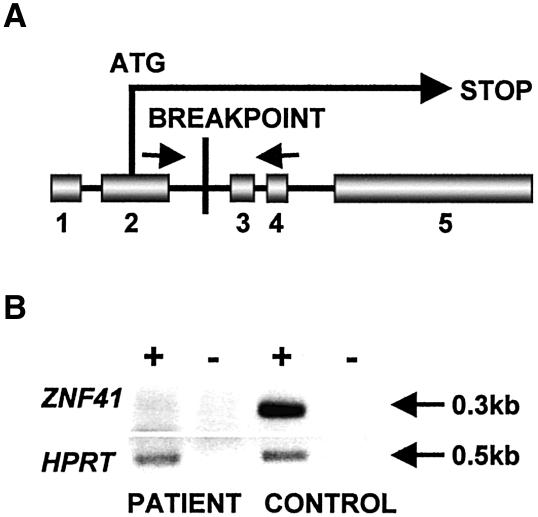
In both breakpoint regions identified in this patient, fully annotated genome sequences are available; therefore, a thorough in silico search for genes and other potential links between genotype and phenotype was possible. Both chromosomes are disrupted within LINE1 repetitive elements (L1ME on X and L1 on 7). On chromosome X, the breakpoint clearly interrupts the coding sequence of the zinc-finger 41 gene (ZNF41 [MIM 314995]). The breakpoint lies within intron 2 (notation is based on the presence of five exons, one of which is composed strictly of 5′ UTR), thereby truncating the predicted ZNF41 protein (GenBank accession number NP_009061) just after amino acid 24 (fig. 3A). Analysis of patient metaphase chromosomes confirmed skewed X inactivation (data not shown). Correspondingly, full-length ZNF41 transcripts were not present in the patient cell line, whereas transcripts were easily detected in control cell lines (fig. 3B). HPRT, however, was present at normal levels in the patient. Data shown are representative examples from several independent experiments.
Figure 3.
A, Schematic diagram (not to scale) indicating the exon-intron structure of ZNF41 relative to the breakpoint location. Arrows indicate location of primers used for RT-PCR analysis. B, RT-PCR analysis by use of ZNF41 primers spanning the breakpoint. ZNF41 product corresponds to nucleotides 407–702 of GenBank accession number NM_007130. Amplification of HPRT served as a control. A plus sign (+) indicates with reverse transcriptase; a minus sign (−) indicates mock reaction.
The chromosome 7 breakpoint does not disrupt any known genes, and, on the basis of both UCSC “Golden Path” (April 2003 freeze) and NIX (UK Human Genome Mapping Project Resource Centre) analyses, there are no ESTs bridging the breakpoint region. The closest documented spliced ESTs or mRNA sequences (GenBank accession numbers AW661872 and BC041841) map ∼96 kb and ∼132 kb to either side of the breakpoint. Further studies, therefore, focused on the X chromosome, where an obvious candidate had been identified.
Identification of ZNF41 Mutations in Two Families with XLMR
Given the absence of this potentially important zinc-finger protein in the investigated patient with translocation, we considered ZNF41 to be a candidate disease gene in families presenting with XLMR. We therefore screened the ZNF41 coding sequence and neighboring intronic sequences for disease-causing mutations in 210 unrelated patients with XLMR. We confirmed the presence of two SNPs (recorded in the GenBank Entrez SNP database), both present in ∼1.5% of the patients, and identified four novel nucleotide exchanges (for a complete description of SNPs identified, see table 4). Of particular interest, we identified one novel amino acid exchange and one intronic splice-site mutation that were not found in >400 control X chromosomes.
Table 4.
Summary of Sequence Alterations Identified in Screening ZNF41 DNA in a Panel of Patients with MRX [Note]
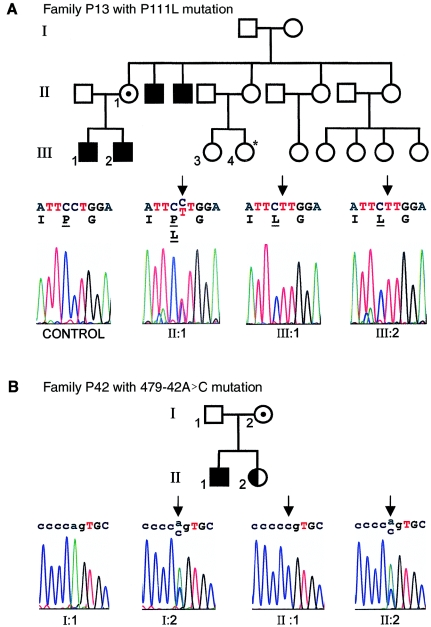
In family P13 (with MRX) (depicted in fig. 4A), the 738C→T (GenBank accession number NM_007130) alteration results in a proline-to-leucine exchange at amino acid 111 (P111L [GenBank accession number NP_009061]) (see fig. 4A). This alteration was absent in the control individuals screened. The affected amino acid lies in the linker region between the repressor domain and the zinc-finger domain. In light of the unique structure of proline, it is plausible that the tertiary structure of the protein is modified or destabilized by this exchange, which could have significant functional consequences. DNA from nonnuclear family members is not available for further studies, but sequence analysis of DNA from the only brother of the patient, who is also mentally disabled, confirmed the presence of the same missense mutation. Both affected boys in this family presented with MRX. The index patient had delayed early milestones: he was first walking freely at age 17 mo, and, at age 5 years 8 mo, he was functioning at an intellectual level of age 3 years (as measured by Brunet-Lezine’s test). He exhibited language retardation, avoided social contact, and was hyperactive. He had no dysmorphic features or additional neurological abnormalities. His older brother has a similar history. The parents are unrelated, and both of the mother's brothers are affected, but additional clinical data on other family members are not available. Analysis of DNA from the mother confirmed that she carries the same mutation on one of her X chromosomes (fig. 4A).
Figure 4.
A, Pedigree of family P13, with sequence corresponding to the proline→leucine mutation (left to right): unrelated control individual, mother (II:1), index patient (III:2), and brother of the index patient (III:1). For the potentially affected female cousin (individual III-4) (indicated with an asterisk [*]), no clinical data are available. Affected nucleotides are indicated with black arrows. B, Pedigree for family P42, with sequence chromatograms indicating the splice-site mutation in affected individuals (left to right): father (I:1), mother (I:2), index patient (II:1), and mildly affected sister (II:2). Uppercase letters indicate coding sequence; affected nucleotides are indicated with black arrows.
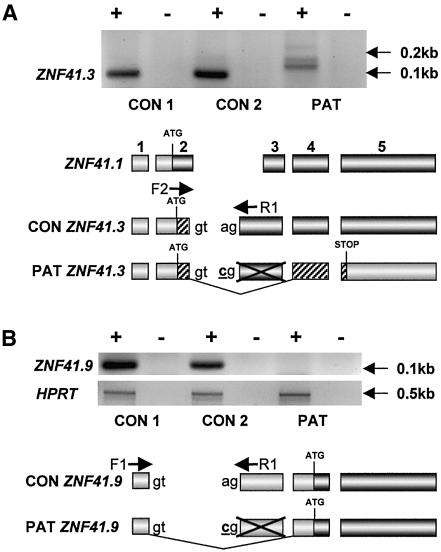
In family P42 (with MRX) (fig. 4B), we detected an adenine-to-cytidine intronic mutation (479-42A→C [GenBank accession number NM_007130]) (fig. 4B [lower panel]) that results in aberrant splicing of ZNF41. More precisely, two documented ZNF41 transcripts—variants 41.3 and 41.6 (corresponding to GenBank accession numbers AJ010018 and AJ010021, respectively; see also Rosati et al. [1999])—and one novel ZNF41 transcript, variant 41.9 described below, contain 5′ extensions of exon 3 (referred to as “exon 2” by Rosati et al. [1999] but now updated in NCBI) that make use of the consensus splice site that is mutated in family P42. Our investigations of the resulting splicing defects in the patient cell line focused primarily on transcript variant 41.3, since variant 41.6 is expressed at low levels (see Rosati et al. [1999]) and could not be readily detected in lymphoblastoid cell lines. As depicted in fig. 5A, transcript variant ZNF41.3 can be amplified in control cell lines but is absent in the patient cell line. Moreover, exclusively in the patient cell line, larger misspliced transcripts are present (fig. 5A [top panel]). Analysis of the predominant transcript variant ZNF41.1 indicated no difference between patients and control individuals (data not shown).
Figure 5.
A, RT-PCR for ZNF41.3 in male control (CON 1), female control (CON 2), and patient (PAT) lymphoblastoid cell lines. The schematic diagram below the cell lines (not to scale) indicates the exonic structure of both normal and predicted aberrant ZNF41.3 transcripts, with respect to the predominant transcript variant ZNF41.1, and the respective locations of primers F2 and R1 (specific for variants 41.3 and 41.6) used for amplification. Shaded regions indicate predicted coding sequence; patterns differentiate between reading frames. The adenine→cytidine intronic splice-site mutation in the patient is underlined and indicated in bold typeface. B, RT-PCR for the novel transcript variant ZNF41.9 in the same samples. As in section A, the schematic diagram indicates the intronic mutation, the structure of ZNF41.9 relative to the predominant transcript ZNF41.1, the location of primers F1 and R1 used for amplification, and the predicted coding sequences of the resulting transcript variants. Amplification of HPRT served as a control. A plus sign (+) indicates with reverse transcriptase; a minus sign (−) indicates mock reaction.
It is interesting that we have also identified a novel splice variant of ZNF41 (designated “41.9” and depicted in fig. 5B) that lacks exon 2 and therefore, like variant ZNF41.4 (Rosati et al. 1999), has a predicted ORF that does not contain the transcriptional repressor domain. Unlike variant ZNF41.4, however, this variant contains the 5′ extension of exon 3 that is present in variant ZNF41.3, and, like variant ZNF41.3, ZNF41.9 is absent in the patient cell line (fig. 5B). This splice-site mutation may not directly affect the coding sequence of variant ZNF41.9, given that the first methionine in the known mRNA sequence lies in exon 4; however, in variant ZNF41.3, the intron 2 acceptor splice site follows predicted amino acid 20, and subsequent skipping of exon 3 would result in the presence of a premature stop codon in exon 5 (fig. 5A). Like other RNAs with premature termination codons, such a transcript would likely be targeted for nonsense-mediated mRNA decay (for review, see Hentze and Kulozik [1999]). This is supported by the fact that we did not detect transcripts lacking exon 3 in the patient cell line when using external primers (data not shown). In any case, it is clear that both normal variants ZNF41.3 and ZNF41.9 are absent specifically in the patient, and this probably has functional consequences.
Like the proline-to-leucine amino acid exchange, this mutation was not found in other screened patients, nor was it detected in the control individuals. The patient has a diagnosis of mild MR. He was born at term (by Cesarean section), with a birth weight of 3,000 g (10th–25th percentile) and a length of 51 cm (50th percentile). He walked at age 12–13 mo and reached early milestones within the normal time frame; however, he exhibited a severe language delay. He first made two-word associations at age 3 years and was first speaking in simple phrases at age 4 years 6 mo. At age 8 years, he was 135 cm tall (90th percentile) and had a head circumference of 53 cm (75th percentile). He had no additional dysmorphic or neurological symptoms, and results of screening for fragile X were negative. At age 10 years 3 mo, the patient was unable to read, and he had essentially no acquisition of mathematics skills. He had mood disorders, with both hyperactivity and aggressiveness, and he required special education. At that time, his verbal IQ score was 83, and performance IQ was 59 (as measured by Wechsler’s scale). Cerebral MRI results were normal. The younger sister of this patient recently received a diagnosis of a borderline form of MR. She also had school difficulties associated with impulsive behavior and hyperactivity. Sequencing confirmed that she is heterozygous for the splice-site mutation (see fig. 4B). The father does not carry the mutation; it was inherited from the mother, who is also a carrier (fig. 4B). We have no clinical data suggesting that the mother is affected, but we never directly addressed that question.
ZNF41 Is Expressed in Multiple Tissues, Including Fetal and Adult Brain
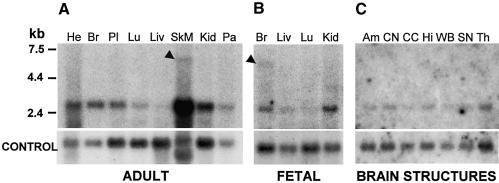
Little is known about the physiological role of the ZNF41 gene; we therefore investigated its expression pattern in both adult and fetal tissues. A predominant transcript was observed at ∼3 kb. The transcript is present in all adult tissues tested, with relatively low expression in lung and liver (fig. 6A). ZNF41 is also transcribed in all fetal tissues tested and shows a similar pattern (fig. 6B). High-stringency hybridization of the ZNF41 cDNA to an adult multiple-brain tissue northern blot also confirmed ubiquitous expression (fig. 6C). It is interesting that a larger transcript of ∼6 kb was detected at significant levels, specifically in fetal brain. The presence of a transcript of the same size was also observed at very low levels in adult skeletal muscle; however, owing to the high levels of RNA present in this sample, no conclusions can be drawn about the tissue specificity of this transcript in the adult. There is currently no data in GenBank to suggest that this band represents cross-hybridization with a closely related gene. Another transcript of ∼2 kb in length was observed exclusively in adult heart. Expression of both the expected 3-kb transcripts and the uncharacterized 6-kb transcript in fetal brain supports a role for the ZNF41 gene in early cognitive development.
Figure 6.
Northern blot hybridization of ZNF41, by use of a probe corresponding to nucleotides 621–1099 of ZNF41 transcript variant 1. A, Adult tissues (left to right): heart, brain, placenta, lung, liver, skeletal muscle, kidney, and pancreas. B, Fetal tissues (left to right): brain, lung, liver, and kidney. C, Adult brain structures (left to right): amygdala, caudate nucleus, corpus callosum, hippocampus, whole brain, substantia nigra, and thalamus. Black arrowheads highlight the presence of a novel 6-kb transcript. Actin (A and C) or GAPDH (B) served as controls for RNA loading.
Discussion
We have shown that the KRAB/FPB gene-family member ZNF41 is disrupted in a female patient with severe MR who carries a de novo reciprocal translocation t(X;7)(p11.3;q11.21). ZNF41 is subject to X inactivation (Rosati et al. 1999); therefore, preferential inactivation of the normal X chromosome results in absence of ZNF41 transcripts in the patient, which strongly suggests that loss of functional ZNF41 causes the observed mental disability. The absence of a disrupted gene on chromosome 7 further supports this hypothesis. In addition, we identified two potentially disease-causing ZNF41 sequence alterations in unrelated families with XLMR. One of these mutations results in aberrant splicing of two ZNF41 isoforms; the other results in exchange of a proline for a leucine residue, which may affect the protein structure. Together, these results strongly suggest that functional alterations in ZNF41 affect cognitive development.
Krueppel-type zinc-finger genes are evolutionarily conserved transcriptional regulators characterized by their Cys2 His2 zinc binding motif, which is typically responsible for sequence-specific DNA binding. ZNF41, absent in our patient, is a member of the subfamily of Krueppel-type zinc-finger proteins harboring a highly conserved N-terminal domain known as the Krueppel-associated box (KRAB). Although the specific functions of ZNF41 are not fully understood, various related genes play an established and important role in human development and disease (Ladomery and Dellaire 2002). In another female patient with severe MRX and a balanced translocation, the disorder probably resulted from the disruption of a related X-chromosomal zinc-finger gene (Lossi et al. 2002). The X-chromosome breakpoint was located just upstream of the Krueppel-like factor 8 (KLF8 [MIM 300286]), also known as the “ZNF741 gene,” and it was confirmed that KLF8 transcripts were absent in the patient cell line. The Wilms tumor suppressor gene WT1 (MIM 194070), which harbors four Krueppel-type zinc fingers, has been implicated in several urogenital developmental disorders, including WAGR syndrome (MIM 194072), which is associated with MR (Call et al. 1990; Rose et al. 1990; Gessler et al. 1992). Of particular relevance, however, are the biochemical studies that highlight the links between the highly conserved KRAB/ZFP subfamily of zinc-finger proteins and chromatin remodeling. Many disorders, several of which are associated with MR, have been linked to defects in processes that govern chromatin structural modification (Hendrich and Bickmore 2001), suggesting that chromatin structural regulation may play a specific and important role in pathways critical for mental function.
Repression of transcription by a KRAB/ZFP requires binding of the corepressor KAP-1 (also known as “TIF1β” and “KRIP-1” [MIM 601742]) (Friedman et al. 1996; Kim et al. 1996; Moosmann et al. 1996; Peng et al. 2000a, 2000b). KAP-1 is a molecular scaffold that coordinates gene-specific silencing by recruiting both heterochromatin-associated proteins (Ryan et al. 1999) and by interacting with the novel histone H3 Lys9–specific methyltransferase SETDB1 (MIM 604396) (Schultz et al. 2002). It is interesting that, within the primary sequence of SETDB1, Schultz et al. (2002) identified a methyl CpG–binding domain that is related to the domain found in the methyl CpG binding protein MeCP2, which is mutated both in patients with Rett syndrome (MIM 312750) (Amir et al. 1999) and in patients with MRX (Couvert et al. 2001; Yntema et al. 2002). Like ZNF41, MECP2 is ubiquitously expressed; yet loss of functional protein results in a neurological phenotype. Although the mechanism by which MECP2 mutations cause MR is not clear, it is well established that MeCP2 binds to methylated CpGs and represses transcription (for review, see Ballestar and Wolffe [2001]), and it has recently been shown that MeCP2 associates with an unidentified methyltransferase that specifically methylates Lys9 of histone H3 (Fuks et al. 2003), as does the KAP-1/KRAB/ZFP binding partner SETDB1.
Further characterization of the KRAB/KAP-1 repressor module has indicated that, in addition to the KRAB domain, a bipartite domain of the plant homeodomain (PHD) finger and a bromodomain, located within the C-terminal portion of KAP-1, are also required for effective gene silencing (Schultz et al. 2002). It is interesting that the point mutations in that study were modeled after naturally occurring mutations in the PHD finger of the human ATRX gene, which has been implicated in both X-linked α-thalassemia/MR syndrome (MIM 301040) (Gibbons et al. 1995) and Juberg-Marsidi syndrome (MIM 309590), which is also associated with MR (Villard et al. 1996a). In light of the fact that the PHD finger of KAP-1 is required for KRAB/ZFP-associated transcriptional repression, the association between the PHD-containing ATRX and MR is especially interesting. ATRX mutations, like KAP-1 mutations, probably inhibit the PHD-dependent formation of multiprotein complexes required for chromatin remodeling. It is therefore conceivable that ATRX mutations and loss of KRAB/ZFP/KAP-1 function lead to MR via similar defects in regulation of chromatin structure.
It may be of significance that many human genes belonging to the KRAB/ZFP family have no obvious orthologue in the mouse. In a recent study on a conserved cluster of these genes on chromosome 19, it was ascertained that, for 21 functional human KRAB/ZFPs, there are only 10 corresponding mouse genes. Shannon et al. (2003) suggest that duplication events occurring after human-rodent evolutionary divergence are responsible for this phenomenon, and they put forward the hypothesis that sequence and functional variations in these duplicated proteins play a role in establishing species-specific traits (Shannon et al. 2003). In light of this, it is interesting that in silico analysis of both publicly available EST sequences and alignment of fully annotated human and mouse genomic sequence (Genome Browser [April 2003 freeze and February 2003 freeze, respectively]) suggest that ZNF41 is one of the KRAB zinc-finger genes for which there is no mouse orthologue. It is enticing to propose that this subset of KRAB/ZFPs may be specifically important for some of the psychological and cognitive traits that distinguish humans from lower mammals.
KRAB zinc-finger proteins are indeed involved in various aspects of physical and intellectual human development and are therefore excellent candidate genes for MR. Since several other potential causes have been ruled out, it is very likely that loss of functional ZNF41 causes the MR observed in our patient with translocation. The identification of two potentially disease-causing ZNF41 mutations in unrelated families with MRX further supports a role for this transcription factor in cognition. The presence of a leucine instead of a proline at ZNF41.1 amino acid position 111 (P111L) may cause structural changes that interfere with normal function of the protein. Perhaps of even greater significance is the mutation found in a second family that disrupts normal splicing of ZNF41. We have shown that two specific ZNF41 transcript variants that make use of the relevant splice site are absent in the patient cell line, suggesting that loss of function of these variants causes the phenotype in the index patient and in his mildly affected sister. It is noteworthy that there are several documented examples of X-linked cognitive disorders that exhibit wide intrafamilial phenotypic variability, sometimes presenting as behavior disorders or mild cognitive impairment in carrier females. In particular, this applies in the case of the L1CAM mutation in a large family described by Ruiz et al. (1995) in which some carrier females partially express the phenotype although others are normal. Similarly, RSK2 mutations, implicated in Coffin-Lowry syndrome (MIM 303600), are associated with a range of mild cognitive deficits in carrier females (Simensen et al. 2002). Also, mutations in the creatine-transporter gene SLC6A8 (MIM 300036), recently implicated in MR, result in various learning disabilities and behavior problems in heterozygous females (Salomons et al. 2001; Hahn et al. 2002). It is interesting that, like the ZNF41 mutation we identified in family P42, one of these familial SLC6A8 mutations is also associated with aberrant splicing (Hahn et al. 2002). It is clear that aberrant splicing can have a range of consequences for cognitive function. Indeed, such mutations in several genes—including both L1CAM and RSK2 (Finckh et al. 2000; Delaunoy et al. 2001) and ATRX (Villard et al. 1996b; Fichera et al. 2001)—have highlighted the link between splicing defects and various forms of MR.
It is also important to recognize that the frequency of causative mutations in any single MRX gene identified to date is relatively low. For example, a mutation in ARHGEF6 (MIM 300267), one of the 13 known genes implicated in monogenic forms of MR, was found in only 1 of 119 families with MRX screened (Kutsche et al. 2000). Alterations in ZNF41 are associated with MR in 2 of 210 unrelated patients screened, and, in addition, our patient with translocation has no functional ZNF41. We conclude that ZNF41 is a good candidate gene for MRX. Further studies will shed more light on the mechanisms by which mutations in the ZNF41 gene result in dysfunction of the brain.
Acknowledgments
We sincerely thank the members of the families and their clinicians, for participation in this study; V. des Portes, for help acquiring additional clinical details; S. Freier and H. Madle, for help with cell culture; R. Sudbrak, for ICRF YAC clones; and RZPD, for distributing clones. This work was supported by the German Human Genome Program (grant number 01KW99087), by the National Genome Research Network (project number 01GR0105), and by the 5th EU Framework, RTD Project-QLRT-2001-01810.
Electronic-Database Information
Accession numbers and URLs for data presented herein are as follows:
- Entrez SNP http://www.ncbi.nih.gov/entrez/query.fcgi?db=snp (for SNP 1192G→A reference sequence rs2498490 and SNP 1351T→G reference sequence rs2498170)
- GenBank, http://www.ncbi.nlm.nih.gov/Genbank/ (for RP13-83I24 [accession number AL590283], ZNF41.1 [accession number NM_007130], ZNF41.6 [accession number AJ010021], ZNF41.3 [accession number AJ010018], RP13-83I24 [accession number AL590283], RP13-479F17 [accession number AL590223], RP11-196D18 [accession number AC069285], ZNF41 protein [accession number NP_009061], spliced ESTs [accession number AW661872], mRNA sequences [accession number BC041841], RP11-30G10 [accession number AQ003212], RP4-733D15 [accession number AL023623], RP11-75A9 [accession number AL139811], RP1-71L16 [accession number AL022165], RP1-250J21 [accession number AL162583], RP3-484G21 [accession number AL163597], RP11-553I6 [accession number AL359698], RP1-306D1 [accession number Z83822], RP1-659F15 [accession number AL096791], RP11-571E6 [accession number AL591503], RP11-479F17 [accession number AL590223], RP11-83I24 [accession number AL590283], RP1-230G1 [accession number AQ489515], RP1-212G6 [accession number AL009172], RP3-473A14 [accession number AL031967], and RP3-393P12 [accession number AL022578])
- Integrated X Chromosome Database, http://ixdb.molgen.mpg.de/
- National Center for Disease Control and Prevention, http://www.cdc.gov/
- Online Mendelian Inheritance in Man (OMIM), http://www.ncbi.nlm.nih.gov/Omim/ (for FMR1, ARX, Partington syndrome, MECP2, AGTR2, FGD1, RSK2, ATRX, L1CAM, FMR2, ZNF41, KLF8, WT1, WAGR syndrome, KAP-1 [or TIF1β or KRIP-1], SETDB1, Rett syndrome, X-linked α-thalassemia/MR syndrome, Juberg-Marsidi syndrome, Coffin-Lowry syndrome, SLC6A8, and ARHGEF6)
- RZPD, http://www.rzpd.de/
- Whitehead Institute, http://www-genome.wi.mit.edu/cgi-bin/contig/phys_map
- UCSC Human Genome Bioinformatics, http://genome.ucsc.edu/
- UK Human Genome Mapping Project Resource Centre, http://www.hgmp.mrc.ac.uk/ (for NIX analysis)
- Wellcome Trust Sanger Institute, http://www.sanger.ac.uk/
References
- Amir RE, Van den Veyver IB, Wan M, Tran CQ, Francke U, Zoghbi HY (1999) Rett syndrome is caused by mutations in X-linked MECP2, encoding methyl-CpG-binding protein 2. Nat Genet 23:185–188 10.1038/13810 [DOI] [PubMed] [Google Scholar]
- Ballestar E, Wolffe AP (2001) Methyl-CpG-binding proteins: targeting specific gene repression. Eur J Biochem 268:1–6 10.1046/j.1432-1327.2001.01869.x [DOI] [PubMed] [Google Scholar]
- Bienvenu T, Poirier K, Friocourt G, Bahi N, Beaumont D, Fauchereau F, Ben Jeema L, Zemni R, Vinet MC, Francis F, Couvert P, Gomot M, Moraine C, van Bokhoven H, Kalscheuer V, Frints S, Gecz J, Ohzaki K, Chaabouni H, Fryns JP, Desportes V, Beldjord C, Chelly J (2002) ARX, a novel Prd-class-homeobox gene highly expressed in the telencephalon, is mutated in X-linked mental retardation. Hum Mol Genet 11:981–991 10.1093/hmg/11.8.981 [DOI] [PubMed] [Google Scholar]
- Brewer CM, Fredericks BJ, Pont JM, Stephenson JB, Tolmie JL (1996) X-linked hydrocephalus masquerading as spina bifida and destructive porencephaly in successive generations in one family. Dev Med Child Neurol 38:359–363 [DOI] [PubMed] [Google Scholar]
- Call K, Glaser T, Ito C, Buckler AJ, Pelletier J, Haber DA, Rose EA, Kral A, Yeger H, Lewis WH, Jones C, Housman DE (1990) Isolation and characterization of a zinc finger polypeptide gene at the human chromosome 11 Wilms’ tumor locus. Cell 60:509–520 [DOI] [PubMed] [Google Scholar]
- Chelly J, Mandel JL (2001) Monogenic causes of X-linked mental retardation. Nat Rev Genet 2:669–680 10.1038/35088558 [DOI] [PubMed] [Google Scholar]
- Couvert P, Bienvenu T, Aquaviva C, Poirier K, Moraine C, Gendrot C, Verloes A, Andres C, Le Fevre AC, Souville I, Steffann J, des Portes V, Ropers HH, Yntema HG, Fryns JP, Briault S, Chelly J, Cherif B (2001) MECP2 is highly mutated in X-linked mental retardation. Hum Mol Genet 10:941–946 10.1093/hmg/10.9.941 [DOI] [PubMed] [Google Scholar]
- Delaunoy J, Abidi F, Zeniou M, Jacquot S, Merienne K, Pannetier S, Schmitt M, Schwartz C, Hanauer A (2001) Mutations in the X-linked RSK2 gene (RPS6KA3) in patients with Coffin-Lowry syndrome. Hum Mutat 17:103–116 [DOI] [PubMed] [Google Scholar]
- Fichera M, Silengo M, Spalletta A, Giudice ML, Romano C, Ragusa A (2001) Prenatal diagnosis of ATR-X syndrome in a fetus with a new G>T splicing mutation in the XNP/ATR-X gene. Prenat Diagn 21:747–751 10.1002/pd.142 [DOI] [PubMed] [Google Scholar]
- Finckh U, Schroder J, Ressler B, Veske A, Gal A (2000) Spectrum and detection rate of L1CAM mutations in isolated and familial cases with clinically suspected L1-disease. Am J Med Genet 92:40–46 [DOI] [PubMed] [Google Scholar]
- Friedman JR, Fredericks WJ, Jensen DE, Speicher DW, Huang XP, Neilson EG, Rauscher FJ 3rd (1996) KAP-1, a novel corepressor for the highly conserved KRAB repression domain. Genes Dev 10:2067–2078 [DOI] [PubMed] [Google Scholar]
- Frints SG, Froyen G, Marynen P, Fryns JP (2002) X-linked mental retardation: vanishing boundaries between non-specific (MRX) and syndromic (MRXS) forms. Clin Genet 62:423–432 10.1034/j.1399-0004.2002.620601.x [DOI] [PubMed] [Google Scholar]
- Fryns JP, Spaepen A, Cassiman JJ, van den Berghe H (1991) X linked complicated spastic paraplegia, MASA syndrome, and X linked hydrocephalus owing to congenital stenosis of the aqueduct of Sylvius: variable expression of the same mutation at Xq28. J Med Genet 28:429–431 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fuks F, Hurd PJ, Wolf D, Nan X, Bird AP, Kouzarides T (2003) The methyl-CpG-binding protein MeCP2 links DNA methylation to histone methylation. J Biol Chem 278:4035–4040 10.1074/jbc.M210256200 [DOI] [PubMed] [Google Scholar]
- Gessler M, Konig A, Bruns GAP (1992) The genomic organization and expression of the WT1 gene. Genomics 12:807–813 [DOI] [PubMed] [Google Scholar]
- Gibbons RJ, Picketts DJ, Villard L, Higgs DR (1995) Mutations in a putative global transcriptional regulator cause X-linked mental retardation with α-thalassemia (ATR-X syndrome). Cell 80:837–845 [DOI] [PubMed] [Google Scholar]
- Hahn KA, Salomons GS, Tackels-Horne D, Wood TC, Taylor HA, Schroer RJ, Lubs HA, Jakobs C, Olson RL, Holden KR, Stevenson RE, Schwartz CE (2002) X-linked mental retardation with seizures and carrier manifestations is caused by a mutation in the creatine-transporter gene (SLC6A8) located in Xq28. Am J Hum Genet 70:1349–1356 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanauer A, Young ID (2002) Coffin-Lowry syndrome: clinical and molecular features. J Med Genet 39:705–713 10.1136/jmg.39.10.705 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hendrich B, Bickmore W (2001) Human diseases with underlying defects in chromatin structure and modification. Hum Mol Genet 10:2233–2242 10.1093/hmg/10.20.2233 [DOI] [PubMed] [Google Scholar]
- Hentze MW, Kulozik AE (1999) A perfect message: RNA surveillance and nonsense-mediated decay. Cell 96:307–310 [DOI] [PubMed] [Google Scholar]
- Jin P, Warren ST (2003) New insights into fragile X syndrome: from molecules to neurobehaviors. Trends Biochem Sci 28:152–158 10.1016/S0968-0004(03)00033-1 [DOI] [PubMed] [Google Scholar]
- Jouet M, Rosenthal A, Armstrong G, MacFarlane J, Stevenson R, Paterson J, Metzenberg A, Ionasescu V, Temple K, Kenwrick S (1994) X-linked spastic paraplegia (SPG1), MASA syndrome and X-linked hydrocephalus result from mutations in the L1 gene. Nat Genet 7:402–407 [DOI] [PubMed] [Google Scholar]
- Kim SS, Chen YM, O’Leary E, Witzgall R, Vidal M, Bonventre JV (1996) A novel member of the RING finger family, KRIP-1, associates with the KRAB-A transcriptional repressor domain of zinc finger proteins. Proc Natl Acad Sci USA 93:15299–15304 10.1073/pnas.93.26.15299 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kutsche K, Yntema H, Brandt A, Jantke I, Nothwang HG, Orth U, Boavida MG, David D, Chelly J, Fryns JP, Moraine C, Ropers HH, Hamel BC, van Bokhoven H, Gal A (2000) Mutations in ARHGEF6, encoding a guanine nucleotide exchange factor for Rho GTPases, in patients with X-linked mental retardation. Nat Genet 26:247–250 10.1038/80002 [DOI] [PubMed] [Google Scholar]
- Ladomery M, Dellaire G (2002) Multifunctional zinc finger proteins in development and disease. Ann Hum Genet 66:331–342 10.1046/j.1469-1809.2002.00121.x [DOI] [PubMed] [Google Scholar]
- Lebel RR, May M, Pouls S, Lubs HA, Stevenson RE, Schwartz CE (2002) Non-syndromic X-linked mental retardation associated with a missense mutation (P312L) in the FGD1 gene. Clin Genet 61:139–145 10.1034/j.1399-0004.2002.610209.x [DOI] [PubMed] [Google Scholar]
- Lossi AM, Laugier-Anfossi F, Depetris D, Gecz J, Gedeon A, Kooy F, Schwartz C, Mattei MG, Croquette MF, Villard L (2002) Abnormal expression of the KLF8 (ZNF741) gene in a female patient with an X;autosome translocation t(X;21)(p11.2;q22.3) and non-syndromic mental retardation. J Med Genet 39:113–117 10.1136/jmg.39.2.113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moosmann P, Georgiev O, Le Douarin B, Bourquin JP, Schaffner W (1996) Transcriptional repression by RING finger protein TIF1 β that interacts with the KRAB repressor domain of KOX1. Nucleic Acids Res 24:4859–4867 10.1093/nar/24.24.4859 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pasteris NG, Cadle A, Logie LJ, Porteous ME, Schwartz CE, Stevenson RE, Glover TW, Wilroy RS, Gorski JL (1994) Isolation and characterization of the faciogenital dysplasia (Aarskog-Scott syndrome) gene: a putative Rho/Rac guanine nucleotide exchange factor. Cell 79:669–678 [DOI] [PubMed] [Google Scholar]
- Peng H, Begg GE, Harper SL, Friedman JR, Speicher DW, Rauscher FJ 3rd (2000a) Biochemical analysis of the Kruppel-associated box (KRAB) transcriptional repression domain. J Biol Chem 275:18000–18010 10.1074/jbc.M001499200 [DOI] [PubMed] [Google Scholar]
- Peng H, Begg GE, Schultz DC, Friedman JR, Jensen DE, Speicher DW, Rauscher FJ 3rd (2000b) Reconstitution of the KRAB-KAP-1 repressor complex: a model system for defining the molecular anatomy of RING-B box-coiled-coil domain-mediated protein-protein interactions. J Mol Biol 295:1139–1162 10.1006/jmbi.1999.3402 [DOI] [PubMed] [Google Scholar]
- Ropers HH, Hoeltzenbein M, Kalscheuer V, Yntema H, Hamel B, Fryns JP, Chelly J, Partington M, Gecz J, Moraine C (2003) Nonsyndromic X-linked mental retardation: where are the missing mutations? Trends Genet 19:316–320 10.1016/S0168-9525(03)00113-6 [DOI] [PubMed] [Google Scholar]
- Rosati M, Franz A, Matarazzo MR, Grimaldi G (1999) Coding region intron/exon organization, alternative splicing, and X-chromosome inactivation of the KRAB/FPB-domain-containing human zinc finger gene ZNF41. Cytogenet Cell Genet 85:291–296 [DOI] [PubMed] [Google Scholar]
- Rose EA, Glaser T, Jones C, Smith CL, Lewis WH, Call KM, Minden M, Champagne E, Bonetta L, Yeger H, Housman DE (1990) Complete physical map of the WAGR region of 11p13 localizes a candidate Wilms’ tumor gene. Cell 60:495–508 [DOI] [PubMed] [Google Scholar]
- Ruiz JC, Cuppens H, Legius E, Fryns JP, Glover T, Marynen P, Cassiman JJ (1995) Mutations in L1-CAM in two families with X linked complicated spastic paraplegia, MASA syndrome, and HSAS. J Med Genet 32:549–552 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ryan RF, Schultz DC, Ayyanathan K, Singh PB, Friedman JR, Fredericks WJ, Rauscher FJ 3rd (1999) KAP-1 corepressor protein interacts and colocalizes with heterochromatic and euchromatic HP1 proteins: a potential role for Kruppel-associated box-zinc finger proteins in heterochromatin-mediated gene silencing. Mol Cell Biol 19:4366–4378 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salomons GS, van Dooren SJM, Verhoeven NM, Cecil KM, Ball WS, Degrauw TJ, Jakobs C (2001) X-linked creatine-transporter gene (SLC6A8) defect: a new creatine-deficiency syndrome. Am J Hum Genet 68:1497–1500 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidt M, DuSart D (1992) Functional disomies for the X chromosome influence the cell selection and hence the X inactivation pattern in females with balanced X-autosome translocations: a review of 122 cases. Am J Med Genet 42:161–169 [DOI] [PubMed] [Google Scholar]
- Schultz DC, Ayyanathan K, Negorev D, Maul GG, Rauscher FJ 3rd (2002) SETDB1: a novel KAP-1-associated histone H3, lysine 9-specific methyltransferase that contributes to HP1-mediated silencing of euchromatic genes by KRAB zinc-finger proteins. Genes Dev 16:919–932 10.1101/gad.973302 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shannon M, Hamilton AT, Gordon L, Branscomb E, Stubbs L (2003) Differential expansion of zinc-finger transcription factor Loci in homologous human and mouse gene clusters. Genome Res 13:1097–1110 10.1101/gr.963903 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siebert PD, Chenchik A, Kellogg DE, Lukyanov KA, Lukyanov SA (1995) An improved PCR method for walking in uncloned genomic DNA. Nucleic Acids Res 23:1087–1088 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simensen RJ, Abidi F, Collins JS, Schwartz CE, Stevenson RE (2002) Cognitive function in Coffin-Lowry syndrome. Clin Genet 61:299–304 10.1034/j.1399-0004.2002.610410.x [DOI] [PubMed] [Google Scholar]
- Stromme P, Mangelsdorf ME, Shaw MA, Lower KM, Lewis SM, Bruyere H, Lutcherath V, Gedeon AK, Wallace RH, Scheffer IE, Turner G, Partington M, Frints SG, Fryns JP, Sutherland GR, Mulley JC, Gecz J (2002) Mutations in the human ortholog of Aristaless cause X-linked mental retardation and epilepsy. Nat Genet 30:441–445 10.1038/ng862 [DOI] [PubMed] [Google Scholar]
- Trivier E, De Cesare D, Jacquot S, Pannetier S, Zackai E, Young I, Mandel JL, Sassone-Corsi P, Hanauer A (1996) Mutations in the kinase Rsk-2 associated with Coffin-Lowry syndrome. Nature 384:567–570 10.1038/384567a0 [DOI] [PubMed] [Google Scholar]
- Vervoort VS, Beachem MA, Edwards PS, Ladd S, Miller KE, de Mollerat X, Clarkson K, DuPont B, Schwartz CE, Stevenson RE, Boyd E, Srivastava AK (2002) AGTR2 mutations in X-linked mental retardation. Science 296:2401–2403 [DOI] [PubMed] [Google Scholar]
- Villard L, Gecz J, Mattei JF, Fontes M, Saugier-Veber P, Munnich A, Lyonnet S (1996a) XNP mutation in a large family with Juberg-Marsidi syndrome. Nat Genet 12:359–360 [DOI] [PubMed] [Google Scholar]
- Villard L, Toutain A, Lossi A-M, Gecz J, Houdayer C, Moraine C, Fontès M (1996b) Splicing mutation in the ATR-X gene can lead to a dysmorphic mental retardation phenotype without α-thalassemia. Am J Hum Genet 58:499–505 [PMC free article] [PubMed] [Google Scholar]
- Warburton D (1991) De novo balanced chromosome rearrangements and extra marker chromosomes identified at prenatal diagnosis: clinical significance and distribution of breakpoints. Am J Hum Genet 49:995–1013 [PMC free article] [PubMed] [Google Scholar]
- Wirth J, Nothwang HG, van der Maarel S, Menzel C, Borck G, Lopez-Pajares I, Brondum-Nielsen K, Tommerup N, Bugge M, Ropers HH, Haaf T (1999) Systematic characterisation of disease associated balanced chromosome rearrangements by FISH: cytogenetically and genetically anchored YACs identify microdeletions and candidate regions for mental retardation genes. J Med Genet 36:271–278 [PMC free article] [PubMed] [Google Scholar]
- Yntema HG, Oudakker AR, Kleefstra T, Hamel BC, van Bokhoven H, Chelly J, Kalscheuer VM, Fryns JP, Raynaud M, Moizard MP, Moraine C (2002) In-frame deletion in MECP2 causes mild nonspecific mental retardation. Am J Med Genet 107:81–83 10.1002/ajmg.10085 [DOI] [PubMed] [Google Scholar]