Abstract
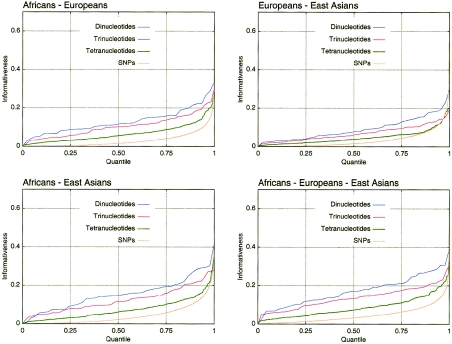
Inference of individual ancestry is useful in various applications, such as admixture mapping and structured-association mapping. Using information-theoretic principles, we introduce a general measure, the informativeness for assignment (In), applicable to any number of potential source populations, for determining the amount of information that multiallelic markers provide about individual ancestry. In a worldwide human microsatellite data set, we identify markers of highest informativeness for inference of regional ancestry and for inference of population ancestry within regions; these markers, which are listed in online-only tables in our article, can be useful both in testing for and in controlling the influence of ancestry on case-control genetic association studies. Markers that are informative in one collection of source populations are generally informative in others. Informativeness of random dinucleotides, the most informative class of microsatellites, is five to eight times that of random single-nucleotide polymorphisms (SNPs), but 2%–12% of SNPs have higher informativeness than the median for dinucleotides. Our results can aid in decisions about the type, quantity, and specific choice of markers for use in studies of ancestry.
Introduction
Inference of individual ancestry from genetic markers is helpful in diverse situations, including admixture and association mapping, forensics, prediction of medical risks, wildlife management, and studies of dispersal, gene flow, and evolutionary history (Shriver et al. 1997; Davies et al. 1999; Primmer et al. 2000; Manel et al. 2002; Bamshad et al. 2003; Campbell et al. 2003; Ziv and Burchard 2003). Statistical methods for ancestry inference use multilocus genotypes and population allele frequencies, either specified in advance or estimated during the inference process, to assign populations of origin to individuals (Smouse et al. 1982; Paetkau et al. 1995; Rannala and Mountain 1997; Cornuet et al. 1999; Pritchard et al. 2000; Guinand et al. 2002).
Because use of highly informative markers can reduce the amount of genotyping required for ancestry inference, it is desirable to measure the extent to which specific markers contribute to this inference. Several approaches have previously been used for measuring these locus contributions (table 1). However, despite their various features in specific scenarios, all of these measures are either difficult to compute, not designed specifically for estimating marker information content, or not applicable to sets with many potential source populations.
Table 1.
Measures of Marker Information Content[Note]
| Criterion | Description | Features | Limitations |
| Absolute allele frequency difference (δ) | |p11-p21| | Is related to amount of linkage disequilibrium in an admixture model (Chakraborty and Weiss 1988); is related to probability of correct assignment in a multilocus no-admixture model (Risch et al. 2002); is related to Fisher information curvature criterion for K=2 (eq. [18]); is related to ORCA for K=2 (eq. [11]) | Requires that only two populations be possible sources; does not take into account all available information about allele frequencies (Stephens et al. 1999; Campbell et al. 2003); statistical features do not apply to the Shriver et al. (1997) multiallelic extension of δ |
| Fst | Excess in probability of identity of alleles from the same population compared with randomly chosen alleles (Excoffier 2001, for example) | Is related, for biallelic markers, to the quotient of expected posterior and prior variance of ancestry in a population equally admixed from two sourcesa (McKeigue 1998; Molokhia et al. 2003) | Performs only slightly better than random markers (Rosenberg et al. 2001) |
| Expected heterozygosityb |
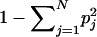 (bias correction can be applied in estimation from data) (bias correction can be applied in estimation from data) |
Performs better than random markers (Rosenberg et al. (2001) and fig. 3) | Measures the amount of variation but not the differences across populations |
| Number of allelesb | N | Performs better than random markers (Rosenberg et al. 2001) | Measures the amount of variation but not the differences across populations; is useful only for multiallelic markers that have variation in number of alleles |
| Fisher information curvature criterion | Reciprocal of the largest eigenvalue of the information matrix for maximum-likelihood estimation of ancestry coefficients (Gomulkiewicz et al. 1990; Millar 1991) | Enables predictions about approximate variances of ancestry estimates (see “Number of Markers” subsection of the “Theory” section); information matrix is additive across loci that are independent within populations | Depends on unknown ancestry coefficients and requires computation for many possible parameter values; largest eigenvalue gives an upper bound that might not be generally applicable across the parameter space |
| Pairwise Kullback-Leibler divergencec |
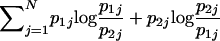 (Brenner 1998; Smith et al. 2001; Anderson and Thompson 2002) (Brenner 1998; Smith et al. 2001; Anderson and Thompson 2002) |
Provides a natural measure, for K=2, of average potential for assignment of an allele to one population compared with the other; has a natural multilocus extension; enables measurement of contributions of specific alleles | Requires that only two populations be possible sources; has upwardly biased estimates in small samples |
| Informativeness for assignment (In) | Equation (4) | Provides a natural measure of potential for assignment of an allele to one population compared with the “average” population; has a natural multilocus extension; enables measurement of contributions of specific alleles or populations; performs better than random or highly heterozygous markers (fig. 3) | Has upwardly biased estimates in small samples |
| Informativeness for ancestry coefficients (Ia) | Equation (14) | Provides a natural measure of potential for assignment of an allele to a point on the set of all possible ancestry coefficient vectors; has a natural multilocus extension; enables measurement of contributions of specific alleles | Has upwardly biased estimates in small samples; is difficult to compute in samples with populations of equal sample size |
| Optimal rate of correct assignment (ORCA) | Equation (10) | Gives the probability of correct assignment of an allele using the decision rule with lowest risk; has a natural multilocus extension (eq. [12]); enables measurement of contributions of specific alleles | Has upwardly biased estimates in small samples |
Note.— Notation is defined in the “Theory” section. All criteria apply to multiallelic loci in any number of populations, except where specified.
A multiallelic statistic related to this ratio was suggested by Molokhia et al. (2003).
Can also be calculated by using average values of the statistic across populations rather than by using values for the whole collection of populations.
A similar statistic based on genotype frequencies was suggested by Shriver et al. (1997). Some authors multiply by a factor of 1/2 in the formula for this statistic.
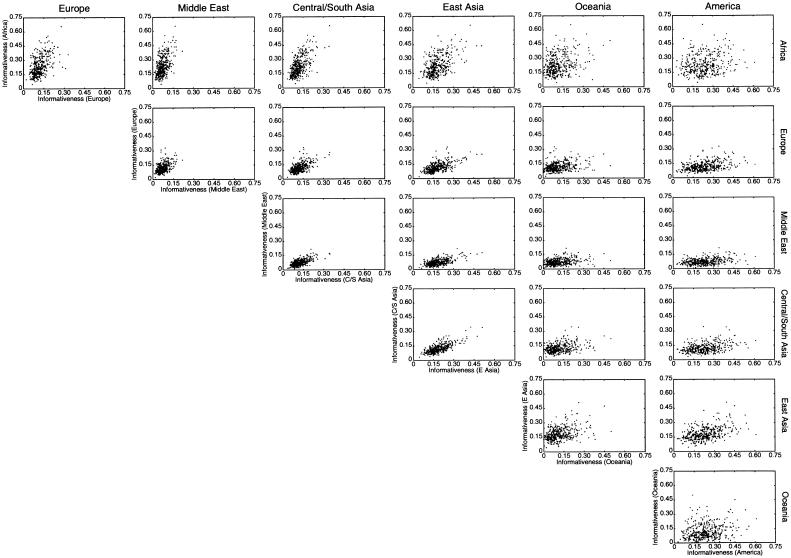
Here, using information-theoretic and decision-theoretic approaches, we introduce new criteria: the informativeness for assignment, the optimal rate of correct assignment, and the informativeness for ancestry coefficients. The choice of statistic for use in identifying markers for ancestry inference depends on the inference algorithm that is being used (table 1). The new statistics, as convenient and statistically motivated general measures applicable to any number of alleles and populations, may be useful both in admixed and in multisource human groups, such as those that have formed in the Western Hemisphere by the intermixing of Africans, Native Americans, and Europeans. We first define the statistics, consider their relationships with δ and Fst (two criteria that are often used to measure marker information content), and study the number of markers needed for inference. We demonstrate that the criteria are highly correlated and proceed using only the informativeness for assignment, or, simply, the informativeness. Informativeness rankings of loci in human microsatellite data are found to be robust, and use of markers of highest informativeness is observed to reduce the number of markers needed for inference of population structure. We consider the relationships of informativeness values in different subsets of the human population, and the relative informativeness of microsatellites and SNPs. Tables A, B, C, D, and E (online only) provide lists of informativeness ranks for various sets of source populations.
Table A.
Informativeness (In) for Distinguishing Groups within Data Sets[Note]
|
In Rank for Data Set |
||||||||||||
| Locus Name | Alternate Name | Repeat Size | World-52 | World-5 | World-7 | Africa | Europe | Middle East | Central/South Asia | East Asia | Oceania | America |
| D10S1208 | ATA5A04 | 3 | 95 | 61 | 73 | 174 | 239 | 212 | 124 | 32 | 211 | 114 |
| D10S1221 | ATA21A03 | 3 | 71 | 165 | 128 | 152 | 56 | 45 | 20 | 137 | 172 | 89 |
| D10S1222 | ATA22D02 | 3 | 135 | 109 | 137 | 167 | 26 | 123 | 106 | 140 | 21 | 325 |
| D10S1225 | ATA24F10 | 3 | 303 | 178 | 204 | 104 | 273 | 82 | 296 | 352 | 285 | 269 |
| D10S1230 | ATA29C03 | 3 | 197 | 149 | 152 | 217 | 179 | 225 | 235 | 345 | 140 | 119 |
| D10S1239 | GATA64A09 | 4 | 291 | 347 | 308 | 349 | 299 | 371 | 263 | 158 | 64 | 231 |
| D10S1248 | GGAA23C05 | 4 | 273 | 256 | 269 | 211 | 322 | 257 | 342 | 271 | 89 | 42 |
| D10S1412 | ATA31G11 | 3 | 258 | 167 | 159 | 306 | 116 | 245 | 304 | 261 | 351 | 359 |
| D10S1423 | GATA70E11 | 4 | 327 | 350 | 341 | 340 | 169 | 367 | 256 | 297 | 131 | 189 |
| D10S1425 | GATA71C09 | 4 | 196 | 153 | 190 | 264 | 287 | 110 | 90 | 326 | 233 | 158 |
| D10S1426 | GATA73E11 | 4 | 363 | 377 | 374 | 373 | 124 | 316 | 341 | 253 | 279 | 212 |
| D10S1430 | GATA84C01 | 4 | 259 | 346 | 347 | 312 | 267 | 74 | 255 | 70 | 265 | 29 |
| D10S1432 | GATA87G01 | 4 | 365 | 351 | 345 | 366 | 360 | 290 | 335 | 319 | 230 | 245 |
| D10S1435 | GATA88F09 | 4 | 313 | 348 | 356 | 226 | 271 | 293 | 262 | 293 | 320 | 141 |
| D10S189 | AFM063XF4 | 2 | 274 | 239 | 219 | 348 | 342 | 315 | 366 | 348 | 174 | 78 |
| D10S212 | AFM198ZB4 | 2 | 229 | 244 | 264 | 103 | 148 | 321 | 258 | 231 | 342 | 93 |
| D10S2327 | GGAT1A4 | 4 | 190 | 148 | 166 | 96 | 294 | 363 | 257 | 320 | 3 | 163 |
| D10S2470 | GATA115E01 | 4 | 224 | 205 | 229 | 367 | 367 | 163 | 226 | 237 | 121 | 203 |
| D10S677 | GGAA2F11 | 4 | 150 | 174 | 171 | 147 | 21 | 139 | 157 | 188 | 364 | 137 |
| D11S1304 | UT2095 | 4 | 36 | 38 | 53 | 112 | 254 | 108 | 31 | 80 | 78 | 184 |
| D11S1392 | GATA6B09 | 4 | 348 | 296 | 314 | 324 | 309 | 128 | 274 | 305 | 332 | 360 |
| D11S1981 | GATA48E02 | 4 | 186 | 240 | 244 | 327 | 156 | 164 | 173 | 122 | 126 | 45 |
| D11S1984 | GGAA17G05 | 4 | 24 | 93 | 79 | 30 | 42 | 33 | 13 | 100 | 110 | 60 |
| D11S1986 | GGAA7G08 | 4 | 11 | 108 | 94 | 17 | 11 | 38 | 3 | 6 | 97 | 23 |
| D11S1993 | ATA1B07 | 3 | 68 | 34 | 34 | 183 | 138 | 73 | 97 | 219 | 75 | 53 |
| D11S1998 | GATA23E06 | 4 | 242 | 294 | 287 | 240 | 363 | 372 | 298 | 104 | 239 | 221 |
| D11S1999 | GATA23F06 | 4 | 130 | 128 | 127 | 272 | 131 | 243 | 188 | 142 | 112 | 275 |
| D11S2000 | GATA28D01 | 4 | 3 | 45 | 43 | 12 | 7 | 3 | 2 | 1 | 20 | 28 |
| D11S2002 | GATA30G01 | 4 | 256 | 324 | 277 | 346 | 172 | 303 | 96 | 170 | 219 | 164 |
| D11S2006 | GATA46A12 | 4 | 376 | 371 | 371 | 227 | 357 | 327 | 265 | 341 | 143 | 374 |
| D11S2362 | ATA33B03 | 3 | 156 | 96 | 118 | 119 | 193 | 287 | 239 | 276 | 296 | 339 |
| D11S2363 | GATA12F04 | 4 | 62 | 150 | 133 | 76 | 23 | 218 | 61 | 39 | 92 | 30 |
| D11S2365 | GATA63F09 | 4 | 332 | 319 | 322 | 186 | 282 | 362 | 355 | 266 | 250 | 174 |
| D11S2371 | GATA90D07 | 4 | 342 | 340 | 353 | 233 | 288 | 283 | 234 | 265 | 142 | 368 |
| D11S4459 | ATA9B04 | 3 | 222 | 90 | 97 | 321 | 291 | 118 | 360 | 372 | 300 | 371 |
| D11S4463 | GATA117D01 | 4 | 316 | 365 | 358 | 291 | 65 | 342 | 250 | 304 | 350 | 219 |
| D11S4464 | GATA64D03 | 4 | 279 | 334 | 337 | 235 | 49 | 260 | 198 | 211 | 155 | 129 |
| D11S969 | AFM205VF10 | 2 | 159 | 80 | 98 | 90 | 228 | 213 | 317 | 172 | 275 | 300 |
| D12S1042 | ATA27A06 | 3 | 133 | 134 | 134 | 79 | 152 | 205 | 305 | 163 | 120 | 63 |
| D12S1045 | ATA29A06 | 3 | 92 | 52 | 49 | 142 | 64 | 78 | 170 | 200 | 102 | 349 |
| D12S1052 | GATA26D02 | 4 | 278 | 274 | 282 | 320 | 13 | 248 | 323 | 268 | 55 | 252 |
| D12S1064 | GATA63D12 | 4 | 194 | 242 | 231 | 91 | 293 | 289 | 192 | 229 | 269 | 80 |
| D12S1294 | GATA73H09 | 4 | 143 | 200 | 220 | 143 | 69 | 100 | 76 | 212 | 164 | 98 |
| D12S1300 | GATA85A04 | 4 | 309 | 259 | 235 | 278 | 374 | 129 | 297 | 311 | 302 | 153 |
| D12S1301 | GATA91H06 | 4 | 141 | 41 | 63 | 195 | 277 | 182 | 351 | 310 | 304 | 356 |
| D12S1638 | AFMB002VD5 | 2 | 271 | 207 | 212 | 288 | 317 | 336 | 89 | 279 | 205 | 277 |
| D12S2070 | ATA25F09 | 3 | 56 | 12 | 13 | 41 | 107 | 214 | 107 | 328 | 303 | 297 |
| D12S2078 | GATA32F05 | 4 | 241 | 213 | 226 | 215 | 161 | 185 | 208 | 208 | 61 | 226 |
| D12S269 | MFD259 | 2 | 67 | 53 | 62 | 65 | 38 | 250 | 111 | 99 | 241 | 104 |
| D12S297 | UT5029 | 4 | 52 | 99 | 67 | 144 | 61 | 25 | 51 | 33 | 44 | 39 |
| D12S372 | GATA4H03 | 4 | 334 | 359 | 346 | 341 | 266 | 322 | 352 | 215 | 254 | 56 |
| D12S373 | GATA6C01 | 4 | 321 | 234 | 232 | 358 | 346 | 208 | 291 | 351 | 186 | 227 |
| D12S395 | GATA4H01 | 4 | 221 | 281 | 275 | 332 | 203 | 314 | 131 | 90 | 326 | 58 |
| D13S1493 | GGAA29H03 | 4 | 217 | 201 | 211 | 151 | 119 | 255 | 197 | 120 | 162 | 211 |
| D13S1807 | GATA11C08 | 4 | 235 | 245 | 261 | 284 | 298 | 341 | 316 | 95 | 127 | 351 |
| D13S285 | AFM309VA9 | 2 | 17 | 120 | 120 | 36 | 18 | 13 | 46 | 8 | 8 | 4 |
| D13S317 | GATA7G10 | 4 | 123 | 84 | 117 | 120 | 183 | 294 | 168 | 275 | 10 | 157 |
| D13S779 | ATA26D07 | 3 | 290 | 169 | 195 | 351 | 364 | 332 | 301 | 256 | 124 | 358 |
| D13S787 | GATA23C03 | 4 | 312 | 355 | 357 | 337 | 349 | 175 | 47 | 260 | 372 | 59 |
| D13S793 | GATA43H03 | 4 | 243 | 147 | 153 | 222 | 205 | 133 | 364 | 221 | 144 | 150 |
| D13S796 | GATA51B02 | 4 | 286 | 270 | 263 | 319 | 247 | 368 | 370 | 187 | 298 | 99 |
| D13S800 | GATA64F08 | 4 | 266 | 271 | 289 | 294 | 211 | 296 | 236 | 130 | 346 | 61 |
| D13S894 | GATA86H01 | 4 | 299 | 310 | 306 | 243 | 224 | 273 | 354 | 238 | 369 | 265 |
| D13S895 | GGAA22G01 | 4 | 249 | 290 | 281 | 210 | 110 | 112 | 139 | 189 | 331 | 244 |
| D14S1007 | AFMB002ZF1 | 2 | 6 | 8 | 3 | 7 | 125 | 114 | 14 | 117 | 1 | 314 |
| D14S1280 | GATA31B09 | 4 | 289 | 220 | 250 | 301 | 353 | 329 | 333 | 274 | 242 | 292 |
| D14S1426 | GATA136B01 | 4 | 57 | 23 | 38 | 296 | 166 | 86 | 67 | 18 | 294 | 217 |
| D14S1434 | GATA168F06 | 4 | 282 | 285 | 295 | 309 | 324 | 320 | 224 | 182 | 203 | 288 |
| D14S587 | GGAA10C09 | 4 | 166 | 194 | 201 | 97 | 122 | 158 | 108 | 107 | 57 | 229 |
| D14S588 | GGAA4A12 | 4 | 85 | 71 | 59 | 224 | 127 | 61 | 126 | 112 | 209 | 68 |
| D14S592 | ATA19H08 | 3 | 132 | 154 | 154 | 72 | 175 | 187 | 223 | 60 | 198 | 140 |
| D14S599 | ATA29G03 | 3 | 98 | 31 | 32 | 205 | 207 | 154 | 163 | 199 | 122 | 353 |
| D14S606 | GATA30A03 | 4 | 328 | 275 | 270 | 369 | 340 | 258 | 84 | 335 | 354 | 242 |
| D14S608 | GATA43H01 | 4 | 120 | 152 | 160 | 161 | 153 | 24 | 154 | 89 | 137 | 90 |
| D14S617 | GGAA21G11 | 4 | 146 | 30 | 54 | 84 | 144 | 209 | 196 | 136 | 291 | 287 |
| D14S742 | GATA74E02 | 4 | 345 | 364 | 362 | 201 | 290 | 120 | 307 | 285 | 213 | 329 |
| D15S128 | AFM273YF9 | 2 | 43 | 101 | 90 | 39 | 50 | 39 | 81 | 29 | 111 | 207 |
| D15S1507 | GATA151F03 | 4 | 276 | 193 | 249 | 277 | 240 | 270 | 315 | 360 | 228 | 192 |
| D15S1515 | GATA197B10 | 4 | 177 | 191 | 180 | 74 | 233 | 92 | 133 | 302 | 229 | 52 |
| D15S165 | AFM248VC5 | 2 | 14 | 29 | 26 | 3 | 46 | 12 | 9 | 157 | 194 | 79 |
| D15S642 | GATA27A03 | 4 | 31 | 158 | 110 | 192 | 2 | 238 | 75 | 59 | 197 | 106 |
| D15S643 | GATA50G06 | 4 | 113 | 251 | 206 | 110 | 12 | 49 | 32 | 43 | 216 | 82 |
| D15S652 | ATA24A08 | 3 | 41 | 68 | 57 | 108 | 79 | 122 | 59 | 47 | 63 | 40 |
| D15S655 | ATA28G05 | 3 | 244 | 137 | 149 | 133 | 191 | 276 | 344 | 356 | 310 | 295 |
| D15S659 | GATA63A03 | 4 | 162 | 184 | 207 | 136 | 88 | 149 | 79 | 49 | 276 | 162 |
| D15S816 | GATA73F01 | 4 | 304 | 277 | 291 | 157 | 285 | 356 | 336 | 299 | 322 | 346 |
| D15S818 | GATA85D02 | 4 | 366 | 354 | 355 | 283 | 344 | 364 | 359 | 353 | 317 | 88 |
| D15S822 | GATA88H02 | 4 | 22 | 162 | 112 | 102 | 27 | 99 | 10 | 4 | 22 | 72 |
| D16S2616 | ATA41E04 | 3 | 193 | 113 | 132 | 137 | 112 | 351 | 270 | 250 | 214 | 271 |
| D16S2621 | GATA71F09 | 4 | 117 | 78 | 101 | 285 | 189 | 288 | 217 | 179 | 71 | 311 |
| D16S2624 | GATA81D12 | 4 | 251 | 253 | 253 | 305 | 314 | 354 | 251 | 242 | 237 | 323 |
| D16S3253 | GATA22F09 | 4 | 203 | 198 | 191 | 314 | 311 | 324 | 43 | 67 | 375 | 319 |
| D16S3396 | ATA55A11 | 3 | 211 | 276 | 276 | 229 | 133 | 249 | 202 | 88 | 38 | 115 |
| D16S3401 | 16PTEL06 | 2 | 16 | 22 | 17 | 57 | 31 | 9 | 42 | 13 | 73 | 66 |
| D16S403 | AFM049XD2 | 2 | 102 | 55 | 58 | 164 | 158 | 186 | 95 | 134 | 101 | 6 |
| D16S422 | AFM249XC5 | 2 | 8 | 70 | 41 | 24 | 37 | 35 | 17 | 14 | 28 | 74 |
| D16S516 | AFM350VD1 | 2 | 129 | 176 | 157 | 69 | 221 | 70 | 99 | 176 | 207 | 296 |
| D16S539 | GATA11C06 | 4 | 311 | 301 | 292 | 318 | 280 | 345 | 347 | 162 | 215 | 222 |
| D16S748 | ATA3A07 | 3 | 88 | 54 | 46 | 93 | 93 | 60 | 38 | 144 | 361 | 342 |
| D16S753 | GGAA3G05 | 4 | 214 | 225 | 239 | 168 | 243 | 140 | 164 | 267 | 50 | 191 |
| D16S764 | GATA42E11 | 4 | 369 | 322 | 330 | 377 | 358 | 226 | 280 | 355 | 181 | 248 |
| D16S769 | GATA71H05 | 4 | 374 | 356 | 366 | 287 | 256 | 298 | 374 | 366 | 202 | 179 |
| D17S1290 | GATA49C09 | 4 | 124 | 177 | 189 | 87 | 104 | 85 | 53 | 31 | 104 | 154 |
| D17S1294 | GGAA9D03 | 4 | 231 | 336 | 333 | 15 | 264 | 308 | 190 | 295 | 220 | 145 |
| D17S1298 | GAAT2C03 | 4 | 215 | 230 | 222 | 37 | 352 | 335 | 345 | 364 | 337 | 304 |
| D17S1299 | GATA25A04 | 4 | 347 | 368 | 367 | 313 | 84 | 228 | 321 | 292 | 293 | 177 |
| D17S1301 | GATA28D11 | 4 | 318 | 333 | 336 | 335 | 331 | 326 | 373 | 152 | 163 | 173 |
| D17S1308 | GTAT1A05 | 4 | 294 | 254 | 196 | 307 | 268 | 347 | 230 | 325 | 377 | 327 |
| D17S2180 | ATC6A06 | 3 | 302 | 265 | 260 | 350 | 296 | 338 | 357 | 294 | 114 | 352 |
| D17S2193 | ATA43A10Z | 3 | 109 | 48 | 65 | 19 | 310 | 107 | 132 | 222 | 357 | 305 |
| D17S2195 | ATA58A02 | 3 | 101 | 49 | 47 | 67 | 341 | 127 | 118 | 98 | 19 | 235 |
| D17S2196 | GATA185H04 | 4 | 152 | 112 | 136 | 204 | 251 | 177 | 117 | 165 | 159 | 263 |
| D17S784 | AFM044XG3 | 2 | 75 | 69 | 95 | 78 | 95 | 251 | 204 | 131 | 66 | 116 |
| D17S974 | GATA8C04 | 4 | 284 | 237 | 254 | 326 | 302 | 311 | 331 | 249 | 133 | 70 |
| D18S1357 | ATA7D07 | 3 | 176 | 173 | 193 | 124 | 307 | 146 | 273 | 92 | 225 | 110 |
| D18S1364 | GATA7E12 | 4 | 118 | 146 | 147 | 228 | 73 | 330 | 172 | 83 | 81 | 2 |
| D18S1370 | ATA45G06 | 3 | 184 | 139 | 139 | 184 | 201 | 97 | 244 | 287 | 234 | 213 |
| D18S1371 | GATA177C03 | 4 | 356 | 264 | 274 | 371 | 320 | 323 | 290 | 324 | 200 | 284 |
| D18S1376 | GATA185C06 | 4 | 373 | 353 | 348 | 297 | 246 | 265 | 363 | 344 | 260 | 340 |
| D18S1390 | 18QTEL11 | 2 | 134 | 131 | 121 | 59 | 89 | 138 | 207 | 283 | 301 | 259 |
| D18S535 | GATA13 | 4 | 216 | 82 | 99 | 376 | 283 | 306 | 167 | 149 | 5 | 334 |
| D18S542 | GATA11A06 | 4 | 72 | 56 | 37 | 27 | 63 | 200 | 161 | 96 | 267 | 155 |
| D18S843 | ACT1A01 | 3 | 346 | 311 | 321 | 310 | 157 | 307 | 349 | 340 | 243 | 313 |
| D18S851 | GATA6D09 | 4 | 262 | 258 | 247 | 220 | 313 | 169 | 242 | 308 | 53 | 350 |
| D18S858 | ATA23G05 | 3 | 245 | 175 | 185 | 196 | 220 | 84 | 248 | 233 | 368 | 161 |
| D18S877 | GATA64H04 | 4 | 297 | 314 | 332 | 279 | 117 | 235 | 195 | 259 | 249 | 109 |
| D19S1034 | GATA21G05 | 4 | 263 | 208 | 216 | 189 | 177 | 223 | 353 | 255 | 328 | 366 |
| D19S245 | MFD235 | 4 | 360 | 374 | 377 | 271 | 262 | 193 | 340 | 282 | 345 | 148 |
| D19S246 | MFD232 | 4 | 151 | 226 | 215 | 163 | 80 | 55 | 243 | 76 | 16 | 139 |
| D19S254 | MFD238 | 4 | 219 | 145 | 155 | 276 | 155 | 217 | 136 | 155 | 247 | 188 |
| D19S433 | GGAA2A03 | 4 | 83 | 127 | 107 | 45 | 168 | 117 | 41 | 64 | 193 | 50 |
| D19S559 | UT7544 | 4 | 295 | 273 | 285 | 317 | 328 | 179 | 299 | 181 | 116 | 283 |
| D19S586 | GATA23B01 | 4 | 336 | 284 | 305 | 261 | 343 | 151 | 271 | 337 | 274 | 131 |
| D19S589 | GATA29B01 | 4 | 362 | 373 | 373 | 370 | 208 | 359 | 343 | 168 | 218 | 240 |
| D19S591 | GATA44F10 | 4 | 213 | 211 | 198 | 282 | 305 | 334 | 153 | 262 | 338 | 84 |
| D19S714 | GATA66B04 | 4 | 264 | 238 | 240 | 129 | 270 | 236 | 150 | 207 | 255 | 279 |
| D1S1589 | ATA4E02 | 3 | 65 | 4 | 11 | 166 | 62 | 274 | 272 | 133 | 257 | 372 |
| D1S1594 | GATA22D12 | 4 | 367 | 293 | 298 | 262 | 355 | 220 | 306 | 369 | 248 | 298 |
| D1S1596 | GATA26G09 | 4 | 370 | 303 | 325 | 360 | 335 | 355 | 361 | 374 | 169 | 181 |
| D1S1597 | GATA27E01 | 4 | 238 | 219 | 251 | 138 | 258 | 192 | 283 | 206 | 180 | 168 |
| D1S1609 | GATA50F11 | 4 | 115 | 121 | 150 | 134 | 100 | 11 | 152 | 52 | 231 | 175 |
| D1S1612 | GGAA3A07 | 4 | 48 | 140 | 93 | 231 | 70 | 46 | 48 | 78 | 189 | 49 |
| D1S1627 | ATA25E07 | 3 | 204 | 229 | 227 | 214 | 184 | 216 | 267 | 257 | 138 | 151 |
| D1S1653 | GATA43A04 | 4 | 314 | 370 | 361 | 265 | 145 | 346 | 358 | 236 | 95 | 101 |
| D1S1660 | GATA48B01 | 4 | 275 | 255 | 259 | 339 | 57 | 299 | 266 | 280 | 362 | 107 |
| D1S1665 | GATA61A06 | 4 | 268 | 282 | 279 | 130 | 265 | 189 | 182 | 186 | 281 | 336 |
| D1S1677 | GGAA22G10 | 4 | 277 | 298 | 294 | 172 | 130 | 271 | 268 | 278 | 80 | 247 |
| D1S1679 | GGAA5F09 | 4 | 170 | 248 | 237 | 127 | 173 | 26 | 40 | 66 | 34 | 132 |
| D1S1728 | GATA109 | 4 | 307 | 227 | 169 | 274 | 323 | 268 | 356 | 376 | 165 | 291 |
| D1S2134 | GATA72H07 | 4 | 69 | 157 | 92 | 33 | 106 | 337 | 93 | 164 | 117 | 124 |
| D1S235 | AFM203YG9 | 2 | 4 | 5 | 5 | 14 | 98 | 75 | 30 | 17 | 37 | 289 |
| D1S2682 | AFMA272XC9 | 2 | 218 | 195 | 184 | 242 | 190 | 134 | 135 | 365 | 188 | 108 |
| D1S3462 | ATA29C07 | 3 | 208 | 263 | 241 | 223 | 312 | 318 | 327 | 91 | 273 | 322 |
| D1S3669 | GATA29A05 | 4 | 144 | 79 | 106 | 175 | 210 | 173 | 113 | 121 | 283 | 94 |
| D1S3720 | ATA47D07 | 3 | 182 | 88 | 84 | 250 | 178 | 352 | 201 | 220 | 358 | 299 |
| D1S3721 | GATA129H04 | 4 | 27 | 135 | 108 | 122 | 15 | 15 | 24 | 7 | 47 | 22 |
| D1S468 | AFM280WE5 | 2 | 106 | 33 | 45 | 190 | 87 | 231 | 160 | 127 | 195 | 364 |
| D1S518 | GATA7C01 | 4 | 103 | 42 | 30 | 199 | 235 | 95 | 199 | 204 | 373 | 261 |
| D1S534 | GATA12A07 | 4 | 7 | 44 | 40 | 2 | 36 | 105 | 37 | 16 | 11 | 26 |
| D1S549 | GATA4H09 | 4 | 240 | 223 | 218 | 359 | 143 | 195 | 252 | 180 | 321 | 33 |
| D1S551 | GATA6A05 | 4 | 308 | 206 | 238 | 254 | 375 | 309 | 319 | 258 | 313 | 143 |
| D20S103 | AFM077XD3 | 2 | 87 | 74 | 56 | 121 | 334 | 339 | 288 | 217 | 299 | 369 |
| D20S1143 | GATA129B03 | 4 | 339 | 305 | 320 | 286 | 206 | 130 | 215 | 349 | 185 | 199 |
| D20S159 | UT1307 | 4 | 34 | 180 | 168 | 9 | 66 | 7 | 45 | 12 | 100 | 18 |
| D20S164 | UT1772 | 4 | 93 | 62 | 35 | 156 | 192 | 204 | 302 | 361 | 108 | 278 |
| D20S201 | GATA8B01 | 4 | 168 | 138 | 148 | 202 | 187 | 31 | 328 | 169 | 87 | 91 |
| D20S451 | UT254 | 4 | 128 | 246 | 230 | 42 | 128 | 83 | 189 | 167 | 147 | 44 |
| D20S477 | GATA29F06 | 4 | 140 | 224 | 175 | 66 | 275 | 116 | 109 | 185 | 86 | 118 |
| D20S478 | GATA42A03 | 4 | 114 | 114 | 135 | 131 | 113 | 157 | 55 | 111 | 324 | 120 |
| D20S480 | GATA45B10 | 4 | 326 | 357 | 365 | 323 | 339 | 348 | 222 | 148 | 262 | 122 |
| D20S481 | GATA47F05 | 4 | 160 | 116 | 161 | 125 | 108 | 196 | 114 | 246 | 263 | 180 |
| D20S482 | GATA51D03 | 4 | 344 | 369 | 372 | 363 | 212 | 278 | 278 | 197 | 48 | 230 |
| D20S851 | AFMA218YB5 | 2 | 26 | 13 | 14 | 280 | 120 | 10 | 5 | 71 | 106 | 9 |
| D21S1411 | UT1355 | 4 | 84 | 87 | 119 | 83 | 35 | 54 | 15 | 150 | 65 | 166 |
| D21S1432 | GATA11C12 | 4 | 181 | 189 | 194 | 116 | 278 | 30 | 179 | 286 | 125 | 127 |
| D21S1437 | GGAA3C07 | 4 | 108 | 171 | 187 | 111 | 90 | 63 | 178 | 84 | 52 | 13 |
| D21S1440 | ATA27F01 | 3 | 239 | 103 | 123 | 148 | 372 | 340 | 324 | 363 | 151 | 250 |
| D21S1446 | GATA70B08 | 4 | 142 | 155 | 162 | 26 | 160 | 291 | 162 | 93 | 167 | 186 |
| D21S2052 | GATA129D11 | 4 | 179 | 235 | 223 | 95 | 97 | 156 | 225 | 103 | 24 | 215 |
| D21S2055 | GATA188F04 | 4 | 2 | 9 | 10 | 13 | 8 | 8 | 6 | 2 | 2 | 10 |
| D22S1045 | ATA37D06 | 3 | 175 | 163 | 170 | 170 | 222 | 34 | 228 | 248 | 152 | 87 |
| D22S1169 | AFMB337ZH9 | 2 | 32 | 18 | 16 | 28 | 60 | 155 | 91 | 118 | 232 | 14 |
| D22S345 | MFD313 | 2 | 121 | 124 | 125 | 176 | 223 | 59 | 121 | 129 | 266 | 17 |
| D22S532 | UT7136 | 4 | 368 | 330 | 350 | 352 | 329 | 374 | 177 | 323 | 305 | 253 |
| D22S683 | GATA11B12 | 4 | 1 | 3 | 4 | 1 | 4 | 6 | 1 | 3 | 49 | 194 |
| D22S686 | GGAA10F06 | 4 | 153 | 269 | 257 | 43 | 126 | 269 | 130 | 228 | 68 | 239 |
| D22S689 | GATA21F03 | 4 | 183 | 268 | 273 | 64 | 319 | 28 | 110 | 28 | 339 | 343 |
| D2S1328 | GATA27A12 | 4 | 169 | 143 | 116 | 260 | 286 | 277 | 151 | 210 | 184 | 256 |
| D2S1334 | GATA4D07 | 4 | 29 | 185 | 173 | 5 | 30 | 4 | 23 | 11 | 94 | 27 |
| D2S1352 | ATA27D04 | 3 | 260 | 216 | 208 | 266 | 316 | 153 | 322 | 273 | 221 | 195 |
| D2S1353 | ATA27H09 | 3 | 127 | 132 | 156 | 197 | 150 | 72 | 74 | 57 | 98 | 201 |
| D2S1356 | ATA4F03 | 3 | 20 | 2 | 2 | 89 | 164 | 62 | 211 | 101 | 173 | 113 |
| D2S1360 | GATA11H10 | 4 | 66 | 156 | 158 | 68 | 10 | 69 | 122 | 195 | 4 | 243 |
| D2S1363 | GATA23D03 | 4 | 199 | 123 | 111 | 255 | 196 | 102 | 320 | 239 | 287 | 65 |
| D2S1384 | GATA52A04 | 4 | 188 | 179 | 177 | 325 | 338 | 233 | 183 | 72 | 268 | 152 |
| D2S1391 | GATA65C03 | 4 | 300 | 300 | 283 | 342 | 159 | 280 | 127 | 331 | 314 | 262 |
| D2S1394 | GATA69E12 | 4 | 375 | 376 | 376 | 343 | 315 | 275 | 362 | 296 | 253 | 214 |
| D2S1399 | GGAA20G04 | 4 | 45 | 119 | 102 | 126 | 32 | 76 | 18 | 19 | 13 | 16 |
| D2S1400 | GGAA20G10 | 4 | 59 | 15 | 18 | 77 | 295 | 64 | 303 | 370 | 261 | 324 |
| D2S1776 | GATA71D01 | 4 | 296 | 295 | 284 | 221 | 219 | 125 | 166 | 330 | 316 | 280 |
| D2S1780 | GATA72G11 | 4 | 247 | 218 | 245 | 106 | 250 | 206 | 277 | 316 | 347 | 344 |
| D2S1788 | GATA86E02 | 4 | 49 | 172 | 165 | 29 | 22 | 1 | 19 | 15 | 84 | 55 |
| D2S1790 | GATA88G05 | 4 | 131 | 170 | 181 | 22 | 28 | 77 | 102 | 160 | 171 | 220 |
| D2S2944 | GATA30E06 | 4 | 287 | 366 | 363 | 92 | 44 | 37 | 200 | 203 | 235 | 156 |
| D2S2952 | GATA116B01 | 4 | 112 | 83 | 115 | 182 | 114 | 160 | 63 | 44 | 99 | 335 |
| D2S2968 | GATA178G09 | 4 | 293 | 186 | 179 | 338 | 289 | 222 | 332 | 313 | 336 | 375 |
| D2S2972 | GATA176C01 | 4 | 232 | 221 | 217 | 139 | 118 | 121 | 294 | 235 | 161 | 273 |
| D2S2986 | 2QTEL47 | 2 | 18 | 28 | 6 | 40 | 132 | 201 | 52 | 128 | 286 | 121 |
| D2S405 | GATA8F07 | 4 | 255 | 190 | 209 | 316 | 292 | 181 | 187 | 322 | 307 | 205 |
| D2S410 | GATA4E11 | 4 | 110 | 126 | 122 | 44 | 103 | 119 | 35 | 116 | 145 | 208 |
| D2S427 | GATA12H10 | 4 | 100 | 25 | 33 | 60 | 325 | 301 | 264 | 339 | 246 | 331 |
| D2S434 | GATA4G12 | 4 | 338 | 321 | 328 | 290 | 356 | 190 | 275 | 300 | 135 | 196 |
| D2S441 | GATA8F03 | 4 | 53 | 19 | 15 | 48 | 147 | 207 | 175 | 110 | 270 | 303 |
| D3S1262 | AFM059XA9 | 2 | 37 | 36 | 48 | 56 | 111 | 126 | 65 | 35 | 32 | 216 |
| D3S1311 | AFM254VE1 | 2 | 15 | 32 | 28 | 117 | 41 | 22 | 22 | 34 | 7 | 264 |
| D3S1560 | AFM217XD6 | 2 | 58 | 91 | 83 | 46 | 92 | 36 | 60 | 48 | 96 | 81 |
| D3S1744 | GATA3C02 | 4 | 254 | 345 | 351 | 304 | 142 | 240 | 129 | 56 | 323 | 77 |
| D3S1746 | GATA8F01 | 4 | 40 | 133 | 126 | 6 | 39 | 40 | 26 | 27 | 136 | 35 |
| D3S1763 | GATA3H01 | 4 | 220 | 231 | 246 | 165 | 94 | 302 | 194 | 138 | 365 | 282 |
| D3S1764 | GATA4A10 | 4 | 198 | 202 | 182 | 238 | 255 | 113 | 240 | 147 | 201 | 223 |
| D3S1766 | GATA6F06 | 4 | 354 | 367 | 370 | 364 | 198 | 361 | 144 | 321 | 366 | 112 |
| D3S1768 | GATA8B05 | 4 | 90 | 118 | 85 | 198 | 139 | 263 | 193 | 109 | 272 | 54 |
| D3S2387 | GATA22G12 | 4 | 9 | 60 | 52 | 23 | 19 | 2 | 12 | 9 | 88 | 21 |
| D3S2398 | GATA6G12 | 4 | 335 | 309 | 293 | 375 | 137 | 239 | 220 | 298 | 208 | 234 |
| D3S2403 | GGAA4B09 | 4 | 138 | 130 | 124 | 249 | 109 | 17 | 16 | 153 | 25 | 348 |
| D3S2409 | ATA10H11 | 3 | 96 | 27 | 19 | 206 | 337 | 310 | 205 | 312 | 371 | 377 |
| D3S2418 | ATA22E01 | 3 | 145 | 67 | 60 | 135 | 259 | 93 | 261 | 264 | 41 | 354 |
| D3S2427 | GATA22F11 | 4 | 5 | 14 | 20 | 8 | 20 | 21 | 7 | 5 | 26 | 62 |
| D3S2432 | GATA27C08 | 4 | 230 | 344 | 340 | 253 | 202 | 135 | 83 | 159 | 335 | 25 |
| D3S2460 | GATA68F07 | 4 | 292 | 209 | 186 | 303 | 181 | 56 | 186 | 373 | 292 | 290 |
| D3S3038 | GATA73D01 | 4 | 248 | 280 | 280 | 181 | 47 | 350 | 149 | 314 | 284 | 135 |
| D3S3039 | GATA7F05 | 4 | 324 | 375 | 375 | 347 | 252 | 136 | 289 | 166 | 85 | 76 |
| D3S3045 | GATA84B12 | 4 | 97 | 37 | 36 | 99 | 300 | 253 | 64 | 106 | 176 | 306 |
| D3S3630 | AFMB296ZF5 | 2 | 105 | 73 | 80 | 63 | 154 | 178 | 279 | 105 | 196 | 48 |
| D3S3644 | AFMB318YF1 | 2 | 331 | 278 | 265 | 114 | 326 | 365 | 325 | 362 | 353 | 228 |
| D3S4523 | ATA34G06 | 3 | 61 | 92 | 76 | 160 | 86 | 247 | 184 | 86 | 29 | 126 |
| D3S4529 | GATA128C02 | 4 | 337 | 287 | 300 | 269 | 348 | 58 | 312 | 291 | 259 | 318 |
| D3S4545 | GATA164B08 | 4 | 21 | 72 | 74 | 38 | 14 | 16 | 34 | 26 | 45 | 142 |
| D4S1625 | GATA107 | 4 | 280 | 360 | 360 | 308 | 52 | 369 | 212 | 190 | 212 | 64 |
| D4S1627 | GATA7D01 | 4 | 329 | 331 | 304 | 322 | 241 | 360 | 350 | 243 | 240 | 147 |
| D4S1629 | GATA8A05 | 4 | 250 | 160 | 174 | 140 | 366 | 300 | 238 | 368 | 177 | 370 |
| D4S1644 | GATA11E09 | 4 | 306 | 304 | 299 | 207 | 151 | 234 | 309 | 342 | 343 | 309 |
| D4S1647 | GATA2F11 | 4 | 227 | 260 | 266 | 149 | 303 | 51 | 281 | 254 | 363 | 20 |
| D4S1652 | GATA5B02 | 4 | 325 | 352 | 334 | 299 | 376 | 131 | 245 | 306 | 356 | 281 |
| D4S2361 | ATA2A03 | 3 | 165 | 81 | 87 | 82 | 281 | 172 | 241 | 175 | 183 | 347 |
| D4S2366 | GATA22G05 | 4 | 174 | 144 | 143 | 105 | 236 | 353 | 246 | 183 | 118 | 170 |
| D4S2367 | GATA24H01 | 4 | 317 | 272 | 252 | 361 | 350 | 272 | 365 | 334 | 256 | 206 |
| D4S2368 | GATA27G03 | 4 | 343 | 288 | 312 | 365 | 345 | 264 | 369 | 315 | 62 | 130 |
| D4S2394 | ATA26B08 | 3 | 126 | 75 | 72 | 86 | 195 | 57 | 142 | 198 | 141 | 302 |
| D4S2397 | ATA27C07 | 3 | 149 | 76 | 88 | 200 | 135 | 227 | 249 | 357 | 132 | 308 |
| D4S2417 | GATA42H02 | 4 | 323 | 217 | 228 | 344 | 272 | 219 | 311 | 371 | 308 | 232 |
| D4S2431 | GGAA19H07 | 4 | 76 | 196 | 167 | 80 | 48 | 81 | 29 | 42 | 178 | 46 |
| D4S2623 | GATA62A12 | 4 | 54 | 105 | 105 | 128 | 40 | 145 | 69 | 20 | 123 | 3 |
| D4S2632 | GATA72G09 | 4 | 28 | 125 | 82 | 62 | 43 | 5 | 8 | 94 | 113 | 123 |
| D4S3243 | GATA10G07 | 4 | 252 | 122 | 130 | 345 | 218 | 199 | 231 | 354 | 325 | 363 |
| D4S3248 | GATA28F03 | 4 | 285 | 308 | 301 | 311 | 260 | 144 | 337 | 173 | 245 | 36 |
| D4S3360 | 4PTEL04 | 2 | 200 | 151 | 172 | 75 | 105 | 161 | 140 | 317 | 359 | 365 |
| D4S403 | AFM157XG3 | 2 | 38 | 21 | 23 | 191 | 140 | 98 | 25 | 113 | 330 | 209 |
| D4S408 | AFM165XC11 | 2 | 70 | 86 | 71 | 257 | 332 | 312 | 27 | 69 | 51 | 169 |
| D5S1456 | GATA11A11 | 4 | 265 | 283 | 288 | 88 | 297 | 65 | 284 | 156 | 166 | 272 |
| D5S1457 | GATA21D04 | 4 | 223 | 279 | 278 | 178 | 136 | 80 | 148 | 201 | 349 | 255 |
| D5S1462 | GATA3H06 | 4 | 207 | 323 | 310 | 298 | 333 | 241 | 237 | 23 | 226 | 149 |
| D5S1470 | GATA7C06 | 4 | 154 | 261 | 258 | 225 | 200 | 71 | 158 | 68 | 12 | 24 |
| D5S1480 | ATA23A10 | 3 | 99 | 107 | 89 | 247 | 185 | 168 | 116 | 38 | 344 | 249 |
| D5S1501 | GATA52A12 | 4 | 33 | 77 | 55 | 51 | 1 | 96 | 80 | 54 | 223 | 51 |
| D5S1505 | GATA62A04 | 4 | 267 | 320 | 335 | 187 | 141 | 188 | 218 | 126 | 39 | 160 |
| D5S1725 | GATA89G08 | 4 | 272 | 232 | 243 | 251 | 234 | 197 | 254 | 192 | 115 | 310 |
| D5S211 | MFD154 | 2 | 50 | 65 | 31 | 185 | 186 | 79 | 120 | 36 | 182 | 83 |
| D5S2488 | ATA20G07 | 3 | 270 | 317 | 316 | 241 | 149 | 237 | 169 | 209 | 74 | 134 |
| D5S2500 | GATA67D03 | 4 | 209 | 307 | 317 | 256 | 188 | 297 | 57 | 85 | 139 | 8 |
| D5S2501 | GATA68A03 | 4 | 283 | 315 | 326 | 245 | 215 | 331 | 269 | 132 | 290 | 197 |
| D5S2505 | GATA84E11 | 4 | 91 | 212 | 213 | 81 | 134 | 174 | 39 | 21 | 82 | 12 |
| D5S2845 | GATA134B03 | 4 | 261 | 257 | 255 | 293 | 123 | 103 | 155 | 184 | 190 | 178 |
| D5S2849 | GATA145D10 | 4 | 234 | 222 | 236 | 289 | 276 | 304 | 176 | 225 | 355 | 260 |
| D5S408 | AFM164XB8 | 2 | 78 | 111 | 104 | 31 | 29 | 132 | 58 | 124 | 105 | 182 |
| D5S816 | GATA2H09 | 4 | 173 | 210 | 242 | 239 | 257 | 109 | 72 | 123 | 14 | 270 |
| D5S817 | GATA3E10 | 4 | 372 | 338 | 323 | 334 | 373 | 284 | 367 | 318 | 329 | 338 |
| D5S820 | GATA6E05 | 4 | 163 | 166 | 178 | 171 | 213 | 305 | 123 | 125 | 103 | 330 |
| D6S1006 | ATC4D09 | 3 | 178 | 58 | 50 | 263 | 361 | 377 | 308 | 375 | 374 | 274 |
| D6S1009 | GATA32B03 | 4 | 167 | 110 | 114 | 268 | 171 | 202 | 171 | 30 | 312 | 267 |
| D6S1017 | GGAT3H10 | 4 | 116 | 66 | 81 | 236 | 244 | 252 | 260 | 251 | 77 | 43 |
| D6S1021 | ATA11D10 | 3 | 147 | 136 | 151 | 107 | 304 | 137 | 73 | 174 | 348 | 276 |
| D6S1027 | ATA22G07 | 3 | 60 | 26 | 29 | 123 | 261 | 148 | 229 | 139 | 93 | 337 |
| D6S1031 | ATA28B11 | 3 | 107 | 181 | 141 | 158 | 204 | 18 | 77 | 178 | 33 | 7 |
| D6S1040 | GATA23F08 | 4 | 353 | 299 | 307 | 295 | 232 | 344 | 292 | 338 | 340 | 333 |
| D6S1051 | GATA61E03 | 4 | 350 | 361 | 359 | 232 | 351 | 191 | 339 | 232 | 334 | 236 |
| D6S1053 | GATA64D02 | 4 | 333 | 329 | 324 | 354 | 231 | 89 | 300 | 213 | 60 | 193 |
| D6S1056 | GATA68H04 | 4 | 212 | 104 | 131 | 252 | 165 | 104 | 213 | 161 | 306 | 159 |
| D6S1277 | GATA81B01 | 4 | 253 | 266 | 272 | 258 | 121 | 111 | 143 | 281 | 319 | 57 |
| D6S1959 | GATA29A01 | 4 | 315 | 291 | 311 | 356 | 308 | 53 | 247 | 154 | 168 | 238 |
| D6S2410 | GATA11E02 | 4 | 319 | 312 | 319 | 213 | 72 | 292 | 334 | 307 | 206 | 202 |
| D6S2436 | GATA165G02 | 4 | 191 | 228 | 233 | 153 | 197 | 224 | 214 | 102 | 251 | 233 |
| D6S2439 | GATA163B10 | 4 | 171 | 214 | 224 | 230 | 54 | 152 | 85 | 81 | 150 | 144 |
| D6S2522 | 6QTEL54 | 2 | 377 | 362 | 368 | 368 | 377 | 376 | 377 | 377 | 367 | 345 |
| D6S305 | AFM242ZG5 | 2 | 51 | 11 | 24 | 177 | 17 | 94 | 119 | 74 | 43 | 105 |
| D6S474 | GATA31 | 4 | 322 | 363 | 364 | 329 | 227 | 171 | 318 | 343 | 27 | 37 |
| D6S942 | UT654 | 4 | 358 | 349 | 344 | 275 | 330 | 198 | 203 | 367 | 311 | 224 |
| D7S1799 | GATA23F05 | 4 | 281 | 292 | 290 | 188 | 194 | 221 | 314 | 119 | 277 | 341 |
| D7S1802 | GATA41G07 | 4 | 359 | 325 | 342 | 259 | 368 | 262 | 253 | 288 | 156 | 373 |
| D7S1804 | GATA43C11 | 4 | 39 | 100 | 103 | 16 | 24 | 42 | 4 | 40 | 72 | 47 |
| D7S1808 | GGAA3F06 | 4 | 55 | 35 | 27 | 70 | 102 | 184 | 210 | 234 | 327 | 246 |
| D7S1818 | GATA24D12 | 4 | 355 | 342 | 327 | 292 | 301 | 366 | 310 | 332 | 83 | 321 |
| D7S1824 | GATA32C12 | 4 | 137 | 233 | 202 | 58 | 71 | 282 | 62 | 230 | 107 | 111 |
| D7S2204 | GATA73D10 | 4 | 82 | 236 | 200 | 100 | 6 | 91 | 44 | 37 | 315 | 11 |
| D7S2477 | AFMB035XB9 | 2 | 35 | 6 | 9 | 20 | 77 | 43 | 104 | 53 | 9 | 316 |
| D7S2846 | GATA31A10 | 4 | 371 | 327 | 354 | 355 | 229 | 357 | 346 | 359 | 18 | 198 |
| D7S3046 | GATA118G10 | 4 | 104 | 262 | 225 | 50 | 5 | 88 | 82 | 45 | 295 | 32 |
| D7S3047 | GATA119B03 | 4 | 351 | 341 | 352 | 234 | 279 | 176 | 293 | 336 | 278 | 138 |
| D7S3051 | GATA137H02 | 4 | 180 | 192 | 203 | 159 | 162 | 211 | 141 | 79 | 217 | 146 |
| D7S3056 | GATA24F03 | 4 | 330 | 297 | 286 | 328 | 180 | 256 | 276 | 290 | 352 | 361 |
| D7S3058 | GATA30D09 | 4 | 164 | 204 | 205 | 216 | 242 | 328 | 147 | 63 | 130 | 31 |
| D7S3061 | GGAA6D03 | 4 | 81 | 117 | 113 | 101 | 83 | 101 | 49 | 77 | 258 | 5 |
| D7S3070 | GATA189C06 | 4 | 246 | 326 | 329 | 180 | 318 | 210 | 105 | 65 | 146 | 183 |
| D7S559 | MFD265 | 2 | 155 | 161 | 176 | 109 | 76 | 87 | 287 | 171 | 69 | 92 |
| D7S817 | GATA13G11 | 4 | 195 | 115 | 140 | 315 | 115 | 246 | 219 | 347 | 191 | 165 |
| D7S820 | GATA3F01 | 4 | 185 | 249 | 262 | 146 | 78 | 230 | 125 | 114 | 282 | 257 |
| D7S821 | GATA5D08 | 4 | 257 | 286 | 267 | 300 | 129 | 143 | 92 | 191 | 175 | 210 |
| D8S1048 | UT7129 | 4 | 352 | 358 | 349 | 330 | 284 | 319 | 338 | 244 | 154 | 176 |
| D8S1108 | GATA50D10 | 4 | 341 | 372 | 369 | 267 | 217 | 295 | 285 | 270 | 109 | 86 |
| D8S1110 | GATA8G10 | 4 | 172 | 182 | 163 | 61 | 82 | 286 | 286 | 218 | 90 | 268 |
| D8S1113 | GGAA8G07 | 4 | 157 | 43 | 69 | 150 | 74 | 358 | 146 | 224 | 187 | 315 |
| D8S1128 | GATA21C12 | 4 | 73 | 97 | 100 | 169 | 3 | 242 | 156 | 194 | 236 | 190 |
| D8S1132 | GATA26E03 | 4 | 205 | 197 | 214 | 281 | 96 | 229 | 232 | 108 | 134 | 15 |
| D8S1136 | GATA41A01 | 4 | 301 | 302 | 297 | 218 | 370 | 343 | 326 | 346 | 288 | 117 |
| D8S1179 | GATA7G07 | 4 | 206 | 313 | 309 | 336 | 170 | 68 | 33 | 75 | 119 | 71 |
| D8S1477 | GGAA20C10 | 4 | 94 | 141 | 142 | 25 | 68 | 44 | 50 | 135 | 148 | 332 |
| D8S2324 | GATA14E09 | 4 | 310 | 337 | 338 | 203 | 336 | 267 | 216 | 241 | 30 | 96 |
| D8S261 | AFM123XG5 | 2 | 42 | 46 | 39 | 34 | 34 | 50 | 66 | 141 | 149 | 128 |
| D8S262 | AFM127XH2 | 2 | 189 | 51 | 77 | 113 | 362 | 285 | 206 | 309 | 376 | 376 |
| D8S373 | UT721 | 4 | 210 | 252 | 248 | 333 | 67 | 142 | 191 | 143 | 59 | 97 |
| D8S503 | AFM193XH4 | 2 | 79 | 63 | 51 | 94 | 75 | 52 | 137 | 284 | 238 | 367 |
| D8S560 | AFMA127YE5 | 2 | 12 | 1 | 1 | 4 | 167 | 14 | 36 | 24 | 36 | 362 |
| D8S592 | GATA6B02 | 4 | 305 | 247 | 268 | 331 | 354 | 370 | 181 | 272 | 252 | 167 |
| D9S1118 | GATA71E08 | 4 | 74 | 85 | 86 | 55 | 101 | 141 | 98 | 61 | 6 | 19 |
| D9S1120 | GATA81C04 | 4 | 192 | 142 | 146 | 353 | 237 | 183 | 259 | 301 | 309 | 73 |
| D9S1121 | GATA87E02 | 4 | 77 | 57 | 61 | 162 | 85 | 29 | 68 | 151 | 56 | 133 |
| D9S1122 | GATA89A11 | 4 | 361 | 332 | 339 | 357 | 327 | 259 | 371 | 263 | 271 | 258 |
| D9S1779 | AFM026TG9 | 2 | 13 | 17 | 12 | 32 | 146 | 66 | 11 | 55 | 128 | 218 |
| D9S1825 | AFMB029XG1 | 2 | 63 | 47 | 68 | 10 | 9 | 170 | 71 | 25 | 17 | 320 |
| D9S1838 | AFMB303ZG9 | 2 | 44 | 40 | 66 | 173 | 16 | 41 | 115 | 51 | 341 | 67 |
| D9S1871 | AFM345TA9 | 2 | 10 | 10 | 8 | 11 | 59 | 19 | 86 | 46 | 157 | 200 |
| D9S2157 | ATA59H06 | 3 | 23 | 64 | 42 | 49 | 55 | 165 | 101 | 146 | 199 | 103 |
| D9S2169 | GATA62F03 | 4 | 225 | 203 | 221 | 53 | 369 | 349 | 282 | 358 | 289 | 251 |
| D9S301 | GATA7D12 | 4 | 187 | 183 | 210 | 270 | 81 | 124 | 221 | 87 | 76 | 204 |
| D9S910 | ATA18A07 | 3 | 119 | 98 | 70 | 35 | 225 | 281 | 138 | 245 | 360 | 241 |
| D9S922 | GATA21F05 | 4 | 125 | 50 | 75 | 54 | 347 | 333 | 94 | 327 | 46 | 254 |
| D9S925 | GATA27A11 | 4 | 89 | 59 | 64 | 154 | 182 | 67 | 103 | 82 | 318 | 100 |
| D9S930 | GATA48D07 | 4 | 269 | 289 | 302 | 212 | 238 | 150 | 185 | 177 | 15 | 171 |
| D9S934 | GATA64G07 | 4 | 340 | 343 | 343 | 374 | 230 | 194 | 227 | 240 | 333 | 187 |
| D9S938 | GGAA22E01 | 4 | 64 | 7 | 7 | 194 | 248 | 261 | 78 | 73 | 158 | 301 |
| F13A1-D6S | SE30 | 4 | 30 | 16 | 21 | 193 | 91 | 32 | 70 | 289 | 42 | 172 |
| NA-D10S-1 | GATA121A08 | 4 | 298 | 318 | 318 | 244 | 174 | 159 | 233 | 226 | 224 | 317 |
| NA-D10S-2 | ATA103C06 | 3 | 136 | 164 | 138 | 115 | 274 | 215 | 145 | 205 | 129 | 38 |
| NA-D11S-1 | ATA34E08 | 3 | 158 | 187 | 144 | 141 | 245 | 166 | 329 | 303 | 70 | 294 |
| NA-D12S-1 | GATA49D12 | 4 | 47 | 95 | 96 | 85 | 53 | 20 | 21 | 10 | 204 | 102 |
| NA-D12S-2 | PAH | 4 | 202 | 250 | 234 | 248 | 209 | 203 | 174 | 58 | 192 | 312 |
| NA-D13S-1 | ATA5A09 | 3 | 122 | 39 | 44 | 179 | 51 | 313 | 348 | 252 | 54 | 285 |
| NA-D14S-1 | GATA193A07 | 4 | 80 | 243 | 188 | 132 | 99 | 27 | 28 | 193 | 23 | 1 |
| NA-D15S-1 | GATA50C03 | 4 | 228 | 267 | 271 | 219 | 226 | 325 | 134 | 223 | 31 | 95 |
| NA-D16S-1 | GATA138C05 | 4 | 111 | 106 | 109 | 98 | 214 | 23 | 165 | 115 | 179 | 69 |
| NA-D17S-1 | ATA78D02 | 3 | 148 | 89 | 78 | 246 | 321 | 232 | 159 | 227 | 222 | 355 |
| NA-D18S-1 | GATA178F11 | 4 | 288 | 316 | 313 | 145 | 249 | 162 | 295 | 145 | 297 | 185 |
| NA-D18S-2 | ATA82B02 | 3 | 25 | 24 | 25 | 21 | 58 | 106 | 56 | 97 | 67 | 41 |
| NA-D1S-1 | ATA79C10 | 3 | 161 | 129 | 145 | 118 | 199 | 266 | 100 | 216 | 170 | 266 |
| NA-D1S-2 | GATA124C08 | 4 | 357 | 335 | 315 | 273 | 253 | 375 | 376 | 329 | 244 | 225 |
| NA-D1S-3 | GATA133A08 | 4 | 349 | 306 | 296 | 362 | 371 | 279 | 330 | 277 | 210 | 136 |
| NA-D1S-4 | ATA42G12 | 3 | 201 | 241 | 256 | 73 | 269 | 244 | 313 | 196 | 280 | 125 |
| NA-D1S-5 | GATA124F08 | 4 | 364 | 339 | 331 | 372 | 306 | 373 | 375 | 269 | 227 | 326 |
| NA-D22S-1 | GATA198B05 | 4 | 139 | 215 | 199 | 237 | 45 | 167 | 112 | 41 | 160 | 85 |
| NA-D4S-1 | GATA70E01 | 4 | 86 | 168 | 192 | 52 | 25 | 48 | 87 | 62 | 91 | 34 |
| NA-D5S-1 | ATA52D02 | 3 | 19 | 20 | 22 | 18 | 216 | 115 | 54 | 22 | 35 | 293 |
| NA-D6S-1 | GATA184A08 | 4 | 46 | 94 | 91 | 47 | 33 | 47 | 88 | 50 | 79 | 75 |
| NA-D7S-1 | GATA104 | 4 | 237 | 159 | 164 | 71 | 365 | 180 | 180 | 333 | 264 | 328 |
| NA-D8S-1 | GATA151F02 | 4 | 233 | 199 | 197 | 209 | 163 | 90 | 128 | 214 | 40 | 307 |
| NA-D8S-2 | GAAT1A4 | 4 | 236 | 188 | 183 | 155 | 176 | 147 | 368 | 247 | 58 | 237 |
| NA-D9S-1 | GATA187D09 | 4 | 320 | 328 | 303 | 302 | 263 | 317 | 209 | 202 | 370 | 286 |
| TPO-D2S | SRA | 4 | 226 | 102 | 129 | 208 | 359 | 254 | 372 | 350 | 153 | 357 |
Table B.
Informativeness (In) for Distinguishing Pairs of Regions[Note]
|
In Rank for Region Pair |
|||||||||||||||||||||||
| Locus Name | Alternate Name | RepeatSize | 1-2 | 1-3 | 1-4 | 1-5 | 1-6 | 1-7 | 2-3 | 2-4 | 2-5 | 2-6 | 2-7 | 3-4 | 3-5 | 3-6 | 3-7 | 4-5 | 4-6 | 4-7 | 5-6 | 5-7 | 6-7 |
| D10S1208 | ATA5A04 | 3 | 33 | 53 | 57 | 44 | 5 | 102 | 125 | 47 | 205 | 30 | 365 | 66 | 230 | 19 | 352 | 104 | 19 | 295 | 42 | 326 | 67 |
| D10S1221 | ATA21A03 | 3 | 99 | 96 | 149 | 162 | 225 | 331 | 211 | 101 | 15 | 88 | 98 | 58 | 12 | 102 | 119 | 28 | 165 | 101 | 136 | 50 | 230 |
| D10S1222 | ATA22D02 | 3 | 105 | 72 | 91 | 66 | 62 | 31 | 64 | 213 | 254 | 251 | 111 | 190 | 145 | 232 | 156 | 126 | 202 | 127 | 290 | 61 | 271 |
| D10S1225 | ATA24F10 | 3 | 205 | 202 | 216 | 268 | 53 | 113 | 302 | 353 | 340 | 127 | 179 | 341 | 369 | 121 | 204 | 321 | 80 | 143 | 83 | 176 | 365 |
| D10S1230 | ATA29C03 | 3 | 49 | 110 | 97 | 145 | 91 | 79 | 89 | 170 | 61 | 277 | 309 | 159 | 167 | 274 | 205 | 53 | 226 | 230 | 154 | 85 | 352 |
| D10S1239 | GATA64A09 | 4 | 191 | 169 | 228 | 297 | 359 | 336 | 194 | 144 | 140 | 285 | 203 | 207 | 146 | 263 | 176 | 237 | 303 | 261 | 338 | 366 | 355 |
| D10S1248 | GGAA23C05 | 4 | 232 | 247 | 177 | 288 | 139 | 350 | 193 | 234 | 322 | 252 | 335 | 245 | 289 | 192 | 325 | 142 | 249 | 266 | 119 | 285 | 122 |
| D10S1412 | ATA31G11 | 3 | 220 | 200 | 217 | 182 | 102 | 101 | 25 | 92 | 300 | 138 | 104 | 19 | 160 | 64 | 54 | 349 | 171 | 156 | 182 | 165 | 291 |
| D10S1423 | GATA70E11 | 4 | 265 | 290 | 321 | 342 | 374 | 367 | 147 | 253 | 176 | 276 | 256 | 244 | 198 | 350 | 236 | 266 | 287 | 282 | 226 | 327 | 284 |
| D10S1425 | GATA71C09 | 4 | 309 | 344 | 336 | 91 | 332 | 254 | 307 | 312 | 120 | 183 | 222 | 370 | 66 | 239 | 211 | 51 | 186 | 255 | 19 | 64 | 89 |
| D10S1426 | GATA73E11 | 4 | 299 | 357 | 286 | 351 | 356 | 377 | 261 | 143 | 305 | 345 | 325 | 213 | 345 | 337 | 366 | 326 | 304 | 310 | 365 | 356 | 354 |
| D10S1430 | GATA84C01 | 4 | 313 | 222 | 300 | 337 | 228 | 358 | 183 | 258 | 309 | 262 | 343 | 171 | 251 | 224 | 363 | 303 | 321 | 374 | 188 | 351 | 311 |
| D10S1432 | GATA87G01 | 4 | 210 | 251 | 205 | 228 | 350 | 326 | 48 | 333 | 364 | 239 | 373 | 199 | 286 | 314 | 370 | 371 | 274 | 369 | 258 | 364 | 310 |
| D10S1435 | GATA88F09 | 4 | 306 | 317 | 310 | 265 | 343 | 299 | 271 | 289 | 336 | 361 | 274 | 232 | 294 | 370 | 300 | 324 | 372 | 299 | 356 | 246 | 324 |
| D10S189 | AFM063XF4 | 2 | 92 | 116 | 34 | 159 | 199 | 281 | 314 | 243 | 274 | 210 | 301 | 117 | 340 | 270 | 298 | 100 | 79 | 177 | 322 | 149 | 224 |
| D10S212 | AFM198ZB4 | 2 | 272 | 159 | 209 | 190 | 111 | 246 | 296 | 304 | 231 | 145 | 331 | 320 | 211 | 146 | 333 | 296 | 181 | 296 | 298 | 172 | 184 |
| D10S2327 | GGAT1A4 | 4 | 25 | 34 | 39 | 137 | 54 | 107 | 230 | 249 | 143 | 242 | 257 | 222 | 228 | 236 | 335 | 194 | 136 | 228 | 48 | 161 | 203 |
| D10S2470 | GATA115E01 | 4 | 253 | 345 | 341 | 326 | 331 | 223 | 197 | 138 | 142 | 149 | 173 | 276 | 248 | 255 | 147 | 136 | 178 | 168 | 363 | 20 | 59 |
| D10S677 | GGAA2F11 | 4 | 322 | 218 | 344 | 304 | 333 | 106 | 84 | 311 | 133 | 229 | 35 | 185 | 104 | 181 | 63 | 125 | 265 | 54 | 345 | 26 | 101 |
| D11S1304 | UT2095 | 4 | 97 | 114 | 106 | 40 | 92 | 117 | 36 | 90 | 25 | 187 | 126 | 91 | 23 | 219 | 95 | 17 | 215 | 98 | 6 | 84 | 70 |
| D11S1392 | GATA6B09 | 4 | 181 | 237 | 188 | 224 | 241 | 234 | 276 | 302 | 321 | 377 | 302 | 319 | 352 | 371 | 286 | 343 | 370 | 304 | 319 | 298 | 302 |
| D11S1981 | GATA48E02 | 4 | 334 | 286 | 276 | 332 | 268 | 210 | 134 | 226 | 258 | 288 | 224 | 115 | 196 | 258 | 261 | 116 | 222 | 181 | 175 | 71 | 175 |
| D11S1984 | GGAA17G05 | 4 | 154 | 124 | 225 | 129 | 164 | 231 | 44 | 42 | 8 | 84 | 68 | 52 | 21 | 97 | 152 | 6 | 31 | 152 | 11 | 224 | 57 |
| D11S1986 | GGAA7G08 | 4 | 152 | 119 | 138 | 119 | 170 | 105 | 54 | 73 | 71 | 140 | 47 | 14 | 60 | 125 | 56 | 69 | 158 | 48 | 127 | 105 | 58 |
| D11S1993 | ATA1B07 | 3 | 183 | 252 | 196 | 52 | 169 | 9 | 122 | 82 | 49 | 92 | 22 | 60 | 22 | 85 | 5 | 64 | 113 | 10 | 113 | 82 | 38 |
| D11S1998 | GATA23E06 | 4 | 292 | 316 | 273 | 309 | 325 | 240 | 356 | 22 | 145 | 375 | 228 | 36 | 168 | 376 | 214 | 360 | 299 | 140 | 196 | 53 | 282 |
| D11S1999 | GATA23F06 | 4 | 213 | 196 | 202 | 60 | 285 | 76 | 223 | 350 | 64 | 348 | 41 | 214 | 44 | 297 | 46 | 29 | 309 | 30 | 106 | 291 | 75 |
| D11S2000 | GATA28D01 | 4 | 43 | 19 | 41 | 127 | 93 | 56 | 5 | 106 | 199 | 77 | 86 | 16 | 188 | 57 | 86 | 190 | 94 | 90 | 66 | 19 | 55 |
| D11S2002 | GATA30G01 | 4 | 200 | 167 | 221 | 362 | 368 | 369 | 189 | 261 | 158 | 178 | 141 | 179 | 101 | 153 | 179 | 147 | 164 | 180 | 344 | 323 | 332 |
| D11S2006 | GATA46A12 | 4 | 288 | 315 | 320 | 350 | 357 | 307 | 215 | 370 | 360 | 355 | 376 | 278 | 368 | 321 | 343 | 368 | 344 | 368 | 340 | 335 | 344 |
| D11S2362 | ATA33B03 | 3 | 48 | 17 | 36 | 19 | 77 | 116 | 174 | 351 | 190 | 248 | 137 | 111 | 336 | 312 | 203 | 107 | 208 | 118 | 170 | 106 | 193 |
| D11S2363 | GATA12F04 | 4 | 115 | 30 | 67 | 77 | 118 | 108 | 11 | 77 | 139 | 181 | 83 | 144 | 227 | 234 | 141 | 152 | 185 | 145 | 251 | 235 | 204 |
| D11S2365 | GATA63F09 | 4 | 328 | 338 | 332 | 354 | 302 | 230 | 374 | 368 | 338 | 333 | 159 | 367 | 365 | 342 | 191 | 351 | 364 | 187 | 349 | 206 | 338 |
| D11S2371 | GATA90D07 | 4 | 311 | 341 | 345 | 322 | 371 | 308 | 346 | 307 | 285 | 274 | 330 | 371 | 267 | 323 | 360 | 183 | 297 | 309 | 266 | 162 | 245 |
| D11S4459 | ATA9B04 | 3 | 36 | 65 | 23 | 42 | 75 | 69 | 37 | 268 | 195 | 164 | 150 | 174 | 209 | 216 | 85 | 205 | 151 | 116 | 309 | 168 | 238 |
| D11S4463 | GATA117D01 | 4 | 352 | 375 | 338 | 346 | 362 | 366 | 176 | 191 | 234 | 360 | 299 | 84 | 357 | 361 | 328 | 92 | 288 | 201 | 342 | 309 | 313 |
| D11S4464 | GATA64D03 | 4 | 368 | 302 | 365 | 310 | 348 | 290 | 61 | 366 | 332 | 353 | 246 | 221 | 238 | 195 | 227 | 315 | 318 | 242 | 250 | 333 | 219 |
| D11S969 | AFM205VF10 | 2 | 46 | 83 | 51 | 53 | 39 | 136 | 129 | 254 | 144 | 108 | 198 | 346 | 217 | 142 | 287 | 170 | 99 | 241 | 50 | 338 | 93 |
| D12S1042 | ATA27A06 | 3 | 303 | 272 | 231 | 260 | 159 | 157 | 96 | 206 | 182 | 143 | 81 | 63 | 117 | 131 | 37 | 115 | 159 | 70 | 287 | 33 | 54 |
| D12S1045 | ATA29A06 | 3 | 71 | 131 | 43 | 27 | 67 | 34 | 71 | 225 | 77 | 97 | 60 | 99 | 17 | 35 | 15 | 34 | 68 | 26 | 359 | 132 | 221 |
| D12S1052 | GATA26D02 | 4 | 338 | 353 | 363 | 374 | 361 | 207 | 331 | 212 | 294 | 317 | 153 | 356 | 353 | 333 | 157 | 347 | 295 | 111 | 351 | 55 | 87 |
| D12S1064 | GATA63D12 | 4 | 184 | 157 | 190 | 187 | 144 | 243 | 348 | 315 | 212 | 156 | 263 | 318 | 183 | 143 | 226 | 146 | 103 | 216 | 254 | 367 | 309 |
| D12S1294 | GATA73H09 | 4 | 290 | 343 | 278 | 121 | 234 | 259 | 279 | 305 | 122 | 279 | 101 | 215 | 86 | 290 | 167 | 75 | 270 | 102 | 104 | 66 | 181 |
| D12S1300 | GATA85A04 | 4 | 227 | 294 | 299 | 300 | 177 | 363 | 291 | 222 | 99 | 13 | 294 | 361 | 199 | 37 | 356 | 202 | 21 | 360 | 79 | 376 | 153 |
| D12S1301 | GATA91H06 | 4 | 151 | 225 | 104 | 82 | 290 | 52 | 140 | 322 | 68 | 133 | 112 | 95 | 35 | 150 | 73 | 37 | 57 | 82 | 59 | 3 | 34 |
| D12S1638 | AFMB002VD5 | 2 | 196 | 186 | 260 | 235 | 137 | 121 | 264 | 117 | 308 | 155 | 187 | 155 | 305 | 130 | 239 | 300 | 127 | 119 | 190 | 118 | 205 |
| D12S2070 | ATA25F09 | 3 | 263 | 295 | 210 | 99 | 100 | 40 | 267 | 210 | 23 | 123 | 3 | 27 | 10 | 68 | 1 | 32 | 115 | 3 | 32 | 39 | 2 |
| D12S2078 | GATA32F05 | 4 | 218 | 298 | 304 | 183 | 258 | 202 | 151 | 248 | 226 | 151 | 266 | 347 | 141 | 166 | 250 | 195 | 166 | 276 | 121 | 201 | 113 |
| D12S269 | MFD259 | 2 | 104 | 98 | 105 | 71 | 57 | 27 | 200 | 140 | 246 | 182 | 56 | 43 | 186 | 100 | 34 | 128 | 109 | 47 | 86 | 59 | 110 |
| D12S297 | UT5029 | 4 | 294 | 348 | 342 | 196 | 73 | 49 | 63 | 46 | 38 | 29 | 6 | 353 | 124 | 55 | 25 | 88 | 33 | 18 | 202 | 98 | 248 |
| D12S372 | GATA4H03 | 4 | 307 | 288 | 347 | 352 | 342 | 353 | 253 | 246 | 359 | 364 | 164 | 74 | 325 | 346 | 139 | 323 | 333 | 226 | 373 | 212 | 229 |
| D12S373 | GATA6C01 | 4 | 373 | 333 | 353 | 295 | 150 | 337 | 297 | 229 | 209 | 87 | 268 | 315 | 147 | 87 | 337 | 96 | 63 | 325 | 233 | 129 | 96 |
| D12S395 | GATA4H01 | 4 | 360 | 359 | 366 | 238 | 183 | 340 | 130 | 346 | 150 | 115 | 298 | 314 | 69 | 69 | 289 | 76 | 77 | 315 | 278 | 284 | 222 |
| D13S1493 | GGAA29H03 | 4 | 214 | 183 | 243 | 214 | 117 | 247 | 368 | 232 | 227 | 160 | 219 | 211 | 185 | 137 | 174 | 286 | 150 | 269 | 101 | 331 | 123 |
| D13S1807 | GATA11C08 | 4 | 283 | 367 | 270 | 287 | 324 | 127 | 196 | 338 | 329 | 315 | 145 | 161 | 254 | 325 | 99 | 328 | 240 | 71 | 184 | 43 | 256 |
| D13S285 | AFM309VA9 | 2 | 50 | 58 | 35 | 65 | 154 | 124 | 108 | 83 | 87 | 172 | 306 | 32 | 74 | 140 | 305 | 145 | 123 | 270 | 39 | 137 | 182 |
| D13S317 | GATA7G10 | 4 | 233 | 310 | 269 | 49 | 235 | 61 | 212 | 236 | 171 | 240 | 93 | 307 | 85 | 271 | 52 | 84 | 257 | 29 | 130 | 40 | 32 |
| D13S779 | ATA26D07 | 3 | 285 | 240 | 277 | 178 | 278 | 181 | 273 | 328 | 147 | 283 | 75 | 375 | 193 | 242 | 127 | 151 | 260 | 103 | 110 | 136 | 48 |
| D13S787 | GATA23C03 | 4 | 323 | 266 | 266 | 290 | 319 | 328 | 337 | 176 | 373 | 350 | 356 | 182 | 377 | 328 | 371 | 281 | 312 | 286 | 259 | 362 | 328 |
| D13S793 | GATA43H03 | 4 | 78 | 64 | 82 | 118 | 119 | 253 | 372 | 374 | 358 | 54 | 154 | 377 | 374 | 71 | 188 | 355 | 44 | 164 | 28 | 152 | 288 |
| D13S796 | GATA51B02 | 4 | 310 | 253 | 306 | 303 | 213 | 314 | 293 | 309 | 326 | 128 | 293 | 297 | 308 | 77 | 252 | 252 | 85 | 281 | 99 | 243 | 317 |
| D13S800 | GATA64F08 | 4 | 314 | 331 | 316 | 267 | 180 | 291 | 243 | 354 | 272 | 241 | 312 | 283 | 265 | 226 | 295 | 241 | 230 | 308 | 61 | 361 | 133 |
| D13S894 | GATA86H01 | 4 | 371 | 293 | 313 | 234 | 254 | 277 | 126 | 128 | 111 | 221 | 215 | 286 | 215 | 339 | 291 | 138 | 342 | 260 | 212 | 353 | 294 |
| D13S895 | GGAA22G01 | 4 | 195 | 257 | 233 | 195 | 313 | 228 | 100 | 174 | 186 | 194 | 288 | 137 | 229 | 326 | 282 | 223 | 248 | 300 | 211 | 373 | 234 |
| D14S1007 | AFMB002ZF1 | 2 | 38 | 50 | 79 | 34 | 157 | 5 | 57 | 12 | 1 | 9 | 16 | 11 | 1 | 6 | 11 | 5 | 43 | 45 | 140 | 21 | 28 |
| D14S1280 | GATA31B09 | 4 | 147 | 175 | 130 | 213 | 242 | 135 | 311 | 339 | 320 | 297 | 332 | 310 | 337 | 296 | 293 | 299 | 253 | 327 | 276 | 144 | 174 |
| D14S1426 | GATA136B01 | 4 | 212 | 213 | 161 | 134 | 267 | 18 | 347 | 183 | 164 | 154 | 21 | 177 | 150 | 164 | 22 | 91 | 73 | 35 | 53 | 5 | 3 |
| D14S1434 | GATA168F06 | 4 | 325 | 369 | 297 | 221 | 252 | 273 | 299 | 284 | 218 | 280 | 247 | 153 | 190 | 218 | 223 | 235 | 339 | 280 | 207 | 185 | 298 |
| D14S587 | GGAA10C09 | 4 | 224 | 340 | 162 | 229 | 151 | 196 | 181 | 180 | 168 | 205 | 139 | 71 | 182 | 188 | 137 | 153 | 275 | 99 | 81 | 167 | 88 |
| D14S588 | GGAA4A12 | 4 | 15 | 29 | 18 | 104 | 116 | 143 | 19 | 125 | 73 | 80 | 169 | 305 | 262 | 114 | 172 | 106 | 108 | 178 | 21 | 80 | 102 |
| D14S592 | ATA19H08 | 3 | 173 | 219 | 136 | 154 | 120 | 100 | 93 | 61 | 251 | 55 | 176 | 178 | 175 | 129 | 182 | 248 | 100 | 213 | 57 | 205 | 199 |
| D14S599 | ATA29G03 | 3 | 58 | 45 | 85 | 150 | 44 | 109 | 363 | 120 | 48 | 250 | 12 | 257 | 48 | 231 | 10 | 33 | 124 | 4 | 34 | 56 | 13 |
| D14S606 | GATA30A03 | 4 | 225 | 176 | 183 | 258 | 208 | 271 | 333 | 369 | 256 | 177 | 262 | 327 | 187 | 141 | 247 | 193 | 134 | 257 | 317 | 355 | 341 |
| D14S608 | GATA43H01 | 4 | 276 | 371 | 244 | 205 | 221 | 174 | 27 | 169 | 331 | 299 | 50 | 17 | 111 | 200 | 80 | 200 | 229 | 7 | 165 | 14 | 63 |
| D14S617 | GGAA21G11 | 4 | 158 | 142 | 89 | 108 | 1 | 134 | 173 | 165 | 248 | 12 | 213 | 149 | 221 | 4 | 194 | 191 | 16 | 186 | 4 | 193 | 31 |
| D14S742 | GATA74E02 | 4 | 289 | 260 | 257 | 289 | 329 | 332 | 249 | 221 | 374 | 376 | 375 | 279 | 288 | 330 | 354 | 222 | 317 | 311 | 361 | 372 | 366 |
| D15S128 | AFM273YF9 | 2 | 45 | 95 | 69 | 20 | 149 | 71 | 18 | 69 | 125 | 212 | 90 | 112 | 71 | 169 | 76 | 78 | 210 | 78 | 186 | 121 | 244 |
| D15S1507 | GATA151F03 | 4 | 130 | 133 | 83 | 59 | 52 | 164 | 339 | 336 | 196 | 231 | 367 | 324 | 204 | 230 | 358 | 230 | 236 | 376 | 187 | 321 | 272 |
| D15S1515 | GATA197B10 | 4 | 83 | 74 | 92 | 155 | 128 | 300 | 290 | 173 | 184 | 303 | 51 | 338 | 271 | 347 | 83 | 216 | 355 | 65 | 330 | 114 | 107 |
| D15S165 | AFM248VC5 | 2 | 44 | 55 | 77 | 4 | 11 | 42 | 35 | 29 | 36 | 60 | 76 | 6 | 20 | 41 | 65 | 25 | 81 | 120 | 232 | 138 | 194 |
| D15S642 | GATA27A03 | 4 | 51 | 59 | 125 | 141 | 314 | 225 | 86 | 100 | 5 | 95 | 79 | 151 | 8 | 158 | 97 | 13 | 183 | 163 | 105 | 247 | 269 |
| D15S643 | GATA50G06 | 4 | 138 | 105 | 111 | 306 | 288 | 280 | 128 | 86 | 194 | 99 | 121 | 141 | 210 | 176 | 215 | 157 | 84 | 130 | 171 | 170 | 345 |
| D15S652 | ATA24A08 | 3 | 192 | 118 | 180 | 180 | 230 | 93 | 91 | 54 | 103 | 101 | 14 | 88 | 140 | 66 | 30 | 112 | 89 | 32 | 133 | 18 | 19 |
| D15S655 | ATA28G05 | 3 | 145 | 134 | 109 | 90 | 40 | 126 | 74 | 148 | 69 | 23 | 300 | 157 | 78 | 33 | 327 | 163 | 60 | 354 | 230 | 197 | 90 |
| D15S659 | GATA63A03 | 4 | 244 | 276 | 245 | 259 | 305 | 140 | 236 | 167 | 314 | 204 | 119 | 293 | 299 | 229 | 128 | 285 | 126 | 158 | 134 | 86 | 36 |
| D15S816 | GATA73F01 | 4 | 264 | 216 | 242 | 277 | 216 | 285 | 172 | 265 | 152 | 331 | 339 | 260 | 195 | 318 | 365 | 118 | 340 | 356 | 141 | 143 | 289 |
| D15S818 | GATA85D02 | 4 | 341 | 300 | 328 | 370 | 320 | 330 | 146 | 375 | 339 | 334 | 311 | 240 | 282 | 313 | 256 | 310 | 314 | 279 | 331 | 217 | 268 |
| D15S822 | GATA88H02 | 4 | 180 | 146 | 340 | 246 | 309 | 137 | 278 | 38 | 46 | 186 | 10 | 22 | 26 | 152 | 6 | 44 | 231 | 17 | 218 | 141 | 94 |
| D16S2616 | ATA41E04 | 3 | 85 | 71 | 86 | 80 | 41 | 47 | 332 | 294 | 222 | 147 | 110 | 308 | 240 | 162 | 105 | 288 | 154 | 117 | 236 | 203 | 361 |
| D16S2621 | GATA71F09 | 4 | 126 | 66 | 48 | 3 | 161 | 32 | 255 | 188 | 29 | 309 | 77 | 142 | 36 | 253 | 91 | 58 | 307 | 161 | 78 | 240 | 148 |
| D16S2624 | GATA81D12 | 4 | 304 | 238 | 294 | 203 | 330 | 262 | 295 | 310 | 57 | 268 | 131 | 374 | 50 | 295 | 123 | 38 | 311 | 134 | 143 | 313 | 132 |
| D16S3253 | GATA22F09 | 4 | 79 | 80 | 133 | 231 | 145 | 182 | 298 | 126 | 203 | 339 | 217 | 119 | 244 | 352 | 199 | 160 | 209 | 108 | 176 | 150 | 220 |
| D16S3396 | ATA55A11 | 3 | 256 | 232 | 153 | 218 | 289 | 173 | 216 | 207 | 303 | 321 | 273 | 184 | 279 | 280 | 207 | 294 | 281 | 204 | 327 | 230 | 231 |
| D16S3401 | 16PTEL06 | 2 | 13 | 39 | 32 | 89 | 101 | 110 | 30 | 13 | 81 | 4 | 43 | 236 | 184 | 14 | 60 | 67 | 9 | 21 | 13 | 54 | 22 |
| D16S403 | AFM049XD2 | 2 | 236 | 224 | 181 | 198 | 82 | 179 | 26 | 52 | 178 | 46 | 132 | 56 | 92 | 26 | 42 | 277 | 4 | 58 | 2 | 65 | 52 |
| D16S422 | AFM249XC5 | 2 | 95 | 112 | 176 | 38 | 141 | 74 | 31 | 44 | 2 | 189 | 17 | 72 | 9 | 174 | 24 | 10 | 213 | 52 | 40 | 322 | 100 |
| D16S516 | AFM350VD1 | 2 | 149 | 209 | 248 | 147 | 238 | 237 | 87 | 99 | 60 | 76 | 103 | 239 | 49 | 190 | 107 | 61 | 205 | 123 | 108 | 175 | 150 |
| D16S539 | GATA11C06 | 4 | 250 | 323 | 362 | 367 | 251 | 357 | 224 | 58 | 189 | 316 | 146 | 298 | 268 | 287 | 224 | 197 | 234 | 238 | 131 | 325 | 149 |
| D16S748 | ATA3A07 | 3 | 109 | 168 | 93 | 116 | 9 | 14 | 133 | 27 | 30 | 21 | 49 | 9 | 45 | 13 | 39 | 171 | 105 | 86 | 125 | 31 | 211 |
| D16S753 | GGAA3G05 | 4 | 291 | 215 | 166 | 85 | 162 | 233 | 259 | 88 | 113 | 216 | 276 | 110 | 116 | 183 | 326 | 177 | 246 | 252 | 314 | 200 | 159 |
| D16S764 | GATA42E11 | 4 | 335 | 299 | 302 | 363 | 370 | 261 | 326 | 376 | 318 | 305 | 178 | 288 | 322 | 315 | 243 | 259 | 278 | 141 | 366 | 182 | 270 |
| D16S769 | GATA71H05 | 4 | 343 | 306 | 339 | 301 | 341 | 359 | 361 | 377 | 317 | 358 | 341 | 372 | 366 | 375 | 318 | 301 | 366 | 336 | 374 | 208 | 280 |
| D17S1290 | GATA49C09 | 4 | 156 | 150 | 156 | 171 | 88 | 175 | 109 | 85 | 276 | 175 | 138 | 116 | 318 | 180 | 113 | 287 | 219 | 75 | 205 | 81 | 144 |
| D17S1294 | GGAA9D03 | 4 | 209 | 164 | 179 | 254 | 236 | 278 | 110 | 348 | 267 | 324 | 323 | 158 | 321 | 336 | 340 | 264 | 327 | 321 | 357 | 358 | 376 |
| D17S1298 | GAAT2C03 | 4 | 84 | 100 | 129 | 97 | 152 | 172 | 306 | 19 | 43 | 273 | 355 | 70 | 82 | 310 | 367 | 211 | 360 | 340 | 328 | 227 | 375 |
| D17S1299 | GATA25A04 | 4 | 320 | 376 | 368 | 319 | 328 | 372 | 70 | 119 | 262 | 293 | 327 | 366 | 341 | 329 | 372 | 346 | 329 | 371 | 282 | 300 | 342 |
| D17S1301 | GATA28D11 | 4 | 208 | 178 | 322 | 266 | 365 | 338 | 323 | 274 | 266 | 253 | 368 | 238 | 243 | 220 | 375 | 238 | 320 | 373 | 237 | 268 | 295 |
| D17S1308 | GTAT1A05 | 4 | 129 | 203 | 155 | 255 | 195 | 356 | 106 | 321 | 177 | 36 | 223 | 258 | 106 | 25 | 234 | 86 | 14 | 247 | 177 | 328 | 265 |
| D17S2180 | ATC6A06 | 3 | 248 | 198 | 301 | 285 | 337 | 147 | 171 | 121 | 96 | 201 | 148 | 218 | 165 | 275 | 150 | 271 | 322 | 135 | 358 | 89 | 141 |
| D17S2193 | ATA43A10Z | 3 | 22 | 10 | 15 | 67 | 15 | 26 | 50 | 235 | 311 | 192 | 129 | 90 | 181 | 251 | 200 | 246 | 117 | 125 | 73 | 72 | 172 |
| D17S2195 | ATA58A02 | 3 | 23 | 23 | 40 | 87 | 72 | 78 | 111 | 187 | 180 | 37 | 258 | 227 | 231 | 51 | 237 | 275 | 45 | 224 | 63 | 148 | 50 |
| D17S2196 | GATA185H04 | 4 | 284 | 305 | 259 | 96 | 292 | 84 | 338 | 208 | 100 | 176 | 54 | 281 | 105 | 238 | 69 | 40 | 223 | 55 | 31 | 104 | 71 |
| D17S784 | AFM044XG3 | 2 | 123 | 125 | 42 | 93 | 200 | 58 | 317 | 158 | 134 | 207 | 134 | 122 | 89 | 266 | 161 | 54 | 147 | 172 | 45 | 4 | 104 |
| D17S974 | GATA8C04 | 4 | 223 | 182 | 186 | 279 | 261 | 266 | 362 | 245 | 323 | 267 | 162 | 306 | 362 | 221 | 220 | 338 | 135 | 176 | 122 | 255 | 66 |
| D18S1357 | ATA7D07 | 3 | 235 | 258 | 218 | 339 | 233 | 112 | 178 | 147 | 211 | 72 | 204 | 351 | 303 | 170 | 154 | 226 | 116 | 153 | 146 | 45 | 65 |
| D18S1364 | GATA7E12 | 4 | 28 | 21 | 11 | 110 | 112 | 185 | 318 | 362 | 306 | 180 | 200 | 268 | 348 | 197 | 222 | 244 | 173 | 173 | 153 | 276 | 125 |
| D18S1370 | ATA45G06 | 3 | 112 | 201 | 117 | 114 | 83 | 274 | 51 | 203 | 154 | 105 | 174 | 128 | 76 | 44 | 230 | 93 | 49 | 193 | 191 | 67 | 114 |
| D18S1371 | GATA177C03 | 4 | 332 | 362 | 253 | 341 | 303 | 184 | 364 | 252 | 354 | 257 | 151 | 266 | 376 | 273 | 165 | 350 | 216 | 237 | 268 | 127 | 129 |
| D18S1376 | GATA185C06 | 4 | 286 | 326 | 279 | 278 | 311 | 335 | 284 | 219 | 349 | 214 | 342 | 294 | 258 | 237 | 332 | 262 | 254 | 349 | 246 | 371 | 356 |
| D18S1390 | 18QTEL11 | 2 | 93 | 106 | 94 | 74 | 174 | 129 | 119 | 55 | 80 | 163 | 66 | 259 | 95 | 133 | 79 | 141 | 143 | 76 | 160 | 178 | 208 |
| D18S535 | GATA13 | 4 | 254 | 180 | 280 | 126 | 31 | 205 | 244 | 334 | 232 | 57 | 182 | 65 | 121 | 30 | 187 | 201 | 35 | 183 | 162 | 97 | 30 |
| D18S542 | GATA11A06 | 4 | 72 | 42 | 65 | 167 | 96 | 186 | 272 | 260 | 126 | 3 | 136 | 241 | 136 | 3 | 180 | 85 | 1 | 128 | 16 | 196 | 64 |
| D18S843 | ACT1A01 | 3 | 367 | 360 | 360 | 237 | 301 | 346 | 357 | 344 | 183 | 254 | 352 | 326 | 142 | 252 | 355 | 89 | 197 | 339 | 102 | 286 | 340 |
| D18S851 | GATA6D09 | 4 | 204 | 177 | 134 | 131 | 295 | 219 | 169 | 130 | 181 | 247 | 88 | 233 | 220 | 320 | 132 | 278 | 251 | 122 | 208 | 179 | 318 |
| D18S858 | ATA23G05 | 3 | 247 | 207 | 262 | 256 | 106 | 212 | 353 | 345 | 172 | 73 | 259 | 247 | 98 | 47 | 232 | 189 | 52 | 215 | 84 | 180 | 226 |
| D18S877 | GATA64H04 | 4 | 280 | 227 | 258 | 166 | 171 | 323 | 334 | 255 | 343 | 264 | 371 | 237 | 312 | 256 | 362 | 240 | 243 | 365 | 292 | 352 | 290 |
| D19S1034 | GATA21G05 | 4 | 202 | 246 | 203 | 146 | 210 | 158 | 164 | 298 | 280 | 296 | 165 | 204 | 156 | 222 | 133 | 242 | 277 | 94 | 288 | 87 | 251 |
| D19S245 | MFD235 | 4 | 365 | 373 | 361 | 325 | 304 | 373 | 373 | 356 | 372 | 356 | 350 | 368 | 363 | 360 | 369 | 345 | 361 | 372 | 368 | 292 | 326 |
| D19S246 | MFD232 | 4 | 262 | 261 | 330 | 273 | 227 | 286 | 17 | 129 | 119 | 169 | 241 | 203 | 224 | 156 | 316 | 109 | 128 | 324 | 129 | 171 | 98 |
| D19S254 | MFD238 | 4 | 187 | 319 | 308 | 333 | 133 | 292 | 136 | 141 | 233 | 59 | 63 | 349 | 335 | 60 | 181 | 337 | 61 | 126 | 69 | 74 | 25 |
| D19S433 | GGAA2A03 | 4 | 63 | 38 | 84 | 115 | 107 | 169 | 83 | 60 | 148 | 161 | 100 | 165 | 192 | 104 | 126 | 208 | 133 | 97 | 225 | 173 | 186 |
| D19S559 | UT7544 | 4 | 229 | 280 | 171 | 211 | 260 | 180 | 191 | 199 | 282 | 347 | 189 | 292 | 349 | 327 | 262 | 375 | 301 | 220 | 269 | 225 | 198 |
| D19S586 | GATA23B01 | 4 | 266 | 254 | 222 | 271 | 215 | 313 | 300 | 343 | 376 | 327 | 275 | 339 | 367 | 343 | 264 | 376 | 332 | 222 | 285 | 249 | 147 |
| D19S589 | GATA29B01 | 4 | 326 | 268 | 349 | 359 | 326 | 374 | 209 | 316 | 348 | 326 | 359 | 277 | 313 | 348 | 322 | 372 | 337 | 348 | 275 | 340 | 333 |
| D19S591 | GATA44F10 | 4 | 182 | 195 | 140 | 151 | 280 | 264 | 322 | 313 | 179 | 190 | 92 | 290 | 293 | 288 | 129 | 178 | 167 | 81 | 294 | 63 | 164 |
| D19S714 | GATA66B04 | 4 | 300 | 235 | 287 | 311 | 165 | 263 | 266 | 194 | 325 | 68 | 195 | 289 | 355 | 88 | 307 | 282 | 53 | 258 | 80 | 257 | 151 |
| D1S1589 | ATA4E02 | 3 | 8 | 18 | 9 | 37 | 35 | 2 | 150 | 263 | 127 | 35 | 115 | 164 | 148 | 42 | 71 | 108 | 40 | 66 | 12 | 1 | 12 |
| D1S1594 | GATA22D12 | 4 | 329 | 336 | 274 | 282 | 287 | 241 | 352 | 153 | 292 | 371 | 194 | 96 | 256 | 356 | 159 | 364 | 283 | 323 | 243 | 345 | 180 |
| D1S1596 | GATA26G09 | 4 | 336 | 287 | 314 | 321 | 373 | 199 | 369 | 318 | 284 | 359 | 211 | 270 | 266 | 332 | 241 | 356 | 356 | 190 | 348 | 163 | 187 |
| D1S1597 | GATA27E01 | 4 | 118 | 92 | 96 | 149 | 113 | 118 | 285 | 320 | 341 | 365 | 221 | 335 | 276 | 358 | 164 | 341 | 357 | 200 | 270 | 220 | 285 |
| D1S1609 | GATA50F11 | 4 | 193 | 153 | 198 | 249 | 189 | 92 | 141 | 215 | 173 | 287 | 120 | 296 | 213 | 298 | 89 | 213 | 310 | 51 | 203 | 11 | 80 |
| D1S1612 | GGAA3A07 | 4 | 141 | 144 | 142 | 244 | 277 | 216 | 143 | 196 | 14 | 47 | 23 | 325 | 30 | 96 | 48 | 7 | 66 | 15 | 308 | 128 | 127 |
| D1S1627 | ATA25E07 | 3 | 305 | 231 | 184 | 76 | 204 | 162 | 226 | 57 | 67 | 152 | 233 | 94 | 43 | 126 | 249 | 113 | 258 | 332 | 238 | 310 | 235 |
| D1S1653 | GATA43A04 | 4 | 317 | 279 | 343 | 348 | 355 | 349 | 62 | 278 | 257 | 286 | 278 | 332 | 310 | 351 | 274 | 331 | 353 | 291 | 370 | 339 | 370 |
| D1S1660 | GATA48B01 | 4 | 293 | 212 | 214 | 241 | 196 | 309 | 345 | 209 | 192 | 96 | 354 | 205 | 179 | 61 | 373 | 172 | 78 | 307 | 109 | 311 | 106 |
| D1S1665 | GATA61A06 | 4 | 171 | 187 | 137 | 177 | 217 | 251 | 34 | 256 | 297 | 363 | 353 | 212 | 158 | 354 | 357 | 217 | 375 | 361 | 261 | 266 | 359 |
| D1S1677 | GGAA22G10 | 4 | 255 | 185 | 303 | 243 | 298 | 255 | 239 | 267 | 242 | 328 | 245 | 138 | 109 | 285 | 177 | 279 | 358 | 202 | 297 | 259 | 263 |
| D1S1679 | GGAA5F09 | 4 | 136 | 184 | 185 | 222 | 222 | 284 | 168 | 200 | 293 | 103 | 317 | 273 | 304 | 139 | 311 | 253 | 110 | 306 | 148 | 316 | 213 |
| D1S1728 | GATA109 | 4 | 70 | 61 | 191 | 240 | 299 | 226 | 234 | 96 | 78 | 43 | 143 | 81 | 114 | 45 | 168 | 206 | 140 | 271 | 223 | 283 | 236 |
| D1S2134 | GATA72H07 | 4 | 128 | 115 | 152 | 227 | 275 | 170 | 82 | 91 | 32 | 91 | 27 | 127 | 27 | 73 | 17 | 8 | 88 | 9 | 335 | 186 | 250 |
| D1S235 | AFM203YG9 | 2 | 3 | 2 | 1 | 1 | 4 | 10 | 47 | 3 | 9 | 120 | 135 | 20 | 29 | 90 | 198 | 49 | 176 | 245 | 23 | 125 | 152 |
| D1S2682 | AFMA272XC9 | 2 | 133 | 108 | 213 | 313 | 125 | 177 | 227 | 295 | 121 | 198 | 155 | 242 | 100 | 157 | 122 | 169 | 163 | 151 | 95 | 119 | 330 |
| D1S3462 | ATA29C07 | 3 | 278 | 320 | 285 | 160 | 187 | 305 | 206 | 11 | 27 | 79 | 226 | 101 | 58 | 124 | 338 | 166 | 282 | 338 | 354 | 122 | 191 |
| D1S3669 | GATA29A05 | 4 | 127 | 154 | 116 | 130 | 69 | 77 | 238 | 132 | 229 | 111 | 161 | 50 | 178 | 101 | 140 | 229 | 82 | 77 | 27 | 154 | 40 |
| D1S3720 | ATA47D07 | 3 | 41 | 24 | 131 | 283 | 79 | 128 | 139 | 16 | 59 | 114 | 152 | 75 | 72 | 53 | 271 | 204 | 69 | 167 | 52 | 83 | 27 |
| D1S3721 | GATA129H04 | 4 | 172 | 126 | 139 | 176 | 197 | 122 | 73 | 33 | 107 | 126 | 44 | 54 | 119 | 127 | 55 | 110 | 71 | 34 | 152 | 94 | 126 |
| D1S468 | AFM280WE5 | 2 | 189 | 308 | 154 | 197 | 127 | 7 | 45 | 9 | 146 | 81 | 8 | 45 | 205 | 173 | 7 | 129 | 212 | 6 | 35 | 2 | 42 |
| D1S518 | GATA7C01 | 4 | 56 | 84 | 29 | 31 | 8 | 63 | 58 | 17 | 53 | 22 | 39 | 77 | 64 | 27 | 68 | 168 | 62 | 93 | 144 | 219 | 247 |
| D1S534 | GATA12A07 | 4 | 163 | 255 | 211 | 25 | 121 | 38 | 56 | 145 | 45 | 86 | 57 | 85 | 18 | 82 | 33 | 12 | 38 | 19 | 71 | 126 | 99 |
| D1S549 | GATA4H09 | 4 | 203 | 297 | 232 | 330 | 85 | 375 | 22 | 162 | 216 | 102 | 201 | 131 | 298 | 91 | 292 | 231 | 101 | 227 | 91 | 233 | 43 |
| D1S551 | GATA6A05 | 4 | 357 | 332 | 354 | 317 | 166 | 312 | 371 | 204 | 263 | 135 | 225 | 250 | 283 | 151 | 217 | 348 | 87 | 251 | 114 | 155 | 39 |
| D20S103 | AFM077XD3 | 2 | 125 | 68 | 78 | 47 | 168 | 99 | 246 | 30 | 12 | 67 | 70 | 51 | 14 | 54 | 74 | 31 | 67 | 206 | 64 | 79 | 192 |
| D20S1143 | GATA129B03 | 4 | 234 | 244 | 226 | 247 | 291 | 272 | 263 | 326 | 345 | 270 | 362 | 352 | 326 | 278 | 344 | 284 | 279 | 346 | 220 | 349 | 261 |
| D20S159 | UT1307 | 4 | 161 | 161 | 157 | 276 | 186 | 239 | 10 | 319 | 117 | 325 | 163 | 59 | 134 | 262 | 186 | 70 | 341 | 129 | 94 | 109 | 92 |
| D20S164 | UT1772 | 4 | 110 | 173 | 224 | 169 | 219 | 35 | 105 | 28 | 3 | 65 | 4 | 200 | 15 | 81 | 13 | 16 | 145 | 11 | 352 | 16 | 86 |
| D20S201 | GATA8B01 | 4 | 124 | 121 | 169 | 292 | 76 | 311 | 88 | 272 | 116 | 289 | 37 | 224 | 157 | 281 | 66 | 134 | 238 | 64 | 67 | 153 | 23 |
| D20S451 | UT254 | 4 | 148 | 170 | 150 | 75 | 214 | 159 | 121 | 105 | 91 | 259 | 269 | 78 | 61 | 212 | 218 | 187 | 331 | 318 | 293 | 360 | 346 |
| D20S477 | GATA29F06 | 4 | 221 | 265 | 264 | 220 | 246 | 260 | 229 | 257 | 37 | 44 | 96 | 304 | 84 | 107 | 124 | 60 | 64 | 147 | 181 | 293 | 218 |
| D20S478 | GATA42A03 | 4 | 6 | 9 | 47 | 5 | 71 | 53 | 43 | 34 | 365 | 330 | 261 | 82 | 170 | 372 | 294 | 90 | 345 | 221 | 256 | 199 | 278 |
| D20S480 | GATA45B10 | 4 | 370 | 370 | 374 | 327 | 367 | 288 | 304 | 347 | 298 | 372 | 248 | 373 | 301 | 373 | 296 | 308 | 368 | 294 | 274 | 275 | 274 |
| D20S481 | GATA47F05 | 4 | 252 | 149 | 199 | 112 | 104 | 83 | 301 | 327 | 130 | 109 | 180 | 246 | 81 | 172 | 225 | 155 | 86 | 197 | 15 | 177 | 61 |
| D20S482 | GATA51D03 | 4 | 349 | 337 | 305 | 360 | 315 | 348 | 329 | 360 | 278 | 314 | 369 | 328 | 292 | 345 | 377 | 228 | 343 | 375 | 329 | 299 | 353 |
| D20S851 | AFMA218YB5 | 2 | 74 | 82 | 73 | 281 | 99 | 96 | 116 | 142 | 41 | 19 | 34 | 104 | 37 | 15 | 47 | 20 | 5 | 23 | 20 | 17 | 5 |
| D21S1411 | UT1355 | 4 | 245 | 262 | 239 | 206 | 240 | 94 | 131 | 93 | 198 | 118 | 91 | 196 | 180 | 223 | 57 | 43 | 102 | 50 | 199 | 9 | 14 |
| D21S1432 | GATA11C12 | 4 | 160 | 228 | 143 | 192 | 156 | 146 | 142 | 241 | 214 | 188 | 252 | 188 | 137 | 202 | 228 | 127 | 201 | 278 | 126 | 76 | 297 |
| D21S1437 | GGAA3C07 | 4 | 55 | 67 | 127 | 101 | 300 | 111 | 76 | 116 | 124 | 167 | 364 | 87 | 155 | 203 | 306 | 148 | 256 | 256 | 115 | 57 | 146 |
| D21S1440 | ATA27F01 | 3 | 14 | 20 | 5 | 23 | 28 | 48 | 103 | 197 | 283 | 368 | 284 | 40 | 306 | 306 | 244 | 329 | 363 | 352 | 273 | 324 | 320 |
| D21S1446 | GATA70B08 | 4 | 64 | 171 | 141 | 88 | 32 | 139 | 6 | 68 | 243 | 202 | 347 | 192 | 172 | 76 | 216 | 258 | 93 | 329 | 151 | 305 | 197 |
| D21S2052 | GATA129D11 | 4 | 217 | 277 | 252 | 298 | 218 | 244 | 190 | 114 | 167 | 78 | 191 | 183 | 242 | 185 | 266 | 180 | 83 | 240 | 96 | 277 | 166 |
| D21S2055 | GATA188F04 | 4 | 12 | 56 | 33 | 84 | 55 | 21 | 4 | 20 | 47 | 17 | 19 | 68 | 103 | 36 | 28 | 139 | 27 | 25 | 43 | 36 | 11 |
| D22S1045 | ATA37D06 | 3 | 73 | 73 | 100 | 233 | 122 | 67 | 159 | 43 | 149 | 269 | 305 | 93 | 144 | 233 | 221 | 207 | 193 | 137 | 149 | 38 | 293 |
| D22S1169 | AFMB337ZH9 | 2 | 18 | 22 | 26 | 30 | 7 | 64 | 153 | 65 | 62 | 2 | 102 | 333 | 73 | 2 | 112 | 55 | 3 | 95 | 24 | 78 | 78 |
| D22S345 | MFD313 | 2 | 188 | 160 | 220 | 223 | 153 | 115 | 137 | 122 | 97 | 292 | 40 | 216 | 79 | 246 | 32 | 144 | 276 | 41 | 112 | 68 | 84 |
| D22S532 | UT7136 | 4 | 351 | 311 | 357 | 376 | 338 | 318 | 327 | 361 | 312 | 278 | 360 | 359 | 261 | 225 | 374 | 295 | 268 | 345 | 323 | 211 | 173 |
| D22S683 | GATA11B12 | 4 | 19 | 12 | 7 | 9 | 12 | 4 | 1 | 168 | 128 | 70 | 33 | 2 | 128 | 75 | 27 | 68 | 24 | 27 | 87 | 15 | 18 |
| D22S686 | GGAA10F06 | 4 | 169 | 189 | 170 | 133 | 192 | 257 | 21 | 136 | 102 | 312 | 370 | 34 | 54 | 201 | 308 | 62 | 285 | 359 | 234 | 254 | 336 |
| D22S689 | GATA21F03 | 4 | 261 | 234 | 249 | 226 | 229 | 224 | 99 | 269 | 237 | 217 | 324 | 210 | 245 | 208 | 208 | 254 | 232 | 253 | 214 | 231 | 201 |
| D2S1328 | GATA27A12 | 4 | 318 | 347 | 337 | 105 | 38 | 296 | 235 | 50 | 11 | 5 | 167 | 309 | 41 | 11 | 280 | 39 | 13 | 317 | 169 | 319 | 91 |
| D2S1334 | GATA4D07 | 4 | 176 | 165 | 167 | 272 | 203 | 217 | 14 | 297 | 138 | 335 | 181 | 73 | 129 | 265 | 166 | 66 | 354 | 121 | 103 | 100 | 111 |
| D2S1352 | ATA27D04 | 3 | 103 | 122 | 122 | 152 | 202 | 194 | 268 | 189 | 265 | 116 | 292 | 80 | 302 | 112 | 313 | 156 | 137 | 235 | 72 | 216 | 305 |
| D2S1353 | ATA27H09 | 3 | 140 | 120 | 194 | 111 | 126 | 37 | 344 | 218 | 287 | 213 | 74 | 229 | 315 | 240 | 104 | 181 | 162 | 63 | 304 | 91 | 135 |
| D2S1356 | ATA4F03 | 3 | 67 | 26 | 24 | 21 | 6 | 16 | 3 | 10 | 31 | 24 | 1 | 284 | 135 | 72 | 9 | 46 | 32 | 1 | 137 | 7 | 7 |
| D2S1360 | GATA11H10 | 4 | 197 | 271 | 256 | 33 | 239 | 145 | 177 | 36 | 33 | 222 | 114 | 243 | 39 | 304 | 155 | 4 | 199 | 104 | 76 | 263 | 283 |
| D2S1363 | GATA23D03 | 4 | 185 | 158 | 197 | 208 | 146 | 248 | 375 | 161 | 114 | 10 | 123 | 121 | 131 | 8 | 111 | 114 | 7 | 277 | 55 | 112 | 60 |
| D2S1384 | GATA52A04 | 4 | 258 | 135 | 230 | 143 | 129 | 249 | 161 | 238 | 89 | 200 | 207 | 28 | 46 | 250 | 93 | 143 | 200 | 259 | 156 | 278 | 105 |
| D2S1391 | GATA65C03 | 4 | 239 | 229 | 265 | 355 | 354 | 265 | 280 | 202 | 191 | 275 | 140 | 303 | 234 | 305 | 125 | 215 | 293 | 114 | 350 | 202 | 232 |
| D2S1394 | GATA69E12 | 4 | 348 | 356 | 373 | 349 | 347 | 342 | 309 | 293 | 366 | 366 | 345 | 295 | 361 | 353 | 329 | 377 | 373 | 344 | 367 | 343 | 362 |
| D2S1399 | GGAA20G04 | 4 | 144 | 143 | 208 | 366 | 148 | 132 | 7 | 87 | 136 | 131 | 25 | 136 | 191 | 93 | 40 | 175 | 119 | 44 | 54 | 29 | 49 |
| D2S1400 | GGAA20G10 | 4 | 4 | 5 | 2 | 2 | 3 | 3 | 29 | 231 | 123 | 215 | 85 | 61 | 113 | 206 | 77 | 159 | 284 | 142 | 376 | 288 | 350 |
| D2S1776 | GATA71D01 | 4 | 364 | 365 | 367 | 343 | 358 | 245 | 102 | 230 | 166 | 311 | 82 | 175 | 246 | 244 | 134 | 162 | 319 | 138 | 219 | 253 | 155 |
| D2S1780 | GATA72G11 | 4 | 87 | 69 | 52 | 103 | 155 | 165 | 308 | 276 | 362 | 318 | 315 | 285 | 356 | 357 | 285 | 330 | 374 | 293 | 316 | 290 | 258 |
| D2S1788 | GATA86E02 | 4 | 114 | 147 | 54 | 232 | 175 | 98 | 49 | 109 | 228 | 209 | 199 | 147 | 208 | 196 | 88 | 83 | 129 | 68 | 204 | 103 | 214 |
| D2S1790 | GATA88G05 | 4 | 122 | 87 | 103 | 92 | 124 | 81 | 38 | 179 | 132 | 166 | 216 | 272 | 171 | 259 | 231 | 135 | 235 | 223 | 272 | 303 | 252 |
| D2S2944 | GATA30E06 | 4 | 201 | 264 | 288 | 314 | 334 | 354 | 270 | 237 | 342 | 266 | 321 | 337 | 375 | 284 | 351 | 370 | 300 | 341 | 318 | 357 | 358 |
| D2S2952 | GATA116B01 | 4 | 53 | 93 | 75 | 50 | 48 | 22 | 219 | 259 | 302 | 211 | 190 | 360 | 260 | 171 | 102 | 250 | 142 | 80 | 118 | 92 | 329 |
| D2S2968 | GATA178G09 | 4 | 69 | 249 | 49 | 79 | 68 | 75 | 12 | 247 | 277 | 271 | 237 | 7 | 139 | 132 | 118 | 367 | 323 | 305 | 332 | 332 | 377 |
| D2S2972 | GATA176C01 | 4 | 62 | 79 | 74 | 139 | 281 | 103 | 205 | 63 | 324 | 197 | 319 | 163 | 360 | 283 | 310 | 307 | 255 | 244 | 291 | 271 | 176 |
| D2S2986 | 2QTEL47 | 2 | 11 | 8 | 13 | 35 | 45 | 36 | 104 | 6 | 16 | 16 | 24 | 21 | 28 | 12 | 19 | 30 | 12 | 22 | 174 | 174 | 351 |
| D2S405 | GATA8F07 | 4 | 166 | 148 | 135 | 106 | 60 | 201 | 208 | 287 | 264 | 153 | 322 | 317 | 309 | 134 | 304 | 227 | 130 | 347 | 222 | 234 | 267 |
| D2S410 | GATA4E11 | 4 | 164 | 145 | 123 | 144 | 147 | 95 | 214 | 118 | 104 | 223 | 29 | 69 | 87 | 187 | 36 | 218 | 273 | 67 | 239 | 184 | 77 |
| D2S427 | GATA12H10 | 4 | 7 | 6 | 6 | 11 | 10 | 59 | 40 | 372 | 351 | 237 | 240 | 46 | 203 | 159 | 345 | 309 | 187 | 205 | 263 | 111 | 117 |
| D2S434 | GATA4G12 | 4 | 345 | 296 | 251 | 334 | 286 | 232 | 254 | 277 | 377 | 351 | 264 | 64 | 330 | 299 | 149 | 325 | 330 | 333 | 303 | 258 | 276 |
| D2S441 | GATA8F03 | 4 | 134 | 141 | 101 | 153 | 2 | 17 | 265 | 31 | 90 | 6 | 26 | 23 | 112 | 1 | 20 | 267 | 8 | 42 | 9 | 23 | 296 |
| D3S1262 | AFM059XA9 | 2 | 35 | 25 | 38 | 62 | 16 | 55 | 182 | 193 | 56 | 42 | 147 | 311 | 55 | 65 | 171 | 23 | 28 | 84 | 68 | 115 | 95 |
| D3S1311 | AFM254VE1 | 2 | 42 | 47 | 46 | 8 | 26 | 60 | 28 | 49 | 7 | 53 | 184 | 10 | 2 | 40 | 114 | 1 | 120 | 272 | 74 | 34 | 210 |
| D3S1560 | AFM217XD6 | 2 | 21 | 57 | 31 | 64 | 64 | 131 | 20 | 15 | 65 | 52 | 253 | 67 | 107 | 165 | 272 | 225 | 74 | 351 | 70 | 269 | 108 |
| D3S1744 | GATA3C02 | 4 | 363 | 354 | 333 | 369 | 308 | 360 | 282 | 216 | 368 | 227 | 295 | 302 | 372 | 272 | 281 | 322 | 242 | 254 | 193 | 270 | 255 |
| D3S1746 | GATA8F01 | 4 | 178 | 152 | 144 | 123 | 43 | 152 | 90 | 113 | 110 | 34 | 220 | 109 | 99 | 39 | 195 | 179 | 54 | 275 | 159 | 301 | 160 |
| D3S1763 | GATA3H01 | 4 | 302 | 248 | 275 | 157 | 143 | 86 | 184 | 266 | 185 | 142 | 72 | 312 | 273 | 276 | 175 | 203 | 192 | 113 | 307 | 252 | 369 |
| D3S1764 | GATA4A10 | 4 | 77 | 51 | 102 | 188 | 178 | 183 | 260 | 296 | 244 | 282 | 130 | 173 | 216 | 213 | 100 | 283 | 292 | 169 | 260 | 221 | 266 |
| D3S1766 | GATA6F06 | 4 | 298 | 334 | 351 | 328 | 296 | 362 | 145 | 330 | 299 | 344 | 340 | 334 | 370 | 359 | 349 | 354 | 350 | 355 | 343 | 341 | 327 |
| D3S1768 | GATA8B05 | 4 | 80 | 36 | 70 | 138 | 134 | 149 | 242 | 363 | 55 | 122 | 71 | 169 | 31 | 106 | 53 | 27 | 118 | 83 | 311 | 142 | 188 |
| D3S2387 | GATA22G12 | 4 | 27 | 41 | 19 | 61 | 49 | 80 | 39 | 40 | 98 | 146 | 142 | 5 | 68 | 94 | 110 | 210 | 111 | 231 | 56 | 130 | 74 |
| D3S2398 | GATA6G12 | 4 | 321 | 123 | 207 | 170 | 244 | 347 | 41 | 227 | 245 | 255 | 372 | 76 | 125 | 83 | 255 | 332 | 214 | 328 | 229 | 274 | 307 |
| D3S2403 | GGAA4B09 | 4 | 32 | 62 | 121 | 98 | 61 | 85 | 24 | 18 | 141 | 171 | 183 | 107 | 174 | 194 | 145 | 87 | 104 | 59 | 244 | 242 | 240 |
| D3S2409 | ATA10H11 | 3 | 106 | 102 | 59 | 57 | 19 | 13 | 262 | 21 | 22 | 14 | 7 | 41 | 16 | 9 | 4 | 19 | 36 | 14 | 195 | 190 | 325 |
| D3S2418 | ATA22E01 | 3 | 29 | 44 | 14 | 46 | 181 | 50 | 46 | 72 | 253 | 61 | 168 | 30 | 249 | 59 | 151 | 305 | 20 | 194 | 25 | 218 | 33 |
| D3S2427 | GATA22F11 | 4 | 20 | 31 | 22 | 41 | 51 | 23 | 65 | 102 | 115 | 62 | 62 | 79 | 122 | 50 | 50 | 81 | 70 | 39 | 26 | 69 | 24 |
| D3S2432 | GATA27C08 | 4 | 287 | 304 | 311 | 331 | 349 | 320 | 175 | 292 | 288 | 291 | 244 | 223 | 319 | 316 | 312 | 290 | 280 | 198 | 277 | 166 | 319 |
| D3S2460 | GATA68F07 | 4 | 107 | 89 | 168 | 201 | 270 | 275 | 281 | 306 | 210 | 56 | 160 | 255 | 189 | 52 | 130 | 263 | 92 | 189 | 158 | 147 | 195 |
| D3S3038 | GATA73D01 | 4 | 237 | 263 | 241 | 189 | 212 | 218 | 277 | 331 | 206 | 249 | 291 | 348 | 154 | 279 | 258 | 79 | 204 | 207 | 286 | 368 | 343 |
| D3S3039 | GATA7F05 | 4 | 350 | 349 | 327 | 372 | 372 | 370 | 324 | 364 | 333 | 357 | 296 | 329 | 354 | 340 | 331 | 256 | 349 | 249 | 324 | 354 | 316 |
| D3S3045 | GATA84B12 | 4 | 146 | 129 | 60 | 117 | 29 | 6 | 292 | 26 | 175 | 38 | 9 | 12 | 241 | 38 | 12 | 154 | 139 | 38 | 88 | 30 | 158 |
| D3S3630 | AFMB296ZF5 | 2 | 10 | 7 | 28 | 24 | 50 | 104 | 80 | 41 | 281 | 83 | 172 | 114 | 264 | 110 | 189 | 209 | 175 | 288 | 38 | 156 | 167 |
| D3S3644 | AFMB318YF1 | 2 | 238 | 233 | 178 | 230 | 184 | 317 | 365 | 95 | 259 | 117 | 308 | 55 | 280 | 168 | 334 | 220 | 55 | 331 | 198 | 375 | 299 |
| D3S4523 | ATA34G06 | 3 | 88 | 107 | 173 | 132 | 263 | 161 | 218 | 94 | 63 | 139 | 5 | 193 | 91 | 198 | 14 | 158 | 266 | 13 | 255 | 6 | 128 |
| D3S4529 | GATA128C02 | 4 | 153 | 151 | 119 | 275 | 257 | 150 | 319 | 198 | 239 | 337 | 205 | 316 | 323 | 355 | 276 | 293 | 334 | 343 | 300 | 250 | 259 |
| D3S4545 | GATA164B08 | 4 | 155 | 139 | 112 | 22 | 191 | 51 | 115 | 59 | 35 | 195 | 45 | 105 | 42 | 267 | 64 | 26 | 203 | 49 | 47 | 264 | 73 |
| D4S1625 | GATA107 | 4 | 315 | 327 | 282 | 296 | 366 | 294 | 310 | 341 | 268 | 367 | 285 | 271 | 233 | 374 | 275 | 269 | 371 | 289 | 362 | 330 | 312 |
| D4S1627 | GATA7D01 | 4 | 219 | 156 | 283 | 357 | 318 | 371 | 330 | 332 | 155 | 110 | 316 | 251 | 130 | 103 | 299 | 221 | 160 | 362 | 313 | 337 | 253 |
| D4S1629 | GATA8A05 | 4 | 37 | 28 | 72 | 36 | 188 | 28 | 343 | 184 | 247 | 323 | 239 | 102 | 329 | 369 | 267 | 57 | 262 | 112 | 360 | 295 | 246 |
| D4S1644 | GATA11E09 | 4 | 251 | 292 | 263 | 174 | 245 | 209 | 258 | 108 | 170 | 235 | 250 | 287 | 214 | 308 | 284 | 233 | 346 | 287 | 347 | 346 | 372 |
| D4S1647 | GATA2F11 | 4 | 362 | 282 | 319 | 168 | 232 | 310 | 220 | 239 | 162 | 220 | 279 | 170 | 206 | 193 | 170 | 176 | 211 | 217 | 235 | 58 | 228 |
| D4S1652 | GATA5B02 | 4 | 358 | 374 | 359 | 291 | 306 | 343 | 180 | 171 | 202 | 228 | 357 | 181 | 102 | 205 | 254 | 124 | 228 | 290 | 341 | 363 | 321 |
| D4S2361 | ATA2A03 | 3 | 135 | 138 | 63 | 142 | 105 | 68 | 192 | 233 | 344 | 193 | 53 | 125 | 285 | 191 | 82 | 272 | 168 | 46 | 155 | 51 | 131 |
| D4S2366 | GATA22G05 | 4 | 61 | 32 | 99 | 100 | 78 | 114 | 274 | 280 | 95 | 51 | 185 | 330 | 159 | 74 | 219 | 149 | 72 | 191 | 241 | 236 | 243 |
| D4S2367 | GATA24H01 | 4 | 273 | 321 | 290 | 207 | 163 | 368 | 107 | 53 | 42 | 18 | 280 | 363 | 153 | 78 | 368 | 167 | 76 | 364 | 216 | 334 | 215 |
| D4S2368 | GATA27G03 | 4 | 375 | 368 | 358 | 377 | 336 | 268 | 294 | 335 | 355 | 256 | 265 | 282 | 300 | 294 | 309 | 224 | 190 | 284 | 280 | 107 | 134 |
| D4S2394 | ATA26B08 | 3 | 143 | 127 | 128 | 113 | 87 | 133 | 360 | 78 | 94 | 15 | 149 | 103 | 83 | 16 | 143 | 140 | 15 | 124 | 30 | 256 | 68 |
| D4S2397 | ATA27C07 | 3 | 96 | 109 | 81 | 16 | 56 | 73 | 240 | 323 | 54 | 144 | 177 | 228 | 34 | 182 | 146 | 35 | 125 | 199 | 93 | 377 | 254 |
| D4S2417 | GATA42H02 | 4 | 377 | 364 | 329 | 269 | 185 | 123 | 335 | 146 | 193 | 157 | 65 | 198 | 200 | 161 | 90 | 340 | 180 | 110 | 168 | 60 | 348 |
| D4S2431 | GGAA19H07 | 4 | 257 | 206 | 132 | 109 | 103 | 193 | 155 | 8 | 75 | 104 | 230 | 24 | 90 | 105 | 273 | 102 | 146 | 319 | 264 | 232 | 279 |
| D4S2623 | GATA62A12 | 4 | 54 | 52 | 110 | 202 | 70 | 153 | 157 | 123 | 86 | 27 | 328 | 235 | 143 | 28 | 348 | 101 | 22 | 268 | 100 | 140 | 37 |
| D4S2632 | GATA72G09 | 4 | 90 | 88 | 189 | 148 | 205 | 222 | 33 | 5 | 21 | 50 | 125 | 18 | 32 | 48 | 84 | 15 | 48 | 100 | 172 | 110 | 212 |
| D4S3243 | GATA10G07 | 4 | 16 | 54 | 27 | 81 | 47 | 25 | 67 | 151 | 118 | 203 | 192 | 29 | 126 | 113 | 94 | 212 | 272 | 209 | 185 | 207 | 239 |
| D4S3248 | GATA28F03 | 4 | 340 | 303 | 352 | 340 | 344 | 227 | 312 | 340 | 270 | 336 | 122 | 267 | 278 | 302 | 158 | 320 | 335 | 148 | 299 | 265 | 216 |
| D4S3360 | 4PTEL04 | 2 | 139 | 166 | 165 | 73 | 231 | 65 | 72 | 285 | 187 | 349 | 124 | 262 | 138 | 249 | 103 | 119 | 315 | 85 | 135 | 88 | 163 |
| D4S403 | AFM157XG3 | 2 | 47 | 77 | 25 | 6 | 23 | 12 | 233 | 175 | 17 | 39 | 118 | 35 | 11 | 32 | 78 | 18 | 50 | 150 | 161 | 181 | 130 |
| D4S408 | AFM165XC11 | 2 | 60 | 99 | 148 | 70 | 42 | 203 | 132 | 25 | 20 | 25 | 128 | 97 | 33 | 23 | 163 | 21 | 34 | 184 | 111 | 192 | 103 |
| D5S1456 | GATA11A11 | 4 | 246 | 220 | 261 | 318 | 182 | 327 | 313 | 178 | 363 | 191 | 338 | 140 | 328 | 207 | 290 | 319 | 239 | 330 | 124 | 359 | 145 |
| D5S1457 | GATA21D04 | 4 | 216 | 285 | 229 | 356 | 323 | 171 | 101 | 205 | 163 | 346 | 229 | 106 | 342 | 364 | 144 | 123 | 290 | 233 | 325 | 47 | 233 |
| D5S1462 | GATA3H06 | 4 | 179 | 241 | 187 | 284 | 351 | 221 | 85 | 97 | 151 | 301 | 238 | 118 | 257 | 331 | 350 | 298 | 352 | 298 | 346 | 241 | 260 |
| D5S1470 | GATA7C06 | 4 | 199 | 197 | 268 | 185 | 279 | 321 | 269 | 67 | 109 | 90 | 271 | 195 | 151 | 122 | 259 | 164 | 221 | 302 | 189 | 90 | 178 |
| D5S1480 | ATA23A10 | 3 | 215 | 221 | 201 | 179 | 97 | 156 | 55 | 124 | 76 | 71 | 84 | 133 | 40 | 67 | 70 | 14 | 29 | 36 | 33 | 134 | 257 |
| D5S1501 | GATA52A12 | 4 | 108 | 76 | 80 | 95 | 20 | 154 | 2 | 1 | 66 | 7 | 157 | 162 | 194 | 34 | 238 | 65 | 37 | 149 | 44 | 336 | 119 |
| D5S1505 | GATA62A04 | 4 | 274 | 181 | 250 | 316 | 255 | 345 | 203 | 373 | 353 | 308 | 334 | 269 | 281 | 322 | 253 | 339 | 298 | 322 | 295 | 308 | 200 |
| D5S1725 | GATA89G08 | 4 | 66 | 33 | 68 | 128 | 247 | 142 | 252 | 352 | 370 | 184 | 346 | 168 | 291 | 115 | 364 | 358 | 122 | 297 | 147 | 306 | 170 |
| D5S211 | MFD154 | 2 | 150 | 46 | 147 | 135 | 194 | 66 | 15 | 51 | 44 | 224 | 11 | 4 | 13 | 99 | 3 | 45 | 198 | 20 | 97 | 146 | 46 |
| D5S2488 | ATA20G07 | 3 | 259 | 210 | 247 | 274 | 321 | 213 | 165 | 223 | 260 | 306 | 218 | 264 | 263 | 335 | 265 | 313 | 316 | 239 | 305 | 294 | 300 |
| D5S2500 | GATA67D03 | 4 | 308 | 307 | 324 | 280 | 226 | 365 | 247 | 288 | 346 | 206 | 351 | 342 | 297 | 184 | 353 | 273 | 156 | 334 | 157 | 315 | 196 |
| D5S2501 | GATA68A03 | 4 | 346 | 352 | 346 | 299 | 363 | 195 | 354 | 154 | 286 | 342 | 243 | 143 | 317 | 363 | 197 | 268 | 359 | 162 | 364 | 116 | 183 |
| D5S2505 | GATA84E11 | 4 | 137 | 111 | 107 | 165 | 190 | 220 | 160 | 177 | 157 | 322 | 283 | 219 | 162 | 341 | 192 | 214 | 313 | 155 | 163 | 99 | 202 |
| D5S2845 | GATA134B03 | 4 | 279 | 318 | 291 | 375 | 207 | 289 | 288 | 155 | 208 | 100 | 297 | 248 | 259 | 119 | 270 | 219 | 112 | 208 | 138 | 160 | 112 |
| D5S2849 | GATA145D10 | 4 | 331 | 243 | 292 | 161 | 198 | 301 | 328 | 244 | 84 | 263 | 197 | 280 | 67 | 243 | 138 | 97 | 267 | 146 | 257 | 93 | 118 |
| D5S408 | AFM164XB8 | 2 | 86 | 70 | 50 | 39 | 86 | 54 | 8 | 70 | 137 | 165 | 94 | 38 | 80 | 135 | 169 | 137 | 138 | 212 | 289 | 237 | 227 |
| D5S816 | GATA2H09 | 4 | 344 | 328 | 158 | 225 | 172 | 151 | 376 | 127 | 213 | 258 | 133 | 154 | 166 | 277 | 184 | 103 | 325 | 179 | 116 | 32 | 306 |
| D5S817 | GATA3E10 | 4 | 230 | 223 | 289 | 217 | 272 | 282 | 202 | 281 | 291 | 260 | 358 | 145 | 152 | 177 | 268 | 302 | 308 | 358 | 333 | 365 | 363 |
| D5S820 | GATA6E05 | 4 | 57 | 91 | 98 | 102 | 262 | 72 | 166 | 251 | 174 | 340 | 107 | 263 | 235 | 366 | 160 | 182 | 365 | 72 | 245 | 117 | 157 |
| D6S1006 | ATC4D09 | 3 | 117 | 194 | 45 | 13 | 14 | 43 | 117 | 271 | 50 | 31 | 78 | 3 | 7 | 5 | 23 | 98 | 51 | 160 | 221 | 369 | 367 |
| D6S1009 | GATA32B03 | 4 | 268 | 278 | 317 | 302 | 65 | 250 | 52 | 48 | 201 | 32 | 80 | 256 | 252 | 21 | 120 | 304 | 25 | 165 | 10 | 135 | 76 |
| D6S1017 | GGAT3H10 | 4 | 17 | 13 | 10 | 43 | 21 | 191 | 241 | 290 | 335 | 284 | 144 | 350 | 270 | 301 | 121 | 306 | 296 | 91 | 145 | 131 | 51 |
| D6S1021 | ATA11D10 | 3 | 81 | 63 | 53 | 29 | 84 | 163 | 127 | 185 | 88 | 290 | 212 | 201 | 236 | 289 | 142 | 173 | 252 | 219 | 194 | 46 | 237 |
| D6S1027 | ATA22G07 | 3 | 9 | 11 | 8 | 12 | 17 | 20 | 201 | 337 | 108 | 219 | 95 | 194 | 93 | 167 | 72 | 82 | 194 | 79 | 179 | 213 | 217 |
| D6S1031 | ATA28B11 | 3 | 31 | 85 | 61 | 173 | 136 | 215 | 23 | 149 | 39 | 58 | 202 | 130 | 118 | 84 | 297 | 41 | 18 | 236 | 107 | 344 | 162 |
| D6S1040 | GATA23F08 | 4 | 194 | 191 | 204 | 294 | 294 | 276 | 355 | 192 | 207 | 226 | 344 | 291 | 250 | 264 | 361 | 192 | 264 | 283 | 372 | 133 | 179 |
| D6S1051 | GATA61E03 | 4 | 330 | 275 | 295 | 345 | 327 | 351 | 320 | 291 | 352 | 265 | 286 | 354 | 371 | 282 | 279 | 365 | 294 | 263 | 315 | 289 | 368 |
| D6S1053 | GATA64D02 | 4 | 281 | 179 | 372 | 315 | 201 | 376 | 275 | 89 | 361 | 304 | 314 | 15 | 351 | 324 | 277 | 257 | 141 | 370 | 279 | 329 | 190 |
| D6S1056 | GATA68H04 | 4 | 269 | 140 | 307 | 361 | 58 | 279 | 123 | 181 | 315 | 85 | 116 | 92 | 149 | 109 | 45 | 249 | 23 | 115 | 18 | 157 | 10 |
| D6S1277 | GATA81B01 | 4 | 228 | 366 | 296 | 262 | 135 | 256 | 42 | 74 | 235 | 244 | 267 | 230 | 197 | 136 | 242 | 174 | 196 | 316 | 139 | 261 | 264 |
| D6S1959 | GATA29A01 | 4 | 353 | 361 | 323 | 338 | 317 | 188 | 359 | 303 | 337 | 370 | 127 | 364 | 339 | 365 | 135 | 314 | 367 | 139 | 321 | 215 | 171 |
| D6S2410 | GATA11E02 | 4 | 369 | 363 | 375 | 371 | 352 | 242 | 350 | 357 | 328 | 295 | 196 | 313 | 334 | 319 | 248 | 366 | 326 | 171 | 339 | 151 | 207 |
| D6S2436 | GATA165G02 | 4 | 65 | 90 | 160 | 158 | 345 | 88 | 207 | 80 | 271 | 232 | 232 | 132 | 332 | 293 | 196 | 255 | 291 | 170 | 281 | 124 | 115 |
| D6S2439 | GATA163B10 | 4 | 175 | 259 | 236 | 181 | 243 | 198 | 120 | 201 | 230 | 82 | 235 | 217 | 218 | 189 | 190 | 184 | 114 | 264 | 92 | 279 | 81 |
| D6S2522 | 6QTEL54 | 2 | 319 | 330 | 254 | 373 | 377 | 322 | 377 | 365 | 240 | 310 | 377 | 344 | 295 | 344 | 376 | 133 | 259 | 377 | 377 | 223 | 287 |
| D6S305 | AFM242ZG5 | 2 | 75 | 40 | 44 | 54 | 36 | 11 | 118 | 134 | 250 | 69 | 58 | 135 | 202 | 123 | 49 | 342 | 65 | 28 | 36 | 13 | 8 |
| D6S474 | GATA31 | 4 | 374 | 335 | 255 | 242 | 360 | 334 | 325 | 137 | 220 | 354 | 289 | 345 | 320 | 362 | 302 | 327 | 328 | 285 | 334 | 307 | 364 |
| D6S942 | UT654 | 4 | 316 | 314 | 325 | 257 | 256 | 333 | 232 | 217 | 310 | 332 | 363 | 323 | 237 | 269 | 319 | 111 | 188 | 267 | 371 | 370 | 371 |
| D7S1799 | GATA23F05 | 4 | 301 | 217 | 192 | 308 | 266 | 211 | 222 | 98 | 215 | 261 | 260 | 254 | 219 | 245 | 321 | 243 | 271 | 353 | 252 | 194 | 301 |
| D7S1802 | GATA41G07 | 4 | 241 | 250 | 309 | 323 | 264 | 236 | 370 | 282 | 296 | 362 | 303 | 340 | 343 | 368 | 303 | 334 | 338 | 250 | 306 | 287 | 349 |
| D7S1804 | GATA43C11 | 4 | 222 | 269 | 215 | 293 | 115 | 176 | 92 | 182 | 223 | 208 | 28 | 37 | 314 | 145 | 59 | 131 | 144 | 31 | 89 | 42 | 20 |
| D7S1808 | GGAA3F06 | 4 | 337 | 239 | 151 | 26 | 132 | 29 | 156 | 79 | 10 | 119 | 15 | 49 | 5 | 128 | 8 | 11 | 161 | 24 | 58 | 226 | 83 |
| D7S1818 | GATA24D12 | 4 | 355 | 313 | 356 | 358 | 369 | 303 | 179 | 214 | 367 | 369 | 193 | 139 | 277 | 349 | 116 | 317 | 377 | 159 | 369 | 198 | 249 |
| D7S1824 | GATA32C12 | 4 | 243 | 256 | 350 | 156 | 160 | 200 | 167 | 107 | 79 | 93 | 117 | 253 | 120 | 149 | 185 | 48 | 96 | 131 | 310 | 282 | 322 |
| D7S2204 | GATA73D10 | 4 | 132 | 190 | 223 | 216 | 248 | 258 | 77 | 24 | 153 | 141 | 209 | 25 | 132 | 160 | 193 | 122 | 106 | 234 | 90 | 159 | 168 |
| D7S2477 | AFMB035XB9 | 2 | 68 | 104 | 124 | 193 | 80 | 39 | 154 | 7 | 28 | 1 | 99 | 26 | 75 | 7 | 92 | 120 | 2 | 73 | 7 | 10 | 1 |
| D7S2846 | GATA31A10 | 4 | 376 | 377 | 376 | 364 | 276 | 361 | 213 | 359 | 319 | 281 | 361 | 362 | 358 | 261 | 283 | 359 | 286 | 301 | 215 | 229 | 185 |
| D7S3046 | GATA118G10 | 4 | 119 | 163 | 163 | 239 | 249 | 287 | 97 | 112 | 92 | 41 | 320 | 180 | 169 | 79 | 339 | 234 | 107 | 357 | 209 | 296 | 169 |
| D7S3047 | GATA119B03 | 4 | 297 | 342 | 281 | 324 | 340 | 252 | 367 | 355 | 369 | 352 | 210 | 336 | 373 | 367 | 209 | 374 | 336 | 188 | 337 | 222 | 335 |
| D7S3051 | GATA137H02 | 4 | 198 | 230 | 206 | 264 | 123 | 166 | 250 | 190 | 165 | 121 | 287 | 150 | 255 | 111 | 288 | 105 | 170 | 232 | 85 | 187 | 138 |
| D7S3056 | GATA24F03 | 4 | 296 | 242 | 200 | 251 | 271 | 352 | 187 | 110 | 224 | 234 | 307 | 322 | 272 | 235 | 269 | 239 | 184 | 182 | 206 | 267 | 339 |
| D7S3058 | GATA30D09 | 4 | 267 | 236 | 318 | 186 | 140 | 283 | 66 | 104 | 160 | 75 | 282 | 226 | 226 | 63 | 324 | 199 | 98 | 312 | 167 | 158 | 72 |
| D7S3061 | GGAA6D03 | 4 | 211 | 193 | 193 | 252 | 90 | 229 | 69 | 64 | 188 | 98 | 64 | 113 | 94 | 58 | 61 | 73 | 132 | 43 | 75 | 120 | 35 |
| D7S3070 | GATA189C06 | 4 | 271 | 312 | 312 | 312 | 310 | 269 | 245 | 286 | 304 | 300 | 227 | 321 | 284 | 311 | 233 | 276 | 348 | 218 | 326 | 239 | 331 |
| D7S559 | MFD265 | 2 | 190 | 130 | 164 | 120 | 158 | 208 | 199 | 250 | 217 | 272 | 166 | 249 | 176 | 307 | 106 | 165 | 237 | 132 | 164 | 62 | 156 |
| D7S817 | GATA13G11 | 4 | 295 | 274 | 355 | 365 | 346 | 97 | 315 | 264 | 106 | 245 | 38 | 357 | 108 | 227 | 62 | 121 | 250 | 37 | 320 | 8 | 26 |
| D7S820 | GATA3F01 | 4 | 333 | 309 | 298 | 199 | 237 | 125 | 287 | 220 | 197 | 225 | 106 | 301 | 232 | 317 | 148 | 247 | 218 | 109 | 217 | 204 | 286 |
| D7S821 | GATA5D08 | 4 | 165 | 226 | 174 | 245 | 284 | 364 | 53 | 299 | 269 | 89 | 188 | 134 | 274 | 108 | 257 | 297 | 131 | 214 | 120 | 280 | 308 |
| D8S1048 | UT7129 | 4 | 354 | 372 | 315 | 347 | 297 | 316 | 358 | 39 | 330 | 246 | 336 | 129 | 364 | 291 | 323 | 311 | 324 | 246 | 312 | 342 | 304 |
| D8S1108 | GATA50D10 | 4 | 347 | 350 | 364 | 335 | 364 | 341 | 98 | 342 | 375 | 302 | 304 | 261 | 338 | 309 | 341 | 373 | 347 | 337 | 284 | 302 | 357 |
| D8S1110 | GATA8G10 | 4 | 89 | 60 | 126 | 210 | 179 | 270 | 124 | 279 | 295 | 196 | 89 | 166 | 225 | 155 | 75 | 335 | 169 | 107 | 200 | 145 | 120 |
| D8S1113 | GGAA8G07 | 4 | 231 | 188 | 219 | 163 | 130 | 120 | 138 | 228 | 74 | 20 | 208 | 167 | 53 | 10 | 246 | 117 | 6 | 243 | 3 | 77 | 4 |
| D8S1128 | GATA21C12 | 4 | 98 | 192 | 195 | 18 | 250 | 141 | 60 | 14 | 18 | 106 | 30 | 365 | 56 | 199 | 115 | 42 | 153 | 144 | 8 | 49 | 161 |
| D8S1132 | GATA26E03 | 4 | 270 | 245 | 120 | 191 | 66 | 297 | 336 | 186 | 347 | 63 | 333 | 146 | 333 | 62 | 346 | 316 | 59 | 248 | 51 | 314 | 97 |
| D8S1136 | GATA41A01 | 4 | 277 | 291 | 271 | 212 | 293 | 304 | 225 | 314 | 200 | 373 | 249 | 191 | 223 | 338 | 240 | 188 | 362 | 274 | 231 | 272 | 262 |
| D8S1179 | GATA7G07 | 4 | 177 | 301 | 212 | 248 | 376 | 206 | 79 | 103 | 289 | 238 | 326 | 186 | 350 | 334 | 210 | 357 | 261 | 262 | 210 | 238 | 225 |
| D8S1477 | GGAA20C10 | 4 | 162 | 205 | 172 | 136 | 173 | 90 | 75 | 32 | 52 | 150 | 158 | 44 | 63 | 148 | 162 | 80 | 195 | 89 | 353 | 27 | 143 |
| D8S2324 | GATA14E09 | 4 | 356 | 346 | 334 | 353 | 353 | 293 | 204 | 76 | 356 | 294 | 231 | 108 | 359 | 257 | 320 | 362 | 225 | 342 | 197 | 317 | 177 |
| D8S261 | AFM123XG5 | 2 | 34 | 15 | 16 | 32 | 114 | 62 | 257 | 66 | 70 | 179 | 36 | 86 | 52 | 154 | 29 | 95 | 121 | 60 | 62 | 209 | 69 |
| D8S262 | AFM127XH2 | 2 | 24 | 27 | 12 | 58 | 13 | 19 | 198 | 242 | 225 | 124 | 214 | 172 | 316 | 80 | 206 | 291 | 152 | 273 | 77 | 228 | 154 |
| D8S373 | UT721 | 4 | 206 | 214 | 284 | 368 | 339 | 160 | 303 | 45 | 159 | 159 | 270 | 39 | 164 | 211 | 278 | 292 | 217 | 175 | 228 | 41 | 124 |
| D8S503 | AFM193XH4 | 2 | 142 | 204 | 113 | 51 | 110 | 33 | 163 | 84 | 24 | 45 | 20 | 31 | 24 | 56 | 18 | 50 | 91 | 57 | 183 | 244 | 121 |
| D8S560 | AFMA127YE5 | 2 | 1 | 4 | 3 | 14 | 138 | 1 | 16 | 23 | 93 | 8 | 73 | 187 | 88 | 17 | 44 | 71 | 10 | 56 | 5 | 35 | 9 |
| D8S592 | GATA6B02 | 4 | 167 | 162 | 145 | 164 | 59 | 204 | 341 | 371 | 334 | 199 | 329 | 275 | 311 | 175 | 317 | 318 | 189 | 350 | 249 | 318 | 273 |
| D9S1118 | GATA71E08 | 4 | 121 | 103 | 118 | 184 | 46 | 298 | 144 | 157 | 327 | 33 | 156 | 57 | 307 | 43 | 153 | 270 | 11 | 185 | 17 | 123 | 62 |
| D9S1120 | GATA81C04 | 4 | 366 | 355 | 370 | 344 | 307 | 130 | 316 | 262 | 273 | 313 | 55 | 331 | 287 | 286 | 67 | 196 | 233 | 40 | 296 | 28 | 79 |
| D9S1121 | GATA87E02 | 4 | 159 | 101 | 76 | 55 | 25 | 267 | 32 | 37 | 129 | 74 | 175 | 42 | 77 | 29 | 251 | 63 | 41 | 87 | 37 | 48 | 41 |
| D9S1122 | GATA89A11 | 4 | 361 | 339 | 371 | 329 | 283 | 302 | 366 | 358 | 313 | 307 | 206 | 299 | 269 | 292 | 202 | 280 | 302 | 196 | 355 | 320 | 292 |
| D9S1779 | AFM026TG9 | 2 | 5 | 3 | 17 | 7 | 30 | 8 | 135 | 4 | 26 | 125 | 61 | 1 | 19 | 116 | 43 | 2 | 56 | 33 | 173 | 164 | 136 |
| D9S1825 | AFMB029XG1 | 2 | 131 | 132 | 95 | 94 | 33 | 91 | 68 | 62 | 221 | 48 | 186 | 62 | 222 | 49 | 173 | 261 | 30 | 229 | 22 | 297 | 29 |
| D9S1838 | AFMB303ZG9 | 2 | 100 | 174 | 90 | 45 | 24 | 306 | 94 | 150 | 40 | 137 | 87 | 148 | 70 | 92 | 131 | 47 | 148 | 53 | 46 | 12 | 15 |
| D9S1871 | AFM345TA9 | 2 | 76 | 43 | 66 | 15 | 18 | 89 | 9 | 81 | 13 | 112 | 31 | 8 | 4 | 46 | 21 | 9 | 75 | 61 | 14 | 73 | 16 |
| D9S2157 | ATA59H06 | 3 | 82 | 81 | 64 | 28 | 167 | 30 | 185 | 71 | 6 | 134 | 32 | 13 | 3 | 98 | 31 | 22 | 224 | 96 | 242 | 52 | 140 |
| D9S2169 | GATA62F03 | 4 | 39 | 49 | 30 | 48 | 95 | 168 | 237 | 367 | 371 | 374 | 348 | 300 | 331 | 377 | 359 | 369 | 376 | 367 | 375 | 350 | 360 |
| D9S301 | GATA7D12 | 4 | 260 | 284 | 272 | 175 | 316 | 70 | 186 | 329 | 275 | 341 | 108 | 252 | 201 | 254 | 87 | 245 | 306 | 106 | 178 | 113 | 82 |
| D9S910 | ATA18A07 | 3 | 113 | 113 | 115 | 140 | 108 | 138 | 228 | 163 | 72 | 40 | 46 | 98 | 47 | 20 | 41 | 52 | 26 | 74 | 213 | 188 | 189 |
| D9S922 | GATA21F05 | 4 | 282 | 322 | 182 | 69 | 89 | 57 | 351 | 133 | 101 | 136 | 48 | 120 | 62 | 95 | 38 | 77 | 191 | 8 | 336 | 22 | 44 |
| D9S925 | GATA27A11 | 4 | 94 | 94 | 71 | 86 | 27 | 41 | 114 | 75 | 85 | 64 | 59 | 225 | 133 | 89 | 81 | 150 | 97 | 62 | 132 | 95 | 142 |
| D9S930 | GATA48D07 | 4 | 342 | 324 | 335 | 336 | 312 | 324 | 221 | 317 | 279 | 230 | 310 | 343 | 324 | 209 | 347 | 344 | 155 | 314 | 150 | 210 | 139 |
| D9S934 | GATA64G07 | 4 | 339 | 351 | 348 | 270 | 273 | 344 | 289 | 166 | 219 | 218 | 349 | 231 | 253 | 241 | 336 | 265 | 269 | 326 | 283 | 245 | 347 |
| D9S938 | GGAA22E01 | 4 | 249 | 283 | 293 | 219 | 37 | 45 | 188 | 156 | 58 | 26 | 2 | 274 | 59 | 24 | 2 | 74 | 17 | 2 | 29 | 25 | 6 |
| F13A1-D6S | SE30 | 4 | 168 | 289 | 227 | 17 | 74 | 155 | 13 | 2 | 4 | 11 | 105 | 48 | 6 | 31 | 109 | 3 | 39 | 154 | 1 | 44 | 17 |
| NA-D10S-1 | GATA121A08 | 4 | 275 | 273 | 246 | 250 | 322 | 315 | 195 | 300 | 249 | 243 | 374 | 202 | 275 | 210 | 330 | 312 | 263 | 363 | 180 | 281 | 315 |
| NA-D10S-2 | ATA103C06 | 3 | 52 | 35 | 37 | 83 | 131 | 197 | 321 | 152 | 161 | 130 | 97 | 176 | 127 | 118 | 96 | 132 | 149 | 157 | 301 | 251 | 314 |
| NA-D11S-1 | ATA34E08 | 3 | 40 | 16 | 55 | 200 | 282 | 148 | 112 | 211 | 51 | 170 | 170 | 209 | 25 | 163 | 101 | 24 | 177 | 133 | 302 | 312 | 242 |
| NA-D12S-1 | GATA49D12 | 4 | 157 | 78 | 159 | 204 | 63 | 192 | 162 | 56 | 82 | 28 | 254 | 33 | 57 | 22 | 315 | 161 | 58 | 195 | 41 | 75 | 56 |
| NA-D12S-2 | PAH | 4 | 174 | 86 | 146 | 172 | 193 | 329 | 305 | 131 | 241 | 173 | 109 | 208 | 296 | 217 | 98 | 352 | 227 | 136 | 262 | 139 | 241 |
| NA-D13S-1 | ATA5A09 | 3 | 116 | 117 | 88 | 107 | 220 | 24 | 248 | 115 | 204 | 66 | 42 | 355 | 123 | 86 | 26 | 36 | 42 | 5 | 60 | 102 | 45 |
| NA-D14S-1 | GATA193A07 | 4 | 91 | 137 | 234 | 215 | 335 | 295 | 152 | 35 | 19 | 162 | 69 | 100 | 51 | 204 | 136 | 72 | 244 | 174 | 117 | 262 | 206 |
| NA-D15S-1 | GATA50C03 | 4 | 170 | 172 | 238 | 209 | 224 | 178 | 340 | 283 | 238 | 338 | 255 | 265 | 207 | 303 | 229 | 232 | 289 | 203 | 271 | 273 | 303 |
| NA-D16S-1 | GATA138C05 | 4 | 30 | 37 | 20 | 68 | 81 | 119 | 59 | 160 | 169 | 94 | 234 | 126 | 247 | 144 | 213 | 251 | 95 | 192 | 166 | 169 | 209 |
| NA-D17S-1 | ATA78D02 | 3 | 242 | 208 | 175 | 72 | 253 | 44 | 349 | 139 | 34 | 132 | 13 | 152 | 38 | 147 | 16 | 56 | 90 | 16 | 98 | 183 | 137 |
| NA-D18S-1 | GATA178F11 | 4 | 327 | 281 | 267 | 286 | 211 | 235 | 78 | 301 | 350 | 319 | 318 | 156 | 290 | 179 | 183 | 336 | 305 | 303 | 253 | 248 | 374 |
| NA-D18S-2 | ATA82B02 | 3 | 59 | 48 | 62 | 78 | 34 | 167 | 217 | 240 | 83 | 49 | 52 | 123 | 110 | 18 | 58 | 94 | 46 | 69 | 49 | 37 | 21 |
| NA-D1S-1 | ATA79C10 | 3 | 324 | 358 | 326 | 253 | 259 | 144 | 342 | 308 | 135 | 233 | 18 | 376 | 177 | 247 | 35 | 130 | 182 | 12 | 82 | 24 | 47 |
| NA-D1S-2 | GATA124C08 | 4 | 240 | 325 | 240 | 307 | 265 | 355 | 149 | 164 | 252 | 107 | 290 | 206 | 327 | 214 | 342 | 353 | 207 | 335 | 265 | 374 | 337 |
| NA-D1S-3 | GATA133A08 | 4 | 186 | 199 | 237 | 263 | 142 | 339 | 81 | 159 | 316 | 298 | 236 | 358 | 347 | 260 | 301 | 361 | 247 | 313 | 267 | 347 | 275 |
| NA-D1S-4 | ATA42G12 | 3 | 226 | 211 | 235 | 236 | 206 | 214 | 256 | 324 | 307 | 320 | 171 | 369 | 212 | 248 | 117 | 260 | 245 | 88 | 192 | 96 | 281 |
| NA-D1S-5 | GATA124F08 | 4 | 372 | 270 | 377 | 305 | 375 | 325 | 158 | 349 | 290 | 343 | 313 | 89 | 161 | 228 | 178 | 236 | 369 | 265 | 248 | 214 | 334 |
| NA-D22S-1 | GATA198B05 | 4 | 359 | 329 | 369 | 261 | 223 | 187 | 251 | 224 | 105 | 174 | 67 | 197 | 97 | 120 | 51 | 198 | 179 | 105 | 142 | 108 | 223 |
| NA-D4S-1 | GATA70E01 | 4 | 102 | 97 | 56 | 63 | 109 | 87 | 210 | 275 | 112 | 329 | 272 | 124 | 115 | 300 | 245 | 99 | 351 | 320 | 247 | 195 | 323 |
| NA-D5S-1 | ATA52D02 | 3 | 2 | 1 | 4 | 10 | 22 | 15 | 113 | 135 | 156 | 185 | 337 | 53 | 96 | 178 | 260 | 186 | 206 | 225 | 224 | 101 | 116 |
| NA-D6S-1 | GATA184A08 | 4 | 26 | 14 | 21 | 56 | 94 | 82 | 231 | 172 | 131 | 158 | 251 | 234 | 65 | 117 | 201 | 59 | 157 | 210 | 201 | 189 | 277 |
| NA-D7S-1 | GATA104 | 4 | 101 | 75 | 58 | 124 | 98 | 190 | 95 | 111 | 301 | 168 | 281 | 220 | 344 | 268 | 235 | 289 | 241 | 166 | 227 | 191 | 109 |
| NA-D8S-1 | GATA151F02 | 4 | 120 | 128 | 108 | 194 | 176 | 189 | 148 | 325 | 236 | 236 | 277 | 160 | 239 | 215 | 263 | 185 | 220 | 211 | 123 | 304 | 165 |
| NA-D8S-2 | GAAT1A4 | 4 | 207 | 136 | 114 | 125 | 274 | 238 | 170 | 195 | 261 | 113 | 366 | 47 | 163 | 70 | 314 | 274 | 47 | 366 | 65 | 260 | 85 |
| NA-D9S-1 | GATA187D09 | 4 | 312 | 267 | 331 | 320 | 269 | 319 | 283 | 273 | 255 | 148 | 242 | 83 | 173 | 138 | 212 | 333 | 174 | 292 | 240 | 348 | 373 |
| TPO-D2S | SRA | 4 | 111 | 155 | 87 | 122 | 209 | 46 | 286 | 270 | 357 | 129 | 113 | 189 | 346 | 186 | 108 | 363 | 172 | 92 | 128 | 70 | 53 |
Note.— For each data set (table 4), markers are ranked from most informative (1) to least informative (377). Names for the markers are the same as in supplementary table 4 of Rosenberg et al. (2002). The regions are coded as follows: 1 = Africa; 2 = Europe; 3 = Middle East; 4 = Central/South Asia; 5 = East Asia; 6 = Oceania; 7 = America. Loci are sorted in lexical order.
Table C.
Informativeness (In) for Distinguishing among Three Regions (First of Three Parts)[Note]
|
In Rank for Set of Three Regions |
|||||||||||||||||
| Locus Name | Alternate Name | RepeatSize | 1-2-3 | 1-2-4 | 1-2-5 | 1-2-6 | 1-2-7 | 1-3-4 | 1-3-5 | 1-3-6 | 1-3-7 | 1-4-5 | 1-4-6 | 1-4-7 | 1-5-6 | 1-5-7 | 1-6-7 |
| D10S1208 | ATA5A04 | 3 | 39 | 35 | 49 | 11 | 120 | 46 | 54 | 8 | 138 | 52 | 10 | 126 | 11 | 96 | 32 |
| D10S1221 | ATA21A03 | 3 | 101 | 132 | 75 | 147 | 192 | 126 | 55 | 163 | 204 | 113 | 205 | 214 | 196 | 207 | 293 |
| D10S1222 | ATA22D02 | 3 | 87 | 108 | 118 | 116 | 66 | 85 | 73 | 91 | 63 | 80 | 102 | 62 | 102 | 40 | 67 |
| D10S1225 | ATA24F10 | 3 | 208 | 222 | 272 | 99 | 160 | 233 | 288 | 103 | 176 | 268 | 95 | 158 | 112 | 187 | 147 |
| D10S1230 | ATA29C03 | 3 | 62 | 63 | 77 | 102 | 108 | 99 | 148 | 132 | 117 | 96 | 116 | 116 | 121 | 100 | 152 |
| D10S1239 | GATA64A09 | 4 | 196 | 207 | 231 | 308 | 272 | 214 | 229 | 304 | 263 | 285 | 328 | 306 | 354 | 354 | 368 |
| D10S1248 | GGAA23C05 | 4 | 234 | 202 | 296 | 208 | 332 | 213 | 294 | 187 | 340 | 227 | 191 | 298 | 176 | 336 | 215 |
| D10S1412 | ATA31G11 | 3 | 185 | 200 | 223 | 141 | 126 | 172 | 202 | 108 | 96 | 237 | 158 | 148 | 151 | 137 | 144 |
| D10S1423 | GATA70E11 | 4 | 294 | 308 | 294 | 336 | 324 | 319 | 312 | 365 | 325 | 329 | 355 | 347 | 349 | 367 | 363 |
| D10S1425 | GATA71C09 | 4 | 337 | 339 | 157 | 301 | 278 | 356 | 132 | 328 | 273 | 137 | 309 | 287 | 109 | 130 | 240 |
| D10S1426 | GATA73E11 | 4 | 329 | 302 | 348 | 358 | 371 | 323 | 367 | 367 | 375 | 350 | 349 | 362 | 372 | 376 | 377 |
| D10S1430 | GATA84C01 | 4 | 273 | 319 | 342 | 284 | 361 | 266 | 300 | 249 | 350 | 332 | 292 | 370 | 285 | 369 | 326 |
| D10S1432 | GATA87G01 | 4 | 201 | 219 | 270 | 302 | 331 | 231 | 262 | 336 | 346 | 258 | 311 | 324 | 311 | 325 | 350 |
| D10S1435 | GATA88F09 | 4 | 317 | 320 | 310 | 354 | 315 | 311 | 306 | 361 | 324 | 303 | 359 | 318 | 320 | 286 | 337 |
| D10S189 | AFM063XF4 | 2 | 113 | 62 | 164 | 173 | 228 | 66 | 196 | 207 | 256 | 90 | 103 | 164 | 236 | 218 | 248 |
| D10S212 | AFM198ZB4 | 2 | 222 | 251 | 239 | 189 | 295 | 193 | 199 | 149 | 268 | 222 | 176 | 274 | 187 | 233 | 195 |
| D10S2327 | GGAT1A4 | 4 | 35 | 37 | 83 | 72 | 110 | 47 | 116 | 87 | 140 | 115 | 77 | 124 | 88 | 127 | 122 |
| D10S2470 | GATA115E01 | 4 | 307 | 298 | 276 | 267 | 225 | 354 | 330 | 330 | 235 | 311 | 302 | 241 | 353 | 166 | 197 |
| D10S677 | GGAA2F11 | 4 | 260 | 343 | 292 | 323 | 116 | 279 | 241 | 274 | 105 | 301 | 340 | 132 | 333 | 112 | 182 |
| D11S1304 | UT2095 | 4 | 96 | 89 | 35 | 101 | 109 | 111 | 30 | 120 | 103 | 32 | 115 | 102 | 22 | 71 | 76 |
| D11S1392 | GATA6B09 | 4 | 210 | 183 | 232 | 275 | 250 | 220 | 270 | 294 | 272 | 236 | 273 | 251 | 273 | 255 | 266 |
| D11S1981 | GATA48E02 | 4 | 303 | 311 | 337 | 297 | 255 | 268 | 308 | 276 | 259 | 283 | 266 | 220 | 282 | 208 | 236 |
| D11S1984 | GGAA17G05 | 4 | 122 | 159 | 66 | 136 | 152 | 153 | 71 | 151 | 192 | 92 | 125 | 216 | 84 | 202 | 142 |
| D11S1986 | GGAA7G08 | 4 | 128 | 143 | 121 | 170 | 90 | 114 | 100 | 152 | 87 | 127 | 169 | 91 | 146 | 104 | 107 |
| D11S1993 | ATA1B07 | 3 | 206 | 177 | 87 | 165 | 21 | 197 | 59 | 164 | 11 | 99 | 163 | 20 | 105 | 24 | 28 |
| D11S1998 | GATA23E06 | 4 | 319 | 238 | 287 | 352 | 273 | 254 | 291 | 352 | 271 | 327 | 323 | 232 | 310 | 232 | 312 |
| D11S1999 | GATA23F06 | 4 | 192 | 216 | 89 | 300 | 82 | 200 | 63 | 277 | 76 | 73 | 287 | 70 | 147 | 117 | 130 |
| D11S2000 | GATA28D01 | 4 | 23 | 38 | 106 | 63 | 41 | 23 | 88 | 48 | 36 | 93 | 68 | 43 | 98 | 39 | 42 |
| D11S2002 | GATA30G01 | 4 | 191 | 234 | 273 | 272 | 275 | 202 | 245 | 261 | 287 | 290 | 280 | 299 | 370 | 373 | 376 |
| D11S2006 | GATA46A12 | 4 | 302 | 322 | 350 | 353 | 345 | 321 | 362 | 353 | 342 | 357 | 358 | 349 | 362 | 348 | 359 |
| D11S2362 | ATA33B03 | 3 | 29 | 47 | 46 | 93 | 95 | 22 | 31 | 82 | 90 | 28 | 84 | 81 | 49 | 63 | 116 |
| D11S2363 | GATA12F04 | 4 | 49 | 72 | 104 | 127 | 103 | 38 | 68 | 94 | 93 | 78 | 112 | 107 | 122 | 118 | 150 |
| D11S2365 | GATA63F09 | 4 | 343 | 338 | 353 | 330 | 240 | 348 | 364 | 339 | 248 | 363 | 344 | 246 | 342 | 276 | 318 |
| D11S2371 | GATA90D07 | 4 | 335 | 333 | 335 | 343 | 333 | 360 | 329 | 364 | 356 | 319 | 363 | 341 | 346 | 308 | 341 |
| D11S4459 | ATA9B04 | 3 | 40 | 30 | 48 | 71 | 59 | 30 | 60 | 99 | 56 | 37 | 56 | 45 | 85 | 50 | 95 |
| D11S4463 | GATA117D01 | 4 | 357 | 345 | 343 | 372 | 359 | 335 | 373 | 376 | 371 | 297 | 353 | 331 | 368 | 361 | 367 |
| D11S4464 | GATA64D03 | 4 | 323 | 373 | 359 | 370 | 326 | 345 | 314 | 308 | 301 | 353 | 365 | 319 | 323 | 330 | 323 |
| D11S969 | AFM205VF10 | 2 | 59 | 49 | 59 | 46 | 117 | 76 | 89 | 65 | 166 | 67 | 44 | 137 | 34 | 133 | 77 |
| D12S1042 | ATA27A06 | 3 | 287 | 283 | 277 | 207 | 163 | 239 | 242 | 186 | 116 | 240 | 197 | 145 | 249 | 141 | 112 |
| D12S1045 | ATA29A06 | 3 | 103 | 69 | 51 | 80 | 47 | 87 | 32 | 62 | 31 | 21 | 49 | 21 | 74 | 28 | 62 |
| D12S1052 | GATA26D02 | 4 | 353 | 353 | 363 | 357 | 248 | 370 | 375 | 368 | 242 | 375 | 366 | 222 | 375 | 236 | 241 |
| D12S1064 | GATA63D12 | 4 | 180 | 189 | 195 | 158 | 246 | 179 | 183 | 144 | 225 | 178 | 134 | 224 | 189 | 261 | 243 |
| D12S1294 | GATA73H09 | 4 | 321 | 304 | 188 | 287 | 231 | 307 | 180 | 297 | 269 | 168 | 281 | 223 | 165 | 161 | 246 |
| D12S1300 | GATA85A04 | 4 | 267 | 265 | 242 | 92 | 330 | 322 | 302 | 138 | 367 | 298 | 133 | 365 | 201 | 371 | 263 |
| D12S1301 | GATA91H06 | 4 | 172 | 140 | 101 | 224 | 81 | 146 | 87 | 251 | 81 | 76 | 157 | 60 | 142 | 12 | 79 |
| D12S1638 | AFMB002VD5 | 2 | 190 | 209 | 244 | 164 | 170 | 217 | 252 | 157 | 180 | 266 | 173 | 163 | 200 | 177 | 172 |
| D12S2070 | ATA25F09 | 3 | 298 | 252 | 111 | 152 | 19 | 225 | 75 | 117 | 18 | 108 | 129 | 19 | 73 | 43 | 8 |
| D12S2078 | GATA32F05 | 4 | 251 | 266 | 219 | 231 | 247 | 331 | 227 | 258 | 264 | 245 | 254 | 275 | 203 | 211 | 206 |
| D12S269 | MFD259 | 2 | 104 | 112 | 119 | 86 | 44 | 92 | 101 | 63 | 30 | 88 | 78 | 32 | 57 | 35 | 44 |
| D12S297 | UT5029 | 4 | 293 | 273 | 151 | 77 | 22 | 359 | 220 | 96 | 42 | 216 | 100 | 50 | 156 | 93 | 106 |
| D12S372 | GATA4H03 | 4 | 309 | 332 | 360 | 363 | 303 | 295 | 354 | 354 | 293 | 367 | 360 | 338 | 371 | 342 | 340 |
| D12S373 | GATA6C01 | 4 | 366 | 361 | 322 | 175 | 346 | 351 | 283 | 153 | 352 | 281 | 139 | 356 | 241 | 300 | 198 |
| D12S395 | GATA4H01 | 4 | 348 | 368 | 281 | 237 | 352 | 371 | 234 | 199 | 348 | 263 | 221 | 360 | 251 | 324 | 288 |
| D13S1493 | GGAA29H03 | 4 | 203 | 231 | 217 | 161 | 236 | 208 | 204 | 143 | 211 | 251 | 166 | 266 | 134 | 264 | 167 |
| D13S1807 | GATA11C08 | 4 | 326 | 294 | 313 | 335 | 169 | 317 | 327 | 350 | 165 | 305 | 308 | 138 | 305 | 143 | 256 |
| D13S285 | AFM309VA9 | 2 | 50 | 40 | 62 | 113 | 138 | 32 | 53 | 111 | 147 | 61 | 104 | 129 | 81 | 101 | 161 |
| D13S317 | GATA7G10 | 4 | 275 | 258 | 137 | 256 | 96 | 303 | 119 | 278 | 80 | 125 | 276 | 57 | 145 | 44 | 57 |
| D13S779 | ATA26D07 | 3 | 279 | 299 | 225 | 296 | 175 | 276 | 218 | 270 | 186 | 220 | 288 | 193 | 222 | 181 | 159 |
| D13S787 | GATA23C03 | 4 | 305 | 280 | 339 | 342 | 348 | 258 | 328 | 324 | 351 | 292 | 316 | 313 | 312 | 344 | 343 |
| D13S793 | GATA43H03 | 4 | 86 | 103 | 145 | 75 | 180 | 84 | 151 | 76 | 179 | 143 | 73 | 183 | 76 | 192 | 230 |
| D13S796 | GATA51B02 | 4 | 296 | 321 | 323 | 221 | 322 | 290 | 307 | 171 | 298 | 308 | 192 | 311 | 216 | 312 | 311 |
| D13S800 | GATA64F08 | 4 | 325 | 327 | 301 | 250 | 320 | 336 | 297 | 259 | 321 | 293 | 245 | 317 | 178 | 323 | 221 |
| D13S894 | GATA86H01 | 4 | 339 | 341 | 282 | 310 | 304 | 312 | 268 | 311 | 303 | 260 | 304 | 295 | 266 | 297 | 301 |
| D13S895 | GGAA22G01 | 4 | 205 | 210 | 203 | 258 | 254 | 246 | 231 | 318 | 270 | 224 | 295 | 273 | 278 | 270 | 280 |
| D14S1007 | AFMB002ZF1 | 2 | 47 | 45 | 7 | 35 | 7 | 56 | 13 | 43 | 5 | 24 | 86 | 18 | 94 | 9 | 17 |
| D14S1280 | GATA31B09 | 4 | 167 | 153 | 213 | 228 | 195 | 158 | 233 | 243 | 196 | 194 | 212 | 189 | 255 | 162 | 177 |
| D14S1426 | GATA136B01 | 4 | 233 | 190 | 176 | 236 | 33 | 187 | 174 | 239 | 29 | 149 | 183 | 31 | 149 | 19 | 14 |
| D14S1434 | GATA168F06 | 4 | 352 | 328 | 268 | 299 | 302 | 329 | 266 | 283 | 305 | 257 | 301 | 301 | 260 | 250 | 290 |
| D14S587 | GGAA10C09 | 4 | 272 | 195 | 230 | 215 | 210 | 218 | 273 | 216 | 234 | 203 | 208 | 168 | 168 | 216 | 168 |
| D14S588 | GGAA4A12 | 4 | 12 | 13 | 39 | 51 | 76 | 24 | 84 | 79 | 107 | 55 | 65 | 85 | 68 | 105 | 118 |
| D14S592 | ATA19H08 | 3 | 175 | 138 | 193 | 96 | 140 | 166 | 195 | 145 | 148 | 169 | 122 | 142 | 116 | 142 | 143 |
| D14S599 | ATA29G03 | 3 | 60 | 71 | 88 | 82 | 46 | 72 | 76 | 75 | 38 | 86 | 67 | 33 | 63 | 108 | 26 |
| D14S606 | GATA30A03 | 4 | 209 | 215 | 248 | 220 | 265 | 185 | 216 | 185 | 247 | 214 | 186 | 249 | 272 | 303 | 291 |
| D14S608 | GATA43H01 | 4 | 292 | 260 | 286 | 279 | 158 | 262 | 244 | 272 | 191 | 230 | 250 | 109 | 221 | 111 | 154 |
| D14S617 | GGAA21G11 | 4 | 148 | 122 | 159 | 10 | 165 | 109 | 158 | 4 | 164 | 118 | 7 | 140 | 4 | 126 | 16 |
| D14S742 | GATA74E02 | 4 | 283 | 269 | 321 | 350 | 351 | 272 | 296 | 335 | 338 | 274 | 317 | 315 | 330 | 347 | 361 |
| D15S128 | AFM273YF9 | 2 | 51 | 46 | 33 | 108 | 49 | 78 | 44 | 126 | 66 | 38 | 124 | 64 | 87 | 47 | 132 |
| D15S1507 | GATA151F03 | 4 | 154 | 130 | 114 | 115 | 233 | 124 | 117 | 113 | 224 | 83 | 98 | 206 | 75 | 152 | 156 |
| D15S1515 | GATA197B10 | 4 | 88 | 93 | 129 | 157 | 149 | 93 | 153 | 172 | 163 | 146 | 171 | 165 | 188 | 229 | 199 |
| D15S165 | AFM248VC5 | 2 | 46 | 43 | 10 | 28 | 42 | 49 | 10 | 26 | 39 | 12 | 39 | 61 | 14 | 15 | 40 |
| D15S642 | GATA27A03 | 4 | 58 | 88 | 30 | 139 | 115 | 96 | 29 | 177 | 123 | 74 | 229 | 184 | 217 | 222 | 302 |
| D15S643 | GATA50G06 | 4 | 136 | 139 | 228 | 185 | 193 | 125 | 222 | 193 | 213 | 208 | 152 | 191 | 287 | 281 | 325 |
| D15S652 | ATA24A08 | 3 | 152 | 175 | 183 | 201 | 56 | 150 | 165 | 135 | 57 | 177 | 190 | 76 | 205 | 77 | 70 |
| D15S655 | ATA28G05 | 3 | 142 | 150 | 116 | 61 | 201 | 137 | 111 | 59 | 193 | 128 | 72 | 199 | 93 | 131 | 90 |
| D15S659 | GATA63A03 | 4 | 268 | 250 | 293 | 280 | 174 | 282 | 292 | 299 | 181 | 280 | 258 | 194 | 274 | 178 | 137 |
| D15S816 | GATA73F01 | 4 | 245 | 268 | 252 | 283 | 317 | 243 | 254 | 263 | 314 | 241 | 274 | 312 | 232 | 258 | 276 |
| D15S818 | GATA85D02 | 4 | 312 | 348 | 367 | 344 | 347 | 305 | 350 | 332 | 329 | 366 | 335 | 340 | 356 | 340 | 333 |
| D15S822 | GATA88H02 | 4 | 174 | 225 | 163 | 249 | 73 | 199 | 126 | 234 | 49 | 226 | 312 | 118 | 292 | 189 | 201 |
| D16S2616 | ATA41E04 | 3 | 83 | 91 | 109 | 74 | 71 | 75 | 94 | 66 | 54 | 95 | 71 | 63 | 78 | 75 | 85 |
| D16S2621 | GATA71F09 | 4 | 109 | 101 | 15 | 216 | 65 | 64 | 11 | 156 | 43 | 11 | 162 | 65 | 32 | 21 | 109 |
| D16S2624 | GATA81D12 | 4 | 291 | 315 | 191 | 319 | 253 | 294 | 166 | 313 | 227 | 188 | 333 | 247 | 277 | 265 | 270 |
| D16S3253 | GATA22F09 | 4 | 82 | 109 | 171 | 167 | 154 | 106 | 182 | 175 | 149 | 183 | 159 | 150 | 191 | 203 | 179 |
| D16S3396 | ATA55A11 | 3 | 252 | 203 | 265 | 311 | 243 | 191 | 258 | 290 | 218 | 204 | 265 | 188 | 291 | 210 | 258 |
| D16S3401 | 16PTEL06 | 2 | 20 | 17 | 42 | 5 | 34 | 39 | 86 | 31 | 60 | 59 | 23 | 41 | 40 | 88 | 51 |
| D16S403 | AFM049XD2 | 2 | 198 | 186 | 216 | 90 | 191 | 184 | 190 | 74 | 118 | 212 | 40 | 131 | 31 | 153 | 97 |
| D16S422 | AFM249XC5 | 2 | 93 | 117 | 17 | 129 | 36 | 138 | 22 | 134 | 40 | 43 | 179 | 78 | 60 | 90 | 100 |
| D16S516 | AFM350VD1 | 2 | 166 | 179 | 123 | 169 | 185 | 235 | 127 | 238 | 201 | 167 | 249 | 218 | 180 | 204 | 227 |
| D16S539 | GATA11C06 | 4 | 289 | 282 | 307 | 289 | 289 | 355 | 351 | 295 | 331 | 354 | 290 | 348 | 281 | 370 | 281 |
| D16S748 | ATA3A07 | 3 | 140 | 85 | 81 | 23 | 24 | 108 | 109 | 18 | 26 | 135 | 45 | 39 | 52 | 34 | 38 |
| D16S753 | GGAA3G05 | 4 | 259 | 212 | 156 | 234 | 284 | 181 | 137 | 189 | 274 | 139 | 199 | 236 | 161 | 183 | 200 |
| D16S764 | GATA42E11 | 4 | 340 | 347 | 354 | 355 | 267 | 314 | 352 | 356 | 282 | 333 | 346 | 243 | 376 | 289 | 328 |
| D16S769 | GATA71H05 | 4 | 341 | 357 | 336 | 360 | 363 | 341 | 332 | 357 | 354 | 324 | 362 | 359 | 357 | 328 | 352 |
| D17S1290 | GATA49C09 | 4 | 162 | 156 | 210 | 134 | 179 | 155 | 213 | 137 | 170 | 195 | 138 | 157 | 158 | 174 | 136 |
| D17S1294 | GGAA9D03 | 4 | 173 | 201 | 250 | 266 | 290 | 174 | 259 | 266 | 288 | 239 | 256 | 280 | 289 | 315 | 324 |
| D17S1298 | GAAT2C03 | 4 | 98 | 80 | 61 | 153 | 194 | 102 | 77 | 179 | 209 | 124 | 203 | 205 | 150 | 139 | 226 |
| D17S1299 | GATA25A04 | 4 | 320 | 331 | 320 | 334 | 368 | 375 | 361 | 351 | 377 | 362 | 354 | 376 | 319 | 364 | 371 |
| D17S1301 | GATA28D11 | 4 | 214 | 271 | 257 | 305 | 336 | 259 | 251 | 291 | 339 | 289 | 356 | 364 | 313 | 317 | 356 |
| D17S1308 | GTAT1A05 | 4 | 165 | 161 | 206 | 109 | 268 | 192 | 217 | 122 | 307 | 191 | 113 | 291 | 240 | 337 | 314 |
| D17S2180 | ATC6A06 | 3 | 232 | 275 | 236 | 294 | 189 | 252 | 240 | 305 | 172 | 304 | 345 | 201 | 339 | 194 | 234 |
| D17S2193 | ATA43A10Z | 3 | 13 | 23 | 60 | 37 | 25 | 12 | 47 | 37 | 37 | 49 | 22 | 25 | 35 | 36 | 27 |
| D17S2195 | ATA58A02 | 3 | 22 | 29 | 64 | 34 | 74 | 28 | 72 | 40 | 89 | 77 | 43 | 88 | 61 | 92 | 52 |
| D17S2196 | GATA185H04 | 4 | 318 | 278 | 161 | 270 | 112 | 292 | 169 | 296 | 115 | 136 | 282 | 114 | 133 | 84 | 149 |
| D17S784 | AFM044XG3 | 2 | 137 | 73 | 108 | 205 | 87 | 71 | 91 | 212 | 94 | 50 | 121 | 69 | 114 | 18 | 91 |
| D17S974 | GATA8C04 | 4 | 225 | 217 | 298 | 269 | 242 | 205 | 290 | 248 | 246 | 277 | 216 | 234 | 254 | 280 | 196 |
| D18S1357 | ATA7D07 | 3 | 241 | 226 | 299 | 196 | 183 | 263 | 326 | 241 | 168 | 296 | 219 | 159 | 269 | 160 | 128 |
| D18S1364 | GATA7E12 | 4 | 30 | 27 | 99 | 83 | 124 | 20 | 105 | 90 | 132 | 71 | 74 | 106 | 120 | 190 | 140 |
| D18S1370 | ATA45G06 | 3 | 146 | 119 | 124 | 94 | 206 | 157 | 149 | 89 | 265 | 112 | 76 | 208 | 111 | 172 | 166 |
| D18S1371 | GATA177C03 | 4 | 359 | 303 | 355 | 315 | 213 | 309 | 370 | 325 | 215 | 328 | 278 | 231 | 318 | 228 | 202 |
| D18S1376 | GATA185C06 | 4 | 308 | 276 | 308 | 307 | 337 | 299 | 303 | 317 | 347 | 282 | 314 | 345 | 308 | 341 | 358 |
| D18S1390 | 18QTEL11 | 2 | 92 | 75 | 74 | 144 | 98 | 107 | 74 | 142 | 101 | 82 | 132 | 100 | 130 | 113 | 178 |
| D18S535 | GATA13 | 4 | 220 | 290 | 204 | 62 | 229 | 206 | 152 | 33 | 207 | 196 | 50 | 235 | 79 | 151 | 39 |
| D18S542 | GATA11A06 | 4 | 64 | 77 | 125 | 19 | 145 | 58 | 107 | 11 | 141 | 100 | 13 | 133 | 65 | 186 | 104 |
| D18S843 | ACT1A01 | 3 | 372 | 369 | 297 | 328 | 374 | 369 | 267 | 319 | 370 | 262 | 303 | 366 | 242 | 320 | 348 |
| D18S851 | GATA6D09 | 4 | 189 | 170 | 174 | 268 | 184 | 164 | 179 | 288 | 195 | 170 | 248 | 175 | 239 | 191 | 307 |
| D18S858 | ATA23G05 | 3 | 253 | 277 | 246 | 128 | 257 | 245 | 208 | 95 | 243 | 259 | 119 | 239 | 139 | 231 | 181 |
| D18S877 | GATA64H04 | 4 | 276 | 281 | 261 | 247 | 353 | 244 | 226 | 232 | 336 | 215 | 227 | 346 | 225 | 304 | 296 |
| D19S1034 | GATA21G05 | 4 | 207 | 198 | 202 | 244 | 182 | 222 | 193 | 244 | 184 | 185 | 241 | 160 | 215 | 129 | 207 |
| D19S245 | MFD235 | 4 | 377 | 370 | 370 | 362 | 377 | 373 | 368 | 360 | 376 | 361 | 352 | 375 | 345 | 366 | 360 |
| D19S246 | MFD232 | 4 | 216 | 289 | 243 | 232 | 291 | 288 | 274 | 211 | 313 | 265 | 220 | 320 | 218 | 269 | 214 |
| D19S254 | MFD238 | 4 | 236 | 236 | 285 | 117 | 196 | 334 | 347 | 147 | 279 | 342 | 145 | 260 | 184 | 256 | 119 |
| D19S433 | GGAA2A03 | 4 | 45 | 53 | 98 | 104 | 114 | 54 | 83 | 73 | 111 | 105 | 106 | 117 | 125 | 140 | 145 |
| D19S559 | UT7544 | 4 | 257 | 204 | 251 | 291 | 221 | 240 | 285 | 302 | 251 | 244 | 267 | 207 | 275 | 217 | 237 |
| D19S586 | GATA23B01 | 4 | 270 | 257 | 309 | 276 | 307 | 257 | 309 | 279 | 304 | 288 | 264 | 284 | 270 | 301 | 253 |
| D19S589 | GATA29B01 | 4 | 311 | 351 | 364 | 349 | 375 | 320 | 340 | 348 | 359 | 373 | 357 | 374 | 350 | 375 | 370 |
| D19S591 | GATA44F10 | 4 | 224 | 188 | 196 | 251 | 187 | 198 | 221 | 289 | 206 | 173 | 231 | 166 | 268 | 169 | 260 |
| D19S714 | GATA66B04 | 4 | 277 | 301 | 326 | 176 | 271 | 274 | 321 | 166 | 291 | 317 | 153 | 286 | 204 | 302 | 211 |
| D1S1589 | ATA4E02 | 3 | 16 | 10 | 20 | 13 | 6 | 15 | 34 | 16 | 6 | 18 | 12 | 5 | 9 | 1 | 3 |
| D1S1594 | GATA22D12 | 4 | 342 | 292 | 316 | 345 | 260 | 278 | 313 | 338 | 245 | 312 | 294 | 285 | 293 | 291 | 255 |
| D1S1596 | GATA26G09 | 4 | 344 | 337 | 331 | 368 | 261 | 308 | 315 | 355 | 260 | 344 | 367 | 238 | 364 | 246 | 269 |
| D1S1597 | GATA27E01 | 4 | 116 | 121 | 178 | 195 | 156 | 113 | 164 | 178 | 133 | 157 | 167 | 141 | 163 | 156 | 170 |
| D1S1609 | GATA50F11 | 4 | 171 | 199 | 229 | 239 | 125 | 189 | 223 | 233 | 100 | 242 | 237 | 95 | 246 | 79 | 110 |
| D1S1612 | GGAA3A07 | 4 | 147 | 158 | 95 | 145 | 107 | 161 | 136 | 203 | 130 | 119 | 193 | 105 | 295 | 225 | 233 |
| D1S1627 | ATA25E07 | 3 | 265 | 229 | 146 | 245 | 244 | 204 | 108 | 220 | 217 | 130 | 232 | 219 | 164 | 168 | 220 |
| D1S1653 | GATA43A04 | 4 | 271 | 330 | 328 | 338 | 334 | 328 | 333 | 349 | 327 | 355 | 364 | 350 | 369 | 363 | 375 |
| D1S1660 | GATA48B01 | 4 | 281 | 253 | 259 | 218 | 340 | 221 | 225 | 158 | 337 | 235 | 175 | 307 | 212 | 313 | 238 |
| D1S1665 | GATA61A06 | 4 | 164 | 154 | 197 | 253 | 263 | 162 | 185 | 268 | 281 | 166 | 240 | 248 | 223 | 235 | 285 |
| D1S1677 | GGAA22G10 | 4 | 229 | 293 | 271 | 318 | 274 | 236 | 201 | 282 | 226 | 286 | 341 | 277 | 303 | 277 | 304 |
| D1S1679 | GGAA5F09 | 4 | 159 | 167 | 208 | 156 | 266 | 190 | 232 | 188 | 289 | 213 | 177 | 283 | 210 | 282 | 259 |
| D1S1728 | GATA109 | 4 | 79 | 128 | 138 | 130 | 159 | 116 | 147 | 123 | 157 | 233 | 238 | 242 | 288 | 252 | 272 |
| D1S2134 | GATA72H07 | 4 | 105 | 135 | 126 | 180 | 99 | 127 | 112 | 161 | 85 | 104 | 185 | 92 | 290 | 201 | 257 |
| D1S235 | AFM203YG9 | 2 | 4 | 1 | 1 | 3 | 11 | 1 | 1 | 3 | 16 | 1 | 1 | 6 | 1 | 5 | 12 |
| D1S2682 | AFMA272XC9 | 2 | 132 | 191 | 211 | 154 | 168 | 169 | 188 | 118 | 139 | 267 | 160 | 190 | 166 | 195 | 235 |
| D1S3462 | ATA29C07 | 3 | 297 | 233 | 140 | 197 | 298 | 285 | 181 | 218 | 341 | 207 | 263 | 334 | 233 | 238 | 262 |
| D1S3669 | GATA29A05 | 4 | 150 | 126 | 148 | 88 | 122 | 131 | 163 | 93 | 124 | 145 | 81 | 90 | 64 | 110 | 50 |
| D1S3720 | ATA47D07 | 3 | 31 | 58 | 127 | 58 | 119 | 63 | 122 | 38 | 127 | 219 | 90 | 149 | 127 | 171 | 56 |
| D1S3721 | GATA129H04 | 4 | 145 | 147 | 177 | 187 | 106 | 135 | 162 | 167 | 91 | 165 | 135 | 82 | 192 | 144 | 157 |
| D1S468 | AFM280WE5 | 2 | 212 | 142 | 179 | 138 | 15 | 196 | 239 | 208 | 20 | 182 | 170 | 17 | 124 | 11 | 25 |
| D1S518 | GATA7C01 | 4 | 54 | 31 | 29 | 14 | 40 | 42 | 42 | 13 | 50 | 39 | 15 | 53 | 27 | 57 | 45 |
| D1S534 | GATA12A07 | 4 | 187 | 176 | 52 | 112 | 64 | 216 | 43 | 127 | 53 | 41 | 105 | 46 | 47 | 32 | 73 |
| D1S549 | GATA4H09 | 4 | 202 | 220 | 279 | 120 | 305 | 255 | 331 | 140 | 365 | 300 | 126 | 337 | 169 | 358 | 169 |
| D1S551 | GATA6A05 | 4 | 362 | 352 | 333 | 217 | 321 | 340 | 325 | 206 | 308 | 352 | 188 | 323 | 207 | 298 | 153 |
| D20S103 | AFM077XD3 | 2 | 95 | 76 | 28 | 119 | 93 | 53 | 21 | 81 | 74 | 40 | 111 | 110 | 90 | 74 | 158 |
| D20S1143 | GATA129B03 | 4 | 249 | 242 | 280 | 278 | 308 | 261 | 280 | 287 | 311 | 256 | 285 | 302 | 280 | 293 | 299 |
| D20S159 | UT1307 | 4 | 117 | 162 | 199 | 230 | 198 | 140 | 206 | 209 | 208 | 179 | 224 | 185 | 198 | 230 | 180 |
| D20S164 | UT1772 | 4 | 129 | 149 | 55 | 126 | 18 | 203 | 80 | 159 | 32 | 121 | 213 | 36 | 259 | 31 | 83 |
| D20S201 | GATA8B01 | 4 | 123 | 169 | 200 | 155 | 150 | 156 | 212 | 146 | 169 | 229 | 144 | 195 | 128 | 292 | 96 |
| D20S451 | UT254 | 4 | 151 | 141 | 103 | 211 | 188 | 145 | 92 | 213 | 187 | 122 | 230 | 202 | 182 | 165 | 251 |
| D20S477 | GATA29F06 | 4 | 238 | 245 | 147 | 159 | 202 | 277 | 200 | 217 | 222 | 192 | 198 | 230 | 247 | 266 | 267 |
| D20S478 | GATA42A03 | 4 | 7 | 15 | 13 | 66 | 43 | 25 | 16 | 98 | 68 | 16 | 120 | 80 | 42 | 22 | 108 |
| D20S480 | GATA45B10 | 4 | 373 | 374 | 351 | 377 | 328 | 376 | 345 | 377 | 334 | 360 | 375 | 343 | 338 | 329 | 334 |
| D20S481 | GATA47F05 | 4 | 218 | 249 | 173 | 160 | 166 | 186 | 123 | 155 | 155 | 161 | 128 | 155 | 71 | 99 | 94 |
| D20S482 | GATA51D03 | 4 | 354 | 346 | 356 | 341 | 373 | 333 | 357 | 347 | 373 | 338 | 338 | 367 | 351 | 362 | 362 |
| D20S851 | AFMA218YB5 | 2 | 90 | 86 | 122 | 48 | 61 | 83 | 113 | 46 | 55 | 111 | 28 | 49 | 104 | 98 | 19 |
| D21S1411 | UT1355 | 4 | 256 | 241 | 224 | 227 | 130 | 265 | 235 | 262 | 110 | 172 | 210 | 115 | 257 | 58 | 68 |
| D21S1432 | GATA11C12 | 4 | 186 | 151 | 184 | 150 | 178 | 177 | 197 | 190 | 199 | 158 | 151 | 180 | 148 | 138 | 205 |
| D21S1437 | GGAA3C07 | 4 | 65 | 95 | 90 | 186 | 151 | 97 | 96 | 200 | 160 | 129 | 252 | 161 | 190 | 95 | 186 |
| D21S1440 | ATA27F01 | 3 | 14 | 6 | 25 | 57 | 51 | 8 | 35 | 56 | 59 | 15 | 35 | 42 | 39 | 51 | 69 |
| D21S1446 | GATA70B08 | 4 | 77 | 98 | 110 | 70 | 172 | 154 | 135 | 61 | 178 | 138 | 64 | 197 | 69 | 155 | 102 |
| D21S2052 | GATA129D11 | 4 | 246 | 227 | 245 | 168 | 234 | 267 | 301 | 235 | 275 | 278 | 189 | 263 | 230 | 290 | 218 |
| D21S2055 | GATA188F04 | 4 | 17 | 20 | 21 | 7 | 4 | 48 | 64 | 32 | 19 | 68 | 26 | 16 | 44 | 29 | 9 |
| D22S1045 | ATA37D06 | 3 | 74 | 78 | 155 | 143 | 118 | 80 | 167 | 150 | 104 | 174 | 137 | 87 | 170 | 94 | 135 |
| D22S1169 | AFMB337ZH9 | 2 | 21 | 24 | 23 | 1 | 38 | 27 | 25 | 2 | 48 | 23 | 2 | 52 | 6 | 46 | 35 |
| D22S345 | MFD313 | 2 | 168 | 197 | 192 | 219 | 91 | 195 | 173 | 197 | 75 | 211 | 223 | 98 | 179 | 136 | 105 |
| D22S532 | UT7136 | 4 | 349 | 362 | 371 | 340 | 357 | 357 | 353 | 306 | 364 | 371 | 337 | 358 | 361 | 331 | 308 |
| D22S683 | GATA11B12 | 4 | 8 | 11 | 16 | 17 | 5 | 6 | 17 | 12 | 3 | 9 | 5 | 4 | 7 | 2 | 2 |
| D22S686 | GGAA10F06 | 4 | 143 | 166 | 141 | 238 | 279 | 151 | 118 | 202 | 266 | 133 | 222 | 276 | 193 | 223 | 275 |
| D22S689 | GATA21F03 | 4 | 235 | 263 | 255 | 252 | 286 | 249 | 250 | 246 | 249 | 253 | 255 | 262 | 256 | 249 | 242 |
| D2S1328 | GATA27A12 | 4 | 327 | 295 | 78 | 32 | 281 | 352 | 128 | 52 | 318 | 153 | 63 | 335 | 91 | 253 | 151 |
| D2S1334 | GATA4D07 | 4 | 133 | 171 | 212 | 242 | 204 | 148 | 205 | 219 | 197 | 181 | 235 | 174 | 209 | 214 | 184 |
| D2S1352 | ATA27D04 | 3 | 118 | 114 | 160 | 148 | 211 | 123 | 187 | 162 | 221 | 148 | 172 | 204 | 137 | 196 | 249 |
| D2S1353 | ATA27H09 | 3 | 156 | 178 | 167 | 171 | 67 | 173 | 170 | 173 | 73 | 164 | 154 | 67 | 175 | 65 | 81 |
| D2S1356 | ATA4F03 | 3 | 34 | 34 | 31 | 15 | 3 | 29 | 39 | 19 | 7 | 19 | 11 | 3 | 20 | 6 | 1 |
| D2S1360 | GATA11H10 | 4 | 221 | 184 | 58 | 240 | 153 | 271 | 62 | 286 | 194 | 47 | 251 | 173 | 101 | 106 | 231 |
| D2S1363 | GATA23D03 | 4 | 194 | 193 | 194 | 69 | 214 | 168 | 191 | 67 | 189 | 197 | 79 | 272 | 138 | 219 | 141 |
| D2S1384 | GATA52A04 | 4 | 184 | 244 | 170 | 199 | 252 | 152 | 103 | 176 | 151 | 171 | 178 | 256 | 136 | 241 | 160 |
| D2S1391 | GATA65C03 | 4 | 250 | 264 | 291 | 314 | 238 | 273 | 293 | 321 | 223 | 307 | 332 | 226 | 365 | 305 | 322 |
| D2S1394 | GATA69E12 | 4 | 356 | 365 | 368 | 369 | 365 | 368 | 366 | 366 | 358 | 374 | 374 | 368 | 366 | 360 | 369 |
| D2S1399 | GGAA20G04 | 4 | 111 | 172 | 235 | 137 | 85 | 178 | 271 | 121 | 98 | 295 | 161 | 122 | 185 | 157 | 113 |
| D2S1400 | GGAA20G10 | 4 | 3 | 4 | 2 | 4 | 2 | 3 | 2 | 6 | 2 | 2 | 3 | 2 | 2 | 4 | 11 |
| D2S1776 | GATA71D01 | 4 | 350 | 366 | 327 | 366 | 239 | 364 | 337 | 342 | 255 | 339 | 368 | 270 | 335 | 295 | 284 |
| D2S1780 | GATA72G11 | 4 | 81 | 68 | 132 | 181 | 176 | 60 | 125 | 181 | 158 | 89 | 150 | 146 | 154 | 158 | 193 |
| D2S1788 | GATA86E02 | 4 | 127 | 84 | 201 | 192 | 132 | 110 | 215 | 192 | 113 | 132 | 123 | 72 | 231 | 145 | 164 |
| D2S1790 | GATA88G05 | 4 | 97 | 120 | 113 | 140 | 135 | 105 | 110 | 154 | 121 | 98 | 142 | 128 | 129 | 116 | 139 |
| D2S2944 | GATA30E06 | 4 | 242 | 248 | 303 | 293 | 319 | 298 | 339 | 322 | 357 | 336 | 334 | 353 | 337 | 357 | 365 |
| D2S2952 | GATA116B01 | 4 | 76 | 70 | 84 | 79 | 54 | 104 | 102 | 80 | 44 | 81 | 66 | 34 | 53 | 30 | 59 |
| D2S2968 | GATA178G09 | 4 | 119 | 56 | 107 | 100 | 102 | 103 | 160 | 130 | 129 | 87 | 101 | 101 | 113 | 107 | 133 |
| D2S2972 | GATA176C01 | 4 | 75 | 54 | 144 | 190 | 142 | 73 | 168 | 223 | 143 | 141 | 207 | 120 | 248 | 150 | 176 |
| D2S2986 | 2QTEL47 | 2 | 11 | 9 | 11 | 8 | 9 | 10 | 14 | 5 | 8 | 14 | 6 | 8 | 46 | 41 | 82 |
| D2S405 | GATA8F07 | 4 | 160 | 163 | 154 | 105 | 245 | 147 | 161 | 92 | 230 | 134 | 96 | 228 | 103 | 179 | 171 |
| D2S410 | GATA4E11 | 4 | 163 | 145 | 142 | 184 | 77 | 134 | 138 | 160 | 78 | 151 | 174 | 86 | 171 | 123 | 101 |
| D2S427 | GATA12H10 | 4 | 5 | 5 | 12 | 16 | 23 | 5 | 9 | 9 | 45 | 8 | 8 | 23 | 10 | 16 | 23 |
| D2S434 | GATA4G12 | 4 | 322 | 309 | 365 | 339 | 287 | 251 | 338 | 309 | 233 | 321 | 310 | 282 | 317 | 284 | 297 |
| D2S441 | GATA8F03 | 4 | 134 | 99 | 134 | 2 | 20 | 94 | 144 | 1 | 15 | 154 | 4 | 22 | 8 | 27 | 24 |
| D3S1262 | AFM059XA9 | 2 | 37 | 41 | 45 | 22 | 60 | 41 | 36 | 23 | 61 | 34 | 17 | 48 | 38 | 64 | 37 |
| D3S1311 | AFM254VE1 | 2 | 42 | 42 | 6 | 31 | 75 | 33 | 5 | 29 | 62 | 6 | 42 | 89 | 17 | 17 | 58 |
| D3S1560 | AFM217XD6 | 2 | 25 | 21 | 34 | 33 | 97 | 31 | 56 | 77 | 137 | 58 | 46 | 139 | 58 | 121 | 92 |
| D3S1744 | GATA3C02 | 4 | 364 | 355 | 376 | 320 | 360 | 353 | 376 | 326 | 355 | 369 | 313 | 344 | 316 | 359 | 344 |
| D3S1746 | GATA8F01 | 4 | 157 | 155 | 136 | 52 | 181 | 139 | 121 | 51 | 162 | 144 | 54 | 187 | 89 | 185 | 114 |
| D3S1763 | GATA3H01 | 4 | 282 | 305 | 226 | 202 | 133 | 286 | 230 | 230 | 153 | 223 | 214 | 144 | 194 | 146 | 192 |
| D3S1764 | GATA4A10 | 4 | 69 | 92 | 143 | 166 | 131 | 69 | 129 | 133 | 106 | 162 | 180 | 153 | 206 | 197 | 213 |
| D3S1766 | GATA6F06 | 4 | 301 | 336 | 329 | 337 | 358 | 358 | 358 | 344 | 369 | 364 | 350 | 373 | 329 | 368 | 354 |
| D3S1768 | GATA8B05 | 4 | 61 | 87 | 79 | 103 | 101 | 55 | 45 | 78 | 67 | 66 | 99 | 99 | 186 | 154 | 163 |
| D3S2387 | GATA22G12 | 4 | 24 | 16 | 40 | 42 | 57 | 16 | 46 | 47 | 72 | 46 | 36 | 74 | 41 | 82 | 46 |
| D3S2398 | GATA6G12 | 4 | 199 | 284 | 260 | 298 | 372 | 171 | 175 | 180 | 286 | 247 | 253 | 325 | 245 | 288 | 331 |
| D3S2403 | GGAA4B09 | 4 | 38 | 51 | 72 | 68 | 79 | 89 | 93 | 85 | 95 | 102 | 91 | 79 | 106 | 109 | 111 |
| D3S2409 | ATA10H11 | 3 | 106 | 65 | 44 | 20 | 12 | 70 | 28 | 10 | 9 | 36 | 20 | 10 | 55 | 42 | 41 |
| D3S2418 | ATA22E01 | 3 | 28 | 19 | 56 | 81 | 53 | 19 | 67 | 102 | 69 | 42 | 62 | 44 | 82 | 59 | 60 |
| D3S2427 | GATA22F11 | 4 | 18 | 18 | 24 | 30 | 16 | 21 | 38 | 34 | 17 | 25 | 33 | 12 | 28 | 23 | 10 |
| D3S2432 | GATA27C08 | 4 | 300 | 312 | 324 | 333 | 314 | 304 | 342 | 345 | 335 | 330 | 342 | 300 | 340 | 310 | 355 |
| D3S2460 | GATA68F07 | 4 | 115 | 160 | 187 | 132 | 203 | 143 | 171 | 116 | 182 | 209 | 195 | 233 | 227 | 242 | 271 |
| D3S3038 | GATA73D01 | 4 | 255 | 254 | 218 | 246 | 264 | 275 | 219 | 265 | 262 | 190 | 236 | 237 | 244 | 263 | 274 |
| D3S3039 | GATA7F05 | 4 | 360 | 359 | 373 | 373 | 369 | 350 | 372 | 372 | 372 | 356 | 372 | 355 | 373 | 377 | 372 |
| D3S3045 | GATA84B12 | 4 | 141 | 79 | 139 | 43 | 10 | 68 | 155 | 39 | 10 | 103 | 57 | 14 | 70 | 20 | 30 |
| D3S3630 | AFMB296ZF5 | 2 | 10 | 14 | 27 | 25 | 70 | 14 | 24 | 30 | 71 | 33 | 51 | 104 | 19 | 70 | 80 |
| D3S3644 | AFMB318YF1 | 2 | 244 | 181 | 241 | 200 | 306 | 167 | 253 | 214 | 316 | 205 | 130 | 294 | 224 | 326 | 287 |
| D3S4523 | ATA34G06 | 3 | 100 | 127 | 91 | 183 | 48 | 144 | 115 | 215 | 58 | 159 | 257 | 84 | 237 | 68 | 189 |
| D3S4529 | GATA128C02 | 4 | 182 | 164 | 238 | 274 | 197 | 163 | 265 | 280 | 214 | 232 | 262 | 210 | 296 | 234 | 252 |
| D3S4545 | GATA164B08 | 4 | 135 | 116 | 47 | 191 | 63 | 118 | 37 | 198 | 64 | 31 | 164 | 58 | 66 | 49 | 84 |
| D4S1625 | GATA107 | 4 | 338 | 324 | 317 | 371 | 311 | 316 | 317 | 374 | 312 | 309 | 369 | 305 | 363 | 327 | 347 |
| D4S1627 | GATA7D01 | 4 | 204 | 270 | 275 | 241 | 339 | 232 | 246 | 221 | 328 | 323 | 286 | 372 | 343 | 374 | 353 |
| D4S1629 | GATA8A05 | 4 | 44 | 50 | 65 | 177 | 58 | 45 | 57 | 191 | 65 | 45 | 182 | 54 | 153 | 53 | 126 |
| D4S1644 | GATA11E09 | 4 | 274 | 246 | 215 | 264 | 256 | 287 | 237 | 293 | 277 | 228 | 293 | 269 | 263 | 245 | 305 |
| D4S1647 | GATA2F11 | 4 | 331 | 344 | 240 | 285 | 329 | 289 | 224 | 255 | 276 | 231 | 271 | 304 | 229 | 206 | 295 |
| D4S1652 | GATA5B02 | 4 | 363 | 350 | 311 | 321 | 370 | 361 | 281 | 315 | 345 | 294 | 315 | 352 | 322 | 356 | 351 |
| D4S2361 | ATA2A03 | 3 | 138 | 100 | 181 | 118 | 69 | 88 | 178 | 119 | 82 | 126 | 87 | 38 | 115 | 78 | 75 |
| D4S2366 | GATA22G05 | 4 | 63 | 106 | 93 | 64 | 134 | 79 | 85 | 57 | 119 | 120 | 85 | 147 | 117 | 128 | 127 |
| D4S2367 | GATA24H01 | 4 | 288 | 261 | 172 | 98 | 344 | 330 | 248 | 168 | 374 | 246 | 165 | 371 | 214 | 333 | 278 |
| D4S2368 | GATA27G03 | 4 | 374 | 372 | 377 | 346 | 313 | 367 | 371 | 346 | 319 | 365 | 318 | 308 | 358 | 267 | 264 |
| D4S2394 | ATA26B08 | 3 | 153 | 129 | 117 | 55 | 148 | 129 | 106 | 50 | 144 | 123 | 55 | 135 | 72 | 149 | 86 |
| D4S2397 | ATA27C07 | 3 | 99 | 94 | 26 | 73 | 100 | 90 | 23 | 84 | 99 | 17 | 59 | 96 | 25 | 73 | 88 |
| D4S2417 | GATA42H02 | 4 | 376 | 360 | 319 | 257 | 164 | 344 | 311 | 250 | 159 | 318 | 244 | 170 | 234 | 148 | 210 |
| D4S2431 | GGAA19H07 | 4 | 226 | 148 | 133 | 131 | 241 | 141 | 124 | 114 | 237 | 110 | 117 | 215 | 141 | 175 | 185 |
| D4S2623 | GATA62A12 | 4 | 55 | 82 | 115 | 40 | 162 | 86 | 134 | 44 | 175 | 156 | 52 | 186 | 119 | 182 | 74 |
| D4S2632 | GATA72G09 | 4 | 94 | 110 | 71 | 97 | 157 | 119 | 66 | 105 | 134 | 94 | 141 | 196 | 181 | 176 | 228 |
| D4S3243 | GATA10G07 | 4 | 36 | 32 | 68 | 65 | 55 | 43 | 95 | 69 | 47 | 85 | 88 | 66 | 97 | 72 | 66 |
| D4S3248 | GATA28F03 | 4 | 330 | 356 | 345 | 356 | 237 | 338 | 335 | 341 | 239 | 359 | 361 | 240 | 348 | 287 | 294 |
| D4S3360 | 4PTEL04 | 2 | 149 | 173 | 130 | 254 | 94 | 182 | 131 | 245 | 97 | 131 | 259 | 93 | 157 | 80 | 131 |
| D4S403 | AFM157XG3 | 2 | 66 | 39 | 9 | 26 | 31 | 37 | 7 | 28 | 33 | 10 | 19 | 27 | 13 | 13 | 21 |
| D4S408 | AFM165XC11 | 2 | 85 | 83 | 37 | 29 | 139 | 128 | 49 | 35 | 173 | 65 | 48 | 203 | 50 | 163 | 120 |
| D5S1456 | GATA11A11 | 4 | 254 | 255 | 318 | 226 | 327 | 242 | 316 | 225 | 309 | 315 | 233 | 322 | 226 | 349 | 239 |
| D5S1457 | GATA21D04 | 4 | 237 | 230 | 284 | 322 | 218 | 253 | 348 | 343 | 203 | 276 | 306 | 217 | 344 | 193 | 265 |
| D5S1462 | GATA3H06 | 4 | 193 | 182 | 227 | 306 | 232 | 212 | 286 | 340 | 292 | 273 | 330 | 259 | 347 | 272 | 319 |
| D5S1470 | GATA7C06 | 4 | 213 | 218 | 189 | 213 | 292 | 238 | 198 | 226 | 294 | 221 | 279 | 321 | 253 | 244 | 298 |
| D5S1480 | ATA23A10 | 3 | 197 | 206 | 169 | 110 | 155 | 209 | 143 | 109 | 142 | 114 | 92 | 119 | 95 | 173 | 162 |
| D5S1501 | GATA52A12 | 4 | 57 | 44 | 82 | 12 | 141 | 67 | 98 | 20 | 150 | 72 | 24 | 127 | 33 | 164 | 65 |
| D5S1505 | GATA62A04 | 4 | 231 | 288 | 332 | 292 | 341 | 229 | 284 | 271 | 300 | 316 | 283 | 332 | 304 | 346 | 289 |
| D5S1725 | GATA89G08 | 4 | 71 | 102 | 168 | 194 | 199 | 62 | 133 | 139 | 174 | 150 | 148 | 171 | 195 | 200 | 209 |
| D5S211 | MFD154 | 2 | 72 | 134 | 102 | 204 | 26 | 59 | 33 | 104 | 13 | 97 | 187 | 40 | 143 | 97 | 72 |
| D5S2488 | ATA20G07 | 3 | 239 | 259 | 289 | 324 | 251 | 248 | 263 | 323 | 254 | 284 | 319 | 252 | 314 | 273 | 313 |
| D5S2500 | GATA67D03 | 4 | 314 | 326 | 325 | 261 | 367 | 337 | 320 | 254 | 366 | 314 | 242 | 361 | 250 | 353 | 303 |
| D5S2501 | GATA68A03 | 4 | 361 | 342 | 338 | 367 | 277 | 342 | 341 | 373 | 258 | 326 | 371 | 244 | 359 | 239 | 283 |
| D5S2505 | GATA84E11 | 4 | 121 | 131 | 153 | 229 | 227 | 115 | 157 | 222 | 198 | 155 | 200 | 182 | 183 | 188 | 229 |
| D5S2845 | GATA134B03 | 4 | 310 | 285 | 330 | 203 | 316 | 310 | 355 | 210 | 315 | 341 | 206 | 292 | 267 | 321 | 222 |
| D5S2849 | GATA145D10 | 4 | 304 | 313 | 205 | 265 | 294 | 284 | 172 | 240 | 253 | 200 | 260 | 268 | 213 | 213 | 216 |
| D5S408 | AFM164XB8 | 2 | 53 | 55 | 70 | 95 | 62 | 52 | 48 | 83 | 77 | 48 | 82 | 71 | 96 | 54 | 89 |
| D5S816 | GATA2H09 | 4 | 358 | 243 | 288 | 262 | 212 | 234 | 260 | 260 | 216 | 184 | 225 | 169 | 173 | 119 | 224 |
| D5S817 | GATA3E10 | 4 | 228 | 267 | 249 | 273 | 310 | 250 | 209 | 237 | 280 | 269 | 299 | 329 | 297 | 311 | 336 |
| D5S820 | GATA6E05 | 4 | 70 | 81 | 97 | 223 | 72 | 101 | 120 | 256 | 102 | 116 | 246 | 75 | 220 | 89 | 165 |
| D6S1006 | ATC4D09 | 3 | 158 | 97 | 36 | 39 | 78 | 74 | 19 | 25 | 51 | 20 | 29 | 68 | 15 | 33 | 55 |
| D6S1009 | GATA32B03 | 4 | 258 | 256 | 283 | 78 | 208 | 297 | 298 | 71 | 232 | 320 | 94 | 261 | 77 | 262 | 103 |
| D6S1017 | GGAT3H10 | 4 | 15 | 12 | 38 | 38 | 88 | 11 | 41 | 49 | 92 | 26 | 37 | 73 | 30 | 103 | 49 |
| D6S1021 | ATA11D10 | 3 | 67 | 61 | 50 | 122 | 161 | 57 | 50 | 112 | 136 | 44 | 108 | 152 | 67 | 83 | 174 |
| D6S1027 | ATA22G07 | 3 | 9 | 8 | 8 | 24 | 14 | 9 | 12 | 22 | 14 | 7 | 16 | 11 | 12 | 14 | 31 |
| D6S1031 | ATA28B11 | 3 | 43 | 36 | 76 | 84 | 144 | 65 | 150 | 115 | 212 | 109 | 83 | 176 | 152 | 254 | 203 |
| D6S1040 | GATA23F08 | 4 | 219 | 213 | 262 | 271 | 299 | 224 | 275 | 281 | 306 | 270 | 291 | 289 | 332 | 271 | 279 |
| D6S1051 | GATA61E03 | 4 | 315 | 323 | 352 | 325 | 338 | 300 | 349 | 316 | 330 | 349 | 324 | 330 | 341 | 355 | 366 |
| D6S1053 | GATA64D02 | 4 | 247 | 314 | 340 | 277 | 362 | 223 | 299 | 257 | 326 | 346 | 239 | 377 | 284 | 372 | 300 |
| D6S1056 | GATA68H04 | 4 | 200 | 296 | 341 | 114 | 249 | 207 | 249 | 100 | 145 | 340 | 89 | 254 | 92 | 307 | 53 |
| D6S1277 | GATA81B01 | 4 | 264 | 247 | 264 | 212 | 285 | 339 | 304 | 204 | 302 | 272 | 215 | 303 | 177 | 268 | 232 |
| D6S1959 | GATA29A01 | 4 | 368 | 354 | 362 | 365 | 226 | 362 | 363 | 358 | 219 | 348 | 351 | 212 | 336 | 257 | 247 |
| D6S2410 | GATA11E02 | 4 | 371 | 375 | 375 | 359 | 276 | 374 | 374 | 359 | 283 | 377 | 370 | 267 | 367 | 275 | 292 |
| D6S2436 | GATA165G02 | 4 | 89 | 113 | 149 | 243 | 129 | 133 | 176 | 273 | 128 | 186 | 297 | 143 | 294 | 122 | 190 |
| D6S2439 | GATA163B10 | 4 | 211 | 214 | 209 | 182 | 224 | 256 | 243 | 253 | 238 | 210 | 218 | 250 | 202 | 221 | 191 |
| D6S2522 | 6QTEL54 | 2 | 345 | 306 | 344 | 364 | 364 | 301 | 359 | 375 | 368 | 302 | 339 | 354 | 377 | 335 | 357 |
| D6S305 | AFM242ZG5 | 2 | 68 | 66 | 105 | 53 | 30 | 44 | 70 | 54 | 24 | 79 | 38 | 15 | 36 | 10 | 7 |
| D6S474 | GATA31 | 4 | 367 | 325 | 312 | 374 | 354 | 306 | 319 | 371 | 343 | 287 | 348 | 314 | 328 | 319 | 373 |
| D6S942 | UT654 | 4 | 313 | 318 | 306 | 312 | 355 | 326 | 289 | 292 | 344 | 261 | 275 | 333 | 300 | 339 | 335 |
| D7S1799 | GATA23F05 | 4 | 261 | 235 | 304 | 290 | 270 | 211 | 279 | 267 | 267 | 271 | 268 | 264 | 301 | 259 | 273 |
| D7S1802 | GATA41G07 | 4 | 266 | 286 | 305 | 309 | 280 | 293 | 324 | 307 | 285 | 331 | 305 | 281 | 307 | 296 | 310 |
| D7S1804 | GATA43C11 | 4 | 230 | 224 | 274 | 198 | 105 | 215 | 322 | 183 | 131 | 252 | 149 | 108 | 174 | 167 | 64 |
| D7S1808 | GGAA3F06 | 4 | 286 | 223 | 43 | 193 | 32 | 165 | 20 | 174 | 21 | 29 | 146 | 26 | 48 | 45 | 48 |
| D7S1818 | GATA24D12 | 4 | 332 | 358 | 372 | 375 | 297 | 327 | 343 | 369 | 257 | 370 | 376 | 290 | 374 | 316 | 332 |
| D7S1824 | GATA32C12 | 4 | 243 | 279 | 165 | 174 | 200 | 302 | 189 | 195 | 229 | 187 | 204 | 227 | 199 | 220 | 250 |
| D7S2204 | GATA73D10 | 4 | 155 | 157 | 185 | 188 | 217 | 180 | 203 | 224 | 240 | 218 | 217 | 271 | 211 | 243 | 254 |
| D7S2477 | AFMB035XB9 | 2 | 91 | 74 | 92 | 18 | 52 | 100 | 139 | 36 | 52 | 163 | 25 | 59 | 62 | 37 | 6 |
| D7S2846 | GATA31A10 | 4 | 375 | 377 | 374 | 327 | 376 | 377 | 377 | 314 | 361 | 376 | 326 | 369 | 309 | 351 | 309 |
| D7S3046 | GATA118G10 | 4 | 126 | 137 | 166 | 135 | 262 | 160 | 214 | 184 | 295 | 217 | 201 | 293 | 265 | 294 | 268 |
| D7S3047 | GATA119B03 | 4 | 336 | 310 | 349 | 348 | 269 | 332 | 360 | 363 | 278 | 334 | 343 | 253 | 352 | 283 | 330 |
| D7S3051 | GATA137H02 | 4 | 215 | 196 | 222 | 133 | 222 | 219 | 269 | 141 | 241 | 225 | 147 | 209 | 167 | 226 | 146 |
| D7S3056 | GATA24F03 | 4 | 263 | 232 | 263 | 281 | 335 | 237 | 261 | 269 | 317 | 243 | 243 | 279 | 271 | 314 | 339 |
| D7S3058 | GATA30D09 | 4 | 240 | 287 | 221 | 142 | 301 | 280 | 228 | 129 | 310 | 250 | 181 | 316 | 172 | 247 | 175 |
| D7S3061 | GGAA6D03 | 4 | 188 | 185 | 237 | 121 | 173 | 183 | 210 | 97 | 152 | 201 | 127 | 156 | 126 | 224 | 98 |
| D7S3070 | GATA189C06 | 4 | 295 | 300 | 314 | 316 | 283 | 324 | 323 | 329 | 290 | 322 | 336 | 288 | 321 | 299 | 327 |
| D7S559 | MFD265 | 2 | 161 | 174 | 162 | 214 | 207 | 149 | 140 | 201 | 154 | 147 | 196 | 181 | 135 | 135 | 188 |
| D7S817 | GATA13G11 | 4 | 306 | 335 | 300 | 326 | 104 | 346 | 287 | 320 | 108 | 325 | 347 | 103 | 360 | 81 | 99 |
| D7S820 | GATA3F01 | 4 | 324 | 316 | 254 | 282 | 177 | 313 | 255 | 300 | 183 | 255 | 272 | 167 | 252 | 184 | 217 |
| D7S821 | GATA5D08 | 4 | 178 | 187 | 233 | 206 | 282 | 188 | 264 | 227 | 322 | 249 | 228 | 297 | 262 | 332 | 346 |
| D8S1048 | UT7129 | 4 | 370 | 297 | 361 | 313 | 349 | 325 | 369 | 327 | 353 | 343 | 325 | 310 | 324 | 350 | 329 |
| D8S1108 | GATA50D10 | 4 | 333 | 364 | 366 | 361 | 350 | 365 | 356 | 362 | 360 | 372 | 373 | 363 | 355 | 352 | 374 |
| D8S1110 | GATA8G10 | 4 | 78 | 125 | 180 | 149 | 146 | 91 | 159 | 124 | 122 | 199 | 168 | 178 | 208 | 237 | 208 |
| D8S1113 | GGAA8G07 | 4 | 195 | 237 | 182 | 89 | 190 | 201 | 146 | 70 | 185 | 189 | 69 | 198 | 56 | 134 | 36 |
| D8S1128 | GATA21C12 | 4 | 130 | 124 | 22 | 162 | 89 | 226 | 51 | 242 | 167 | 57 | 226 | 177 | 45 | 67 | 204 |
| D8S1132 | GATA26E03 | 4 | 280 | 192 | 266 | 107 | 325 | 170 | 256 | 101 | 323 | 180 | 75 | 257 | 99 | 285 | 173 |
| D8S1136 | GATA41A01 | 4 | 290 | 274 | 234 | 332 | 293 | 281 | 257 | 334 | 299 | 234 | 321 | 296 | 279 | 279 | 321 |
| D8S1179 | GATA7G07 | 4 | 227 | 194 | 253 | 303 | 259 | 260 | 318 | 370 | 261 | 279 | 322 | 258 | 315 | 260 | 316 |
| D8S1477 | GGAA20C10 | 4 | 181 | 146 | 120 | 178 | 136 | 176 | 154 | 196 | 146 | 142 | 194 | 113 | 197 | 76 | 125 |
| D8S2324 | GATA14E09 | 4 | 346 | 329 | 369 | 351 | 318 | 315 | 365 | 337 | 333 | 368 | 327 | 342 | 325 | 343 | 317 |
| D8S261 | AFM123XG5 | 2 | 27 | 26 | 32 | 87 | 28 | 13 | 18 | 72 | 22 | 22 | 61 | 29 | 51 | 69 | 61 |
| D8S262 | AFM127XH2 | 2 | 26 | 22 | 63 | 27 | 35 | 18 | 78 | 21 | 41 | 53 | 18 | 28 | 29 | 48 | 18 |
| D8S373 | UT721 | 4 | 217 | 221 | 290 | 260 | 223 | 228 | 295 | 284 | 228 | 347 | 298 | 213 | 331 | 198 | 225 |
| D8S503 | AFM193XH4 | 2 | 170 | 123 | 53 | 85 | 29 | 142 | 52 | 107 | 35 | 63 | 109 | 47 | 100 | 60 | 63 |
| D8S560 | AFMA127YE5 | 2 | 2 | 2 | 5 | 6 | 1 | 4 | 8 | 24 | 1 | 3 | 14 | 1 | 21 | 3 | 4 |
| D8S592 | GATA6B02 | 4 | 177 | 180 | 220 | 124 | 258 | 159 | 211 | 106 | 250 | 198 | 118 | 255 | 131 | 248 | 187 |
| D9S1118 | GATA71E08 | 4 | 102 | 118 | 198 | 41 | 220 | 98 | 186 | 42 | 205 | 175 | 32 | 225 | 43 | 240 | 115 |
| D9S1120 | GATA81C04 | 4 | 369 | 367 | 358 | 347 | 123 | 372 | 346 | 333 | 114 | 345 | 329 | 111 | 327 | 114 | 148 |
| D9S1121 | GATA87E02 | 4 | 110 | 96 | 100 | 59 | 235 | 82 | 61 | 27 | 231 | 54 | 21 | 162 | 24 | 124 | 71 |
| D9S1122 | GATA89A11 | 4 | 365 | 371 | 357 | 331 | 309 | 366 | 336 | 310 | 297 | 358 | 331 | 309 | 326 | 334 | 320 |
| D9S1779 | AFM026TG9 | 2 | 6 | 7 | 4 | 21 | 8 | 7 | 3 | 15 | 4 | 5 | 27 | 7 | 16 | 7 | 20 |
| D9S1825 | AFMB029XG1 | 2 | 120 | 104 | 128 | 45 | 127 | 95 | 130 | 41 | 125 | 107 | 31 | 125 | 26 | 120 | 22 |
| D9S1838 | AFMB303ZG9 | 2 | 131 | 111 | 54 | 67 | 171 | 136 | 81 | 68 | 236 | 51 | 60 | 154 | 23 | 85 | 54 |
| D9S1871 | AFM345TA9 | 2 | 48 | 64 | 14 | 50 | 50 | 34 | 6 | 17 | 28 | 13 | 34 | 56 | 5 | 38 | 13 |
| D9S2157 | ATA59H06 | 3 | 80 | 60 | 18 | 125 | 27 | 50 | 15 | 110 | 27 | 27 | 140 | 51 | 110 | 25 | 87 |
| D9S2169 | GATA62F03 | 4 | 41 | 33 | 67 | 106 | 147 | 36 | 69 | 131 | 177 | 56 | 110 | 151 | 107 | 132 | 194 |
| D9S301 | GATA7D12 | 4 | 284 | 291 | 247 | 329 | 121 | 291 | 236 | 312 | 109 | 248 | 320 | 123 | 264 | 115 | 129 |
| D9S910 | ATA18A07 | 3 | 107 | 115 | 112 | 76 | 84 | 120 | 97 | 60 | 83 | 101 | 70 | 97 | 140 | 147 | 134 |
| D9S922 | GATA21F05 | 4 | 316 | 228 | 152 | 163 | 83 | 227 | 141 | 148 | 70 | 117 | 143 | 35 | 108 | 26 | 34 |
| D9S925 | GATA27A11 | 4 | 84 | 67 | 85 | 49 | 45 | 81 | 99 | 55 | 46 | 84 | 47 | 37 | 54 | 52 | 33 |
| D9S930 | GATA48D07 | 4 | 334 | 349 | 346 | 317 | 342 | 343 | 344 | 303 | 349 | 351 | 289 | 339 | 302 | 322 | 286 |
| D9S934 | GATA64G07 | 4 | 347 | 334 | 302 | 295 | 366 | 347 | 305 | 301 | 363 | 313 | 307 | 357 | 299 | 318 | 345 |
| D9S938 | GGAA22E01 | 4 | 262 | 262 | 186 | 56 | 17 | 296 | 194 | 58 | 23 | 206 | 53 | 24 | 59 | 61 | 5 |
| F13A1-D6S | SE30 | 4 | 183 | 144 | 19 | 47 | 137 | 230 | 26 | 88 | 171 | 30 | 93 | 172 | 3 | 62 | 47 |
| NA-D10S-1 | GATA121A08 | 4 | 278 | 272 | 278 | 304 | 343 | 264 | 276 | 285 | 332 | 275 | 300 | 328 | 283 | 306 | 342 |
| NA-D10S-2 | ATA103C06 | 3 | 52 | 48 | 80 | 91 | 128 | 40 | 65 | 86 | 120 | 70 | 97 | 136 | 144 | 180 | 219 |
| NA-D11S-1 | ATA34E08 | 3 | 33 | 52 | 73 | 151 | 111 | 35 | 40 | 125 | 86 | 75 | 184 | 121 | 286 | 205 | 245 |
| NA-D12S-1 | GATA49D12 | 4 | 114 | 152 | 150 | 54 | 216 | 112 | 104 | 45 | 202 | 176 | 80 | 200 | 86 | 170 | 93 |
| NA-D12S-2 | PAH | 4 | 144 | 168 | 207 | 209 | 230 | 132 | 177 | 182 | 190 | 202 | 211 | 229 | 228 | 251 | 282 |
| NA-D13S-1 | ATA5A09 | 3 | 124 | 107 | 131 | 123 | 39 | 122 | 114 | 136 | 25 | 69 | 107 | 13 | 123 | 55 | 43 |
| NA-D14S-1 | GATA193A07 | 4 | 112 | 136 | 86 | 233 | 167 | 175 | 145 | 264 | 220 | 193 | 296 | 265 | 261 | 278 | 315 |
| NA-D15S-1 | GATA50C03 | 4 | 179 | 208 | 214 | 248 | 215 | 210 | 207 | 247 | 200 | 238 | 261 | 211 | 258 | 215 | 244 |
| NA-D16S-1 | GATA138C05 | 4 | 32 | 28 | 57 | 44 | 92 | 26 | 79 | 64 | 112 | 62 | 41 | 83 | 80 | 102 | 117 |
| NA-D17S-1 | ATA78D02 | 3 | 248 | 211 | 96 | 235 | 37 | 194 | 82 | 229 | 34 | 91 | 202 | 30 | 160 | 66 | 124 |
| NA-D18S-1 | GATA178F11 | 4 | 285 | 307 | 334 | 288 | 300 | 270 | 310 | 231 | 244 | 310 | 269 | 278 | 276 | 274 | 306 |
| NA-D18S-2 | ATA82B02 | 3 | 56 | 57 | 69 | 36 | 86 | 51 | 58 | 14 | 88 | 60 | 30 | 94 | 37 | 86 | 29 |
| NA-D1S-1 | ATA79C10 | 3 | 355 | 340 | 256 | 286 | 113 | 363 | 277 | 298 | 126 | 264 | 277 | 112 | 219 | 125 | 138 |
| NA-D1S-2 | GATA124C08 | 4 | 269 | 240 | 295 | 225 | 323 | 283 | 334 | 275 | 362 | 299 | 247 | 336 | 298 | 365 | 338 |
| NA-D1S-3 | GATA133A08 | 4 | 176 | 205 | 269 | 210 | 296 | 241 | 282 | 205 | 320 | 291 | 209 | 326 | 238 | 338 | 277 |
| NA-D1S-4 | ATA42G12 | 3 | 223 | 239 | 258 | 255 | 219 | 247 | 238 | 236 | 188 | 254 | 234 | 192 | 235 | 199 | 261 |
| NA-D1S-5 | GATA124F08 | 4 | 328 | 376 | 347 | 376 | 356 | 318 | 272 | 331 | 284 | 337 | 377 | 351 | 334 | 309 | 364 |
| NA-D22S-1 | GATA198B05 | 4 | 351 | 363 | 267 | 259 | 209 | 349 | 247 | 228 | 156 | 306 | 270 | 221 | 243 | 212 | 223 |
| NA-D4S-1 | GATA70E01 | 4 | 108 | 90 | 94 | 172 | 143 | 77 | 90 | 169 | 135 | 64 | 131 | 130 | 118 | 91 | 155 |
| NA-D5S-1 | ATA52D02 | 3 | 1 | 3 | 3 | 9 | 13 | 2 | 4 | 7 | 12 | 4 | 9 | 9 | 18 | 8 | 15 |
| NA-D6S-1 | GATA184A08 | 4 | 19 | 25 | 41 | 60 | 80 | 17 | 27 | 53 | 79 | 35 | 58 | 77 | 83 | 87 | 121 |
| NA-D7S-1 | GATA104 | 4 | 73 | 59 | 135 | 111 | 186 | 61 | 142 | 128 | 161 | 106 | 114 | 134 | 132 | 159 | 123 |
| NA-D8S-1 | GATA151F02 | 4 | 125 | 133 | 175 | 179 | 205 | 130 | 192 | 170 | 210 | 160 | 156 | 179 | 162 | 227 | 183 |
| NA-D8S-2 | GAAT1A4 | 4 | 169 | 165 | 190 | 222 | 288 | 117 | 156 | 165 | 252 | 152 | 136 | 245 | 159 | 209 | 212 |
| NA-D9S-1 | GATA187D09 | 4 | 299 | 317 | 315 | 263 | 312 | 269 | 278 | 252 | 296 | 335 | 284 | 327 | 306 | 345 | 349 |
| TPO-D2S | SRA | 4 | 139 | 105 | 158 | 146 | 68 | 121 | 184 | 194 | 84 | 140 | 155 | 55 | 155 | 56 | 78 |
Note.— For each data set (table 4), markers are ranked from most informative (1) to least informative (377). Names for the markers are the same as in supplementary table 4 of Rosenberg et al. (2002). The regions are coded as follows: 1 = Africa; 2 = Europe; 3 = Middle East; 4 = Central/South Asia; 5 = East Asia; 6 = Oceania; 7 = America. Loci are sorted in lexical order.
Table D.
Informativeness (In) for Distinguishing among Three Regions (Second of Three Parts)[Note]
|
In Rank for Set of Three Regions |
||||||||||||
| Locus Name | Alternate Name | RepeatSize | 2-3-4 | 2-3-5 | 2-3-6 | 2-3-7 | 2-4-5 | 2-4-6 | 2-4-7 | 2-5-6 | 2-5-7 | 2-6-7 |
| D10S1208 | ATA5A04 | 3 | 55 | 216 | 29 | 357 | 115 | 27 | 313 | 59 | 336 | 119 |
| D10S1221 | ATA21A03 | 3 | 91 | 10 | 100 | 116 | 21 | 125 | 92 | 58 | 42 | 145 |
| D10S1222 | ATA22D02 | 3 | 152 | 178 | 229 | 124 | 211 | 244 | 134 | 290 | 126 | 222 |
| D10S1225 | ATA24F10 | 3 | 349 | 360 | 141 | 205 | 353 | 122 | 183 | 163 | 247 | 244 |
| D10S1230 | ATA29C03 | 3 | 149 | 95 | 261 | 241 | 61 | 247 | 270 | 155 | 134 | 345 |
| D10S1239 | GATA64A09 | 4 | 176 | 155 | 278 | 192 | 178 | 282 | 226 | 275 | 258 | 311 |
| D10S1248 | GGAA23C05 | 4 | 253 | 294 | 215 | 330 | 246 | 252 | 310 | 230 | 337 | 241 |
| D10S1412 | ATA31G11 | 3 | 26 | 172 | 87 | 63 | 278 | 154 | 118 | 213 | 171 | 176 |
| D10S1423 | GATA70E11 | 4 | 234 | 196 | 317 | 256 | 234 | 280 | 290 | 254 | 265 | 293 |
| D10S1425 | GATA71C09 | 4 | 357 | 94 | 230 | 216 | 83 | 219 | 238 | 81 | 121 | 171 |
| D10S1426 | GATA73E11 | 4 | 218 | 332 | 343 | 356 | 298 | 322 | 322 | 363 | 358 | 366 |
| D10S1430 | GATA84C01 | 4 | 220 | 274 | 232 | 351 | 313 | 299 | 367 | 278 | 361 | 335 |
| D10S1432 | GATA87G01 | 4 | 167 | 282 | 251 | 361 | 370 | 261 | 373 | 302 | 376 | 337 |
| D10S1435 | GATA88F09 | 4 | 287 | 326 | 365 | 302 | 342 | 368 | 305 | 368 | 302 | 346 |
| D10S189 | AFM063XF4 | 2 | 244 | 333 | 248 | 316 | 212 | 147 | 244 | 286 | 264 | 277 |
| D10S212 | AFM198ZB4 | 2 | 335 | 251 | 174 | 342 | 275 | 193 | 332 | 233 | 275 | 237 |
| D10S2327 | GGAT1A4 | 4 | 258 | 189 | 254 | 307 | 180 | 210 | 255 | 148 | 216 | 268 |
| D10S2470 | GATA115E01 | 4 | 221 | 203 | 217 | 166 | 141 | 164 | 166 | 216 | 81 | 106 |
| D10S677 | GGAA2F11 | 4 | 191 | 112 | 201 | 46 | 151 | 266 | 51 | 257 | 39 | 98 |
| D11S1304 | UT2095 | 4 | 68 | 18 | 181 | 88 | 22 | 194 | 99 | 27 | 64 | 109 |
| D11S1392 | GATA6B09 | 4 | 333 | 349 | 375 | 309 | 343 | 376 | 320 | 369 | 330 | 358 |
| D11S1981 | GATA48E02 | 4 | 153 | 228 | 241 | 220 | 207 | 238 | 188 | 259 | 168 | 228 |
| D11S1984 | GGAA17G05 | 4 | 35 | 11 | 86 | 98 | 6 | 51 | 87 | 11 | 47 | 55 |
| D11S1986 | GGAA7G08 | 4 | 34 | 66 | 128 | 51 | 69 | 141 | 45 | 116 | 66 | 71 |
| D11S1993 | ATA1B07 | 3 | 77 | 33 | 88 | 14 | 60 | 92 | 17 | 73 | 36 | 21 |
| D11S1998 | GATA23E06 | 4 | 57 | 176 | 377 | 246 | 177 | 303 | 161 | 279 | 150 | 326 |
| D11S1999 | GATA23F06 | 4 | 282 | 50 | 326 | 41 | 51 | 345 | 40 | 168 | 99 | 112 |
| D11S2000 | GATA28D01 | 4 | 14 | 129 | 50 | 60 | 188 | 84 | 78 | 103 | 56 | 61 |
| D11S2002 | GATA30G01 | 4 | 230 | 145 | 164 | 182 | 174 | 181 | 184 | 245 | 209 | 235 |
| D11S2006 | GATA46A12 | 4 | 320 | 361 | 338 | 365 | 372 | 358 | 376 | 366 | 374 | 373 |
| D11S2362 | ATA33B03 | 3 | 212 | 267 | 287 | 175 | 187 | 253 | 148 | 229 | 154 | 205 |
| D11S2363 | GATA12F04 | 4 | 38 | 127 | 154 | 87 | 129 | 168 | 101 | 193 | 124 | 159 |
| D11S2365 | GATA63F09 | 4 | 375 | 370 | 351 | 183 | 362 | 360 | 176 | 360 | 223 | 308 |
| D11S2371 | GATA90D07 | 4 | 364 | 299 | 319 | 362 | 257 | 292 | 340 | 303 | 291 | 309 |
| D11S4459 | ATA9B04 | 3 | 135 | 171 | 185 | 106 | 229 | 180 | 154 | 224 | 172 | 199 |
| D11S4463 | GATA117D01 | 4 | 148 | 289 | 355 | 311 | 170 | 330 | 249 | 346 | 299 | 347 |
| D11S4464 | GATA64D03 | 4 | 215 | 258 | 243 | 219 | 349 | 350 | 272 | 333 | 323 | 300 |
| D11S969 | AFM205VF10 | 2 | 268 | 164 | 111 | 234 | 165 | 100 | 225 | 89 | 239 | 126 |
| D12S1042 | ATA27A06 | 3 | 115 | 142 | 121 | 50 | 157 | 149 | 72 | 215 | 70 | 73 |
| D12S1045 | ATA29A06 | 3 | 130 | 37 | 63 | 31 | 65 | 96 | 49 | 164 | 78 | 117 |
| D12S1052 | GATA26D02 | 4 | 337 | 343 | 339 | 160 | 319 | 307 | 135 | 342 | 145 | 170 |
| D12S1064 | GATA63D12 | 4 | 352 | 233 | 168 | 269 | 215 | 140 | 259 | 218 | 308 | 267 |
| D12S1294 | GATA73H09 | 4 | 284 | 122 | 306 | 142 | 120 | 289 | 109 | 175 | 85 | 200 |
| D12S1300 | GATA85A04 | 4 | 327 | 162 | 22 | 336 | 148 | 19 | 336 | 40 | 300 | 110 |
| D12S1301 | GATA91H06 | 4 | 180 | 49 | 131 | 84 | 70 | 90 | 112 | 78 | 26 | 64 |
| D12S1638 | AFMB002VD5 | 2 | 187 | 320 | 139 | 230 | 280 | 132 | 160 | 210 | 221 | 201 |
| D12S2070 | ATA25F09 | 3 | 140 | 16 | 96 | 1 | 30 | 127 | 3 | 36 | 5 | 4 |
| D12S2078 | GATA32F05 | 4 | 275 | 168 | 146 | 254 | 227 | 152 | 279 | 145 | 251 | 163 |
| D12S269 | MFD259 | 2 | 122 | 227 | 136 | 40 | 197 | 136 | 48 | 165 | 88 | 104 |
| D12S297 | UT5029 | 4 | 113 | 61 | 28 | 10 | 43 | 22 | 4 | 60 | 15 | 44 |
| D12S372 | GATA4H03 | 4 | 175 | 344 | 356 | 159 | 345 | 355 | 197 | 374 | 254 | 284 |
| D12S373 | GATA6C01 | 4 | 311 | 190 | 83 | 314 | 164 | 72 | 302 | 167 | 229 | 135 |
| D12S395 | GATA4H01 | 4 | 292 | 105 | 102 | 293 | 140 | 126 | 331 | 172 | 267 | 229 |
| D13S1493 | GGAA29H03 | 4 | 299 | 241 | 179 | 203 | 255 | 162 | 243 | 166 | 276 | 178 |
| D13S1807 | GATA11C08 | 4 | 239 | 279 | 330 | 117 | 348 | 293 | 107 | 296 | 129 | 257 |
| D13S285 | AFM309VA9 | 2 | 60 | 84 | 143 | 290 | 96 | 135 | 278 | 95 | 198 | 221 |
| D13S317 | GATA7G10 | 4 | 280 | 131 | 258 | 65 | 144 | 255 | 54 | 200 | 86 | 74 |
| D13S779 | ATA26D07 | 3 | 353 | 198 | 264 | 114 | 189 | 271 | 93 | 190 | 107 | 96 |
| D13S787 | GATA23C03 | 4 | 242 | 376 | 349 | 372 | 326 | 329 | 326 | 354 | 375 | 362 |
| D13S793 | GATA43H03 | 4 | 377 | 374 | 81 | 191 | 368 | 67 | 178 | 88 | 218 | 148 |
| D13S796 | GATA51B02 | 4 | 323 | 331 | 95 | 274 | 312 | 101 | 292 | 150 | 295 | 269 |
| D13S800 | GATA64F08 | 4 | 306 | 270 | 236 | 303 | 285 | 260 | 330 | 179 | 349 | 243 |
| D13S894 | GATA86H01 | 4 | 188 | 156 | 284 | 245 | 124 | 276 | 224 | 201 | 240 | 266 |
| D13S895 | GGAA22G01 | 4 | 127 | 194 | 242 | 272 | 209 | 218 | 297 | 211 | 320 | 251 |
| D14S1007 | AFMB002ZF1 | 2 | 13 | 1 | 5 | 13 | 1 | 11 | 19 | 3 | 1 | 6 |
| D14S1280 | GATA31B09 | 4 | 342 | 337 | 291 | 324 | 335 | 272 | 341 | 305 | 284 | 271 |
| D14S1426 | GATA136B01 | 4 | 247 | 159 | 178 | 25 | 122 | 113 | 29 | 111 | 10 | 10 |
| D14S1434 | GATA168F06 | 4 | 271 | 217 | 252 | 260 | 242 | 312 | 280 | 262 | 242 | 302 |
| D14S587 | GGAA10C09 | 4 | 138 | 186 | 205 | 147 | 175 | 241 | 128 | 156 | 160 | 154 |
| D14S588 | GGAA4A12 | 4 | 108 | 114 | 66 | 133 | 78 | 79 | 156 | 39 | 101 | 111 |
| D14S592 | ATA19H08 | 3 | 92 | 208 | 76 | 173 | 232 | 69 | 181 | 91 | 226 | 136 |
| D14S599 | ATA29G03 | 3 | 269 | 48 | 266 | 15 | 39 | 189 | 6 | 77 | 23 | 16 |
| D14S606 | GATA30A03 | 4 | 365 | 260 | 193 | 275 | 268 | 191 | 289 | 265 | 313 | 290 |
| D14S608 | GATA43H01 | 4 | 40 | 167 | 223 | 64 | 266 | 267 | 31 | 284 | 54 | 107 |
| D14S617 | GGAA21G11 | 4 | 158 | 256 | 10 | 204 | 239 | 18 | 203 | 16 | 244 | 31 |
| D14S742 | GATA74E02 | 4 | 270 | 335 | 360 | 366 | 314 | 357 | 352 | 376 | 377 | 377 |
| D15S128 | AFM273YF9 | 2 | 46 | 79 | 147 | 67 | 82 | 196 | 73 | 195 | 111 | 188 |
| D15S1507 | GATA151F03 | 4 | 358 | 232 | 265 | 369 | 240 | 268 | 374 | 241 | 339 | 332 |
| D15S1515 | GATA197B10 | 4 | 297 | 253 | 340 | 77 | 204 | 326 | 63 | 304 | 93 | 128 |
| D15S165 | AFM248VC5 | 2 | 9 | 26 | 51 | 68 | 26 | 57 | 76 | 71 | 60 | 91 |
| D15S642 | GATA27A03 | 4 | 96 | 8 | 130 | 86 | 9 | 139 | 110 | 34 | 45 | 139 |
| D15S643 | GATA50G06 | 4 | 120 | 201 | 145 | 176 | 169 | 99 | 130 | 153 | 162 | 189 |
| D15S652 | ATA24A08 | 3 | 66 | 110 | 80 | 18 | 84 | 91 | 14 | 121 | 8 | 12 |
| D15S655 | ATA28G05 | 3 | 128 | 74 | 30 | 295 | 98 | 41 | 328 | 65 | 192 | 102 |
| D15S659 | GATA63A03 | 4 | 245 | 312 | 226 | 126 | 294 | 167 | 139 | 227 | 149 | 92 |
| D15S816 | GATA73F01 | 4 | 252 | 165 | 316 | 352 | 146 | 338 | 353 | 228 | 243 | 342 |
| D15S818 | GATA85D02 | 4 | 298 | 305 | 321 | 283 | 352 | 339 | 317 | 356 | 316 | 329 |
| D15S822 | GATA88H02 | 4 | 48 | 36 | 175 | 11 | 36 | 176 | 12 | 149 | 28 | 63 |
| D16S2616 | ATA41E04 | 3 | 341 | 263 | 183 | 121 | 267 | 166 | 131 | 209 | 173 | 216 |
| D16S2621 | GATA71F09 | 4 | 195 | 32 | 292 | 83 | 42 | 314 | 120 | 119 | 90 | 193 |
| D16S2624 | GATA81D12 | 4 | 356 | 65 | 295 | 148 | 62 | 296 | 155 | 161 | 147 | 190 |
| D16S3253 | GATA22F09 | 4 | 173 | 229 | 352 | 209 | 183 | 259 | 158 | 261 | 197 | 278 |
| D16S3396 | ATA55A11 | 3 | 219 | 292 | 303 | 248 | 292 | 300 | 240 | 337 | 288 | 304 |
| D16S3401 | 16PTEL06 | 2 | 43 | 102 | 7 | 36 | 58 | 5 | 20 | 6 | 58 | 9 |
| D16S403 | AFM049XD2 | 2 | 29 | 104 | 37 | 70 | 185 | 15 | 75 | 14 | 113 | 65 |
| D16S422 | AFM249XC5 | 2 | 36 | 3 | 152 | 20 | 3 | 174 | 25 | 31 | 13 | 68 |
| D16S516 | AFM350VD1 | 2 | 150 | 51 | 124 | 97 | 63 | 129 | 96 | 70 | 96 | 99 |
| D16S539 | GATA11C06 | 4 | 156 | 240 | 313 | 184 | 166 | 257 | 167 | 236 | 232 | 220 |
| D16S748 | ATA3A07 | 3 | 22 | 40 | 20 | 43 | 48 | 38 | 62 | 28 | 29 | 70 |
| D16S753 | GGAA3G05 | 4 | 147 | 136 | 206 | 312 | 131 | 221 | 257 | 231 | 225 | 233 |
| D16S764 | GATA42E11 | 4 | 361 | 345 | 334 | 218 | 336 | 321 | 180 | 355 | 227 | 272 |
| D16S769 | GATA71H05 | 4 | 376 | 366 | 376 | 348 | 351 | 374 | 357 | 371 | 314 | 352 |
| D17S1290 | GATA49C09 | 4 | 100 | 281 | 182 | 125 | 250 | 198 | 100 | 238 | 161 | 155 |
| D17S1294 | GGAA9D03 | 4 | 201 | 271 | 327 | 334 | 287 | 336 | 338 | 343 | 348 | 371 |
| D17S1298 | GAAT2C03 | 4 | 71 | 73 | 310 | 367 | 59 | 269 | 314 | 206 | 215 | 365 |
| D17S1299 | GATA25A04 | 4 | 174 | 272 | 297 | 341 | 274 | 298 | 346 | 301 | 325 | 351 |
| D17S1301 | GATA28D11 | 4 | 305 | 268 | 237 | 375 | 252 | 287 | 372 | 270 | 338 | 331 |
| D17S1308 | GTAT1A05 | 4 | 232 | 139 | 33 | 207 | 158 | 33 | 246 | 104 | 268 | 166 |
| D17S2180 | ATC6A06 | 3 | 165 | 141 | 227 | 167 | 159 | 248 | 150 | 248 | 110 | 168 |
| D17S2193 | ATA43A10Z | 3 | 109 | 215 | 211 | 152 | 288 | 169 | 138 | 184 | 158 | 169 |
| D17S2195 | ATA58A02 | 3 | 163 | 179 | 27 | 206 | 224 | 31 | 223 | 57 | 191 | 54 |
| D17S2196 | GATA185H04 | 4 | 304 | 140 | 210 | 71 | 81 | 183 | 65 | 87 | 71 | 80 |
| D17S784 | AFM044XG3 | 2 | 198 | 125 | 245 | 168 | 89 | 165 | 163 | 118 | 44 | 144 |
| D17S974 | GATA8C04 | 4 | 339 | 363 | 263 | 194 | 333 | 215 | 162 | 256 | 249 | 142 |
| D18S1357 | ATA7D07 | 3 | 260 | 255 | 117 | 186 | 210 | 94 | 175 | 129 | 143 | 103 |
| D18S1364 | GATA7E12 | 4 | 345 | 342 | 194 | 215 | 308 | 177 | 193 | 197 | 270 | 173 |
| D18S1370 | ATA45G06 | 3 | 106 | 101 | 73 | 181 | 143 | 93 | 179 | 152 | 131 | 137 |
| D18S1371 | GATA177C03 | 4 | 325 | 373 | 293 | 163 | 356 | 246 | 191 | 311 | 187 | 174 |
| D18S1376 | GATA185C06 | 4 | 293 | 323 | 239 | 349 | 318 | 229 | 351 | 281 | 372 | 336 |
| D18S1390 | 18QTEL11 | 2 | 133 | 88 | 159 | 78 | 85 | 150 | 70 | 138 | 92 | 150 |
| D18S535 | GATA13 | 4 | 213 | 183 | 39 | 193 | 244 | 45 | 200 | 117 | 170 | 49 |
| D18S542 | GATA11A06 | 4 | 277 | 148 | 1 | 164 | 119 | 2 | 143 | 7 | 167 | 20 |
| D18S843 | ACT1A01 | 3 | 367 | 197 | 270 | 368 | 167 | 237 | 360 | 178 | 309 | 339 |
| D18S851 | GATA6D09 | 4 | 172 | 193 | 280 | 119 | 214 | 232 | 106 | 234 | 135 | 226 |
| D18S858 | ATA23G05 | 3 | 344 | 161 | 71 | 273 | 216 | 77 | 258 | 107 | 222 | 192 |
| D18S877 | GATA64H04 | 4 | 290 | 346 | 272 | 370 | 300 | 258 | 369 | 318 | 373 | 340 |
| D19S1034 | GATA21G05 | 4 | 217 | 209 | 249 | 158 | 276 | 302 | 149 | 309 | 165 | 252 |
| D19S245 | MFD235 | 4 | 374 | 375 | 368 | 371 | 369 | 365 | 370 | 372 | 364 | 364 |
| D19S246 | MFD232 | 4 | 73 | 132 | 120 | 237 | 118 | 131 | 273 | 130 | 195 | 160 |
| D19S254 | MFD238 | 4 | 236 | 277 | 64 | 112 | 261 | 64 | 82 | 98 | 98 | 24 |
| D19S433 | GGAA2A03 | 4 | 85 | 154 | 134 | 96 | 147 | 144 | 77 | 194 | 120 | 152 |
| D19S559 | UT7544 | 4 | 255 | 319 | 337 | 228 | 320 | 323 | 210 | 328 | 250 | 260 |
| D19S586 | GATA23B01 | 4 | 350 | 372 | 347 | 277 | 377 | 343 | 260 | 353 | 310 | 255 |
| D19S589 | GATA29B01 | 4 | 294 | 329 | 342 | 344 | 365 | 348 | 363 | 344 | 365 | 367 |
| D19S591 | GATA44F10 | 4 | 332 | 259 | 234 | 111 | 205 | 187 | 81 | 237 | 94 | 147 |
| D19S714 | GATA66B04 | 4 | 274 | 350 | 90 | 270 | 305 | 73 | 230 | 122 | 269 | 130 |
| D1S1589 | ATA4E02 | 3 | 196 | 138 | 45 | 93 | 126 | 44 | 98 | 23 | 34 | 27 |
| D1S1594 | GATA22D12 | 4 | 203 | 303 | 371 | 190 | 311 | 332 | 245 | 339 | 287 | 264 |
| D1S1596 | GATA26G09 | 4 | 351 | 316 | 363 | 251 | 339 | 362 | 215 | 358 | 246 | 279 |
| D1S1597 | GATA27E01 | 4 | 343 | 317 | 364 | 195 | 347 | 366 | 228 | 357 | 283 | 314 |
| D1S1609 | GATA50F11 | 4 | 250 | 192 | 286 | 92 | 192 | 297 | 74 | 249 | 53 | 140 |
| D1S1612 | GGAA3A07 | 4 | 256 | 21 | 78 | 35 | 16 | 71 | 23 | 42 | 17 | 40 |
| D1S1627 | ATA25E07 | 3 | 118 | 67 | 155 | 253 | 73 | 171 | 253 | 134 | 202 | 217 |
| D1S1653 | GATA43A04 | 4 | 231 | 262 | 307 | 264 | 302 | 324 | 303 | 330 | 319 | 349 |
| D1S1660 | GATA48B01 | 4 | 285 | 207 | 93 | 373 | 198 | 104 | 343 | 135 | 328 | 194 |
| D1S1665 | GATA61A06 | 4 | 132 | 175 | 335 | 327 | 260 | 369 | 359 | 336 | 333 | 374 |
| D1S1677 | GGAA22G10 | 4 | 223 | 187 | 314 | 214 | 273 | 347 | 234 | 321 | 272 | 307 |
| D1S1679 | GGAA5F09 | 4 | 222 | 284 | 115 | 319 | 270 | 110 | 319 | 158 | 340 | 218 |
| D1S1728 | GATA109 | 4 | 142 | 99 | 53 | 165 | 112 | 80 | 186 | 100 | 156 | 122 |
| D1S2134 | GATA72H07 | 4 | 104 | 27 | 68 | 26 | 23 | 83 | 22 | 123 | 46 | 101 |
| D1S235 | AFM203YG9 | 2 | 10 | 14 | 89 | 151 | 14 | 97 | 132 | 21 | 61 | 134 |
| D1S2682 | AFMA272XC9 | 2 | 278 | 126 | 190 | 141 | 171 | 200 | 164 | 136 | 128 | 249 |
| D1S3462 | ATA29C07 | 3 | 49 | 43 | 107 | 288 | 38 | 123 | 222 | 106 | 104 | 172 |
| D1S3669 | GATA29A05 | 4 | 105 | 195 | 112 | 172 | 228 | 95 | 136 | 86 | 203 | 86 |
| D1S3720 | ATA47D07 | 3 | 44 | 64 | 67 | 198 | 75 | 59 | 151 | 56 | 109 | 60 |
| D1S3721 | GATA129H04 | 4 | 37 | 90 | 109 | 38 | 77 | 86 | 32 | 133 | 75 | 82 |
| D1S468 | AFM280WE5 | 2 | 16 | 137 | 113 | 9 | 86 | 109 | 5 | 68 | 7 | 14 |
| D1S518 | GATA7C01 | 4 | 32 | 54 | 24 | 53 | 56 | 29 | 55 | 44 | 68 | 56 |
| D1S534 | GATA12A07 | 4 | 95 | 29 | 70 | 44 | 27 | 55 | 42 | 51 | 57 | 77 |
| D1S549 | GATA4H09 | 4 | 80 | 211 | 92 | 201 | 223 | 111 | 205 | 132 | 230 | 88 |
| D1S551 | GATA6A05 | 4 | 303 | 301 | 156 | 258 | 299 | 108 | 247 | 151 | 234 | 94 |
| D20S103 | AFM077XD3 | 2 | 58 | 12 | 69 | 81 | 15 | 60 | 113 | 22 | 35 | 95 |
| D20S1143 | GATA129B03 | 4 | 336 | 339 | 279 | 358 | 340 | 285 | 362 | 289 | 367 | 316 |
| D20S159 | UT1307 | 4 | 53 | 86 | 231 | 131 | 93 | 342 | 152 | 177 | 116 | 187 |
| D20S164 | UT1772 | 4 | 82 | 9 | 57 | 7 | 5 | 75 | 7 | 67 | 3 | 29 |
| D20S201 | GATA8B01 | 4 | 185 | 130 | 273 | 49 | 138 | 278 | 57 | 162 | 87 | 59 |
| D20S451 | UT254 | 4 | 89 | 81 | 218 | 227 | 125 | 277 | 282 | 255 | 266 | 328 |
| D20S477 | GATA29F06 | 4 | 291 | 56 | 72 | 105 | 53 | 56 | 119 | 66 | 114 | 116 |
| D20S478 | GATA42A03 | 4 | 45 | 242 | 328 | 265 | 181 | 309 | 211 | 341 | 294 | 319 |
| D20S480 | GATA45B10 | 4 | 363 | 304 | 374 | 289 | 315 | 375 | 296 | 345 | 301 | 330 |
| D20S481 | GATA47F05 | 4 | 314 | 124 | 166 | 226 | 172 | 124 | 213 | 64 | 164 | 121 |
| D20S482 | GATA51D03 | 4 | 359 | 308 | 345 | 376 | 277 | 337 | 375 | 329 | 356 | 370 |
| D20S851 | AFMA218YB5 | 2 | 129 | 34 | 19 | 37 | 32 | 13 | 28 | 13 | 9 | 5 |
| D21S1411 | UT1355 | 4 | 143 | 184 | 180 | 74 | 97 | 116 | 68 | 170 | 50 | 45 |
| D21S1432 | GATA11C12 | 4 | 179 | 169 | 188 | 225 | 196 | 201 | 268 | 176 | 183 | 282 |
| D21S1437 | GGAA3C07 | 4 | 94 | 146 | 191 | 323 | 133 | 204 | 307 | 141 | 186 | 240 |
| D21S1440 | ATA27F01 | 3 | 93 | 273 | 331 | 255 | 309 | 364 | 318 | 348 | 321 | 355 |
| D21S1446 | GATA70B08 | 4 | 31 | 143 | 101 | 217 | 222 | 138 | 323 | 222 | 341 | 273 |
| D21S2052 | GATA129D11 | 4 | 159 | 212 | 127 | 231 | 173 | 78 | 207 | 102 | 217 | 132 |
| D21S2055 | GATA188F04 | 4 | 6 | 45 | 16 | 21 | 52 | 17 | 18 | 15 | 18 | 8 |
| D22S1045 | ATA37D06 | 3 | 83 | 150 | 238 | 266 | 135 | 208 | 185 | 198 | 148 | 310 |
| D22S1169 | AFMB337ZH9 | 2 | 184 | 82 | 3 | 107 | 57 | 3 | 83 | 8 | 77 | 22 |
| D22S345 | MFD313 | 2 | 157 | 87 | 271 | 30 | 111 | 275 | 41 | 174 | 63 | 89 |
| D22S532 | UT7136 | 4 | 370 | 314 | 246 | 374 | 334 | 279 | 365 | 319 | 324 | 283 |
| D22S683 | GATA11B12 | 4 | 3 | 78 | 49 | 22 | 100 | 48 | 36 | 94 | 27 | 15 |
| D22S686 | GGAA10F06 | 4 | 39 | 62 | 222 | 320 | 87 | 301 | 361 | 263 | 274 | 363 |
| D22S689 | GATA21F03 | 4 | 183 | 224 | 204 | 262 | 251 | 234 | 309 | 250 | 293 | 263 |
| D2S1328 | GATA27A12 | 4 | 170 | 20 | 8 | 224 | 20 | 8 | 216 | 9 | 103 | 42 |
| D2S1334 | GATA4D07 | 4 | 67 | 92 | 244 | 132 | 102 | 351 | 153 | 191 | 123 | 203 |
| D2S1352 | ATA27D04 | 3 | 162 | 288 | 123 | 325 | 225 | 128 | 274 | 131 | 282 | 253 |
| D2S1353 | ATA27H09 | 3 | 283 | 325 | 250 | 103 | 247 | 211 | 71 | 291 | 115 | 141 |
| D2S1356 | ATA4F03 | 3 | 11 | 41 | 31 | 3 | 28 | 25 | 1 | 38 | 2 | 2 |
| D2S1360 | GATA11H10 | 4 | 110 | 39 | 269 | 139 | 18 | 199 | 90 | 101 | 102 | 215 |
| D2S1363 | GATA23D03 | 4 | 240 | 158 | 12 | 135 | 121 | 10 | 189 | 26 | 119 | 33 |
| D2S1384 | GATA52A04 | 4 | 98 | 60 | 221 | 127 | 116 | 207 | 231 | 154 | 208 | 167 |
| D2S1391 | GATA65C03 | 4 | 279 | 243 | 312 | 150 | 226 | 290 | 142 | 292 | 181 | 230 |
| D2S1394 | GATA69E12 | 4 | 322 | 368 | 366 | 350 | 373 | 372 | 355 | 373 | 368 | 376 |
| D2S1399 | GGAA20G04 | 4 | 28 | 103 | 77 | 27 | 132 | 119 | 33 | 99 | 41 | 38 |
| D2S1400 | GGAA20G10 | 4 | 63 | 91 | 192 | 72 | 154 | 256 | 114 | 267 | 144 | 232 |
| D2S1776 | GATA71D01 | 4 | 161 | 185 | 274 | 109 | 186 | 325 | 126 | 271 | 142 | 191 |
| D2S1780 | GATA72G11 | 4 | 321 | 365 | 348 | 317 | 350 | 353 | 316 | 349 | 344 | 323 |
| D2S1788 | GATA86E02 | 4 | 86 | 200 | 203 | 120 | 149 | 172 | 125 | 251 | 175 | 223 |
| D2S1790 | GATA88G05 | 4 | 146 | 135 | 196 | 196 | 150 | 213 | 227 | 183 | 233 | 224 |
| D2S2944 | GATA30E06 | 4 | 307 | 364 | 285 | 346 | 354 | 288 | 342 | 323 | 362 | 350 |
| D2S2952 | GATA116B01 | 4 | 312 | 283 | 197 | 143 | 291 | 186 | 147 | 203 | 190 | 259 |
| D2S2968 | GATA178G09 | 4 | 18 | 147 | 150 | 129 | 325 | 305 | 283 | 314 | 297 | 334 |
| D2S2972 | GATA176C01 | 4 | 124 | 340 | 235 | 322 | 269 | 205 | 267 | 285 | 327 | 246 |
| D2S2986 | 2QTEL47 | 2 | 15 | 17 | 11 | 16 | 13 | 9 | 10 | 33 | 40 | 75 |
| D2S405 | GATA8F07 | 4 | 296 | 280 | 153 | 318 | 254 | 157 | 344 | 223 | 298 | 270 |
| D2S410 | GATA4E11 | 4 | 131 | 100 | 212 | 32 | 145 | 233 | 43 | 212 | 72 | 79 |
| D2S427 | GATA12H10 | 4 | 111 | 235 | 176 | 267 | 360 | 236 | 239 | 300 | 237 | 202 |
| D2S434 | GATA4G12 | 4 | 189 | 362 | 333 | 199 | 367 | 346 | 306 | 365 | 317 | 322 |
| D2S441 | GATA8F03 | 4 | 50 | 111 | 2 | 24 | 107 | 6 | 26 | 5 | 25 | 25 |
| D3S1262 | AFM059XA9 | 2 | 267 | 63 | 60 | 162 | 44 | 47 | 122 | 46 | 105 | 85 |
| D3S1311 | AFM254VE1 | 2 | 17 | 2 | 40 | 115 | 4 | 70 | 196 | 24 | 49 | 143 |
| D3S1560 | AFM217XD6 | 2 | 19 | 70 | 82 | 229 | 76 | 42 | 254 | 45 | 196 | 115 |
| D3S1744 | GATA3C02 | 4 | 300 | 371 | 256 | 300 | 355 | 239 | 287 | 277 | 331 | 291 |
| D3S1746 | GATA8F01 | 4 | 90 | 93 | 35 | 189 | 137 | 39 | 233 | 76 | 235 | 120 |
| D3S1763 | GATA3H01 | 4 | 286 | 239 | 213 | 122 | 221 | 182 | 95 | 214 | 146 | 197 |
| D3S1764 | GATA4A10 | 4 | 264 | 257 | 257 | 123 | 279 | 306 | 170 | 287 | 205 | 250 |
| D3S1766 | GATA6F06 | 4 | 288 | 321 | 344 | 340 | 341 | 356 | 358 | 350 | 357 | 359 |
| D3S1768 | GATA8B05 | 4 | 281 | 44 | 122 | 62 | 55 | 134 | 85 | 140 | 76 | 123 |
| D3S2387 | GATA22G12 | 4 | 12 | 76 | 94 | 110 | 114 | 103 | 165 | 105 | 132 | 108 |
| D3S2398 | GATA6G12 | 4 | 102 | 163 | 151 | 304 | 293 | 250 | 356 | 274 | 342 | 348 |
| D3S2403 | GGAA4B09 | 4 | 25 | 123 | 162 | 144 | 80 | 118 | 86 | 189 | 189 | 209 |
| D3S2409 | ATA10H11 | 3 | 56 | 23 | 15 | 5 | 19 | 20 | 9 | 29 | 12 | 26 |
| D3S2418 | ATA22E01 | 3 | 41 | 222 | 44 | 161 | 238 | 30 | 174 | 54 | 228 | 52 |
| D3S2427 | GATA22F11 | 4 | 64 | 106 | 52 | 56 | 91 | 63 | 50 | 49 | 80 | 23 |
| D3S2432 | GATA27C08 | 4 | 251 | 300 | 305 | 278 | 310 | 283 | 220 | 299 | 245 | 315 |
| D3S2460 | GATA68F07 | 4 | 310 | 230 | 62 | 169 | 256 | 87 | 190 | 120 | 177 | 127 |
| D3S3038 | GATA73D01 | 4 | 340 | 206 | 282 | 286 | 182 | 249 | 277 | 264 | 318 | 317 |
| D3S3039 | GATA7F05 | 4 | 362 | 357 | 357 | 333 | 332 | 363 | 298 | 362 | 353 | 354 |
| D3S3045 | GATA84B12 | 4 | 42 | 223 | 46 | 12 | 127 | 61 | 16 | 85 | 22 | 35 |
| D3S3630 | AFMB296ZF5 | 2 | 61 | 261 | 98 | 171 | 208 | 117 | 198 | 108 | 206 | 162 |
| D3S3644 | AFMB318YF1 | 2 | 145 | 313 | 171 | 345 | 231 | 85 | 308 | 180 | 350 | 247 |
| D3S4523 | ATA34G06 | 3 | 160 | 89 | 184 | 8 | 88 | 192 | 8 | 137 | 4 | 41 |
| D3S4529 | GATA128C02 | 4 | 309 | 302 | 354 | 261 | 259 | 335 | 275 | 320 | 257 | 306 |
| D3S4545 | GATA164B08 | 4 | 65 | 42 | 208 | 61 | 31 | 163 | 46 | 75 | 67 | 81 |
| D4S1625 | GATA107 | 4 | 331 | 275 | 373 | 279 | 295 | 373 | 294 | 364 | 311 | 343 |
| D4S1627 | GATA7D01 | 4 | 334 | 174 | 135 | 326 | 219 | 160 | 350 | 186 | 303 | 256 |
| D4S1629 | GATA8A05 | 4 | 209 | 324 | 353 | 276 | 152 | 291 | 173 | 340 | 285 | 301 |
| D4S1644 | GATA11E09 | 4 | 228 | 214 | 290 | 282 | 191 | 273 | 263 | 269 | 273 | 324 |
| D4S1647 | GATA2F11 | 4 | 229 | 191 | 216 | 210 | 190 | 226 | 252 | 226 | 166 | 274 |
| D4S1652 | GATA5B02 | 4 | 164 | 151 | 225 | 308 | 161 | 227 | 325 | 280 | 351 | 341 |
| D4S2361 | ATA2A03 | 3 | 181 | 310 | 173 | 73 | 317 | 159 | 52 | 207 | 95 | 105 |
| D4S2366 | GATA22G05 | 4 | 326 | 144 | 61 | 208 | 130 | 62 | 201 | 114 | 182 | 146 |
| D4S2367 | GATA24H01 | 4 | 154 | 83 | 41 | 328 | 66 | 34 | 311 | 52 | 211 | 131 |
| D4S2368 | GATA27G03 | 4 | 328 | 338 | 289 | 296 | 321 | 240 | 284 | 307 | 241 | 219 |
| D4S2394 | ATA26B08 | 3 | 178 | 119 | 18 | 179 | 103 | 16 | 141 | 18 | 178 | 47 |
| D4S2397 | ATA27C07 | 3 | 295 | 47 | 167 | 174 | 54 | 148 | 199 | 93 | 199 | 204 |
| D4S2417 | GATA42H02 | 4 | 254 | 247 | 195 | 89 | 245 | 175 | 84 | 173 | 84 | 177 |
| D4S2431 | GGAA19H07 | 4 | 23 | 77 | 99 | 249 | 47 | 88 | 208 | 146 | 169 | 212 |
| D4S2623 | GATA62A12 | 4 | 182 | 116 | 26 | 337 | 92 | 28 | 295 | 55 | 200 | 66 |
| D4S2632 | GATA72G09 | 4 | 7 | 24 | 48 | 99 | 10 | 32 | 79 | 41 | 55 | 113 |
| D4S3243 | GATA10G07 | 4 | 62 | 109 | 144 | 128 | 160 | 223 | 202 | 185 | 194 | 227 |
| D4S3248 | GATA28F03 | 4 | 330 | 290 | 336 | 145 | 316 | 349 | 145 | 327 | 207 | 242 |
| D4S3360 | 4PTEL04 | 2 | 205 | 149 | 283 | 108 | 179 | 341 | 116 | 260 | 136 | 213 |
| D4S403 | AFM157XG3 | 2 | 126 | 13 | 42 | 101 | 24 | 46 | 140 | 37 | 83 | 83 |
| D4S408 | AFM165XC11 | 2 | 52 | 28 | 21 | 146 | 17 | 23 | 133 | 25 | 79 | 84 |
| D5S1456 | GATA11A11 | 4 | 214 | 358 | 200 | 331 | 337 | 217 | 333 | 208 | 370 | 236 |
| D5S1457 | GATA21D04 | 4 | 139 | 234 | 341 | 177 | 153 | 317 | 236 | 306 | 133 | 298 |
| D5S1462 | GATA3H06 | 4 | 84 | 170 | 301 | 281 | 199 | 311 | 250 | 294 | 224 | 286 |
| D5S1470 | GATA7C06 | 4 | 155 | 153 | 125 | 271 | 117 | 143 | 266 | 125 | 176 | 198 |
| D5S1480 | ATA23A10 | 3 | 97 | 53 | 65 | 76 | 40 | 53 | 64 | 50 | 89 | 124 |
| D5S1501 | GATA52A12 | 4 | 1 | 72 | 9 | 149 | 34 | 7 | 89 | 12 | 184 | 46 |
| D5S1505 | GATA62A04 | 4 | 313 | 318 | 323 | 299 | 366 | 320 | 348 | 335 | 355 | 305 |
| D5S1725 | GATA89G08 | 4 | 273 | 336 | 163 | 363 | 371 | 178 | 345 | 246 | 363 | 254 |
| D5S211 | MFD154 | 2 | 4 | 15 | 126 | 4 | 35 | 206 | 11 | 109 | 31 | 36 |
| D5S2488 | ATA20G07 | 3 | 243 | 264 | 324 | 244 | 286 | 318 | 235 | 317 | 281 | 303 |
| D5S2500 | GATA67D03 | 4 | 316 | 328 | 207 | 360 | 329 | 203 | 349 | 253 | 359 | 280 |
| D5S2501 | GATA68A03 | 4 | 246 | 330 | 362 | 247 | 262 | 340 | 204 | 359 | 236 | 276 |
| D5S2505 | GATA84E11 | 4 | 194 | 157 | 332 | 223 | 184 | 313 | 206 | 242 | 179 | 287 |
| D5S2845 | GATA134B03 | 4 | 249 | 254 | 108 | 291 | 213 | 98 | 251 | 142 | 255 | 149 |
| D5S2849 | GATA145D10 | 4 | 319 | 98 | 262 | 178 | 108 | 265 | 172 | 219 | 141 | 195 |
| D5S408 | AFM164XB8 | 2 | 21 | 75 | 105 | 100 | 113 | 142 | 137 | 204 | 137 | 165 |
| D5S816 | GATA2H09 | 4 | 259 | 236 | 299 | 187 | 163 | 281 | 157 | 199 | 97 | 258 |
| D5S817 | GATA3E10 | 4 | 226 | 220 | 214 | 315 | 306 | 286 | 364 | 315 | 366 | 360 |
| D5S820 | GATA6E05 | 4 | 262 | 202 | 346 | 137 | 201 | 354 | 94 | 282 | 122 | 210 |
| D6S1006 | ATC4D09 | 3 | 47 | 22 | 13 | 48 | 79 | 43 | 127 | 53 | 112 | 118 |
| D6S1009 | GATA32B03 | 4 | 81 | 180 | 23 | 95 | 195 | 26 | 104 | 30 | 130 | 39 |
| D6S1017 | GGAT3H10 | 4 | 324 | 306 | 308 | 130 | 338 | 310 | 124 | 276 | 185 | 125 |
| D6S1021 | ATA11D10 | 3 | 171 | 152 | 288 | 185 | 134 | 274 | 221 | 220 | 117 | 265 |
| D6S1027 | ATA22G07 | 3 | 263 | 115 | 199 | 85 | 123 | 224 | 105 | 187 | 125 | 196 |
| D6S1031 | ATA28B11 | 3 | 72 | 57 | 56 | 213 | 41 | 35 | 212 | 63 | 174 | 138 |
| D6S1040 | GATA23F08 | 4 | 315 | 265 | 267 | 364 | 218 | 251 | 321 | 298 | 260 | 261 |
| D6S1051 | GATA61E03 | 4 | 354 | 369 | 300 | 301 | 363 | 295 | 299 | 325 | 326 | 338 |
| D6S1053 | GATA64D02 | 4 | 69 | 356 | 325 | 310 | 284 | 214 | 337 | 334 | 354 | 295 |
| D6S1056 | GATA68H04 | 4 | 141 | 226 | 103 | 79 | 283 | 58 | 129 | 80 | 188 | 37 |
| D6S1277 | GATA81B01 | 4 | 101 | 188 | 177 | 243 | 194 | 212 | 291 | 217 | 279 | 275 |
| D6S1959 | GATA29A01 | 4 | 371 | 359 | 372 | 154 | 344 | 367 | 146 | 367 | 219 | 238 |
| D6S2410 | GATA11E02 | 4 | 366 | 353 | 320 | 222 | 364 | 316 | 182 | 332 | 220 | 239 |
| D6S2436 | GATA165G02 | 4 | 117 | 297 | 260 | 221 | 237 | 235 | 192 | 283 | 231 | 207 |
| D6S2439 | GATA163B10 | 4 | 197 | 221 | 132 | 197 | 230 | 115 | 256 | 127 | 261 | 129 |
| D6S2522 | 6QTEL54 | 2 | 373 | 309 | 350 | 377 | 236 | 308 | 377 | 352 | 335 | 357 |
| D6S305 | AFM242ZG5 | 2 | 125 | 218 | 91 | 59 | 272 | 65 | 44 | 79 | 51 | 17 |
| D6S474 | GATA31 | 4 | 301 | 291 | 359 | 305 | 253 | 331 | 276 | 331 | 296 | 368 |
| D6S942 | UT654 | 4 | 289 | 278 | 311 | 343 | 235 | 270 | 329 | 361 | 371 | 372 |
| D7S1799 | GATA23F05 | 4 | 190 | 219 | 247 | 297 | 202 | 243 | 300 | 266 | 248 | 299 |
| D7S1802 | GATA41G07 | 4 | 355 | 348 | 369 | 332 | 324 | 352 | 293 | 351 | 322 | 361 |
| D7S1804 | GATA43C11 | 4 | 88 | 248 | 187 | 29 | 193 | 197 | 24 | 181 | 52 | 28 |
| D7S1808 | GGAA3F06 | 4 | 87 | 6 | 129 | 6 | 8 | 133 | 13 | 32 | 19 | 30 |
| D7S1818 | GATA24D12 | 4 | 166 | 322 | 358 | 138 | 346 | 370 | 169 | 375 | 252 | 296 |
| D7S1824 | GATA32C12 | 4 | 169 | 108 | 133 | 156 | 64 | 102 | 123 | 139 | 140 | 182 |
| D7S2204 | GATA73D10 | 4 | 24 | 134 | 142 | 188 | 94 | 106 | 177 | 126 | 193 | 175 |
| D7S2477 | AFMB035XB9 | 2 | 20 | 46 | 4 | 91 | 37 | 1 | 66 | 2 | 16 | 3 |
| D7S2846 | GATA31A10 | 4 | 346 | 341 | 277 | 329 | 359 | 304 | 347 | 295 | 334 | 297 |
| D7S3046 | GATA118G10 | 4 | 116 | 128 | 55 | 313 | 136 | 66 | 335 | 97 | 262 | 157 |
| D7S3047 | GATA119B03 | 4 | 372 | 377 | 367 | 233 | 375 | 359 | 218 | 370 | 280 | 327 |
| D7S3051 | GATA137H02 | 4 | 211 | 225 | 118 | 298 | 142 | 137 | 271 | 113 | 253 | 185 |
| D7S3056 | GATA24F03 | 4 | 208 | 237 | 233 | 284 | 217 | 216 | 229 | 243 | 286 | 320 |
| D7S3058 | GATA30D09 | 4 | 134 | 166 | 59 | 285 | 168 | 82 | 281 | 128 | 210 | 114 |
| D7S3061 | GGAA6D03 | 4 | 76 | 121 | 75 | 58 | 109 | 114 | 47 | 110 | 108 | 53 |
| D7S3070 | GATA189C06 | 4 | 308 | 295 | 309 | 238 | 304 | 328 | 241 | 324 | 271 | 313 |
| D7S559 | MFD265 | 2 | 257 | 199 | 298 | 136 | 206 | 264 | 159 | 235 | 139 | 214 |
| D7S817 | GATA13G11 | 4 | 347 | 133 | 253 | 54 | 128 | 262 | 39 | 244 | 21 | 51 |
| D7S820 | GATA3F01 | 4 | 302 | 238 | 296 | 134 | 233 | 228 | 115 | 247 | 163 | 211 |
| D7S821 | GATA5D08 | 4 | 151 | 244 | 97 | 200 | 307 | 121 | 217 | 144 | 259 | 206 |
| D8S1048 | UT7129 | 4 | 136 | 367 | 276 | 347 | 263 | 245 | 269 | 308 | 360 | 318 |
| D8S1108 | GATA50D10 | 4 | 248 | 334 | 304 | 306 | 376 | 334 | 334 | 338 | 346 | 356 |
| D8S1110 | GATA8G10 | 4 | 199 | 249 | 158 | 82 | 327 | 184 | 111 | 232 | 159 | 133 |
| D8S1113 | GGAA8G07 | 4 | 186 | 68 | 17 | 236 | 104 | 14 | 237 | 10 | 118 | 13 |
| D8S1128 | GATA21C12 | 4 | 79 | 30 | 138 | 66 | 25 | 105 | 56 | 20 | 14 | 100 |
| D8S1132 | GATA26E03 | 4 | 261 | 354 | 79 | 353 | 323 | 74 | 301 | 112 | 352 | 158 |
| D8S1136 | GATA41A01 | 4 | 241 | 210 | 361 | 232 | 220 | 371 | 265 | 312 | 263 | 312 |
| D8S1179 | GATA7G07 | 4 | 121 | 293 | 275 | 259 | 297 | 230 | 288 | 268 | 306 | 288 |
| D8S1477 | GGAA20C10 | 4 | 33 | 58 | 140 | 155 | 50 | 146 | 103 | 159 | 65 | 164 |
| D8S2324 | GATA14E09 | 4 | 114 | 351 | 268 | 280 | 322 | 231 | 264 | 293 | 315 | 248 |
| D8S261 | AFM123XG5 | 2 | 112 | 69 | 165 | 42 | 74 | 130 | 53 | 92 | 73 | 78 |
| D8S262 | AFM127XH2 | 2 | 210 | 276 | 104 | 211 | 249 | 145 | 248 | 124 | 238 | 161 |
| D8S373 | UT721 | 4 | 75 | 177 | 189 | 294 | 176 | 158 | 194 | 188 | 152 | 186 |
| D8S503 | AFM193XH4 | 2 | 74 | 25 | 58 | 23 | 33 | 68 | 34 | 48 | 37 | 32 |
| D8S560 | AFMA127YE5 | 2 | 27 | 80 | 6 | 57 | 68 | 4 | 59 | 4 | 62 | 7 |
| D8S592 | GATA6B02 | 4 | 360 | 347 | 202 | 338 | 358 | 220 | 354 | 272 | 347 | 292 |
| D9S1118 | GATA71E08 | 4 | 103 | 298 | 38 | 157 | 282 | 21 | 168 | 43 | 201 | 58 |
| D9S1120 | GATA81C04 | 4 | 338 | 315 | 315 | 45 | 258 | 284 | 37 | 322 | 48 | 93 |
| D9S1121 | GATA87E02 | 4 | 30 | 85 | 43 | 180 | 71 | 49 | 97 | 61 | 106 | 67 |
| D9S1122 | GATA89A11 | 4 | 368 | 327 | 318 | 212 | 330 | 319 | 214 | 347 | 292 | 289 |
| D9S1779 | AFM026TG9 | 2 | 2 | 19 | 110 | 52 | 7 | 54 | 35 | 82 | 59 | 97 |
| D9S1825 | AFMB029XG1 | 2 | 54 | 204 | 47 | 170 | 203 | 36 | 187 | 47 | 256 | 43 |
| D9S1838 | AFMB303ZG9 | 2 | 137 | 52 | 116 | 102 | 49 | 151 | 69 | 69 | 20 | 48 |
| D9S1871 | AFM345TA9 | 2 | 8 | 4 | 54 | 19 | 11 | 89 | 38 | 17 | 24 | 18 |
| D9S2157 | ATA59H06 | 3 | 51 | 7 | 106 | 34 | 12 | 153 | 60 | 72 | 11 | 90 |
| D9S2169 | GATA62F03 | 4 | 329 | 352 | 370 | 355 | 374 | 377 | 368 | 377 | 369 | 375 |
| D9S301 | GATA7D12 | 4 | 276 | 246 | 302 | 90 | 289 | 333 | 117 | 288 | 153 | 153 |
| D9S910 | ATA18A07 | 3 | 144 | 55 | 34 | 33 | 72 | 40 | 58 | 96 | 82 | 69 |
| D9S922 | GATA21F05 | 4 | 200 | 113 | 137 | 47 | 105 | 170 | 30 | 171 | 38 | 50 |
| D9S925 | GATA27A11 | 4 | 123 | 118 | 84 | 75 | 106 | 81 | 61 | 90 | 69 | 72 |
| D9S930 | GATA48D07 | 4 | 318 | 311 | 220 | 335 | 328 | 202 | 324 | 221 | 290 | 231 |
| D9S934 | GATA64G07 | 4 | 235 | 250 | 240 | 354 | 243 | 242 | 339 | 273 | 307 | 344 |
| D9S938 | GGAA22E01 | 4 | 227 | 59 | 25 | 2 | 67 | 24 | 2 | 19 | 6 | 1 |
| F13A1-D6S | SE30 | 4 | 5 | 5 | 14 | 80 | 2 | 12 | 80 | 1 | 33 | 11 |
| NA-D10S-1 | GATA121A08 | 4 | 265 | 269 | 219 | 359 | 296 | 254 | 371 | 240 | 345 | 333 |
| NA-D10S-2 | ATA103C06 | 3 | 238 | 160 | 119 | 94 | 139 | 120 | 108 | 192 | 155 | 184 |
| NA-D11S-1 | ATA34E08 | 3 | 168 | 38 | 172 | 140 | 46 | 190 | 171 | 160 | 157 | 225 |
| NA-D12S-1 | GATA49D12 | 4 | 59 | 71 | 32 | 292 | 90 | 37 | 195 | 35 | 127 | 76 |
| NA-D12S-2 | PAH | 4 | 233 | 286 | 224 | 113 | 271 | 209 | 121 | 252 | 151 | 180 |
| NA-D13S-1 | ATA5A09 | 3 | 266 | 182 | 74 | 39 | 101 | 52 | 27 | 74 | 91 | 34 |
| NA-D14S-1 | GATA193A07 | 4 | 70 | 31 | 186 | 104 | 29 | 173 | 88 | 83 | 74 | 156 |
| NA-D15S-1 | GATA50C03 | 4 | 317 | 252 | 329 | 257 | 248 | 315 | 232 | 313 | 278 | 321 |
| NA-D16S-1 | GATA138C05 | 4 | 99 | 181 | 114 | 202 | 200 | 107 | 219 | 147 | 214 | 181 |
| NA-D17S-1 | ATA78D02 | 3 | 206 | 35 | 157 | 17 | 45 | 112 | 15 | 84 | 32 | 62 |
| NA-D18S-1 | GATA178F11 | 4 | 177 | 296 | 228 | 235 | 357 | 327 | 327 | 326 | 329 | 369 |
| NA-D18S-2 | ATA82B02 | 3 | 202 | 96 | 36 | 55 | 95 | 50 | 67 | 62 | 43 | 19 |
| NA-D1S-1 | ATA79C10 | 3 | 369 | 173 | 255 | 28 | 155 | 222 | 21 | 143 | 30 | 57 |
| NA-D1S-2 | GATA124C08 | 4 | 193 | 285 | 161 | 321 | 290 | 156 | 315 | 205 | 343 | 262 |
| NA-D1S-3 | GATA133A08 | 4 | 216 | 307 | 259 | 250 | 331 | 263 | 261 | 310 | 312 | 281 |
| NA-D1S-4 | ATA42G12 | 3 | 348 | 266 | 294 | 153 | 301 | 294 | 144 | 297 | 180 | 294 |
| NA-D1S-5 | GATA124F08 | 4 | 192 | 213 | 281 | 239 | 281 | 361 | 312 | 316 | 289 | 353 |
| NA-D22S-1 | GATA198B05 | 4 | 237 | 107 | 149 | 69 | 156 | 188 | 91 | 157 | 100 | 151 |
| NA-D4S-1 | GATA70E01 | 4 | 207 | 120 | 322 | 263 | 110 | 344 | 304 | 258 | 212 | 325 |
| NA-D5S-1 | ATA52D02 | 3 | 78 | 117 | 160 | 287 | 162 | 185 | 286 | 202 | 213 | 208 |
| NA-D6S-1 | GATA184A08 | 4 | 225 | 97 | 148 | 240 | 99 | 161 | 242 | 169 | 204 | 245 |
| NA-D7S-1 | GATA104 | 4 | 119 | 287 | 198 | 242 | 264 | 195 | 209 | 239 | 277 | 183 |
| NA-D8S-1 | GATA151F02 | 4 | 204 | 231 | 209 | 268 | 241 | 225 | 262 | 196 | 305 | 234 |
| NA-D8S-2 | GAAT1A4 | 4 | 107 | 205 | 85 | 339 | 265 | 76 | 366 | 115 | 332 | 179 |
| NA-D9S-1 | GATA187D09 | 4 | 224 | 245 | 169 | 252 | 303 | 179 | 285 | 225 | 304 | 285 |
| TPO-D2S | SRA | 4 | 272 | 355 | 170 | 118 | 361 | 155 | 102 | 182 | 138 | 87 |
Note.— For each data set (table 4), markers are ranked from most informative (1) to least informative (377). Names for the markers are the same as in supplementary table 4 of Rosenberg et al. (2002). The regions are coded as follows: 1 = Africa; 2 = Europe; 3 = Middle East; 4 = Central/South Asia; 5 = East Asia; 6 = Oceania; 7 = America. Loci are sorted in lexical order.
Table E.
Informativeness (In) for Distinguishing among Three Regions (Third of Three Parts)[Note]
|
In Rank for Set of Three Regions |
||||||||||||
| Locus Name | Alternate Name | RepeatSize | 3-4-5 | 3-4-6 | 3-4-7 | 3-5-6 | 3-5-7 | 3-6-7 | 4-5-6 | 4-5-7 | 4-6-7 | 5-6-7 |
| D10S1208 | ATA5A04 | 3 | 154 | 20 | 303 | 53 | 343 | 107 | 39 | 273 | 93 | 96 |
| D10S1221 | ATA21A03 | 3 | 14 | 129 | 100 | 58 | 45 | 150 | 102 | 55 | 174 | 147 |
| D10S1222 | ATA22D02 | 3 | 137 | 227 | 150 | 253 | 117 | 241 | 234 | 102 | 215 | 224 |
| D10S1225 | ATA24F10 | 3 | 360 | 121 | 199 | 173 | 258 | 257 | 119 | 202 | 204 | 234 |
| D10S1230 | ATA29C03 | 3 | 105 | 243 | 215 | 220 | 153 | 317 | 150 | 115 | 309 | 211 |
| D10S1239 | GATA64A09 | 4 | 190 | 278 | 217 | 275 | 241 | 305 | 322 | 325 | 345 | 367 |
| D10S1248 | GGAA23C05 | 4 | 233 | 225 | 301 | 197 | 323 | 211 | 188 | 256 | 212 | 149 |
| D10S1412 | ATA31G11 | 3 | 170 | 89 | 73 | 129 | 96 | 99 | 238 | 209 | 225 | 239 |
| D10S1423 | GATA70E11 | 4 | 255 | 336 | 271 | 293 | 268 | 320 | 280 | 310 | 305 | 297 |
| D10S1425 | GATA71C09 | 4 | 63 | 249 | 252 | 76 | 104 | 182 | 55 | 105 | 178 | 47 |
| D10S1426 | GATA73E11 | 4 | 343 | 332 | 348 | 368 | 375 | 373 | 359 | 354 | 353 | 370 |
| D10S1430 | GATA84C01 | 4 | 264 | 261 | 366 | 244 | 357 | 333 | 292 | 373 | 356 | 317 |
| D10S1432 | GATA87G01 | 4 | 326 | 302 | 369 | 307 | 373 | 354 | 310 | 377 | 342 | 336 |
| D10S1435 | GATA88F09 | 4 | 314 | 368 | 312 | 358 | 312 | 355 | 372 | 307 | 352 | 335 |
| D10S189 | AFM063XF4 | 2 | 219 | 157 | 232 | 323 | 293 | 303 | 165 | 162 | 170 | 251 |
| D10S212 | AFM198ZB4 | 2 | 283 | 197 | 330 | 238 | 261 | 237 | 281 | 264 | 248 | 222 |
| D10S2327 | GGAT1A4 | 4 | 229 | 208 | 293 | 172 | 273 | 300 | 118 | 220 | 221 | 136 |
| D10S2470 | GATA115E01 | 4 | 227 | 237 | 161 | 301 | 87 | 125 | 249 | 73 | 101 | 59 |
| D10S677 | GGAA2F11 | 4 | 119 | 223 | 62 | 214 | 44 | 93 | 277 | 40 | 109 | 103 |
| D11S1304 | UT2095 | 4 | 22 | 202 | 88 | 28 | 55 | 108 | 22 | 54 | 104 | 29 |
| D11S1392 | GATA6B09 | 4 | 356 | 373 | 322 | 364 | 336 | 349 | 365 | 333 | 350 | 329 |
| D11S1981 | GATA48E02 | 4 | 147 | 214 | 204 | 233 | 176 | 229 | 186 | 108 | 186 | 128 |
| D11S1984 | GGAA17G05 | 4 | 11 | 63 | 130 | 18 | 95 | 90 | 9 | 81 | 55 | 53 |
| D11S1986 | GGAA7G08 | 4 | 46 | 110 | 47 | 95 | 71 | 69 | 138 | 67 | 71 | 79 |
| D11S1993 | ATA1B07 | 3 | 36 | 84 | 5 | 49 | 6 | 11 | 94 | 31 | 25 | 62 |
| D11S1998 | GATA23E06 | 4 | 213 | 316 | 164 | 284 | 161 | 325 | 299 | 140 | 273 | 191 |
| D11S1999 | GATA23F06 | 4 | 38 | 311 | 37 | 131 | 84 | 95 | 126 | 76 | 85 | 122 |
| D11S2000 | GATA28D01 | 4 | 138 | 60 | 70 | 81 | 68 | 49 | 91 | 62 | 63 | 30 |
| D11S2002 | GATA30G01 | 4 | 118 | 145 | 191 | 215 | 212 | 251 | 237 | 237 | 259 | 349 |
| D11S2006 | GATA46A12 | 4 | 367 | 344 | 363 | 352 | 369 | 359 | 362 | 375 | 368 | 361 |
| D11S2362 | ATA33B03 | 3 | 217 | 253 | 156 | 296 | 211 | 258 | 193 | 119 | 172 | 172 |
| D11S2363 | GATA12F04 | 4 | 186 | 203 | 136 | 266 | 198 | 212 | 215 | 171 | 188 | 246 |
| D11S2365 | GATA63F09 | 4 | 371 | 365 | 195 | 365 | 243 | 324 | 369 | 223 | 324 | 321 |
| D11S2371 | GATA90D07 | 4 | 266 | 338 | 358 | 315 | 298 | 341 | 288 | 246 | 311 | 256 |
| D11S4459 | ATA9B04 | 3 | 212 | 192 | 99 | 267 | 133 | 167 | 229 | 153 | 181 | 259 |
| D11S4463 | GATA117D01 | 4 | 189 | 314 | 249 | 370 | 351 | 356 | 278 | 203 | 298 | 338 |
| D11S4464 | GATA64D03 | 4 | 279 | 242 | 245 | 246 | 278 | 236 | 315 | 312 | 284 | 287 |
| D11S969 | AFM205VF10 | 2 | 232 | 123 | 290 | 103 | 302 | 152 | 72 | 276 | 119 | 101 |
| D12S1042 | ATA27A06 | 3 | 99 | 127 | 49 | 191 | 39 | 43 | 210 | 56 | 68 | 66 |
| D12S1045 | ATA29A06 | 3 | 26 | 52 | 19 | 54 | 18 | 41 | 113 | 41 | 74 | 260 |
| D12S1052 | GATA26D02 | 4 | 362 | 342 | 137 | 356 | 157 | 178 | 350 | 113 | 137 | 126 |
| D12S1064 | GATA63D12 | 4 | 195 | 139 | 236 | 211 | 286 | 254 | 175 | 278 | 247 | 342 |
| D12S1294 | GATA73H09 | 4 | 82 | 290 | 133 | 162 | 100 | 225 | 159 | 78 | 197 | 108 |
| D12S1300 | GATA85A04 | 4 | 242 | 38 | 368 | 80 | 359 | 153 | 65 | 368 | 149 | 206 |
| D12S1301 | GATA91H06 | 4 | 39 | 85 | 76 | 59 | 10 | 55 | 40 | 12 | 28 | 14 |
| D12S1638 | AFMB002VD5 | 2 | 298 | 134 | 180 | 210 | 231 | 202 | 197 | 164 | 159 | 186 |
| D12S2070 | ATA25F09 | 3 | 12 | 73 | 1 | 14 | 2 | 2 | 43 | 5 | 5 | 10 |
| D12S2078 | GATA32F05 | 4 | 191 | 170 | 286 | 134 | 203 | 158 | 142 | 235 | 164 | 117 |
| D12S269 | MFD259 | 2 | 143 | 87 | 35 | 115 | 70 | 66 | 97 | 64 | 75 | 86 |
| D12S297 | UT5029 | 4 | 128 | 41 | 21 | 118 | 65 | 82 | 86 | 43 | 66 | 201 |
| D12S372 | GATA4H03 | 4 | 300 | 330 | 168 | 372 | 219 | 276 | 370 | 263 | 296 | 305 |
| D12S373 | GATA6C01 | 4 | 144 | 72 | 341 | 149 | 221 | 143 | 117 | 190 | 117 | 140 |
| D12S395 | GATA4H01 | 4 | 91 | 95 | 329 | 117 | 226 | 196 | 130 | 257 | 218 | 275 |
| D13S1493 | GGAA29H03 | 4 | 240 | 155 | 221 | 146 | 237 | 141 | 166 | 315 | 182 | 163 |
| D13S1807 | GATA11C08 | 4 | 276 | 292 | 81 | 283 | 93 | 249 | 257 | 86 | 191 | 160 |
| D13S285 | AFM309VA9 | 2 | 69 | 109 | 265 | 78 | 184 | 215 | 76 | 198 | 196 | 102 |
| D13S317 | GATA7G10 | 4 | 98 | 282 | 42 | 183 | 56 | 59 | 179 | 37 | 40 | 48 |
| D13S779 | ATA26D07 | 3 | 235 | 257 | 131 | 202 | 151 | 110 | 187 | 126 | 95 | 72 |
| D13S787 | GATA23C03 | 4 | 338 | 320 | 336 | 343 | 377 | 366 | 314 | 337 | 331 | 339 |
| D13S793 | GATA43H03 | 4 | 377 | 77 | 201 | 96 | 236 | 171 | 64 | 212 | 139 | 127 |
| D13S796 | GATA51B02 | 4 | 306 | 76 | 269 | 123 | 274 | 230 | 103 | 271 | 251 | 238 |
| D13S800 | GATA64F08 | 4 | 274 | 240 | 311 | 182 | 344 | 224 | 167 | 340 | 239 | 171 |
| D13S894 | GATA86H01 | 4 | 208 | 353 | 292 | 297 | 319 | 338 | 269 | 284 | 323 | 316 |
| D13S895 | GGAA22G01 | 4 | 218 | 287 | 283 | 285 | 330 | 309 | 239 | 342 | 283 | 299 |
| D14S1007 | AFMB002ZF1 | 2 | 1 | 12 | 17 | 4 | 1 | 5 | 29 | 8 | 17 | 34 |
| D14S1280 | GATA31B09 | 4 | 341 | 276 | 319 | 308 | 267 | 260 | 286 | 265 | 262 | 194 |
| D14S1426 | GATA136B01 | 4 | 115 | 111 | 26 | 112 | 14 | 8 | 59 | 10 | 7 | 3 |
| D14S1434 | GATA168F06 | 4 | 193 | 258 | 250 | 227 | 208 | 268 | 291 | 248 | 325 | 253 |
| D14S587 | GGAA10C09 | 4 | 160 | 217 | 111 | 152 | 166 | 135 | 176 | 135 | 141 | 105 |
| D14S588 | GGAA4A12 | 4 | 214 | 104 | 169 | 68 | 143 | 122 | 42 | 106 | 111 | 51 |
| D14S592 | ATA19H08 | 3 | 204 | 113 | 194 | 109 | 194 | 180 | 104 | 231 | 165 | 132 |
| D14S599 | ATA29G03 | 3 | 41 | 193 | 6 | 72 | 19 | 17 | 47 | 9 | 9 | 21 |
| D14S606 | GATA30A03 | 4 | 226 | 167 | 270 | 226 | 281 | 273 | 218 | 298 | 278 | 362 |
| D14S608 | GATA43H01 | 4 | 94 | 191 | 36 | 189 | 54 | 96 | 214 | 15 | 51 | 46 |
| D14S617 | GGAA21G11 | 4 | 209 | 11 | 192 | 11 | 215 | 25 | 14 | 201 | 39 | 27 |
| D14S742 | GATA74E02 | 4 | 277 | 334 | 339 | 348 | 371 | 370 | 333 | 345 | 360 | 376 |
| D15S128 | AFM273YF9 | 2 | 73 | 175 | 77 | 155 | 86 | 144 | 183 | 96 | 184 | 208 |
| D15S1507 | GATA151F03 | 4 | 249 | 264 | 372 | 251 | 329 | 328 | 244 | 360 | 336 | 290 |
| D15S1515 | GATA197B10 | 4 | 287 | 362 | 83 | 345 | 122 | 151 | 330 | 97 | 134 | 168 |
| D15S165 | AFM248VC5 | 2 | 15 | 44 | 67 | 43 | 41 | 72 | 85 | 77 | 120 | 197 |
| D15S642 | GATA27A03 | 4 | 10 | 182 | 116 | 44 | 64 | 168 | 68 | 94 | 226 | 210 |
| D15S643 | GATA50G06 | 4 | 173 | 131 | 178 | 204 | 209 | 259 | 136 | 154 | 175 | 249 |
| D15S652 | ATA24A08 | 3 | 111 | 70 | 25 | 102 | 25 | 14 | 110 | 21 | 16 | 25 |
| D15S655 | ATA28G05 | 3 | 108 | 56 | 337 | 86 | 218 | 126 | 144 | 280 | 152 | 170 |
| D15S659 | GATA63A03 | 4 | 312 | 201 | 159 | 237 | 150 | 103 | 171 | 155 | 81 | 60 |
| D15S816 | GATA73F01 | 4 | 176 | 340 | 365 | 249 | 265 | 348 | 216 | 234 | 348 | 202 |
| D15S818 | GATA85D02 | 4 | 311 | 319 | 280 | 333 | 288 | 313 | 337 | 290 | 310 | 300 |
| D15S822 | GATA88H02 | 4 | 30 | 158 | 8 | 114 | 17 | 48 | 190 | 45 | 87 | 146 |
| D16S2616 | ATA41E04 | 3 | 289 | 179 | 122 | 229 | 171 | 221 | 228 | 186 | 240 | 313 |
| D16S2621 | GATA71F09 | 4 | 42 | 268 | 110 | 100 | 92 | 162 | 143 | 161 | 222 | 137 |
| D16S2624 | GATA81D12 | 4 | 53 | 326 | 151 | 159 | 123 | 187 | 172 | 141 | 190 | 177 |
| D16S3253 | GATA22F09 | 4 | 184 | 281 | 144 | 281 | 199 | 286 | 199 | 136 | 180 | 187 |
| D16S3396 | ATA55A11 | 3 | 292 | 273 | 211 | 310 | 251 | 267 | 317 | 251 | 275 | 285 |
| D16S3401 | 16PTEL06 | 2 | 135 | 14 | 41 | 22 | 83 | 18 | 10 | 39 | 12 | 18 |
| D16S403 | AFM049XD2 | 2 | 125 | 8 | 45 | 6 | 60 | 28 | 2 | 83 | 15 | 15 |
| D16S422 | AFM249XC5 | 2 | 8 | 171 | 33 | 37 | 34 | 74 | 53 | 69 | 105 | 106 |
| D16S516 | AFM350VD1 | 2 | 55 | 213 | 114 | 105 | 91 | 138 | 125 | 116 | 147 | 129 |
| D16S539 | GATA11C06 | 4 | 260 | 270 | 247 | 254 | 291 | 240 | 205 | 277 | 219 | 200 |
| D16S748 | ATA3A07 | 3 | 48 | 30 | 53 | 26 | 26 | 57 | 131 | 80 | 153 | 111 |
| D16S753 | GGAA3G05 | 4 | 145 | 199 | 291 | 216 | 240 | 231 | 270 | 230 | 234 | 227 |
| D16S764 | GATA42E11 | 4 | 321 | 315 | 203 | 350 | 257 | 306 | 325 | 174 | 255 | 298 |
| D16S769 | GATA71H05 | 4 | 363 | 377 | 344 | 377 | 318 | 350 | 374 | 294 | 343 | 312 |
| D17S1290 | GATA49C09 | 4 | 282 | 198 | 93 | 258 | 156 | 132 | 259 | 117 | 129 | 134 |
| D17S1294 | GGAA9D03 | 4 | 295 | 333 | 332 | 354 | 363 | 375 | 353 | 348 | 372 | 375 |
| D17S1298 | GAAT2C03 | 4 | 113 | 325 | 349 | 272 | 262 | 374 | 340 | 292 | 373 | 353 |
| D17S1299 | GATA25A04 | 4 | 357 | 347 | 375 | 335 | 368 | 368 | 328 | 365 | 369 | 340 |
| D17S1301 | GATA28D11 | 4 | 243 | 259 | 373 | 255 | 337 | 323 | 287 | 330 | 351 | 286 |
| D17S1308 | GTAT1A05 | 4 | 114 | 26 | 240 | 75 | 234 | 147 | 62 | 253 | 154 | 284 |
| D17S2180 | ATC6A06 | 3 | 222 | 297 | 155 | 294 | 130 | 193 | 348 | 150 | 205 | 196 |
| D17S2193 | ATA43A10Z | 3 | 185 | 177 | 148 | 171 | 146 | 218 | 128 | 127 | 127 | 92 |
| D17S2195 | ATA58A02 | 3 | 251 | 36 | 224 | 67 | 201 | 63 | 63 | 214 | 53 | 58 |
| D17S2196 | GATA185H04 | 4 | 85 | 230 | 69 | 107 | 80 | 100 | 70 | 58 | 89 | 44 |
| D17S784 | AFM044XG3 | 2 | 64 | 195 | 167 | 127 | 46 | 164 | 67 | 33 | 135 | 28 |
| D17S974 | GATA8C04 | 4 | 359 | 196 | 198 | 248 | 280 | 139 | 181 | 238 | 103 | 107 |
| D18S1357 | ATA7D07 | 3 | 303 | 164 | 172 | 213 | 142 | 119 | 163 | 114 | 96 | 76 |
| D18S1364 | GATA7E12 | 4 | 324 | 181 | 209 | 224 | 295 | 186 | 184 | 239 | 148 | 167 |
| D18S1370 | ATA45G06 | 3 | 81 | 54 | 196 | 83 | 129 | 115 | 89 | 121 | 106 | 99 |
| D18S1371 | GATA177C03 | 4 | 365 | 252 | 206 | 328 | 204 | 179 | 293 | 226 | 200 | 165 |
| D18S1376 | GATA185C06 | 4 | 286 | 256 | 356 | 268 | 352 | 339 | 261 | 366 | 347 | 358 |
| D18S1390 | 18QTEL11 | 2 | 136 | 161 | 87 | 135 | 98 | 129 | 158 | 123 | 138 | 195 |
| D18S535 | GATA13 | 4 | 131 | 19 | 174 | 77 | 137 | 35 | 90 | 158 | 33 | 64 |
| D18S542 | GATA11A06 | 4 | 123 | 1 | 160 | 9 | 196 | 36 | 4 | 151 | 20 | 49 |
| D18S843 | ACT1A01 | 3 | 149 | 235 | 361 | 177 | 306 | 340 | 129 | 267 | 315 | 263 |
| D18S851 | GATA6D09 | 4 | 254 | 294 | 140 | 278 | 185 | 293 | 264 | 184 | 268 | 276 |
| D18S858 | ATA23G05 | 3 | 156 | 58 | 230 | 62 | 181 | 148 | 82 | 200 | 151 | 161 |
| D18S877 | GATA64H04 | 4 | 291 | 245 | 364 | 303 | 367 | 331 | 290 | 363 | 328 | 333 |
| D19S1034 | GATA21G05 | 4 | 192 | 246 | 118 | 243 | 116 | 206 | 296 | 109 | 214 | 221 |
| D19S245 | MFD235 | 4 | 368 | 371 | 374 | 375 | 370 | 369 | 373 | 364 | 370 | 352 |
| D19S246 | MFD232 | 4 | 171 | 133 | 327 | 151 | 270 | 172 | 111 | 225 | 156 | 109 |
| D19S254 | MFD238 | 4 | 354 | 75 | 165 | 116 | 178 | 53 | 98 | 139 | 34 | 32 |
| D19S433 | GGAA2A03 | 4 | 200 | 132 | 97 | 186 | 155 | 131 | 203 | 134 | 132 | 198 |
| D19S559 | UT7544 | 4 | 366 | 324 | 266 | 332 | 296 | 289 | 321 | 285 | 264 | 237 |
| D19S586 | GATA23B01 | 4 | 373 | 351 | 257 | 349 | 303 | 262 | 349 | 283 | 244 | 216 |
| D19S589 | GATA29B01 | 4 | 347 | 352 | 343 | 340 | 345 | 361 | 351 | 367 | 363 | 345 |
| D19S591 | GATA44F10 | 4 | 271 | 232 | 101 | 305 | 134 | 194 | 227 | 91 | 130 | 151 |
| D19S714 | GATA66B04 | 4 | 342 | 80 | 305 | 139 | 332 | 166 | 87 | 286 | 124 | 142 |
| D1S1589 | ATA4E02 | 3 | 133 | 46 | 71 | 29 | 23 | 26 | 19 | 17 | 23 | 5 |
| D1S1594 | GATA22D12 | 4 | 288 | 310 | 219 | 311 | 254 | 248 | 301 | 355 | 276 | 255 |
| D1S1596 | GATA26G09 | 4 | 322 | 349 | 227 | 336 | 245 | 288 | 368 | 218 | 265 | 231 |
| D1S1597 | GATA27E01 | 4 | 333 | 366 | 202 | 330 | 227 | 299 | 345 | 260 | 307 | 296 |
| D1S1609 | GATA50F11 | 4 | 239 | 318 | 68 | 274 | 51 | 117 | 274 | 26 | 94 | 41 |
| D1S1612 | GGAA3A07 | 4 | 28 | 100 | 40 | 89 | 52 | 84 | 60 | 19 | 47 | 183 |
| D1S1627 | ATA25E07 | 3 | 54 | 174 | 274 | 119 | 173 | 207 | 233 | 293 | 301 | 281 |
| D1S1653 | GATA43A04 | 4 | 345 | 360 | 307 | 363 | 328 | 365 | 371 | 343 | 366 | 374 |
| D1S1660 | GATA48B01 | 4 | 182 | 74 | 354 | 99 | 340 | 159 | 106 | 304 | 143 | 158 |
| D1S1665 | GATA61A06 | 4 | 169 | 361 | 355 | 286 | 284 | 372 | 323 | 314 | 376 | 330 |
| D1S1677 | GGAA22G10 | 4 | 165 | 312 | 189 | 263 | 190 | 263 | 346 | 254 | 300 | 291 |
| D1S1679 | GGAA5F09 | 4 | 296 | 126 | 323 | 184 | 341 | 233 | 147 | 327 | 207 | 226 |
| D1S1728 | GATA109 | 4 | 141 | 79 | 200 | 98 | 188 | 127 | 202 | 270 | 229 | 258 |
| D1S2134 | GATA72H07 | 4 | 20 | 71 | 18 | 92 | 30 | 77 | 81 | 20 | 78 | 280 |
| D1S235 | AFM203YG9 | 2 | 34 | 99 | 182 | 24 | 103 | 136 | 57 | 156 | 199 | 81 |
| D1S2682 | AFMA272XC9 | 2 | 152 | 165 | 129 | 108 | 106 | 216 | 124 | 133 | 254 | 213 |
| D1S3462 | ATA29C07 | 3 | 79 | 187 | 333 | 154 | 186 | 245 | 303 | 227 | 294 | 220 |
| D1S3669 | GATA29A05 | 4 | 162 | 83 | 105 | 74 | 174 | 85 | 66 | 145 | 58 | 43 |
| D1S3720 | ATA47D07 | 3 | 103 | 49 | 214 | 46 | 149 | 56 | 78 | 148 | 48 | 37 |
| D1S3721 | GATA129H04 | 4 | 92 | 88 | 39 | 142 | 82 | 87 | 101 | 63 | 57 | 121 |
| D1S468 | AFM280WE5 | 2 | 158 | 172 | 10 | 125 | 7 | 29 | 105 | 4 | 35 | 13 |
| D1S518 | GATA7C01 | 4 | 86 | 47 | 82 | 60 | 94 | 79 | 112 | 159 | 114 | 215 |
| D1S534 | GATA12A07 | 4 | 13 | 51 | 28 | 33 | 29 | 61 | 16 | 23 | 36 | 90 |
| D1S549 | GATA4H09 | 4 | 257 | 98 | 254 | 143 | 300 | 112 | 123 | 249 | 92 | 84 |
| D1S551 | GATA6A05 | 4 | 329 | 117 | 255 | 167 | 228 | 104 | 139 | 261 | 83 | 74 |
| D20S103 | AFM077XD3 | 2 | 24 | 55 | 108 | 19 | 40 | 81 | 38 | 95 | 126 | 97 |
| D20S1143 | GATA129B03 | 4 | 340 | 303 | 360 | 289 | 358 | 321 | 284 | 358 | 322 | 294 |
| D20S159 | UT1307 | 4 | 75 | 269 | 127 | 164 | 145 | 161 | 164 | 93 | 157 | 88 |
| D20S164 | UT1772 | 4 | 19 | 105 | 13 | 91 | 3 | 42 | 140 | 3 | 54 | 75 |
| D20S201 | GATA8B01 | 4 | 159 | 266 | 66 | 176 | 108 | 64 | 146 | 103 | 52 | 45 |
| D20S451 | UT254 | 4 | 83 | 239 | 246 | 203 | 213 | 301 | 312 | 332 | 354 | 360 |
| D20S477 | GATA29F06 | 4 | 78 | 96 | 128 | 136 | 170 | 163 | 88 | 169 | 140 | 247 |
| D20S478 | GATA42A03 | 4 | 122 | 346 | 259 | 298 | 255 | 347 | 275 | 187 | 318 | 274 |
| D20S480 | GATA45B10 | 4 | 332 | 375 | 328 | 337 | 322 | 344 | 334 | 318 | 335 | 307 |
| D20S481 | GATA47F05 | 4 | 127 | 142 | 234 | 70 | 168 | 142 | 51 | 179 | 107 | 52 |
| D20S482 | GATA51D03 | 4 | 293 | 358 | 376 | 344 | 362 | 377 | 327 | 346 | 374 | 354 |
| D20S851 | AFMA218YB5 | 2 | 33 | 9 | 34 | 12 | 15 | 4 | 5 | 2 | 4 | 2 |
| D21S1411 | UT1355 | 4 | 107 | 169 | 59 | 239 | 32 | 44 | 114 | 18 | 26 | 17 |
| D21S1432 | GATA11C12 | 4 | 139 | 194 | 241 | 163 | 148 | 280 | 169 | 163 | 299 | 188 |
| D21S1437 | GGAA3C07 | 4 | 148 | 219 | 282 | 170 | 193 | 253 | 192 | 152 | 233 | 98 |
| D21S1440 | ATA27F01 | 3 | 269 | 307 | 278 | 312 | 315 | 319 | 352 | 362 | 361 | 332 |
| D21S1446 | GATA70B08 | 4 | 216 | 94 | 273 | 137 | 253 | 170 | 161 | 329 | 223 | 240 |
| D21S2052 | GATA129D11 | 4 | 228 | 136 | 260 | 169 | 287 | 208 | 93 | 252 | 145 | 169 |
| D21S2055 | GATA188F04 | 4 | 110 | 32 | 30 | 38 | 42 | 10 | 35 | 42 | 10 | 19 |
| D22S1045 | ATA37D06 | 3 | 155 | 210 | 176 | 205 | 124 | 281 | 201 | 104 | 232 | 154 |
| D22S1169 | AFMB337ZH9 | 2 | 84 | 4 | 112 | 8 | 97 | 34 | 7 | 85 | 30 | 55 |
| D22S345 | MFD313 | 2 | 104 | 267 | 38 | 157 | 59 | 80 | 200 | 68 | 91 | 83 |
| D22S532 | UT7136 | 4 | 320 | 247 | 370 | 277 | 327 | 284 | 309 | 311 | 280 | 236 |
| D22S683 | GATA11B12 | 4 | 52 | 29 | 16 | 87 | 22 | 15 | 49 | 13 | 11 | 16 |
| D22S686 | GGAA10F06 | 4 | 49 | 218 | 313 | 180 | 206 | 322 | 222 | 269 | 355 | 318 |
| D22S689 | GATA21F03 | 4 | 246 | 224 | 238 | 241 | 238 | 219 | 251 | 268 | 253 | 225 |
| D2S1328 | GATA27A12 | 4 | 47 | 16 | 316 | 34 | 195 | 88 | 46 | 242 | 100 | 180 |
| D2S1334 | GATA4D07 | 4 | 76 | 286 | 120 | 168 | 132 | 160 | 173 | 89 | 162 | 91 |
| D2S1352 | ATA27D04 | 3 | 210 | 118 | 277 | 141 | 301 | 264 | 107 | 224 | 261 | 209 |
| D2S1353 | ATA27H09 | 3 | 259 | 212 | 85 | 300 | 144 | 155 | 236 | 88 | 112 | 178 |
| D2S1356 | ATA4F03 | 3 | 101 | 67 | 3 | 104 | 11 | 6 | 52 | 1 | 3 | 11 |
| D2S1360 | GATA11H10 | 4 | 25 | 272 | 147 | 122 | 118 | 265 | 54 | 71 | 201 | 205 |
| D2S1363 | GATA23D03 | 4 | 117 | 7 | 175 | 30 | 119 | 37 | 20 | 185 | 60 | 68 |
| D2S1384 | GATA52A04 | 4 | 50 | 205 | 126 | 145 | 113 | 134 | 177 | 258 | 173 | 162 |
| D2S1391 | GATA65C03 | 4 | 261 | 323 | 134 | 318 | 187 | 238 | 318 | 166 | 237 | 277 |
| D2S1394 | GATA69E12 | 4 | 370 | 370 | 350 | 373 | 361 | 367 | 376 | 369 | 375 | 372 |
| D2S1399 | GGAA20G04 | 4 | 183 | 106 | 48 | 93 | 62 | 50 | 99 | 51 | 56 | 40 |
| D2S1400 | GGAA20G10 | 4 | 106 | 220 | 89 | 259 | 127 | 210 | 320 | 195 | 293 | 365 |
| D2S1776 | GATA71D01 | 4 | 215 | 289 | 146 | 265 | 200 | 191 | 272 | 183 | 211 | 212 |
| D2S1780 | GATA72G11 | 4 | 349 | 369 | 302 | 360 | 326 | 335 | 367 | 326 | 338 | 314 |
| D2S1788 | GATA86E02 | 4 | 151 | 173 | 75 | 235 | 114 | 156 | 162 | 84 | 125 | 189 |
| D2S1790 | GATA88G05 | 4 | 179 | 262 | 248 | 257 | 244 | 266 | 232 | 244 | 270 | 303 |
| D2S2944 | GATA30E06 | 4 | 375 | 313 | 359 | 341 | 376 | 360 | 347 | 371 | 357 | 366 |
| D2S2952 | GATA116B01 | 4 | 301 | 168 | 104 | 178 | 131 | 198 | 152 | 112 | 185 | 184 |
| D2S2968 | GATA178G09 | 4 | 142 | 163 | 142 | 208 | 182 | 220 | 358 | 350 | 365 | 368 |
| D2S2972 | GATA176C01 | 4 | 331 | 260 | 279 | 329 | 334 | 282 | 304 | 287 | 246 | 252 |
| D2S2986 | 2QTEL47 | 2 | 23 | 5 | 11 | 36 | 43 | 65 | 41 | 50 | 72 | 268 |
| D2S405 | GATA8F07 | 4 | 307 | 156 | 346 | 230 | 316 | 272 | 209 | 303 | 282 | 279 |
| D2S410 | GATA4E11 | 4 | 109 | 207 | 50 | 187 | 73 | 73 | 276 | 129 | 110 | 143 |
| D2S427 | GATA12H10 | 4 | 201 | 149 | 256 | 225 | 239 | 201 | 265 | 189 | 166 | 150 |
| D2S434 | GATA4G12 | 4 | 302 | 304 | 212 | 331 | 232 | 275 | 341 | 324 | 330 | 308 |
| D2S441 | GATA8F03 | 4 | 116 | 2 | 20 | 3 | 21 | 21 | 12 | 47 | 65 | 63 |
| D3S1262 | AFM059XA9 | 2 | 44 | 62 | 138 | 55 | 112 | 113 | 21 | 72 | 69 | 80 |
| D3S1311 | AFM254VE1 | 2 | 2 | 59 | 143 | 15 | 24 | 121 | 32 | 57 | 216 | 89 |
| D3S1560 | AFM217XD6 | 2 | 126 | 101 | 294 | 101 | 229 | 169 | 83 | 323 | 146 | 116 |
| D3S1744 | GATA3C02 | 4 | 361 | 275 | 288 | 295 | 325 | 304 | 271 | 296 | 288 | 269 |
| D3S1746 | GATA8F01 | 4 | 120 | 45 | 222 | 82 | 205 | 120 | 108 | 282 | 144 | 217 |
| D3S1763 | GATA3H01 | 4 | 280 | 250 | 162 | 299 | 230 | 311 | 252 | 172 | 263 | 344 |
| D3S1764 | GATA4A10 | 4 | 248 | 244 | 125 | 264 | 167 | 205 | 306 | 228 | 281 | 282 |
| D3S1766 | GATA6F06 | 4 | 369 | 364 | 362 | 371 | 372 | 364 | 361 | 370 | 359 | 346 |
| D3S1768 | GATA8B05 | 4 | 35 | 116 | 65 | 113 | 61 | 101 | 127 | 75 | 123 | 223 |
| D3S2387 | GATA22G12 | 4 | 66 | 68 | 123 | 69 | 110 | 86 | 100 | 194 | 113 | 73 |
| D3S2398 | GATA6G12 | 4 | 178 | 140 | 276 | 147 | 246 | 239 | 273 | 331 | 317 | 304 |
| D3S2403 | GGAA4B09 | 4 | 129 | 150 | 94 | 218 | 183 | 204 | 145 | 101 | 118 | 267 |
| D3S2409 | ATA10H11 | 3 | 21 | 17 | 7 | 20 | 13 | 22 | 50 | 30 | 70 | 261 |
| D3S2418 | ATA22E01 | 3 | 207 | 21 | 158 | 51 | 216 | 47 | 24 | 250 | 31 | 35 |
| D3S2427 | GATA22F11 | 4 | 87 | 57 | 46 | 41 | 74 | 19 | 37 | 61 | 19 | 22 |
| D3S2432 | GATA27C08 | 4 | 319 | 306 | 258 | 321 | 290 | 351 | 295 | 204 | 306 | 295 |
| D3S2460 | GATA68F07 | 4 | 244 | 78 | 171 | 110 | 165 | 123 | 155 | 196 | 158 | 182 |
| D3S3038 | GATA73D01 | 4 | 161 | 274 | 264 | 271 | 292 | 329 | 208 | 240 | 292 | 351 |
| D3S3039 | GATA7F05 | 4 | 339 | 356 | 308 | 355 | 365 | 357 | 343 | 316 | 333 | 350 |
| D3S3045 | GATA84B12 | 4 | 150 | 61 | 14 | 88 | 33 | 40 | 132 | 38 | 102 | 85 |
| D3S3630 | AFMB296ZF5 | 2 | 230 | 146 | 225 | 121 | 210 | 175 | 116 | 236 | 242 | 113 |
| D3S3644 | AFMB318YF1 | 2 | 225 | 93 | 310 | 222 | 364 | 291 | 135 | 357 | 227 | 322 |
| D3S4523 | ATA34G06 | 3 | 130 | 236 | 12 | 206 | 5 | 71 | 254 | 7 | 84 | 69 |
| D3S4529 | GATA128C02 | 4 | 337 | 359 | 326 | 347 | 309 | 336 | 329 | 322 | 334 | 302 |
| D3S4545 | GATA164B08 | 4 | 37 | 216 | 64 | 90 | 78 | 105 | 71 | 70 | 88 | 82 |
| D4S1625 | GATA107 | 4 | 270 | 374 | 285 | 353 | 305 | 346 | 366 | 321 | 341 | 347 |
| D4S1627 | GATA7D01 | 4 | 187 | 153 | 342 | 185 | 294 | 255 | 243 | 351 | 303 | 320 |
| D4S1629 | GATA8A05 | 4 | 163 | 308 | 177 | 369 | 321 | 327 | 240 | 147 | 228 | 326 |
| D4S1644 | GATA11E09 | 4 | 250 | 343 | 298 | 306 | 308 | 353 | 336 | 319 | 362 | 371 |
| D4S1647 | GATA2F11 | 4 | 197 | 209 | 190 | 234 | 136 | 217 | 225 | 142 | 249 | 173 |
| D4S1652 | GATA5B02 | 4 | 112 | 222 | 272 | 240 | 269 | 302 | 260 | 299 | 316 | 357 |
| D4S2361 | ATA2A03 | 3 | 268 | 147 | 63 | 198 | 102 | 116 | 174 | 74 | 90 | 93 |
| D4S2366 | GATA22G05 | 4 | 177 | 81 | 226 | 158 | 225 | 176 | 154 | 210 | 160 | 265 |
| D4S2367 | GATA24H01 | 4 | 194 | 91 | 371 | 150 | 333 | 232 | 157 | 338 | 235 | 278 |
| D4S2368 | GATA27G03 | 4 | 297 | 254 | 304 | 304 | 249 | 256 | 250 | 207 | 203 | 152 |
| D4S2394 | ATA26B08 | 3 | 96 | 15 | 135 | 16 | 169 | 51 | 18 | 170 | 42 | 65 |
| D4S2397 | ATA27C07 | 3 | 40 | 162 | 181 | 84 | 159 | 199 | 77 | 211 | 210 | 262 |
| D4S2417 | GATA42H02 | 4 | 265 | 185 | 95 | 193 | 90 | 181 | 230 | 124 | 231 | 199 |
| D4S2431 | GGAA19H07 | 4 | 59 | 97 | 262 | 166 | 207 | 227 | 198 | 241 | 269 | 289 |
| D4S2623 | GATA62A12 | 4 | 140 | 31 | 325 | 64 | 233 | 76 | 56 | 188 | 61 | 67 |
| D4S2632 | GATA72G09 | 4 | 18 | 40 | 80 | 42 | 58 | 89 | 28 | 44 | 97 | 174 |
| D4S3243 | GATA10G07 | 4 | 121 | 151 | 113 | 148 | 128 | 133 | 241 | 221 | 272 | 233 |
| D4S3248 | GATA28F03 | 4 | 313 | 328 | 154 | 313 | 223 | 244 | 344 | 229 | 256 | 270 |
| D4S3360 | 4PTEL04 | 2 | 157 | 296 | 98 | 194 | 111 | 154 | 213 | 100 | 176 | 135 |
| D4S403 | AFM157XG3 | 2 | 9 | 34 | 91 | 21 | 63 | 68 | 45 | 99 | 98 | 139 |
| D4S408 | AFM165XC11 | 2 | 29 | 27 | 170 | 35 | 107 | 97 | 36 | 110 | 108 | 115 |
| D5S1456 | GATA11A11 | 4 | 317 | 221 | 300 | 219 | 349 | 228 | 221 | 361 | 257 | 193 |
| D5S1457 | GATA21D04 | 4 | 224 | 321 | 179 | 361 | 152 | 285 | 282 | 130 | 287 | 203 |
| D5S1462 | GATA3H06 | 4 | 263 | 337 | 318 | 342 | 317 | 342 | 357 | 300 | 327 | 315 |
| D5S1470 | GATA7C06 | 4 | 168 | 183 | 281 | 160 | 189 | 209 | 217 | 191 | 267 | 155 |
| D5S1480 | ATA23A10 | 3 | 27 | 50 | 58 | 31 | 69 | 98 | 11 | 28 | 64 | 118 |
| D5S1501 | GATA52A12 | 4 | 132 | 39 | 193 | 56 | 276 | 102 | 33 | 175 | 80 | 125 |
| D5S1505 | GATA62A04 | 4 | 323 | 327 | 296 | 327 | 313 | 292 | 324 | 341 | 290 | 271 |
| D5S1725 | GATA89G08 | 4 | 316 | 137 | 334 | 195 | 354 | 226 | 195 | 336 | 208 | 214 |
| D5S211 | MFD154 | 2 | 17 | 102 | 4 | 39 | 9 | 16 | 109 | 49 | 43 | 77 |
| D5S2488 | ATA20G07 | 3 | 309 | 341 | 275 | 325 | 307 | 332 | 326 | 302 | 313 | 323 |
| D5S2500 | GATA67D03 | 4 | 327 | 189 | 357 | 217 | 353 | 271 | 194 | 334 | 250 | 229 |
| D5S2501 | GATA68A03 | 4 | 281 | 350 | 183 | 366 | 214 | 279 | 363 | 168 | 260 | 243 |
| D5S2505 | GATA84E11 | 4 | 188 | 329 | 173 | 262 | 163 | 270 | 247 | 149 | 238 | 157 |
| D5S2845 | GATA134B03 | 4 | 267 | 112 | 251 | 165 | 248 | 145 | 149 | 213 | 128 | 133 |
| D5S2849 | GATA145D10 | 4 | 100 | 265 | 153 | 212 | 115 | 157 | 231 | 125 | 163 | 153 |
| D5S408 | AFM164XB8 | 2 | 72 | 107 | 163 | 179 | 162 | 188 | 206 | 215 | 202 | 266 |
| D5S816 | GATA2H09 | 4 | 153 | 300 | 184 | 207 | 105 | 294 | 196 | 87 | 297 | 144 |
| D5S817 | GATA3E10 | 4 | 203 | 226 | 314 | 250 | 304 | 315 | 335 | 372 | 371 | 369 |
| D5S820 | GATA6E05 | 4 | 237 | 367 | 124 | 316 | 177 | 252 | 311 | 111 | 198 | 181 |
| D6S1006 | ATC4D09 | 3 | 16 | 10 | 51 | 7 | 27 | 39 | 96 | 219 | 193 | 363 |
| D6S1009 | GATA32B03 | 4 | 284 | 25 | 157 | 25 | 175 | 45 | 25 | 193 | 62 | 39 |
| D6S1017 | GGAT3H10 | 4 | 325 | 331 | 107 | 270 | 160 | 124 | 267 | 138 | 99 | 78 |
| D6S1021 | ATA11D10 | 3 | 221 | 283 | 188 | 273 | 126 | 246 | 235 | 143 | 266 | 156 |
| D6S1027 | ATA22G07 | 3 | 95 | 184 | 84 | 161 | 99 | 137 | 178 | 128 | 179 | 228 |
| D6S1031 | ATA28B11 | 3 | 70 | 48 | 267 | 97 | 283 | 189 | 48 | 217 | 116 | 204 |
| D6S1040 | GATA23F08 | 4 | 253 | 285 | 335 | 319 | 279 | 296 | 316 | 222 | 271 | 232 |
| D6S1051 | GATA61E03 | 4 | 374 | 317 | 297 | 338 | 331 | 345 | 342 | 320 | 340 | 355 |
| D6S1053 | GATA64D02 | 4 | 234 | 188 | 287 | 339 | 338 | 295 | 223 | 359 | 258 | 273 |
| D6S1056 | GATA68H04 | 4 | 174 | 66 | 74 | 66 | 88 | 24 | 30 | 165 | 18 | 20 |
| D6S1277 | GATA81B01 | 4 | 202 | 176 | 289 | 153 | 242 | 223 | 185 | 279 | 291 | 241 |
| D6S1959 | GATA29A01 | 4 | 351 | 372 | 152 | 362 | 217 | 235 | 360 | 208 | 241 | 244 |
| D6S2410 | GATA11E02 | 4 | 358 | 335 | 213 | 346 | 247 | 277 | 356 | 206 | 245 | 242 |
| D6S2436 | GATA165G02 | 4 | 278 | 279 | 186 | 317 | 222 | 214 | 300 | 176 | 192 | 176 |
| D6S2439 | GATA163B10 | 4 | 223 | 166 | 228 | 192 | 224 | 140 | 137 | 262 | 142 | 114 |
| D6S2522 | 6QTEL54 | 2 | 256 | 322 | 377 | 367 | 347 | 363 | 308 | 305 | 337 | 324 |
| D6S305 | AFM242ZG5 | 2 | 252 | 86 | 43 | 94 | 36 | 20 | 73 | 29 | 13 | 9 |
| D6S474 | GATA31 | 4 | 348 | 354 | 306 | 357 | 335 | 371 | 355 | 328 | 358 | 364 |
| D6S942 | UT654 | 4 | 211 | 248 | 320 | 309 | 346 | 352 | 245 | 289 | 314 | 377 |
| D7S1799 | GATA23F05 | 4 | 241 | 251 | 352 | 269 | 264 | 316 | 283 | 291 | 326 | 264 |
| D7S1802 | GATA41G07 | 4 | 352 | 357 | 299 | 351 | 342 | 362 | 339 | 309 | 339 | 343 |
| D7S1804 | GATA43C11 | 4 | 175 | 130 | 29 | 188 | 79 | 33 | 122 | 36 | 21 | 31 |
| D7S1808 | GGAA3F06 | 4 | 4 | 138 | 9 | 23 | 16 | 30 | 44 | 35 | 49 | 95 |
| D7S1818 | GATA24D12 | 4 | 290 | 355 | 117 | 359 | 172 | 250 | 375 | 197 | 289 | 288 |
| D7S1824 | GATA32C12 | 4 | 93 | 141 | 166 | 199 | 202 | 242 | 134 | 146 | 195 | 327 |
| D7S2204 | GATA73D10 | 4 | 97 | 120 | 185 | 140 | 180 | 174 | 115 | 192 | 171 | 145 |
| D7S2477 | AFMB035XB9 | 2 | 74 | 3 | 72 | 10 | 35 | 3 | 6 | 32 | 2 | 1 |
| D7S2846 | GATA31A10 | 4 | 372 | 295 | 309 | 291 | 311 | 261 | 298 | 308 | 277 | 218 |
| D7S3046 | GATA118G10 | 4 | 198 | 92 | 347 | 144 | 314 | 195 | 180 | 335 | 217 | 230 |
| D7S3047 | GATA119B03 | 4 | 376 | 363 | 216 | 374 | 282 | 337 | 364 | 259 | 312 | 325 |
| D7S3051 | GATA137H02 | 4 | 180 | 135 | 268 | 138 | 275 | 184 | 121 | 199 | 187 | 123 |
| D7S3056 | GATA24F03 | 4 | 294 | 233 | 244 | 261 | 289 | 318 | 224 | 233 | 279 | 311 |
| D7S3058 | GATA30D09 | 4 | 231 | 82 | 324 | 133 | 260 | 118 | 151 | 247 | 122 | 100 |
| D7S3061 | GGAA6D03 | 4 | 80 | 90 | 56 | 61 | 81 | 32 | 84 | 66 | 41 | 54 |
| D7S3070 | GATA189C06 | 4 | 310 | 345 | 253 | 326 | 263 | 326 | 338 | 255 | 329 | 328 |
| D7S559 | MFD265 | 2 | 181 | 291 | 119 | 252 | 101 | 190 | 211 | 107 | 183 | 119 |
| D7S817 | GATA13G11 | 4 | 146 | 271 | 57 | 245 | 28 | 58 | 258 | 14 | 38 | 26 |
| D7S820 | GATA3F01 | 4 | 273 | 293 | 139 | 287 | 197 | 278 | 246 | 173 | 209 | 250 |
| D7S821 | GATA5D08 | 4 | 272 | 122 | 233 | 156 | 299 | 243 | 168 | 274 | 252 | 254 |
| D8S1048 | UT7129 | 4 | 330 | 299 | 284 | 334 | 360 | 334 | 331 | 317 | 308 | 337 |
| D8S1108 | GATA50D10 | 4 | 353 | 339 | 340 | 324 | 350 | 358 | 354 | 353 | 367 | 348 |
| D8S1110 | GATA8G10 | 4 | 258 | 143 | 90 | 201 | 121 | 111 | 220 | 167 | 131 | 164 |
| D8S1113 | GGAA8G07 | 4 | 71 | 6 | 261 | 2 | 135 | 13 | 3 | 160 | 8 | 4 |
| D8S1128 | GATA21C12 | 4 | 61 | 206 | 145 | 52 | 75 | 177 | 34 | 79 | 169 | 57 |
| D8S1132 | GATA26E03 | 4 | 318 | 64 | 295 | 106 | 356 | 149 | 92 | 301 | 115 | 124 |
| D8S1136 | GATA41A01 | 4 | 199 | 348 | 239 | 292 | 256 | 307 | 307 | 272 | 319 | 292 |
| D8S1179 | GATA7G07 | 4 | 346 | 301 | 243 | 320 | 285 | 298 | 289 | 297 | 285 | 257 |
| D8S1477 | GGAA20C10 | 4 | 56 | 152 | 106 | 175 | 76 | 146 | 219 | 59 | 136 | 131 |
| D8S2324 | GATA14E09 | 4 | 336 | 229 | 317 | 280 | 355 | 274 | 263 | 356 | 274 | 245 |
| D8S261 | AFM123XG5 | 2 | 58 | 125 | 52 | 73 | 67 | 62 | 75 | 98 | 73 | 87 |
| D8S262 | AFM127XH2 | 2 | 308 | 108 | 237 | 124 | 259 | 130 | 141 | 281 | 194 | 130 |
| D8S373 | UT721 | 4 | 172 | 180 | 207 | 228 | 164 | 200 | 248 | 132 | 167 | 112 |
| D8S503 | AFM193XH4 | 2 | 31 | 65 | 31 | 47 | 31 | 38 | 95 | 90 | 76 | 179 |
| D8S560 | AFMA127YE5 | 2 | 77 | 13 | 60 | 13 | 49 | 7 | 8 | 46 | 6 | 7 |
| D8S592 | GATA6B02 | 4 | 335 | 200 | 351 | 256 | 339 | 290 | 266 | 352 | 304 | 306 |
| D9S1118 | GATA71E08 | 4 | 247 | 18 | 149 | 48 | 191 | 67 | 15 | 180 | 44 | 38 |
| D9S1120 | GATA81C04 | 4 | 285 | 284 | 44 | 314 | 57 | 92 | 279 | 27 | 67 | 70 |
| D9S1121 | GATA87E02 | 4 | 60 | 33 | 132 | 32 | 109 | 60 | 27 | 65 | 37 | 33 |
| D9S1122 | GATA89A11 | 4 | 304 | 305 | 210 | 322 | 277 | 287 | 332 | 275 | 286 | 334 |
| D9S1779 | AFM026TG9 | 2 | 5 | 53 | 27 | 63 | 48 | 83 | 26 | 22 | 50 | 166 |
| D9S1825 | AFMB029XG1 | 2 | 205 | 35 | 187 | 45 | 235 | 46 | 31 | 288 | 45 | 36 |
| D9S1838 | AFMB303ZG9 | 2 | 57 | 128 | 86 | 65 | 50 | 54 | 79 | 16 | 32 | 12 |
| D9S1871 | AFM345TA9 | 2 | 3 | 42 | 22 | 5 | 8 | 9 | 13 | 34 | 22 | 23 |
| D9S2157 | ATA59H06 | 3 | 7 | 114 | 54 | 50 | 12 | 78 | 160 | 60 | 150 | 138 |
| D9S2169 | GATA62F03 | 4 | 355 | 376 | 367 | 376 | 366 | 376 | 377 | 376 | 377 | 373 |
| D9S301 | GATA7D12 | 4 | 245 | 288 | 92 | 236 | 120 | 114 | 268 | 144 | 133 | 104 |
| D9S910 | ATA18A07 | 3 | 45 | 23 | 55 | 57 | 72 | 52 | 74 | 92 | 77 | 207 |
| D9S922 | GATA21F05 | 4 | 68 | 144 | 23 | 126 | 20 | 31 | 212 | 11 | 29 | 61 |
| D9S925 | GATA27A11 | 4 | 164 | 103 | 78 | 111 | 89 | 94 | 120 | 82 | 82 | 110 |
| D9S930 | GATA48D07 | 4 | 350 | 190 | 345 | 223 | 320 | 234 | 191 | 295 | 189 | 148 |
| D9S934 | GATA64G07 | 4 | 262 | 255 | 331 | 282 | 310 | 343 | 297 | 306 | 344 | 331 |
| D9S938 | GGAA22E01 | 4 | 67 | 22 | 2 | 17 | 4 | 1 | 17 | 6 | 1 | 8 |
| F13A1-D6S | SE30 | 4 | 6 | 28 | 103 | 1 | 37 | 23 | 1 | 48 | 27 | 6 |
| NA-D10S-1 | GATA121A08 | 4 | 299 | 231 | 353 | 232 | 324 | 314 | 253 | 347 | 332 | 272 |
| NA-D10S-2 | ATA103C06 | 3 | 124 | 119 | 102 | 190 | 140 | 165 | 207 | 182 | 230 | 309 |
| NA-D11S-1 | ATA34E08 | 3 | 32 | 178 | 121 | 120 | 85 | 185 | 148 | 122 | 220 | 301 |
| NA-D12S-1 | GATA49D12 | 4 | 62 | 37 | 229 | 27 | 141 | 91 | 61 | 137 | 79 | 50 |
| NA-D12S-2 | PAH | 4 | 328 | 234 | 109 | 276 | 154 | 183 | 302 | 181 | 213 | 235 |
| NA-D13S-1 | ATA5A09 | 3 | 90 | 69 | 24 | 71 | 66 | 27 | 23 | 25 | 14 | 56 |
| NA-D14S-1 | GATA193A07 | 4 | 51 | 215 | 141 | 128 | 138 | 197 | 156 | 177 | 224 | 192 |
| NA-D15S-1 | GATA50C03 | 4 | 238 | 298 | 220 | 290 | 252 | 312 | 294 | 245 | 295 | 310 |
| NA-D16S-1 | GATA138C05 | 4 | 236 | 124 | 205 | 200 | 220 | 192 | 170 | 205 | 155 | 185 |
| NA-D17S-1 | ATA78D02 | 3 | 43 | 115 | 15 | 85 | 38 | 70 | 80 | 53 | 59 | 120 |
| NA-D18S-1 | GATA178F11 | 4 | 305 | 228 | 231 | 260 | 250 | 297 | 319 | 313 | 364 | 341 |
| NA-D18S-2 | ATA82B02 | 3 | 89 | 24 | 61 | 40 | 53 | 12 | 58 | 52 | 24 | 24 |
| NA-D1S-1 | ATA79C10 | 3 | 196 | 238 | 32 | 174 | 47 | 75 | 133 | 24 | 46 | 42 |
| NA-D1S-2 | GATA124C08 | 4 | 334 | 211 | 338 | 279 | 374 | 330 | 285 | 374 | 321 | 359 |
| NA-D1S-3 | GATA133A08 | 4 | 364 | 277 | 321 | 302 | 348 | 308 | 305 | 349 | 302 | 319 |
| NA-D1S-4 | ATA42G12 | 3 | 275 | 263 | 115 | 247 | 125 | 247 | 242 | 118 | 236 | 219 |
| NA-D1S-5 | GATA124F08 | 4 | 166 | 280 | 197 | 231 | 179 | 269 | 313 | 243 | 349 | 293 |
| NA-D22S-1 | GATA198B05 | 4 | 134 | 154 | 79 | 132 | 77 | 109 | 189 | 131 | 168 | 159 |
| NA-D4S-1 | GATA70E01 | 4 | 88 | 309 | 263 | 242 | 192 | 310 | 256 | 232 | 346 | 283 |
| NA-D5S-1 | ATA52D02 | 3 | 102 | 159 | 218 | 181 | 158 | 173 | 226 | 178 | 177 | 141 |
| NA-D6S-1 | GATA184A08 | 4 | 65 | 148 | 223 | 130 | 147 | 213 | 153 | 157 | 243 | 248 |
| NA-D7S-1 | GATA104 | 4 | 315 | 241 | 208 | 288 | 271 | 203 | 262 | 216 | 161 | 175 |
| NA-D8S-1 | GATA151F02 | 4 | 206 | 204 | 235 | 196 | 297 | 222 | 182 | 266 | 206 | 190 |
| NA-D8S-2 | GAAT1A4 | 4 | 167 | 43 | 315 | 79 | 266 | 128 | 69 | 339 | 121 | 94 |
| NA-D9S-1 | GATA187D09 | 4 | 220 | 160 | 242 | 209 | 272 | 283 | 255 | 344 | 320 | 356 |
| TPO-D2S | SRA | 4 | 344 | 186 | 96 | 221 | 139 | 106 | 204 | 120 | 86 | 71 |
Note.— For each data set (table 4), markers are ranked from most informative (1) to least informative (377). Names for the markers are the same as in supplementary table 4 of Rosenberg et al. (2002). The regions are coded as follows: 1 = Africa; 2 = Europe; 3 = Middle East; 4 = Central/South Asia; 5 = East Asia; 6 = Oceania; 7 = America. Loci are sorted in lexical order.
Theory
Consider populations i=1,2,…,K and loci l=1,2,…,L, with K⩾2 and L⩾1. Locus l has alleles j=1,2,…,N(l). The relative frequency for allele j of locus l in population i is p(l)ij; this quantity represents a parametric rather than a sample frequency. The (parametric) average frequency of allele j at locus l is defined as
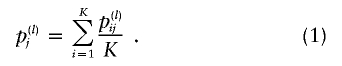 |
We use “log” to denote the natural logarithm, with 0log0=0.
Informativeness for Assignment: the No-Admixture Model
In the no-admixture model, individuals are each assumed to originate from one of K populations. Suppose we are given a random individual, whose (random) population assignment is Q, with Q∈{1,2,…,K}. The probability that the individual belongs to population i is ℙ(Q=i); we assume that each population has the same initial probability of being the source of an unknown individual, so that ℙ(Q=i)=1/K for all i. The (random) genotype of one of the individual’s two alleles at locus l is J(l).
Our aim is to measure the amount of “information” gained about Q from knowledge of J(l) and to compare this quantity across different values of l. This measurement can be performed in a natural way, through use of an information-theoretic framework. If the value of J(l) is unknown, there is uncertainty, or entropy, regarding the value of the random variable Q. Once the value of J(l) is known, the entropy of Q decreases. The reduction in uncertainty about Q due to knowledge of J(l) is the mutual information, In(Q;J(l))=Hn(Q)-Hn(Q|J(l)), where Hn(Q) is the initial entropy of Q, and Hn(Q|J(l)) is the conditional entropy of Q given knowledge of J(l) (the subscript “n” refers to the no-admixture model). Using standard definitions (Cover and Thomas 1991, chapter 2), and leaving off superscripts for convenience, we have
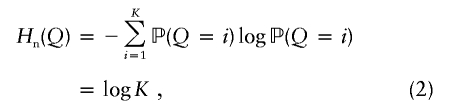 |
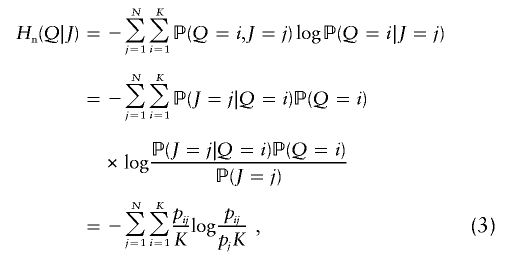 |
and
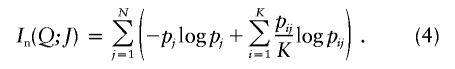 |
We refer to In(Q;J) as the informativeness for assignment. For a given set of populations, the minimal In of 0 occurs when all alleles have equal frequencies in all populations. The maximal value, logK, occurs when N⩾K and no allele is found in more than one population.
This measure of the amount of “information” about the ancestry Q contained in the genotype J also arises from a likelihood approach. The quantity 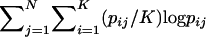 can be viewed as the expected log-likelihood associated with drawing an allele randomly from the set of populations {1,2,…,K}. The term
can be viewed as the expected log-likelihood associated with drawing an allele randomly from the set of populations {1,2,…,K}. The term 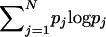 is the expected log-likelihood associated with drawing an allele from a hypothetical “average” population whose allele frequencies equal the mean across the K populations. Thus, equation (4) gives the expected logarithm of the likelihood ratio whose numerator is the likelihood that an allele is assigned to one of the populations and whose denominator is the likelihood that an allele is assigned to the “average” population. When the minimum description length principle (Barron et al. 1998) is used, up to a constant, In also equals the expected reduction, upon observation of J, in the length of the optimal coding of the random variable Q. It gives the average (taken across populations) Kullback-Leibler information (Kullback 1959, p. 6) for distinguishing population-specific allele frequency distributions from the distribution for the “average population.” For K=2, In is similar to a previously proposed statistic based on Kullback-Leibler information (table 1).
is the expected log-likelihood associated with drawing an allele from a hypothetical “average” population whose allele frequencies equal the mean across the K populations. Thus, equation (4) gives the expected logarithm of the likelihood ratio whose numerator is the likelihood that an allele is assigned to one of the populations and whose denominator is the likelihood that an allele is assigned to the “average” population. When the minimum description length principle (Barron et al. 1998) is used, up to a constant, In also equals the expected reduction, upon observation of J, in the length of the optimal coding of the random variable Q. It gives the average (taken across populations) Kullback-Leibler information (Kullback 1959, p. 6) for distinguishing population-specific allele frequency distributions from the distribution for the “average population.” For K=2, In is similar to a previously proposed statistic based on Kullback-Leibler information (table 1).
The expression for In(Q;J) also has a close correspondence with the G statistic obtained from a contingency table, each of whose N columns of K elements gives the relative frequencies of an allele in the K populations. This G statistic is given by (Sokal and Rohlf 1995, p. 737)
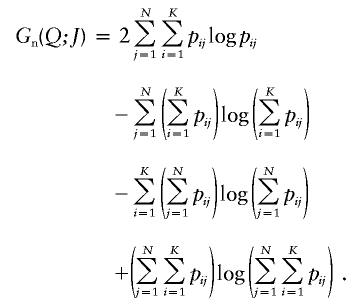 |
Using equation (1) and 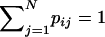 , we can simplify to
, we can simplify to
Thus, for any number of populations and alleles, identifying loci of high informativeness for assignment is equivalent to identifying loci with large values of Gn.
Note that In(Q;J) is a sum over alleles. The contribution of allele j to informativeness for assignment is
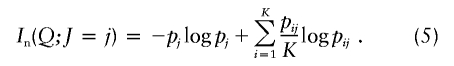 |
For fixed pj, the minimal allelic informativeness of 0 occurs when pij=pj for all i. For fixed pj⩽1/K, the maximum, pjlogK, occurs when pij=Kpj for exactly one value of i and pij=0 for all other values.
Similarly, it is possible to write In(Q;J) as a sum over populations, with the contribution of population i to the informativeness for assignment equaling
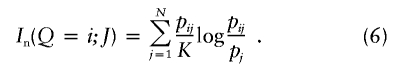 |
Equations (5) and (6) enable calculation of the specific contribution of an allele or population to In. These computations are useful, since alleles at a locus might differ in their importance for assignment of individuals to populations. Populations might also differ in their degree of difference from the “average” population, so that assignment to some populations is easier than assignment to others. Henceforth, we use “informativeness” only in relation to the In statistic (and the Ia statistic, to be defined later), although we use “informative” and “information” more generally, to describe “ability to infer ancestry.” Note that, if the prior assignment of individuals is not uniformly distributed, general priors, ℙ(Q=i)=qi, can be accommodated by replacing ℙ(Q=i) with qi in the derivations of equations (2) and (3) and 1/K with qi in equations (1), (4), (5), and (6).
In can also be extended for use in assignment to populations of multilocus diploid genotypes rather than of single alleles. For convenience, we treat diploid genotypes as being ordered so that (j1,j2) differs from (j2,j1) (an unordered genotype is assigned randomly to one of the possible ordered genotypes). If we assume both Hardy-Weinberg proportions and independence of loci within populations, equation (4) applies with l=1Lpij(l)1pij(l)2 in place of pij, 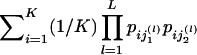 in place of pj, and with the sum taken over all l=1L[N(l)]2 possible multilocus genotypes, {(j(1)1,j(1)2),(j(2)1,j(2)2),…,(j(L)1,j(L)2)}. Although this sum is difficult to evaluate if the number of possible multilocus genotypes is large, it can, in principle, predict the informativeness of multilocus sets; note that informativeness is not additive over loci, since loci that are independent within populations may still contribute to ancestry inference in a correlated manner.
in place of pj, and with the sum taken over all l=1L[N(l)]2 possible multilocus genotypes, {(j(1)1,j(1)2),(j(2)1,j(2)2),…,(j(L)1,j(L)2)}. Although this sum is difficult to evaluate if the number of possible multilocus genotypes is large, it can, in principle, predict the informativeness of multilocus sets; note that informativeness is not additive over loci, since loci that are independent within populations may still contribute to ancestry inference in a correlated manner.
Relationship of In to δ and Fst
For the simplest case in which informativeness is of interest—namely, for K=N=2—it is possible to relate In to δ and to Fst. In the mathematical development, we let δ equal the signed difference between the frequencies of allele 1 in two populations, p11-p21, and, without loss of generality, we assume that p11⩾p21, so that δ∈[0,1]; when applied to data, it is implicit that δ refers to the absolute difference, |p11-p21|. Denoting σ=p11+p21, we must have σ∈[δ,2-δ]. Simplifying equation (4) in terms of δ and σ, we obtain (fig. 1A)
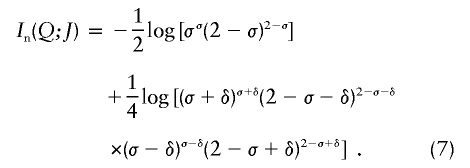 |
As it should be, In is invariant with respect to a transposition of alleles (δ→-δ and σ→2-σ), of populations (δ→-δ), or of both alleles and populations (σ→2-σ).
Figure 1.
Relationship of informativeness for assignment (In) to δ (A), and Fst (B). The plots are based on two alleles and two source populations, and they use equations (7) and (8).
For K=2 and N=2, Fst for a locus (henceforth used interchangeably with F), can be written as (modified from p. 167 of Weir 1996)
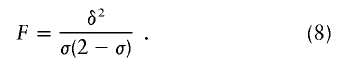 |
Solving equation (8) for δ and using equation (7), we can express In in terms of F and σ (fig. 1B).
For a fixed value of δ or F, a biallelic marker is best able to infer ancestry if one of its alleles is absent in one of the populations: if the random genotype J equals this allele, there is no uncertainty about the origin of an individual. In other words, for a fixed δ (Campbell et al. 2003) or F, the markers with the greatest ability to infer ancestry have a value of σ near one of the extremes. The statistic In captures this aspect of ancestry inference ability (fig. 1; table 2): for a fixed δ, informativeness declines from its maximum at σ=δ to its minimum at σ=1 and then climbs to a second maximum at σ=2-δ; for a fixed F, the minimal informativeness is at σ=1, and the maxima are at σ=2F/(1+F) and at σ=2/(1+F).
Table 2.
Relationship of informativeness for assignment (In), δ, and Fst[Note]
| Quantity (A) | Expression in Terms of σ and Other Quantity (B) | Allowable Values of σ in Terms of A | Allowable Values of A in Terms of B | Maximal Width of the Rangeof A Given the Value of B | Minimal Informativeness forAssignment in Terms of σ and A | Maximal Informativeness forAssignment in Terms of σ and A | Maximal Width of the Range of Informativeness Given the Value of A |
| δ | 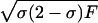 |
[δ,2-δ] | 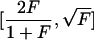 |
.1349 at F≈.0874 |
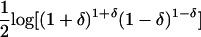 at σ=1 at σ=1
|
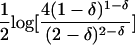 at σ=δ or σ=2-δ at σ=δ or σ=2-δ
|
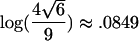 at δ=1/2 at δ=1/2
|
| F | 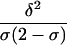 |
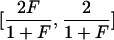 |
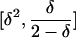 |
5φ-8≈.0902 at δ=2-φ≈.3820 |
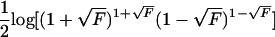 at σ=1 at σ=1
|
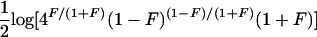 at at 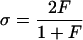 or or 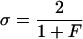
|
.0417 at F≈.4772 |
Note.— In the upper row of the table, A=δ and B=F. In the lower row, A=F and B=δ. 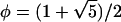 .
.
A comparison of figure 1A and 1B demonstrates that informativeness varies less across values of σ for a fixed F than for a fixed δ; thus, In is more closely related to F than to δ. By considering the difference between the maximal and minimal informativeness over values of σ (table 2), it can be shown that the value of F predicts the value of In to within 0.0417, whereas δ predicts In only to within 0.0849. The mean difference between the upper and lower bounds on In, given the value of F, is 0.0282; the corresponding mean difference between the upper and lower bounds on In, given δ, is more than twice as large, equaling 0.0569 (fig. 2A and 2B).
Figure 2.
Informativeness for assignment (In), δ, and Fst for 8,714 SNPs, based on allele frequency estimates in African Americans and European Americans. A, In vs. δ. B, In vs. Fst. C, Fst vs. δ. Upper and lower bounds for the dependent variable, given the independent variable, are taken from table 2. A red vertical line marks the point of greatest difference between the upper and lower curves. Mean differences between upper and lower curves are 3/4-log2≈0.0569 (A), [16log2+2-6(log2)2-π2]/12≈0.0282 (B), and 2log2-4/3≈0.0530 (C). Spearman rank correlation coefficients between the variables are 0.921, 0.998, and 0.943 in A, B, and C, respectively.
An additional consequence of equation (8) and the requirement that σ∈[δ,2-δ] is that δ can be used to predict F fairly accurately, and vice versa (fig. 2C). This is useful in cases in which only one of these two measures has been reported. Given δ and allowing σ to vary over [δ,2-δ], F ranges from a minimum of δ2, when σ=1, to a maximum of δ/(2-δ), when σ=δ or σ=2-δ. Thus, either δ2 or δ/(2-δ) can be regarded as a substitute for F; for any values of δ and σ, it can be shown that δ/(2-δ)-0.0902⩽F⩽δ2+0.0902. The maximal discrepancy between the two approximations, 0.0902, is attained when δ≈0.3820 (table 2). The mean of the two bounds, or (δ+2δ2-δ3)/(4-2δ), is always within 0.0451 of F. The accuracy of such simple approximations as F≈δ2 and F≈δ/(2-δ) is perhaps somewhat surprising.
Predictions of δ from F are slightly less accurate than the reverse predictions. Given F, as σ ranges over [2F/(1+F),2/(1+F)], δ ranges from a minimum of 2F/(1+F) at σ=1 to a maximum of  at σ=2F/(1+F) or σ=2/(1+F). The maximal width of this range of δ, given F, is 0.1349, and it is attained at F≈0.0874.
at σ=2F/(1+F) or σ=2/(1+F). The maximal width of this range of δ, given F, is 0.1349, and it is attained at F≈0.0874.
Optimal Rate of Correct Assignment
If we use the no-admixture model, another way to measure marker information content is to pursue a decision-theoretic approach. We adopt an assignment rule in which observing that one of the two alleles of a random individual is j leads to assignment to population i with probability dij, where 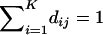 for each j. We also choose a cost function cii′, which gives the penalty for assignment to population i′ when the correct population is i. Under the assumption of a uniform prior on the population of origin, or ℙ(Q=i)=1/K for all i, the aim is to choose an assignment rule, or a set of values of dij, that minimizes the expected value of the loss (Weiss 1961, p. 69), or
for each j. We also choose a cost function cii′, which gives the penalty for assignment to population i′ when the correct population is i. Under the assumption of a uniform prior on the population of origin, or ℙ(Q=i)=1/K for all i, the aim is to choose an assignment rule, or a set of values of dij, that minimizes the expected value of the loss (Weiss 1961, p. 69), or
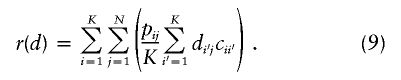 |
If cii′ is taken to equal 0 when i=i′ and 1 otherwise, minimizing equation (9) is equivalent to maximizing the probability of correct assignment, or 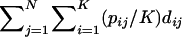 . The optimal rate of correct assignment (ORCA) is the probability of correct assignment when the optimal rule is used. To determine this rule, note that, for allele j, the maximum of the linear function
. The optimal rate of correct assignment (ORCA) is the probability of correct assignment when the optimal rule is used. To determine this rule, note that, for allele j, the maximum of the linear function 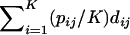 over the set
over the set 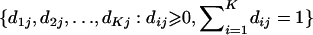 must occur at one of the vertices of the set. Consequently, this maximum occurs when each allele is always assigned to the population in which it is most frequent, and it equals
must occur at one of the vertices of the set. Consequently, this maximum occurs when each allele is always assigned to the population in which it is most frequent, and it equals 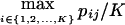 . Adding across alleles, we obtain
. Adding across alleles, we obtain
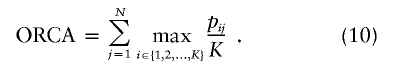 |
Similarly to In, the minimal value of ORCA, 1/K, occurs when all alleles have equal frequencies in all populations, and the maximal value, 1, occurs when N⩾K and no allele is found in more than one population. Also similarly to In, general prior assignment probabilities, ℙ(Q=i)=qi, can be accommodated by replacing 1/K with qi in equations (9) and (10). Note that, for K=N=2 with p11⩾p21, we obtain (p11+p22)/2 for ORCA, or
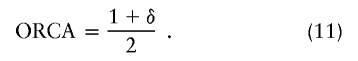 |
Similarly to In, ORCA can be extended for evaluation of sets of many loci. Because the maximal correct assignment probability when (Hardy-Weinberg) diploid genotypes rather than individual alleles are assigned to populations equals 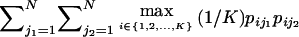 , when multilocus diploid genotypes (at loci that are independent within populations) are assigned, the corresponding probability is
, when multilocus diploid genotypes (at loci that are independent within populations) are assigned, the corresponding probability is
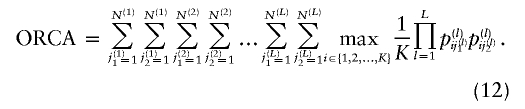 |
Equation (12) can, in principle, predict the probabilities of correct assignment of sets of one or more loci in procedures (Buchanan et al. 1994; Paetkau et al. 1995; Banks et al. 2003) that assign multilocus genotypes to their most likely source populations.
Informativeness for Ancestry Coefficients: the Admixture Model
We have introduced two new measures that can facilitate assignment of individuals to populations. Often, however, a goal of ancestry inference is to estimate “ancestry coefficients” for an individual whose ancestry is from two or more populations (Rannala and Mountain 1997; Pritchard et al. 2000; Anderson and Thompson 2002). Such an individual has a vector of K ancestry coefficients that sum to 1, where the coefficient for population i gives the fraction of the individual’s genome that derives from population i. Ancestry is now a random vector Q rather than a discrete random variable. The mutual information Ia(Q;J) that quantifies the amount of information about Q provided by knowledge of J is of interest. With Q=(Q1,Q2,…,QK), where Qi is the (random) ancestry coefficient for the ith population and 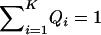 , we have
, we have
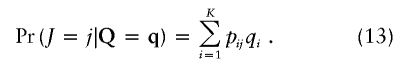 |
Similarly to the no-admixture case, any assumptions could be made for the initial probability distribution of Q. That is, any distribution defined on the set 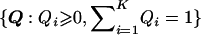 is suitable. For simplicity, we assume that this distribution is uniform: all collections of ancestry coefficients that sum to 1 are a priori equally likely.
is suitable. For simplicity, we assume that this distribution is uniform: all collections of ancestry coefficients that sum to 1 are a priori equally likely.
Using the definition of mutual information for continuous random variables (Cover and Thomas 1991, chapter 9), Ia(Q;J) equals (see the appendix)
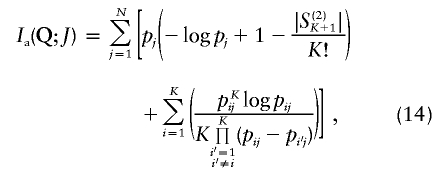 |
where S(2)K+1 is a Stirling number of the first kind (Abramowitz and Stegun 1965, p. 833) (the first six values of |S(2)K+1|, starting at K=2, are 3, 11, 50, 274, 1,764, and 13,068). The quantity Ia is termed the “informativeness for ancestry coefficients.” Ia has a similar multilocus extension to that of In, and, for K=N=2, relationships of Ia to δ and F are qualitatively similar to the corresponding relationships of In to δ and F (not shown). For example, for fixed δ, the maxima of Ia over σ occur at σ=δ and σ=2-δ, and the minimum is at σ=1.
Number of Markers
The statistic Ia suggests a way to prioritize markers for use in inference of ancestry coefficients, but it does not have a simple relationship with the number of markers needed for this inference. However, using a maximum-likelihood approach, it is possible to approximate this number of markers. Equation (13) gives the likelihood of ancestry coefficients (q1,q2,…,qK-1) in a haploid one-locus model, with qK=1-q1-…-qK-1. The expected Fisher information matrix, U, for the likelihood function has dimensions (K-1)×(K-1) and, for each i and i′, the (i,i′)th element equals (Millar 1991, eq. [A.3])
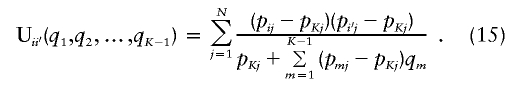 |
Multiplying equation (15) by 2, we can obtain the corresponding value for (Hardy-Weinberg) diploids. When standard maximum-likelihood theory is used (Elandt-Johnson 1971), the variance-covariance matrix of the ancestry coefficient maximum-likelihood estimates is approximated by U-1. For K⩾3, a straightforward transformation of U can enable inclusion of qK in the matrix (Millar 1991); for K=2, the maximum-likelihood estimates  and
and  have equal variances.
have equal variances.
Using equation (15), in the (diploid) case of K=2, we obtain
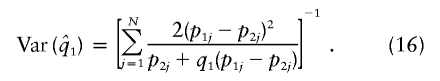 |
For biallelic markers (N=2), equation (16) reduces to
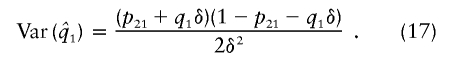 |
If we consider all possible values of q1 and assume that p11⩾p21, the largest value of equation (17) occurs at q1=(1-2p21)/(2δ), producing an upper bound for the approximate variance equal to
 |
Because the information matrix for a set of loci that are independent within populations is the sum of the matrices for the individual loci, the number of independent markers, all with the same value δ, that are required to achieve 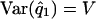 , is
, is
 |
Using equation (19), 35 biallelic markers with δ=0.6 are necessary for achieving an SD of 0.1, in agreement with a previous suggestion of ∼40 such markers (Hoggart et al. 2003).
The number of independent biallelic markers required for accurate estimation of ancestry coefficients in the two-population admixture model (table 3) is considerably larger than the number required for assignment in corresponding no-admixture models (Risch et al. 2002; Campbell et al. 2003). However, our computations assume that estimation of ancestry coefficients occurs by maximum likelihood; other estimation procedures or use of dependencies between markers might reduce the number of markers needed. In addition, since it is based on an upper bound for the variance and not on the general expression, equation (19) might further overestimate the number of markers needed. Note that, because only the upper bound and not the general expression is directly related to δ, markers with high values of In and Ia rather than of δ might often produce smaller variances at values of q1 relevant to individuals under consideration.
Table 3.
Number of Biallelic Markers Required for Achieving a Specified Standard Deviation in Ancestry Coefficient Estimates for a Two-Population Model (eq. [19])
|
No. of Markers Requiredfor an SD of |
||||
| δ | .2 | .1 | .05 | .01 |
| .9 | 4 | 16 | 62 | 1,544 |
| .8 | 5 | 20 | 79 | 1,954 |
| .7 | 7 | 26 | 103 | 2,552 |
| .6 | 9 | 35 | 139 | 3,473 |
| .5 | 13 | 50 | 200 | 5,000 |
| .4 | 20 | 79 | 313 | 7,813 |
| .3 | 35 | 139 | 556 | 13,889 |
| .2 | 79 | 313 | 1,250 | 31,250 |
| .1 | 313 | 1,250 | 5,000 | 125,000 |
Estimation of Informativeness
In, ORCA, and Ia have been defined parametrically, as inherent properties of a marker together with a set of populations. In practice, however, estimates made from data rather than parametric allele frequencies must be used. For a given locus, let the number of copies of allele j observed at the locus in population i equal nij, and let the total number of observations in population i equal ni. A simple estimator of informativeness statistics is the count estimate, in which pij is estimated by nij/ni, and the estimated 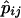 values are inserted in place of the parametric values.
values are inserted in place of the parametric values.
This estimator can produce biased estimates; consider, for example, two samples taken from the same population. Since allele counts in the two samples are likely to differ by chance, markers will have positive estimated In for distinguishing the two samples and estimated ORCA >1/2 when parametric informativeness equals 0 and parametric ORCA is 1/2. This bias is not of major concern when the goal is to compare informativeness estimates for different loci through use of the same sample (Brenner 1998), since a systematic bias affects all loci in a similar manner. In addition, in comparisons of locus informativeness across different samples, sample-specific biases should affect all loci similarly, and the relationship between informativeness estimates of a locus in two samples is preserved even if the estimates are biased.
In, ORCA, and Ia have been defined using frequencies of alleles rather than of diploid genotypes. If alleles within individuals are not independent, so that within-population genotype frequencies do not correspond to Hardy-Weinberg proportions, the definitions can be applied treating J as a random diploid genotype, and the count estimates of genotype frequencies can be used in estimation. We do not consider this issue further.
Data
We consider various subsets (table 4) of a data set of 377 microsatellite markers—45 dinucleotides, 58 trinucleotides, and 274 tetranucleotides—genotyped in 1,056 individuals from 52 human populations (Cann et al. 2002; Rosenberg et al. 2002; Zhivotovsky et al. 2003; Human Diversity Panel Genotypes Web site; Human STRP Screening Sets Web site). Names of regions and regional affiliations of populations are the same as in the article by Rosenberg et al. (2002). At least 50 of the 377 markers are among those reported in table 1 of the article by Collins-Schramm et al. (2002), and at least 212 of them are included in table 1 of the article by Smith et al. (2001).
Table 4.
Data Sets and Spearman Rank Correlation Coefficients of In, ORCA, and Ia
|
Correlation Coefficient |
||||
| Data Set | Description of Groups | In and ORCA | In and Ia | ORCA and Ia |
| World-52 | 52 populations representing seven regions | .920 | … | … |
| World-5 | 5 regional groups (Africa, Eurasia, East Asia, Oceania, America) | .956 | .994 | .956 |
| World-7 | 7 regional groups (Africa, Europe, Middle East, Central/South Asia, East Asia, Oceania, America) | .953 | .990 | .947 |
| Africa | 6 populations (Bantu [Kenya], Mandenka, Yoruba, San, Mbuti Pygmy, Biaka Pygmy) | .945 | .986 | .937 |
| Europe | 8 populations (Orcadian, Adygei, Russian, Basque, French, Italian, Sardinian, Tuscan) | .878 | .973 | .867 |
| Middle East | 4 populations (Mozabite, Bedouin, Druze, Palestinian) | .873 | .994 | .891 |
| Central/South Asia | 9 populations (Balochi, Brahui, Makrani, Sindhi, Pathan, Burusho, Hazara, Uygur, Kalash) | .883 | … | … |
| East Asia | 18 populations (Han, Han [N. China], Dai, Daur, Hezhen, Lahu, Miao, Oroqen, She, Tujia, Tu, Xibo, Yi, Mongola, Naxi, Cambodian, Japanese, Yakut) | .915 | … | … |
| Oceania | 2 populations (Melanesian, Papuan) | .921 | .998 | .940 |
| America | 5 populations (Karitiana, Surui, Colombian, Maya, Pima) | .934 | .989 | .945 |
To compare informativeness for microsatellites and SNPs, we consider SNPs that have been studied in African Americans, European Americans, and East Asians and that were found to have few enough errors for use in analysis of population divergence (Akey et al. 2002; Joshua Akey's Homepage). From the Akey et al. (2002) data, we exclude several types of SNPs: (1) SNPs genotyped by the Whitehead Institute, whose European American sample differed from that used by the other genotyping centers; (2) SNPs with unknown sample sizes or with sample size <40 (20 individuals) in at least one of the three groups; (3) SNPs with unknown, nonunique, or nonautosomal map positions; and (4) SNPs whose frequencies were obtained by DNA pooling or for which one or more of the reported allele frequencies could not be expressed as a rounded quotient of an integer and the reported sample size. The 8,714 SNPs we use were all genotyped by Celera, Motorola, or Orchid.
Statistical Properties of Informativeness
In this section, we demonstrate that the proposed informativeness statistics are, indeed, useful measures. First, we show that the statistics In, ORCA, and Ia produce similar estimates, so that we can proceed using only one statistic, the informativeness for assignment. Second, we demonstrate that the In statistic is robust, in that rankings of locus informativeness do not vary greatly across resamples of the data. Third, we show that the In statistic does indeed measure ability to infer ancestry, in that population structure inference using markers of high informativeness requires fewer markers than inference using markers of low informativeness.
Relationship between In, ORCA, and Ia
For each of the data sets in table 4, we computed In, ORCA, and Ia for each of the loci, using the allele count estimates with equations (4), (10), and (14). For each data set, Spearman rank correlation coefficients (Gibbons 1985, p. 226) of locus In and ORCA, In and Ia, and ORCA and Ia values were computed. Loci for which two or more populations had an identical allele frequency estimate were not used in the latter two calculations, to avoid obtaining denominators of 0 in the computation of Ia. For the World-52, Central/South Asia, and East Asia data sets, in which many populations had the same sample sizes and therefore had numerous opportunities to produce equal allele frequency estimates in two or more populations, there were many such loci, and the correlation coefficients involving Ia were not computed.
Rankings of loci by In, ORCA, and Ia were all highly correlated, with the largest correlations observed between In and Ia (table 4). Thus, for convenience, in the remainder of this article we restrict attention to estimates of In, or, simply, the informativeness, and we assume that all three measures have similar properties.
Locus Informativeness Rankings
For each of the 10 data sets (table 4), markers were ranked from highest to lowest estimated informativeness (table A [online only]). To assess the robustness of these rankings, or the extent to which they are affected by the particular choice of individuals included in the data, we performed bootstrap replicates.
Individuals were resampled with replacement within groups, holding group sample sizes fixed. For each replicate, informativeness was estimated for each locus, and loci were ranked by estimated informativeness. In some replicates, for at least one group and one locus, the resample included only individuals who did not have genotypes at the locus. This situation arose only for data sets in which some groups had small sample sizes (⩽10). In these data sets, it was possible for a resample to consist solely of copies of a few individuals. Thus, if these few individuals had no data at a locus, the resample also had no data. For each data set, excluding these replicates, which were discarded, 1,000 resamples were performed. For data sets in which all groups had larger sample sizes (>10), it was not necessary to discard any replicates.
To assess the variability of In values across bootstrap replicates, for each locus, we computed the ratio of the SD of the bootstrap values of In to the value estimated from the data, and we averaged this quantity across loci. Three statistics were used to compare the locus informativeness rankings that were estimated from the data and those that were obtained in the bootstrap replicates: (1) the mean across replicates of RVd,Vb, where Vd denotes the vector of informativeness ranks based on the data, Vb denotes the vector of ranks based on the bth bootstrap replicate, and R denotes the Spearman rank correlation coefficient; (2) the Kendall coefficient of concordance of the 1,000 bootstrap replicates (Gibbons 1985, p. 250); (3) the mean across loci of the mean across replicates of the absolute deviation between the rank of a locus in the data and its rank in the replicate. This third statistic was also computed using only the 50 loci of highest estimated informativeness.
Although informativeness fluctuated noticeably across replicates for individual loci, rankings in different replicates were highly concordant with each other and were highly correlated with the rankings based on the estimates from the data (table 5). Similar patterns of correlation across bootstrap replicates were observed for all of the data sets. The World-52, World-5, and World-7 data sets, which contained the most data, produced the most robust informativeness ranks and values; the least robust were found for Oceania, the smallest data set.
Table 5.
Robustness of Informativeness Statistics[Note]
| Data Set | No. of Replicates Discarded | Mean Ratio ofBootstrap SD toEstimated In Value | Mean ± SD Spearman Rank Correlation Coefficient | Kendall Coefficient of Concordance of Ranks | Mean ± SD Absolute Deviation of Ranks | Mean ± SD Absolute Deviation of Ranks (50 Loci of Highest In) |
| World-52 | 1,460 | .064 | .977 ± .002 | .974 | 17.53 ± 8.27 | 7.38 ± 4.38 |
| World-5 | 0 | .119 | .978 ± .003 | .959 | 17.24 ± 5.60 | 8.57 ± 4.42 |
| World-7 | 0 | .106 | .980 ± .003 | .964 | 16.44 ± 5.30 | 8.19 ± 4.09 |
| Africa | 27 | .215 | .909 ± .018 | .852 | 34.75 ± 12.57 | 16.13 ± 9.06 |
| Europe | 514 | .274 | .845 ± .021 | .770 | 45.39 ± 17.40 | 17.81 ± 10.92 |
| Middle East | 0 | .317 | .839 ± .020 | .734 | 46.35 ± 15.18 | 22.51 ± 11.47 |
| Central/South Asia | 108 | .223 | .880 ± .017 | .818 | 39.71 ± 15.32 | 16.80 ± 12.27 |
| East Asia | 392 | .170 | .919 ± .009 | .883 | 32.47 ± 13.95 | 13.75 ± 10.13 |
| Oceania | 0 | .683 | .836 ± .034 | .723 | 46.45 ± 15.72 | 21.44 ± 9.74 |
| America | 0 | .180 | .935 ± .012 | .883 | 29.04 ± 10.81 | 13.56 ± 6.28 |
Note.— See the “Locus Informativeness Rankings” subsection of the “Statistical Properties of Informativeness” section for a description of the quantities in the table.
The fluctuation of ranks of individual loci across replicates indicates that exact ranks of loci (such as in tables A, B, C, D, and E [online only]) should be regarded with caution. However, fluctuations were small enough that the markers of highest informativeness usually had low ranks in bootstrap replicates (rightmost column of table 5). Thus, confidence can be placed in general statements such as a locus being “among the most informative markers” in a data set.
Performance of Markers of High Informativeness in Ancestry Inference
One way to test the utility of In as a measurement of the ability of a marker to infer ancestry is to check whether the population structure inferred using the markers of highest In more closely approximates the population structure inferred using all of the markers than does the population structure inferred using the markers of lowest In. Using the computer program structure (Pritchard et al. 2000; available from the Pritchard Lab Web site), with five clusters and the full data of 1,056 individuals, we previously found that the genetically inferred population structure corresponded fairly closely to the five regions in the World-5 data set (Rosenberg et al. 2002). Thus, if In indeed measures ability to infer ancestry, informativeness of a locus in the World-5 data set should correlate well with the contribution of the locus to population structure inference using five clusters.
We therefore ran structure with all 1,056 individuals in the data, using the markers of highest informativeness for the World-5 data set. For various choices of the number of markers, M, five structure runs were performed with the M markers of highest In, and five runs were performed with the M most heterozygous markers (table S4 of Rosenberg et al. [2002]). Expected heterozygosity was used for comparison, because, among several statistics studied in a previous analysis (Rosenberg et al. 2001), it produced the greatest reduction in the number of markers needed for inference. One run was performed with each of 20 random sets of M markers; for each value of M, random sets were chosen independently of the sets that were selected for the other values of M. Five runs were performed with the M markers of lowest In. All structure runs used five clusters, and, as in the study by Rosenberg et al. (2002), they employed the admixture model for individual ancestry (Pritchard et al. 2000), the F model for allele frequency correlations (Falush et al. 2003), and a burn-in period of length 20,000 followed by 10,000 iterations.
The similarity coefficient C (Rosenberg et al. 2002) was used to compare runs with subsets of the markers against 10 runs that employed all 377 markers and were performed by Rosenberg et al. (2002). As in that study, the normalization required in the computation of C was based on the runs that used all of the markers. For each value of M, M<377, each of the 10 runs that used all 377 markers was compared with each of the five runs that used the M markers of highest informativeness, for a total of 50 comparisons. For M=377, the 90 pairwise comparisons of the 10 full-data runs were performed. For each M, the first quartile, median, and third quartile of the distribution of the 50 values were obtained (90 values for M=377). Comparisons to the full-data runs were made in an analogous manner, using the runs based on the least informative, most heterozygous, and random markers. For the random markers and M<377, the similarity coefficient distribution was based on 200 comparisons.
Figure 3 indicates that, in general, fewer loci chosen according to the highest informativeness were required than random loci for inferring a population structure similar to that obtained with all the loci. This pattern was observed especially for small and intermediate values of M; although similarity coefficients at these M often varied considerably across runs, runs based on the markers of highest In generally produced greater similarity coefficients than those based on random or highly heterozygous markers. For larger values of M, the difference in similarity coefficients across criteria was less pronounced, partly because the sets of markers chosen by different criteria had greater overlap than for small values of M. However, runs that used the markers of lowest In produced similarity coefficients that were considerably smaller than those obtained by the other sets of markers. Many more of the markers of lowest In than of those of highest In were required to obtain inferred population structures that were visually similar to that inferred using the full data (fig. 4). Thus, high informativeness is a useful indicator of the ability of a marker to infer ancestry; more dramatically, low informativeness suggests that a locus is not of great utility for inference of ancestry.
Figure 3.
Similarity coefficients for runs based on reduced sets of markers and runs based on the full data. Sets of markers were chosen with each of four methods: highest informativeness, highest expected heterozygosity, random, and lowest informativeness.
Figure 4.
Inferred population structure with five clusters, based on markers of highest and lowest informativeness and plotted using distruct (available from Noah Rosenberg's Homepage). Each individual is represented by a thin vertical line, which is partitioned into five colored segments that represent the individual’s estimated ancestry coefficients in the five clusters. Black lines separate individuals of different populations, which are labeled below the figure. The left-right order of individuals is the same in all plots. The bottom plot is the same as is shown in figure 1 of Rosenberg et al. (2002); each of the other graphs is based on the highest-likelihood run among five runs with the relevant set of loci.
Comparison of Rankings across Data Sets
For pairs of data sets, we computed correlation coefficients of locus informativeness (table 6). Most pairs of rankings had correlations of at least 0.2. Markers that had high informativeness for inference of regional ancestry tended to be informative for inference within several regions. One exception was that informativeness in the America data set was not correlated with informativeness in the World-5 and World-7 data sets.
Table 6.
Pearson (above the Diagonal) and Spearman (below the Diagonal) Correlation Coefficients of Locus Informativeness Values for Pairs of Data Sets
| World-52 | World-5 | World-7 | Africa | Europe | MiddleEast | Central/SouthAsia | EastAsia | Oceania | America | |
| World-52 | … | .835 | .867 | .766 | .640 | .635 | .782 | .728 | .433 | .319 |
| World-5 | .860 | … | .986 | .586 | .356 | .411 | .494 | .402 | .272 | −.007 |
| World-7 | .887 | .987 | … | .605 | .380 | .431 | .526 | .422 | .278 | .024 |
| Africa | .754 | .608 | .621 | … | .505 | .521 | .598 | .521 | .317 | .180 |
| Europe | .626 | .391 | .415 | .490 | … | .500 | .600 | .607 | .310 | .387 |
| Middle East | .579 | .419 | .434 | .480 | .481 | … | .651 | .590 | .324 | .338 |
| Central/South Asia | .746 | .508 | .536 | .542 | .582 | .570 | … | .733 | .368 | .374 |
| East Asia | .685 | .397 | .420 | .455 | .565 | .492 | .670 | … | .402 | .444 |
| Oceania | .379 | .242 | .246 | .276 | .316 | .298 | .306 | .370 | … | .232 |
| America | .286 | −.006 | .023 | .151 | .376 | .297 | .338 | .413 | .231 | … |
The highest correlations for pairs of regions occurred for regions that were geographically proximate, such as Central/South Asia and East Asia. All correlations for pairs of regions, among those that included two of Africa, Europe, Middle East, Central/South Asia, and East Asia, were larger than correlations that involved Oceania or America. The smallest correlation for a pair of regions was between informativeness in Africa and informativeness in America. Larger absolute levels of informativeness in Africa, America, and Oceania (fig. 5) are consistent with the greater observed differentiation among populations in these regions (Rosenberg et al. 2002).
Figure 5.
Correlations of informativeness for pairs of regional data sets
Most loci were ranked poorly in at least one data set (table A [online only]). D11S2000, D16S3401, D16S422, D21S2055, and D3S2427 were the only markers to rank among the 75 most informative in all data sets; note that D21S2055 was one of three loci identified by Zhivotovsky et al. (2003) as unusually variable. D13S285 and D7S1804 were highly informative in all seven regional data sets (rank ⩽75) but were less informative in at least one of the three worldwide data sets (rank >75). Conversely, D14S1007, D1S235, D22S683, D2S1356, D8S560, D9S1779, D9S1871, NA-D18S-2, and NA-D5S-1 were highly informative in the worldwide data sets (rank ⩽25) but were less informative in one or more of the regional data sets (rank >75).
Microsatellites and SNPs
Dinucleotide loci, which show the most variation among the markers in these data (Zhivotovsky et al. 2003), were generally more informative than tetranucleotide loci (table 7), consistent with the generally greater differentiation of dinucleotides across human populations (Ruiz Linares 1999; Rosenberg et al. 2003). Dinucleotides were usually also more informative than trinucleotides, but, in many cases, trinucleotides and tetranucleotides had similar levels of informativeness. However, for the worldwide data sets and for Africa, tetranucleotides were by far the least informative class of microsatellite. For example, although 73% of the loci were tetranucleotides, in the World-7 data set, the 25 loci of highest informativeness included only 7 tetranucleotides. Of the 100 loci of lowest informativeness in this data set, 97 were tetranucleotides.
Table 7.
Informativeness and Repeat Size
|
Mean Informativeness Rank |
P Value (Two-Tailed Mann-Whitney Test) |
|||||
| Data Set | Dinucleotides | Trinucleotides | Tetranucleotides | Dinucleotides vs. Trinucleotides | Dinucleotides vs. Tetranucleotides | Trinucleotides vs. Tetranucleotides |
| World-52 | 94 | 151 | 213 | <.001 | <.001 | <.001 |
| World-5 | 86 | 119 | 221 | .009 | <.001 | <.001 |
| World-7 | 85 | 119 | 221 | .011 | <.001 | <.001 |
| Africa | 107 | 161 | 209 | <.001 | <.001 | .001 |
| Europe | 136 | 201 | 195 | <.001 | .001 | .790 |
| Middle East | 140 | 189 | 197 | .011 | .002 | .564 |
| Central/South Asia | 124 | 205 | 196 | <.001 | <.001 | .603 |
| East Asia | 138 | 206 | 194 | <.001 | .002 | .466 |
| Oceania | 168 | 189 | 192 | .314 | .176 | .789 |
| America | 185 | 232 | 181 | .048 | .865 | <.001 |
To compare informativeness of microsatellites and SNPs, we determined the informativeness of microsatellites for assignment with three source populations: Africans, Europeans, and East Asians. For these groups, we also determined informativeness for each pairwise combination of source populations (tables B and C [online only]). Similarly, we estimated informativeness of SNPs among African Americans, European Americans, and East Asians. Because the individuals and populations in the microsatellite and SNP data sets were not the same, our comparison of microsatellite and SNP informativeness can only be regarded as approximate. Inclusion of some extremely isolated populations in the microsatellite data but not in the SNP data might exaggerate the relative informativeness of microsatellites. However, this effect might be counteracted by a SNP ascertainment procedure that produced greater divergence across populations than is characteristic of randomly chosen SNP markers (J. Akey, personal communication); the microsatellite data likely show little or no such effect (Rosenberg et al. 2002). The small-sample upward bias in informativeness might also impact relative informativeness estimates.
For each set of source populations, randomly chosen microsatellites had greater informativeness than random SNPs (fig. 6). The ratios of median dinucleotide informativeness to median SNP informativeness were 7.8 (Africans vs. Europeans), 6.8 (Africans vs. East Asians), 5.1 (Europeans vs. East Asians), and 5.3 (Africans vs. Europeans vs. East Asians). The ratios of means were 4.3, 3.7, 2.8, and 3.8, respectively, and the 50th percentile of dinucleotide informativeness corresponded to the 96th, 95th, 88th, and 98th percentiles of SNP informativeness.
Figure 6.
Informativeness quantiles for microsatellites and SNPs. For each set of populations, curves follow the same relative order over most of the domain (from top to bottom: dinucleotides, trinucleotides, tetranucleotides, and SNPs). SNPs were genotyped in African Americans and European Americans rather than in Africans and Europeans.
One threshold proposed for declaring a SNP to be highly informative is δ=0.5 (Shriver et al. 1997), a value exceeded by 1.9%, 4.6%, and 2.7% of the SNPs (among those polymorphic in the relevant pair of populations) for African Americans and European Americans, African Americans and East Asians, and European Americans and East Asians, respectively. The value δ=0.5 corresponds to Fst∈[0.250,0.333] and In∈[0.131,0.216] (table 2); for corresponding comparisons, averaging across the three classes of loci, 26.0%, 42.0%, and 12.4% of microsatellites exceed the lower bound of In, and 5.9%, 10.2%, and 1.5% exceed the upper bound.
Discussion
In this article, we have introduced new statistics, In, ORCA, and Ia, for measuring the information provided by loci about ancestry. In, which is highly correlated with ORCA and Ia (table 4), is robust, in that it gives similar results in bootstrap replicates (table 5). The statistic is effective for inference of ancestry, in that population structure is more easily inferred using markers that have high values of In than using those that have low values (figs. 3 and 4). Although it is closely related to δ in the case of biallelic markers in two source populations (figs. 1 and 2; table 2), unlike δ, In captures the dependence of information content on the position of allele frequencies in the unit interval.
Use of markers of highest informativeness is desirable for reduction of genotyping effort in such situations as forensics (Shriver et al. 1997; Lowe et al. 2001), admixture mapping (Dean et al. 1994; McKeigue 1998), and structured-association mapping (Pritchard and Donnelly 2001; Hoggart et al. 2003). In these scenarios, it is desirable to maximize information about individual ancestry at minimal cost. For the case of admixture mapping, the additional constraint that loci must be located in candidate regions of the genome applies; unlike other ancestry inference scenarios, admixture mapping makes use not of the ancestry of an individual as a whole but of particular parts of an individual genome. Thus, ideal marker sets for admixture mapping must have representation in regions of interest as well as high informativeness.
Highly informative markers are also useful in testing for population stratification in case-control genetic association studies (Pritchard and Rosenberg 1999), although the test does not use individual ancestry estimates. The goal is to determine whether cases and controls differ in ancestry to such an extent that an excess number of random markers will, by chance, be associated with disease status. Because they have the greatest potential to differentiate among ancestry groups, the most informative markers offer the greatest power to reject the null hypothesis of no genomewide allele-frequency differences between cases and controls; thus, their use offers a cautious approach in dealing with population stratification. If allele-frequency differences are detected, these markers are ideal for structured-association methods that employ individual ancestry estimates to avoid identifying the associations that result from ancestry differences rather than from true association with disease status (Pritchard and Donnelly 2001). The number of these markers needed for desired precision in estimated ancestry coefficients can be approximated using the maximum-likelihood model.
Consequently, the panels in tables A, B, C, D, and E (online only) can provide a resource for tests of population stratification. For example, in European Americans, the test might use markers that are most informative for distinguishing among various types of European ancestry (table A [online only]); in Hispanic Americans, it might employ markers that are most informative for distinguishing European from Native American ancestry (table B [online only]) or for distinguishing European, Native American, and African ancestry (table C [online only]). Note that panels in tables A–E utilize groups and classifications that might not be identical to those needed in applications: for example, if ancestry inference in African Americans is of interest, the African and European groups in our data do not fully represent the groups from which African Americans have descended. However, we observed that informativeness in one region was often highly correlated with informativeness in another region (table 6; fig. 5). Thus, while the most informative markers in a data set need not be the most informative for use with a different collection of groups, this imperfect panel of markers is likely to be considerably more informative than a random panel. The observed pattern, in which informativeness correlations were highest for neighboring geographic regions, is likely to be a consequence of the correlation of allele frequencies that results from shared ancestry (Ramachandran et al., in press). Populations from neighboring regions typically share ancestors more recently, so that their allele frequencies are more strongly correlated.
Two exceptions to the general pattern of correlation across data sets were Oceania and America, in which informativeness was not very highly correlated with informativeness in other regions. The small correlations likely indicate that many of the markers that are extremely variable in other regions by chance must not have been highly variable in founder groups of Oceania and the Americas. That informativeness patterns across di-, tri-, and tetranucleotides were different in Oceania and America from those of the other data sets suggests that bottlenecks were strong enough to obscure the typical patterns of variation for these three classes of markers.
Thus, to identify a panel of markers that are generally useful for inference of regional ancestry and for population ancestry inference within regions (Hoggart et al. 2003), it is most difficult to find markers that are informative both within continental Eastern Hemisphere regions and within Oceania and the Americas. We have identified a small number of generally informative markers; many more loci will need to be screened if markers that are informative in every region are to be found. Alternatively, a general panel might be assembled by collecting markers useful for inference between specific pairs of groups. Such a procedure may be advantageous, because, unlike sequential accumulation of generally informative markers, it avoids duplication of effort by accounting for the possibility that markers of high informativeness can provide information about ancestry in different ways. A systematic procedure to identify maximally informative sets or loci that are conditionally optimal, given the markers that have already been chosen, might use multilocus In, multilocus ORCA (eq. [12]), or a decision tree (Guinand et al. 2002).
Although random microsatellites are considerably more informative than random SNPs for distinguishing among pairs of populations, and highly informative loci constitute a greater fraction of microsatellites than of SNPs, the right-hand tail of the distribution of SNP informativeness crosses that of microsatellites (fig. 6), suggesting that, if enough SNPs are screened, a set with informativeness comparable to that of the set of the most informative microsatellites can be found. This observation may be less applicable to the problem of distinguishing among K source populations for K>2. For a locus with N alleles, if N⩾K, the informativeness of the locus can potentially be as large as logK, whereas, if N=2, the maximal informativeness is no larger than log2, regardless of the value of K. Because microsatellites in the data of Rosenberg et al. (2002) have an average of 12.4 alleles, for relatively large values of K, microsatellites have greater potential for higher information content than SNPs, most of which are biallelic. Thus, for large K, the relative performance of microsatellites compared with SNPs will likely be greater than is seen in figure 6 for sets of two and three source populations.
Thus, for inferring ancestry among groups such as African Americans, European Americans, and East Asians, for which genomewide SNP allele frequencies have already been obtained (Akey et al. 2002), use of the most informative known SNPs is likely to be most efficient. However, because informativeness for distinguishing among populations such as different Native American groups is not correlated with informativeness for distinguishing among major regional groups (table 6), SNPs chosen by their informativeness in other scenarios will likely be considerably less useful in these populations than randomly chosen microsatellites. Until the most informative SNPs are identified for a set of populations of interest, use of microsatellites, especially dinucleotides, may lead to greater statistical efficiency in inference of ancestry. Of course, technical problems associated with dinucleotides (Ghebranious et al. 2003) might outweigh the efficiency that derives from their use, and factors such as laboratory fixed costs and difficulties in multiplexing might make the application of less informative markers more economical. The decision about which markers to use for inference of ancestry in any particular context should incorporate a combination of economic, technical, and statistical concerns.
Acknowledgments
N.A.R. was supported by an NSF Postdoctoral Fellowship in Biological Informatics. We thank J. Akey for assistance with the SNP data set, P. Calabrese and D. Conti for discussions, and E. Ziv and two reviewers for thoughtful comments on the manuscript.
Appendix: Informativeness for Ancestry Coefficients
Define 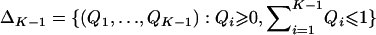 and suppose
and suppose 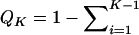 on ΔK-1. The probability density function for Q=(Q1,Q2,…,QK), which we denote by fQ(q), can be regarded as a function defined on ΔK-1. An elementary calculation shows that ΔK-1dQ1…dQK-1=1/(K-1)!, and, therefore,
on ΔK-1. The probability density function for Q=(Q1,Q2,…,QK), which we denote by fQ(q), can be regarded as a function defined on ΔK-1. An elementary calculation shows that ΔK-1dQ1…dQK-1=1/(K-1)!, and, therefore,
Similarly to the discrete no-admixture model, we can apply the continuous-variable analogue of mutual information (Cover and Thomas 1991, chapter 9) to define informativeness in the admixture model, or Ia(Q;J) (the subscript “a” refers to the admixture model). The informativeness for ancestry coefficients is the difference of entropy Ha(Q) and conditional entropy Ha(Q|J). By use of equation (A1) and the definition, the entropy is
The conditional entropy of Q given J is given by
By use of equations (13) and (A1), the following integral can be evaluated:
Setting aij=pij(K-1)!/pj and substituting equations (13), (A1), and (A4) into (A3), we have
If we assume that, for all j, if i≠i′, then pij≠pi′j, by applying the result of Rosenberg and Stong (2003) with K-1 in place of k to the function f(x)=xKlogx/K!, it can be shown that the integral in equation (A5) evaluates to
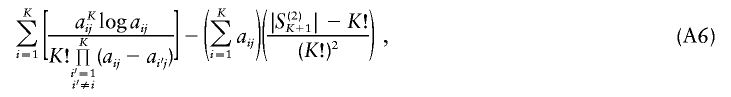 |
where S(2)K+1=(-1)K+1K!(1+2-1+3-1+…+K-1) is a Stirling number of the first kind. Finally, inserting equation (A6) into (A5) and simplifying gives
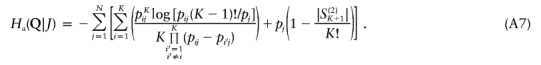 |
The expression for the mutual information (eq. [14]) is obtained by use of Ia(Q;J)=Ha(Q)-Ha(Q|J) with equations (A2) and (A7). Note that the definition of Ia is sensible only if no two populations share the same frequency for any allele. It is appropriate to assume that parametric allele frequencies are unequal in different populations; however, when allele frequencies are estimated from samples of small and equal size, this assumption will often not be met.
Footnotes
This article is dedicated to the memory of Ryk Ward.
Electronic-Database Information
The URLs for data presented herein are as follows:
- Human Diversity Panel Genotypes, Center for Medical Genetics, http://research.marshfieldclinic.org/genetics/Freq/FreqInfo.htm (for microsatellite genotypes)
- Human STRP Screening Sets, Center for Medical Genetics, http://research.marshfieldclinic.org/genetics/sets/combo.html (for Marshfield panel 10)
- Joshua Akey’s Homepage, http://cgi.uc.edu/~jakey/ (for SNP allele frequencies)
- Noah Rosenberg’s Homepage, http://www.cmb.usc.edu/~noahr/distruct.html (for distruct software)
- Pritchard Lab, http://pritch.bsd.uchicago.edu/ (for structure software)
References
- Abramowitz MA, Stegun IA (1965) Handbook of mathematical functions. Dover, New York [Google Scholar]
- Akey JM, Zhang G, Zhang K, Jin L, Shriver MD (2002) Interrogating a high-density SNP map for signatures of natural selection. Genome Res 12:1805–1814 10.1101/gr.631202 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson EC, Thompson EA (2002) A model-based method for identifying species hybrids using multilocus genetic data. Genetics 160:1217–1229 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bamshad MJ, Wooding S, Watkins WS, Ostler CT, Batzer MA, Jorde LB (2003) Human population genetic structure and inference of group membership. Am J Hum Genet 72:578–589 10.1086/368061 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Banks MA, Eichert W, Olsen JB (2003) Which genetic loci have greater population assignment power? Bioinformatics 19:1436–1438 10.1093/bioinformatics/btg172 [DOI] [PubMed] [Google Scholar]
- Barron A, Rissanen J, Yu B (1998) The minimum description length principle in coding and modeling. IEEE Trans Inform Theory 44:2743–2760 10.1109/18.720554 [DOI] [Google Scholar]
- Brenner CH (1998) Difficulties in the estimation of ethnic affiliation. Am J Hum Genet 62:1558–1560 10.1086/301856 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buchanan FC, Adams LJ, Littlejohn RP, Maddox JF, Crawford AM (1994). Determination of evolutionary relationships among sheep breeds using microsatellites. Genomics 22:397–403 10.1006/geno.1994.1401 [DOI] [PubMed] [Google Scholar]
- Campbell D, Duchesne P, Bernatchez L (2003) AFLP utility for population assignment studies: analytical investigation and empirical comparison with microsatellites. Mol Ecol 12:1979–1991 10.1046/j.1365-294X.2003.01856.x [DOI] [PubMed] [Google Scholar]
- Cann HM, de Toma C, Cazes L, Legrand M-F, Morel V, Piouffre L, Bodmer J, et al (2002) A human genome diversity cell line panel. Science 296:261–262 10.1126/science.296.5566.261b [DOI] [PubMed] [Google Scholar]
- Chakraborty R, Weiss KM (1988) Admixture as a tool for finding linked genes and detecting that difference from allelic association between loci. Proc Natl Acad Sci USA 85:9119–9123 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collins-Schramm HE, Phillips CM, Operario DJ, Lee JS, Weber JL, Hanson RL, Knowler WC, et al (2002) Ethnic-difference markers for use in mapping by admixture linkage disequilibrium. Am J Hum Genet 70:737–750 10.1086/339368 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cornuet J-M, Piry S, Luikart G, Estoup A, Solignac M (1999) New methods employing multilocus genotypes to select or exclude populations as origins of individuals. Genetics 153:1989–2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cover TM, Thomas JA (1991) Elements of information theory. Wiley, New York [Google Scholar]
- Davies N, Villablanca FX, Roderick GK (1999) Determining the source of individuals: multilocus genotyping in nonequilibrium population genetics. Trends Ecol Evol 14:17–21 10.1016/S0169-5347(98)01530-4 [DOI] [PubMed] [Google Scholar]
- Dean M, Stephens JC, Winkler C, Lomb DA, Ramsburg M, Boaze R, Stewart C, et al (1994) Polymorphic admixture typing in human ethnic populations. Am J Hum Genet 55:788–808 [PMC free article] [PubMed] [Google Scholar]
- Elandt-Johnson RC (1971) Probability models and statistical methods in genetics. Wiley, New York [Google Scholar]
- Excoffier L (2001) Analysis of population subdivision. In: Balding DJ, Bishop M, Cannings C (eds) Handbook of statistical genetics. John Wiley & Sons, Chichester, UK, pp 271–307 [Google Scholar]
- Falush D, Stephens M, Pritchard JK (2003) Inference of population structure using multilocus genotype data: linked loci and correlated allele frequencies. Genetics 164:1567–1587 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghebranious N, Vaske D, Yu A, Zhao C, Marth G, Weber JL (2003) STRP screening sets for the human genome at 5 cM density. BMC Genomics 4:6 10.1186/1471-2164-4-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gibbons JD (1985) Nonparametric statistical inference. 2nd ed. Marcel Dekker, New York [Google Scholar]
- Gomulkiewicz R, Brodziak JKT, Mangel M (1990) Ranking loci for genetic stock identification by curvature methods. Can J Fish Aquat Sci 47:611–619 [Google Scholar]
- Guinand B, Topchy A, Page KS, Burnham-Curtis MK, Punch WF, Scribner KT (2002) Comparisons of likelihood and machine learning methods of individual classification. J Hered 93:260–269 10.1093/jhered/93.4.260 [DOI] [PubMed] [Google Scholar]
- Hoggart CJ, Parra EJ, Shriver MD, Bonilla C, Kittles RA, Clayton DG, McKeigue PM (2003) Control of confounding of genetic associations in stratified populations. Am J Hum Genet 72:1492–1504 10.1086/375613 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kullback S (1959) Information theory and statistics. Dover, Mineola, New York [Google Scholar]
- Lowe AL, Urquhart A, Foreman LA, Evett IW (2001) Inferring ethnic origin by means of an STR profile. Forensic Sci Int 119:17–22 10.1016/S0379-0738(00)00387-X [DOI] [PubMed] [Google Scholar]
- Manel S, Berthier P, Luikart G (2002) Detecting wildlife poaching: identifying the origin of individuals with Bayesian assignment tests and multilocus genotypes. Conserv Biol 16:650–659 10.1046/j.1523-1739.2002.00576.x [DOI] [Google Scholar]
- McKeigue PM (1998) Mapping genes that underlie ethnic differences in disease risk: methods for detecting linkage in admixed populations, by conditioning on parental admixture. Am J Hum Genet 63:241–251 10.1086/301908 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Millar RB (1991) Selecting loci for genetic stock identification using maximum likelihood, and the connection with curvature methods. Can J Fish Aquat Sci 48:2173–2179 [Google Scholar]
- Molokhia M, Hoggart C, Patrick AL, Shriver M, Parra E, Ye J, Silman AJ, et al (2003) Relation of risk of systemic lupus erythematosus to west African admixture in a Caribbean population. Hum Genet 112:310–318 [DOI] [PubMed] [Google Scholar]
- Paetkau D, Calvert W, Stirling I, Strobeck C (1995) Microsatellite analysis of population structure in Canadian polar bears. Mol Ecol 4:347–354 [DOI] [PubMed] [Google Scholar]
- Primmer CR, Koskinen MT, Piironen J (2000) The one that did not get away: individual assignment using microsatellite data detects a case of fishing competition fraud. Proc R Soc Lond B 267:1699–1704 10.1098/rspb.2000.1197 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pritchard JK, Donnelly P (2001) Case-control studies of association in structured or admixed populations. Theor Popul Biol 60:227–237 10.1006/tpbi.2001.1543 [DOI] [PubMed] [Google Scholar]
- Pritchard JK, Rosenberg NA (1999) Use of unlinked genetic markers to detect population stratification in association studies. Am J Hum Genet 65:220–228 10.1086/302449 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pritchard JK, Stephens M, Donnelly P (2000) Inference of population structure using multilocus genotype data. Genetics 155:945–959 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramachandran S, Rosenberg NA, Zhivotovsky LA, Feldman MW. Robustness of the inference of human population structure: a comparison of X-chromosomal and autosomal microsatellites. Hum Genomics (in press) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rannala B, Mountain JL (1997) Detecting immigration by using multilocus genotypes. Proc Natl Acad Sci USA 94:9197–9201 10.1073/pnas.94.17.9197 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Risch N, Burchard E, Ziv E, Tang H (2002) Categorization of humans in biomedical research: genes, race and disease. Genome Biol 3:comment2007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosenberg N, Stong R (2003) Problem 11039. Amer Math Monthly 110:743 [Google Scholar]
- Rosenberg NA, Burke T, Elo K, Feldman MW, Freidlin PJ, Groenen MAM, Hillel J, et al (2001) Empirical evaluation of genetic clustering methods using multilocus genotypes from 20 chicken breeds. Genetics 159:699–713 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosenberg NA, Pritchard JK, Weber JL, Cann HM, Kidd KK, Zhivotovsky LA, Feldman MW (2002) Genetic structure of human populations. Science 298:2381–2385 10.1126/science.1078311 [DOI] [PubMed] [Google Scholar]
- ——— (2003) Response to comment on “Genetic structure of human populations.” Science 300:1877 [DOI] [PubMed] [Google Scholar]
- Ruiz Linares A (1999) Microsatellites and the reconstruction of the history of human populations. In: Goldstein DB, Schlötterer C (eds) Microsatellites: evolution and applications. Oxford University Press, Oxford, pp 183–197 [Google Scholar]
- Shriver MD, Smith MW, Jin L, Marcini A, Akey JM, Deka R, Ferrell RE (1997) Ethnic-affiliation estimation by use of population-specific DNA markers. Am J Hum Genet 60:957–964 [PMC free article] [PubMed] [Google Scholar]
- Smith MW, Lautenberger JA, Shin HD, Chretien J-P, Shrestha S, Gilbert DA, O’Brien SJ (2001) Markers for mapping by admixture linkage disequilibrium in African American and Hispanic populations. Am J Hum Genet 69:1080–1094 10.1086/323922 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smouse PE, Spielman RS, Park MH (1982) Multiple-locus allocation of individuals to groups as a function of the genetic variation within and differences among human populations. Am Nat 119:445–463 10.1086/283925 [DOI] [Google Scholar]
- Sokal RR, Rohlf FJ (1995) Biometry. 3rd ed. Freeman, New York [Google Scholar]
- Stephens JC, Smith MW, Shin HD, O’Brien SJ (1999) Tracking linkage disequilibrium in admixed populations with MALD using microsatellite loci. In: Goldstein DB, Schlötterer C (eds) Microsatellites: evolution and applications. Oxford University Press, Oxford, pp 211–224 [Google Scholar]
- Weir BS (1996) Genetic data analysis II. Sinauer, Sunderland, MA [Google Scholar]
- Weiss L (1961) Statistical decision theory. McGraw-Hill, New York [Google Scholar]
- Zhivotovsky LA, Rosenberg NA, Feldman MW (2003) Features of evolution and expansion of modern humans, inferred from genomewide microsatellite markers. Am J Hum Genet 72:1171–1186 10.1086/375120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ziv E, Burchard EG (2003) Human population structure and genetic association studies. Pharmacogenomics 4:431–441 [DOI] [PubMed] [Google Scholar]