Abstract
De novo LINE-1 (long interspersed element–1, or L1) retrotransposition events are responsible for ∼1/1,000 disease-causing mutations in humans. Previously, L1.2 was identified as the likely progenitor of a mutagenic insertion in the factor VIII gene in a patient with hemophilia A. It subsequently was shown to be one of a small number of active L1s in the human genome. Here, we demonstrate that L1.2 is present at an intermediate insertion allele frequency in worldwide human populations and that common alleles (L1.2A and L1.2B) exhibit an ∼16-fold difference in their ability to retrotranspose in cultured human HeLa cells. Chimera analysis revealed that two amino acid substitutions (S1259L and I1220M) downstream of the conserved cysteine-rich motif in L1 open reading frame 2 are largely responsible for the observed reduction in L1.2A retrotransposition efficiency. Thus, common L1 alleles can vary widely in their retrotransposition potential. We propose that such allelic heterogeneity can influence the potential L1 mutational load present in an individual genome.
LINE-1 (long interspersed element–1, or L1) is an abundant family of non–long terminal repeat retrotransposons that comprises ∼17% of human DNA (Smit 1996; Lander et al. 2001). The vast majority (>99.8%) of L1s can no longer retrotranspose because they are 5′ truncated, internally rearranged, or mutated (reviewed by Moran and Gilbert 2002). However, the average human genome is estimated to contain ∼60–100 retrotransposition-competent L1s (RC-L1s), and ∼10% of these elements are classified as highly active, or “hot” (Sassaman et al. 1997; Brouha et al. 2003). The majority of RC-L1s are members of the Ta (Transcribed active) subfamily (Skowronski et al. 1988), and many are polymorphic with respect to presence, indicating that they have retrotransposed since the origin of our species (Boissinot et al. 2000; Sheen et al. 2000; Ovchinnikov et al. 2001; Myers et al. 2002).
A consensus RC-L1 is ∼6 kb in length and contains a 5′ UTR, two nonoverlapping ORFs (ORF1 and ORF2), and a 3′ UTR that ends in a poly (A) tail (fig. 1B) (Scott et al. 1987; Dombroski et al. 1991; Moran and Gilbert 2002; Brouha et al. 2003). ORF1 encodes a 40-kDa nucleic acid–binding protein (Holmes et al. 1992; Hohjoh and Singer 1996, 1997). ORF2 encodes a protein with both endonuclease (EN) and reverse transcriptase (RT) activities that also contains a cysteine-rich motif (CX3CX7HX4C) of unknown function (Fanning and Singer 1987; Mathias et al. 1991; Feng et al. 1996). Both proteins are required for retrotransposition (Moran et al. 1996), which likely occurs by a mechanism termed “target site–primed reverse transcription” (TPRT) (Luan et al. 1993; Luan and Eickbush 1995; Feng et al. 1996).
Figure 1.
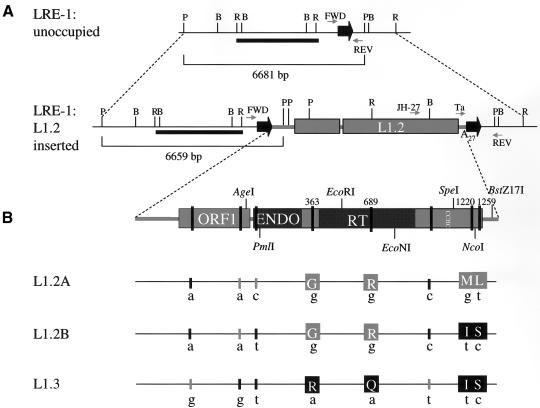
LRE-1 locus and sequence differences between L1.2A, L1.2B, and L1.3. A, The LRE-1 locus has both unoccupied and L1.2 inserted alleles. L1.2 is flanked by 15-bp target site duplications (bold black horizontal arrows) (Dombroski et al. 1991). The positions of the diagnostic restriction sites (P = PstI, B = BamHI, and R = EcoRI) were inferred from the available sequence of the unoccupied (GenBank accession number D87016) and inserted alleles (Dombroski et al. 1991). The thick black line represents the EcoRI probe used in Southern blot analysis by Dombroski et al. (1991). The sizes of the PstI restriction fragments detected from the unoccupied and inserted alleles are indicated under the brackets and would not have been differentiated in the previous analysis (Dombroski et al. 1991). The relative positions of the primers (FWD, REV, Ta, and JH-27) used to determine both the inserted allele frequency and the allele frequencies of L1.2A and L1.2B are designated by gray horizontal arrows. B, A schematic of a full-length RC-L1. The EN and RT domains and the cysteine-rich motif of ORF2 are indicated. Thick vertical lines within the L1 schematic denote the relative positions of sequence differences between the three L1s. Diagnostic DNA and amino acid sequences peculiar to L1.2A, L1.2B, and L1.3 are shown below the schematic. Thick black and gray lines indicate nucleotide differences, whereas black and gray squares denote amino acid sequence differences. Black lines and squares indicate that the designated nucleotide and amino acid match the consensus sequence of a “hot” L1 (Brouha et al. 2003). Gray lines and squares indicate deviations from the consensus sequence. The amino acid numbering convention is based on the sequence of L1.2 (Dombroski et al. 1991). G = glycine; R = arginine; Q = glutamine; M = methionine; I = isoleucine; L = leucine; S = serine. The approximate locations of the restriction sites used to construct the chimeric L1s are shown. For example, JM101L1.2A (M1220I) was created by replacing the 0.25-kb SpeI-NcoI restriction fragment from pJCC48/L1.2A with an analogous fragment from pJCC48/L1.3 (Moran et al. 1996). The 8.1-kb NotI-BstZ17I restriction fragment from the resultant construct then was used to replace the corresponding fragment in JM101L1.3 (Sassaman et al. 1997; Wei et al. 2001). Similar strategies were employed to make the other chimeric constructs noted in tables 4 and 5.
RC-L1 retrotransposition continues to impact the human genome. For example, 14 disease-producing de novo L1 retrotransposition events have been identified in humans (reviewed by Ostertag and Kazazian 2001). RC-L1s also can mobilize sequences derived from both their 5′ and 3′ flanks in cis by a process termed “L1-mediated transduction” (Holmes et al. 1994; Moran et al. 1999; Goodier et al. 2000; Pickeral et al. 2000; Lander et al. 2001). Finally, the RC-L1 encoded proteins also may function in trans, resulting in the mobilization of Alu elements and the formation of processed pseudogenes, which together comprise ∼10% of genomic DNA (Boeke 1997; Esnault et al. 2000; Wei et al. 2001; Dewannieux et al. 2003; Ejima and Yang 2003). Thus, either directly or through the promiscuous mobilization of cellular RNAs, L1 retrotransposition continues to shape the genome.
L1.2 was identified as the likely progenitor of a mutagenic insertion into the factor VIII gene in a patient with hemophilia A (Kazazian et al. 1988; Dombroski et al. 1991). It is a member of the Ta subfamily and resides at a locus designated “LRE-1” on chromosome 22q11.22 (fig. 1A). The initial characterization of L1.2 identified at least two alleles, L1.2A and L1.2B. L1.2A was isolated from a commercial genomic library, and L1.2B was isolated from the mother of the patient. These two elements differ at only 3 nt positions in ORF2 (fig. 1B) (Dombroski et al. 1991).
Previous Southern blot data indicated that L1.2 might be fixed with respect to presence in human DNA (Dombroski et al. 1991). However, we were able to identify only the unoccupied allele of LRE-1 in the human genome working draft sequence (HGWD), indicating that L1.2 may be polymorphic with respect to presence. Indeed, examination of the HGWD indicated that the previous Southern blot analyses could not differentiate between occupied (i.e., inserted) and unoccupied (i.e., empty) alleles of LRE-1, because the PstI restriction fragment in both alleles is similar in size (fig. 1A).
To determine the inserted allele frequency of L1.2, we used a previously developed PCR-based strategy (Sheen et al. 2000; Ovchinnikov et al. 2001; Myers et al. 2002) to assess the presence/absence status of L1.2 in a panel of 80 DNA samples isolated from 20 individuals of African American, Alaskan native, European, or South American descent (fig. 1A). These data then were used to calculate the L1.2 insertion frequency and heterozygosity values for each population group, as well as the average values across all four populations. The average L1.2 insertion frequency across all populations is 0.425, and the average unbiased heterozygosity is 0.492 (table 1). We next extended our analysis to include an additional 365 individuals of unknown ethnic/racial origin. For these 365 individuals, the L1.2 insertion frequency is 0.374, and the unbiased heterozygosity value is 0.469 (table 1). Thus, our data indicate that L1.2 is present at an intermediate insertion frequency in the human population, consistent with the human-specific origin of the L1 Ta subfamily (Boissinot et al. 2000; Sheen et al. 2000).
Table 1.
Insertion Allele Frequency of L1.2 at Locus LRE1
|
No. of Individuals with Genotyped |
||||||
| Populationa | Sample(n)b | Insertion (+) Frequencyc | + / + | + / − | − / − | Unbiased Heterozygosity |
| African Americans | 20 | .325 | 0 | 13 | 7 | .45 |
| Alaskan natives | 20 | .5 | 4 | 12 | 4 | .513 |
| German European | 20 | .4 | 3 | 10 | 7 | .492 |
| South Americans | 20 | .475 | 4 | 11 | 5 | .512 |
| Average | .425 | .492 | ||||
| Additional population | 365 | .374 | 59 | 155 | 151 | .469 |
Populations of individual DNAs used in this study.
Number of individual DNA samples tested for L1.2.
Insertion (+) frequency of L1.2.
Empty alleles are denoted with a (−). +/+ and −/− indicate individuals who are homozygous for presence/absence, respectively. +/− indicates heterozygous individuals. Exact tests using a Markov chain (Guo and Thompson 1992) were used to test for HWE. When a 1% CI was used, none of the populations deviated significantly from HWE.
To assess the respective allele frequencies of L1.2A and B, we used primers JH27 and REV to amplify the LRE-1 locus from 57 individuals who were either heterozygous (+/−; 46 individuals) or homozygous (+/+; 11 individuals) for the insertion (fig. 1A). The resultant PCR products were cloned, and 45 of 68 PCR products yielded high-quality DNA sequence. Diagnostic nucleotides then were used to differentiate L1.2A from L1.2B (fig. 1B; table 2). The allele frequency of L1.2A in these samples is 0.667, whereas that of L1.2B is 0.333. Since the average L1.2 insertion frequency is 0.425 (see table 1 and above), we calculate the overall allele frequencies for L1.2A and L1.2B as 0.283 and 0.142, respectively (table 3). These values are in general agreement with previously published data (Dombroski et al. 1991). Although we characterized only a relatively small number of individual L1.2 alleles, our data also suggest that the L1.2A and L1.2B allele frequencies may differ among population groups.
Table 2.
L1.2A/L1.2B Allele Frequency Determination: Actual Number of Individuals Sequenced, by Genotype[Note]
|
No. of |
No. of Individuals Sequenced |
Frequency of |
|||||||
| Population | + AllelesSequenced | L1.2 AAlleles | L1.2 BAlleles | A/A | A/B | A/− | B/− | L1.2A | L1.2B |
| African Americans | 8 | 3 | 5 | 0 | 0 | 3 | 5 | .375 | .625 |
| Alaskan natives | 12 | 11 | 1 | 2 | 0 | 7 | 1 | .917 | .083 |
| German European | 10 | 5 | 5 | 0 | 0 | 5 | 5 | .5 | .5 |
| South Americans | 15 |
11 |
4 |
2 |
1 |
6 |
3 |
.733 | .267 |
| Total/average | 45 | 30 | 15 | 4 | 1 | 21 | 14 | .667 | .333 |
Note.— Allele frequencies were determined as in table 1. Details regarding the ascertainment of alleles are provided in the text.
Table 3.
L1.2A/ L1.2B Allele Frequency Determination: Predicted Number of Individuals with Given Genotypes if All Individuals Were Sequenced[Note]
|
Predicted No. of Individuals with Genotype |
OverallFrequency of |
||||||||
| Population | A/A | A/B | B/B | A/− | B/− | − / − | L1.2A | L1.2B | (+) |
| African Americans | 0 | 0 | 0 | 4.875 | 8.125 | 7 | .122 | .203 | .325 |
| Alaskan natives | 3.682 | .304 | .014 | 10.668 | 1.332 | 4 | .459 | .042 | .5 |
| German European | .75 | 1.5 | .75 | 5 | 5 | 7 | .2 | .2 | .4 |
| South Americans | 2.537 | 1.391 | .071 | 7.466 | 3.534 | 5 |
.348 | .127 | .475 |
| Total/average | 6.669 | 3.665 | .665 | 28.337 | 17.663 | 23 | .283 | .142 | .425 |
Note.— Allele frequencies were determined as in table 1. Details regarding the ascertainment of alleles are provided in the text.
We recently determined that L1.2B is a “hot” L1 (Brouha et al. 2003), and we demonstrate here that it can retrotranspose at a ∼16-fold higher efficiency than L1.2A in cultured human HeLa cells (table 4). Two of three nucleotide differences between L1.2A and L1.2B result in amino acid changes (I1220M and S1259L) downstream of the conserved cysteine-rich motif (CX3CX7HX4C) of L1 ORF2 (fig. 1B). It is notable that the amino acid substitutions in L1.2A (M1220 and L1259) deviate from the consensus sequence of a “hot” L1 (Brouha et al. 2003). To determine the amino acid(s) responsible for the observed difference in retrotransposition efficiency, we took advantage of unique restriction sites within the L1 sequence to generate L1.2A/L1.2B chimeras (fig. 1B). These chimeras then were tested for their ability to retrotranspose in the transient retrotransposition assay (table 4) (Wei et al. 2000). It is interesting that we found that S1259L is responsible for ∼80% of the difference in retrotransposition activity between L1.2A and L1.2B. By comparison, I1220M affects retrotransposition to a lesser extent, accounting for ∼20% of the difference.
Table 4.
Results of the Cultured Cell Retrotransposition Assay: JM101/L1.2A, JM101/L1.2B, and Associated Chimeras[Note]
|
Mean No. of Colonies per Well ± SD |
% of JM101/L1.2B |
||||||||
| Construct | Trial 1(n=3) | Trial 2(n=3) | Trial 3(n=9) | Trial 4(n=9) | Trial 1(n=3) | Trial 2(n=3) | Trial 3(n=9) | Trial 4(n=9) | Averageof Trials |
| JM101/L1.2B | 142 ± 16 | 49 ± 4.7 | 60 ± 6.7 | 78 ± 12 | 100 | 100 | 100 | 100 | 100 |
| JM101/L1.2A (M1220I/L1259S) | 132 ± 11 | 49 ± 3.5 | 55 ± 6.7 | 67 ± 12 | 93 | 100 | 92 | 86 | 93 |
| JM101/L1.2A (L1259S) | 86 ± 11 | 57 ± 4.0 | 56 ± 9.8 | 64 ± 11 | 61 | 116 | 93 | 93 | 91 |
| JM101/L1.2A (M1220I) | 21 ± 1.5 | 13 ± 4.7 | 17 ± 3.2 | 29 ± 2.3 | 15 | 27 | 28 | 37 | 27 |
| JM101/L1.2A | 8 ± 1.7 | 2 ± 1.0 | 4 ± 2.2 | 5 ± 1.5 | 6 | 4 | 7 | 6 | 6 |
| JM105/L1.3 | 0 | .7 ± 1.2 | .1 ± .3 | .2 ± .7 | 0 | <.01 | 0 | <.01 | <.01 |
Note.— The constructs in tables 4 and 5 were tested in the same trial (i.e., trial 1 = trial 1 in the two tables). Cells were plated in six-well dishes at 5×103 cells/well in all trials except trial 2, in which they were plated at 2×103 cells/well. In each trial, .5 μg of each construct was cotransfected with an equal amount of hrGFP (Stratagene). n = the number of wells scored in each trial. Transfection efficiencies for each construct were determined by flow cytometry (Wei et al. 2000). The proportion of cells expressing GFP averaged 81% and was similar for each of the tested constructs. The number of G418-resistant foci and the standard deviation for each trial is reported. JM105/L1.3 contains a missense mutation in the RT active site and serves as a negative control (Wei et al. 2001).
L1.2A and L1.2B also share two other amino acid substitutions that deviate from the consensus sequence of a “hot” L1 (Brouha et al. 2003). The first substitution, R363G, is located between the L1 ORF2 EN and RT domains, in a putative Myb-like domain (Kubo et al. 2001); the second substitution, Q689R, is located in the L1 RT domain. We hypothesized that these substitutions may affect retrotransposition efficiency, since L1.3, a previously characterized RC-L1 that matches the hot L1 consensus at these positions, retrotransposes in HeLa cells at ∼10–20-fold higher efficiencies than L1.2A (Dombroski et al. 1991, 1993; Sassaman et al. 1997; Wei et al. 2000). To examine the effect of these substitutions on retrotransposition, we created L1.2A/L1.3 chimeras and assayed each of the resultant constructs for their ability to retrotranspose (fig. 1B; table 5). Consistent with the above analysis, our data indicate that I1220M and S1259L are largely responsible for the difference in retrotransposition activity between L1.2A and L1.3. By comparison, R363G is responsible for only <10% of the difference in retrotransposition, whereas Q689R had virtually no effect on retrotransposition (tables 4 and 5).
Table 5.
Results of the Cultured Cell Retrotransposition Assay: JM101/L1.2A, JM101/L1.3, and Associated Chimeras[Note]
|
Mean No. of Coloniesper Well ± SD |
% of JM101/L1.3 |
||||||
| Construct | Trial 1(n=3) | Trial 2(n=3) | Trial 3(n=9) | Trial 1(n=3) | Trial 2(n=3) | Trial 3(n=9) | Averageof Trials |
| JM101/L1.3 | 202 ± 35 | 71 ± 1.5 | 77 ± 6.4 | 100 | 100 | 100 | 100 |
| JM101/L1.3 (Q689R) | 199 ± 36 | 65 ± 5.0 | 77 ± 17 | 99 | 92 | 100 | 97 |
| JM101/L1.3 (R363G) | 148 ± 14 | 67 ± 5.2 | 65 ± 13 | 73 | 94 | 84 | 84 |
| JM101/L1.3 (I1220M) | 124 ± 15 | 54 ± 3.5 | 64 ± 7.5 | 61 | 76 | 83 | 73 |
| JM101/L1.3 (S1259L) | 28 ± 5.6 | 19 ± 5.0 | 23 ± 4.1 | 14 | 27 | 30 | 24 |
| JM101/L1.2A (G363R) | 10 ± 1.0 | 5 ± .6 | 10 ± 3.6 | 5 | 7 | 13 | 8 |
| JM101/L1.3 (I1220M/S1259L) | 12 ± 4.0 | 3 ± 1.5 | 7 ± 3.2 | 6 | 4 | 9 | 6 |
| JM101/L1.2A | 8 ± 1.7 | 2 ± 1.0 | 4 ± 2.2 | 4 | 3 | 5 | 4 |
| JM105/L1.3 | 0 | .7 ± 1.2 | .1 ± .3 | 0 | <.01 | <.01 | <.01 |
Note.— The constructs in tables 4 and 5 were tested in the same trial (i.e., trial 1 = trial 1 in the two tables). Cells were plated in six-well dishes at 5×103 cells/well in all trials except trial 2, in which they were plated at 2×103 cells/well. In each trial, .5 μg of each construct was cotransfected with an equal amount of hrGFP (Stratagene). n = the number of wells scored in each trial. Transfection efficiencies for each construct were determined by flow cytometry (Wei et al. 2000). The proportion of cells expressing GFP averaged 81% and was similar for each of the tested constructs. The number of G418-resistant foci and the standard deviation for each trial is reported. JM105/L1.3 contains a missense mutation in the RT active site and serves as a negative control (Wei et al. 2001).
In sum, we have determined that L1.2 is present at an intermediate insertion frequency in various population groups and have demonstrated that common alleles (L1.2A and L1.2B) exhibit a ∼16-fold difference in their ability to retrotranspose in cultured human HeLa cells. Chimera analysis revealed that two amino acid substitutions (I1220M and S1259L) downstream of the conserved cysteine-rich motif (CX3CX7HX4C) in L1 ORF2 likely are responsible for the observed reduction in L1.2A retrotransposition efficiency. Interestingly, S1259 also is conserved in the canine ORF2-encoded protein (Choi et al. 1999), although it is encoded by a different codon (UCG in human vs. AGU in canine). Similarly, threonine, another nucleophilic amino acid, is present in the active mouse consensus and rat ORF2-encoded proteins. Thus, our data suggest that the amino acid difference (S1259L), and not nucleotide changes that adversely affect L1 RNA, causes most of the observed reduction in L1.2A retrotransposition efficiency.
Future studies are needed to determine how amino acid substitutions downstream of the conserved cysteine-rich motif adversely affect L1 retrotransposition. Preliminary data indicate that retrotransposed sequences derived from L1.3 and L1.2A in cultured human cells are similar in structure and commonly are 5′ truncated (Moran et al. 1996; Gilbert et al. 2002). Thus, it is likely that I1220M and S1259L directly affect retrotransposition efficiency, perhaps by altering template binding, ribonucleoprotein particle transport, and/or initial steps in TPRT. However, it is possible that some of the difference in retrotransposition efficiency we observe reflects the ability of L1.3 and L1.2B to retrotranspose 1.6 kb of sequence (i.e., the length of retrotransposed cDNA required to confer G418-resistance to cells) more efficiently than L1.2A.
Our data suggest that, besides presence/absence dimorphism, allelic heterogeneity also can contribute to differences in L1 retrotransposition potential. For example, if an L1 is polymorphic with respect to presence—and, when present, has two possible alleles (A and B) that differ in their ability to retrotranspose (i.e., A = 1× vs. B = 16× retrotransposition efficiencies)—then any individual in the population may have one of the following six possible genotypes at that specific locus: B16×/B16×, B16×/A1×, B16×/−, A1×/A1×, A1×/−, or −/−. On the basis of our calculated allele frequencies, we would predict that a person of genotype B16×/B16× has the greatest probability of obtaining a new retrotransposition event, with the probability decreasing in the following genotypic order: B16×/B16× (2% of individuals) > B16×/A1× (8% of individuals) > B16×/− (16% of individuals) > A1×/A1× (8% of individuals) > A1×/− (33% of individuals) > −/− (33% of individuals). Similarly, three L1 alleles with different retrotransposition activities would produce 10 potential genotypes, and so on. Indeed, our data predict that both presence/absence dimorphism and L1 allelic heterogeneity could, in principle, influence the L1 mutational load present in an individual’s genome.
Acknowledgments
We thank members of the University of Michigan DNA sequencing core for help with sequencing. We thank Dr. Nicolas Gilbert, Ms. Amy Hulme, Mr. Reid Alisch, and other members of the Moran laboratory for critically evaluating the manuscript. M.A.B is supported, in part, by National Science Foundation grant BCS-0218338 and National Institutes of Health (NIH) grant GM59290. H.H.K. is supported, in part, by NIH grant GM45398. J.V.M. is supported, in part, by NIH grant GM60518 and by a grant from the W. M. Keck Foundation. The University of Michigan Cancer Center helped to defray some of the costs of DNA sequencing.
Electronic-Database Information
The accession number and URL for data presented herein are as follows:
- GenBank, http://www.ncbi.nlm.nih.gov/Genbank/ (for sequence of the LRE-1 unoccupied allele (accession number D87016)
References
- Boeke JD (1997) LINEs and Alus—the poly (A) connection. Nat Genet 16:6–7 [DOI] [PubMed] [Google Scholar]
- Boissinot S, Chevret P, Furano AV (2000) L1 (LINE-1) retrotransposon evolution and amplification in recent human history. Mol Biol Evol 17:915–928 [DOI] [PubMed] [Google Scholar]
- Brouha B, Schustak J, Badge RM, Lutz-Prigge S, Farley AH, Moran JV, Kazazian HH Jr (2003) Hot L1s account for the bulk of retrotransposition in the human population. Proc Natl Acad Sci USA 100:5280–5285 10.1073/pnas.0831042100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi Y, Ishiguro N, Shinagawa M, Kim C-J, Okamoto Y, Minami S, Ogihara K (1999) Molecular structure of canine LINE-1 elements in canine transmissible venereal tumor. Anim Genet 30:51–53 [DOI] [PubMed] [Google Scholar]
- Dewannieux D, Esnault C, Heidmann T (2003) LINE-mediated retrotransposition of marked Alu sequences. Nat Genet 35:41–48 10.1038/ng1223 [DOI] [PubMed] [Google Scholar]
- Dombroski BA, Mathias SL, Nanthakumar E, Scott AF, Kazazian HH Jr (1991) Isolation of an active human transposable element. Science 254:1805–1808 [DOI] [PubMed] [Google Scholar]
- Dombroski BA, Scott AF, Kazazian HH Jr (1993) Two additional potential retrotransposons isolated from a human L1 subfamily that contains an active retrotransposable element. Proc Natl Acad Sci USA 90:6513–6517 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ejima Y, Yang L (2003) Trans mobilization of genomic DNA as a mechanism for retrotransposon-mediated exon shuffling. Hum Mol Genet 12:1321–1328 10.1093/hmg/ddg138 [DOI] [PubMed] [Google Scholar]
- Esnault CJ, Maestre J, Heidmann T (2000) Human LINE-1 retrotransposons generate processed pseudogenes. Nat Genet 24:363–367 10.1038/74184 [DOI] [PubMed] [Google Scholar]
- Fanning T, Singer MF (1987) The LINE-1 DNA sequences in four mammalian orders predict proteins that conserve homologies to retrovirus proteins. Nucleic Acids Res 15:2251–2260 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng Q, Moran JV, Kazazian HH Jr, Boeke JD (1996) Human L1 retrotransposon encodes a conserved endonuclease required for retrotransposition. Cell 87:905–916 [DOI] [PubMed] [Google Scholar]
- Gilbert N, Lutz-Prigge S, Moran JV (2002) Genomic deletions created upon LINE-1 retrotransposition. Cell 110:315–325 [DOI] [PubMed] [Google Scholar]
- Goodier JL, Ostertag, EM, Kazazian HH Jr (2000) Transduction of 3′ flanking sequences is common in L1 retrotransposition. Hum Mol Genet 9:653–657 10.1093/hmg/9.4.653 [DOI] [PubMed] [Google Scholar]
- Guo SW, Thompson EA (1992) A Monte Carlo method for combined segregation and linkage analysis. Am J Hum Genet 51:1111–1126 [PMC free article] [PubMed] [Google Scholar]
- Hohjoh H, Singer MF (1996) Cytoplasmic ribonucleoprotein complexes containing human LINE-1 protein and RNA. EMBO J 15:630–639 [PMC free article] [PubMed] [Google Scholar]
- ——— (1997) Sequence-specific single-strand RNA binding protein encoded by the human LINE-1 retrotransposon. EMBO J 16:6034–6043 10.1093/emboj/16.19.6034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holmes SE, Dombroski BA, Krebs CM, Boehm CD, Kazazian HH Jr (1994) A new retrotransposable human L1 element from the LRE2 locus on chromosome 1q produces a chimaeric insertion. Nat Genet 7:143–148 [DOI] [PubMed] [Google Scholar]
- Holmes SE, Singer MF, Swergold GD (1992) Studies on p40, the leucine zipper motif-containing protein encoded by the first open reading frame of an active human LINE-1 transposable element. J Biol Chem 267:19765–19768 [PubMed] [Google Scholar]
- Kazazian HH Jr, Wong C, Youssoufian H, Scott AF, Phillips DG, Antonarakis SE (1988) Haemophilia A resulting from de novo insertion of L1 sequences represents a novel mechanism for mutation in man. Nature 332:164–166 10.1038/332164a0 [DOI] [PubMed] [Google Scholar]
- Kubo Y, Okazaki S, Anzai T, Fujiwara H (2001) Structural and phylogenetic analysis of TRAS, telomeric repeat-specific non-LTR retrotransposon families in Lepidopteran insects. Mol Biol Evol 18:848–857 [DOI] [PubMed] [Google Scholar]
- Lander ES, Linton LM, Birren B, Nusbaum C, Zody MC, Baldwin J, Devon K, et al (2001) Initial sequencing and analysis of the human genome. Nature 409:860–921 10.1038/35057062 [DOI] [PubMed] [Google Scholar]
- Luan DD, Eickbush TH (1995) RNA template requirements for target DNA-primed reverse transcription by the R2 retrotransposable element. Mol Cell Biol 15:3882–3891 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luan DD, Korman MH, Jakubczak JL, Eickbush TH (1993) Reverse transcription of R2Bm RNA is primed by a nick at the chromosomal target site: a mechanism for non-LTR retrotransposition. Cell 72:595–605 [DOI] [PubMed] [Google Scholar]
- Mathias SL, Scott AF, Kazazian HH Jr, Boeke JD, Gabriel A (1991) Reverse transcriptase encoded by a human transposable element. Science 254:1808–1810 [DOI] [PubMed] [Google Scholar]
- Moran JV, DeBerardinis RJ, Kazazian HH Jr (1999) Exon shuffling by L1 retrotransposition. Science 283:1530–1534 10.1126/science.283.5407.1530 [DOI] [PubMed] [Google Scholar]
- Moran JV, Gilbert N (2002) Mammalian LINE-1 retrotransposons and related elements. In: Craig N, Craggie R, Gellert M, Lambowitz A (eds) Mobile DNA II. ASM Press, Washington, DC, pp 836–869 [Google Scholar]
- Moran JV, Holmes SE, Naas TP, DeBerardinis RJ, Boeke JD, Kazazian HH Jr (1996) High frequency retrotransposition in cultured mammalian cells. Cell 87:917–927 [DOI] [PubMed] [Google Scholar]
- Myers JS, Vincent BJ, Udall H, Watkins WS, Morrish TA, Kilroy GE, Swergold GD, Henke J, Henke L, Moran JV, Jorde LB, Batzer MA (2002) A comprehensive analysis of recently integrated human Ta L1 elements. Am J Hum Genet 71:312–326 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ostertag EM, Kazazian HH Jr (2001) Biology of mammalian L1 retrotransposons. Annu Rev Genet 35:501–538 10.1146/annurev.genet.35.102401.091032 [DOI] [PubMed] [Google Scholar]
- Ovchinnikov I, Troxel AB, Swergold GD (2001) Genomic characterization of recent human LINE-1 insertions: evidence supporting random insertion. Genome Res 11:2050–2058 10.1101/gr.194701 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pickeral OK, Makalowski W, Boguski MS, Boeke JD (2000) Frequent human genomic DNA transduction driven by LINE-1 retrotransposition. Genome Res 10:411–415 10.1101/gr.10.4.411 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sassaman DM, Dombroski BA, Moran JV, Kimberland ML, Naas TP, DeBerardinis RJ, Gabriel A, Swergold GD, Kazazian HH Jr (1997) Many human L1 elements are capable of retrotransposition. Nat Genet 16:37–43 [DOI] [PubMed] [Google Scholar]
- Scott AF, Schmeckpeper BJ, Abdelrazik M, Comey CT, O’Hara B, Rossiter JP, Cooley T, Heath P, Smith KD, Margolet L (1987) Origin of the human L1 elements: proposed progenitor genes deduced from a consensus DNA sequence. Genomics 1:113–125 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheen F, Sherry ST, Risch GM, Robichaux M, Nasidze I, Stoneking M, Batzer MA, Swergold GD (2000) Reading between the LINEs: human genomic variation induced by LINE-1 retrotransposition. Genome Res 10:1496–1508 10.1101/gr.149400 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skowronski J, Fanning TG, Singer MF (1988) Unit-length line-1 transcripts in human teratocarcinoma cells. Mol Cell Biol 8:1385–1397 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smit AFA (1996) The origin of interspersed repeats in the human genome. Curr Opin Genet Dev 6:743–748 10.1016/S0959-437X(96)80030-X [DOI] [PubMed] [Google Scholar]
- Wei W, Gilbert N, Ooi SL, Lawler JF, Ostertag EM, Kazazian HH, Boeke JD, Moran JV (2001) Human L1 retrotransposition: cis preference versus trans complementation. Mol Cell Biol 21:1429–1439 10.1128/MCB.21.4.1429-1439.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei W, Morrish TA, Alisch RS, Moran JV (2000) A transient assay reveals that cultured human cells can accommodate multiple LINE-1 retrotransposition events. Anal Biochem 284:435–438 10.1006/abio.2000.4675 [DOI] [PubMed] [Google Scholar]