Abstract
The signaling molecule cyclic AMP (cAMP) is a ubiquitous second messenger that enables cells to detect and respond to extracellular signals. cAMP is generated by the enzyme adenylyl cyclase, which is activated or inhibited by the Gα subunits of heterotrimeric G proteins in response to ligand-activated G-protein-coupled receptors. Here we identified the unique gene (CAC1) encoding adenylyl cyclase in the opportunistic fungal pathogen Cryptococcus neoformans. The CAC1 gene was disrupted by transformation and homologous recombination. In stark contrast to the situation for Saccharomyces cerevisiae, in which adenylyl cyclase is essential, C. neoformans cac1 mutant strains were viable and had no vegetative growth defect. Furthermore, cac1 mutants maintained the yeast-like morphology of wild-type cells, in contrast to the constitutively filamentous phenotype found upon the loss of adenylyl cyclase in another basidiomycete pathogen, Ustilago maydis. Like C. neoformans mutants lacking the Gα protein Gpa1, cac1 mutants were mating defective and failed to produce two inducible virulence factors: capsule and melanin. As a consequence, cac1 mutant strains were avirulent in animal models of cryptococcal meningitis. Reintroduction of the wild-type CAC1 gene or the addition of exogenous cAMP suppressed cac1 mutant phenotypes. Moreover, the overexpression of adenylyl cyclase restored mating and virulence factor production in gpa1 mutant strains. Physiological studies revealed that the Gα protein Gpa1 and adenylyl cyclase controlled cAMP production in response to glucose, and no cAMP was detectable in extracts from cac1 or gpa1 mutant strains. These findings provide direct evidence that Gpa1 and adenylyl cyclase function in a conserved signal transduction pathway controlling cAMP production, hyphal differentiation, and virulence of this human fungal pathogen.
The conversion of intracellular ATP to cyclic AMP (cAMP), catalyzed by adenylyl cyclase, is a central reaction in eukaryotic signal transduction. The control of cAMP concentration is principally determined by the precise regulation of adenylyl cyclase activity (54). Adenylyl cyclases are either activated or inhibited by interactions with Gα or βγ subunits liberated from heterotrimeric G proteins in response to ligand-activated G-protein-coupled receptors. The molecular basis of G protein activation of adenylyl cyclase was recently determined by X-ray crystallographic analysis of activated Gα subunits with the catalytic domains of the enzyme (55). Here we present studies that illustrate how the regulation of adenylyl cyclase by Gα proteins has been conserved between microorganisms and mammals.
Although the basic catalytic functions of adenylyl cyclases are shared in fungi, the mechanisms of activation and the downstream targets of these enzymes differ among divergent fungal species. In the budding yeast Saccharomyces cerevisiae, the single gene encoding adenylyl cyclase, CYR1, is essential (40, 57). Two partially redundant Ras proteins, Ras1 and Ras2, and the Gα protein Gpa2 activate adenylyl cyclase in response to nutrient conditions (10, 29, 37, 58) and intracellular acidification (10). The target of cAMP, cAMP-dependent protein kinase (PKA), plays central roles in filamentation, sporulation, and stress survival (6, 42, 47).
In contrast to the situation in budding yeast, adenylyl cyclase is not essential in the fission yeast Schizosaccharomyces pombe (39). Mutants lacking adenylyl cyclase (cyr1−) exhibit precocious mating that is no longer repressed by nutrients, as in wild-type cells (39). The Gα protein Gpa2, and not Ras1, plays a central role in regulating adenylyl cyclase in S. pombe (17).
Pathogenic fungi have coopted these conserved signal transduction pathways to regulate their virulence (4, 5, 35). In the corn smut fungus Ustilago maydis, the morphological transitions involved in mating and pathogenicity are dependent upon cAMP signaling. For example, disruption of the adenylyl cyclase gene uac1 results in constitutive filamentation (19). The U. maydis Gα protein Gpa3 activates adenylyl cyclase in response to specific nutritional signals and is required for pathogenesis (21, 27, 46).
Cryptococcus neoformans is an opportunistic human fungal pathogen and an excellent model system for the genetic and molecular dissection of microbial pathogenicity (2). Recently, it was demonstrated that the C. neoformans Gα protein Gpa1 regulates mating and the expression of the inducible virulence factors melanin and capsule. As a consequence, gpa1 mutant strains are attenuated for virulence in animal models of cryptococcal meningitis (3, 15; B. M. Allen, J. G. Kimbrough, J. Heitman, and J. A. Alspaugh, unpublished data). Because exogenous cAMP restores mating and virulence factor production in gpa1 mutant cells, it was proposed that Gpa1 might regulate adenylyl cyclase. Interestingly, Gpa1 belongs to a conserved subgroup of fungal Gα proteins that primarily respond to nutrient deprivation signals (35). We also note that the central role that this conserved Gα protein-cAMP signaling pathway plays in pathogenesis could not have been predicted from studies of model organisms such as budding and fission yeasts.
To investigate the direct role of cAMP signaling in fungal differentiation and microbial pathogenesis, we cloned and disrupted the C. neoformans CAC1 gene, which encodes adenylyl cyclase. Strikingly, cac1 mutants were viable, lacked any detectable cAMP, and maintained the budding growth phenotype of wild-type cells. These findings indicate that adenylyl cyclase and cAMP are dispensable for viability and do not play a major role in determining morphology in this pathogenic basidiomycete under conditions that promote growth as a budding yeast. Mutant strains lacking adenylyl cyclase were sterile, failed to induce capsule or produce melanin, and were avirulent in animal models. Exogenous cAMP or reintroduction of the wild-type gene restored mating and virulence factor production. Genetic epistasis tests support a model in which the Gα protein Gpa1 regulates adenylyl cyclase. Taken together with recent studies on the role of PKA in C. neoformans (15), these findings reveal that the central features of Gα protein regulation of adenylyl cyclase are conserved between unicellular and multicellular eukaryotes. The similarities and differences among cAMP signal transduction pathways in divergent fungi demonstrate how a conserved nutrient-sensing signaling pathway that controls differentiation in nonpathogenic yeasts has been coopted for the control of virulence in pathogenic fungi.
MATERIALS AND METHODS
Strains and media.
The strains used are listed in Table 1. Except for strain JEC20, a serotype D MATa strain used in all of the mating experiments (31), the C. neoformans strains used in these experiments were derived from the serotype A wild-type strain H99 (43). H99-ura5 is a spontaneous 5-fluoroorotic acid-resistant derivative of H99 that was isolated on 5-fluoroorotic acid-containing medium by using the method of Kwon-Chung et al. (33). Strain AAC1 is a serotype A gpa1 mutant strain, and strain AAC3 is a gpa1 GPA1-reconstituted strain (3). RPC3, LCC2, and LCC23 are cac1 mutant strains described in this study. RPC7 and LCC22-1 are cac1 CAC1-reconstituted strains derived from strains RPC3 and LCC22, respectively. Strain AAC17 is a gpa1 mutant strain in which the CAC1 gene was ectopically integrated in multiple copies.
TABLE 1.
Strains used
| Serotype | Strain | Genotype | Reference or source |
|---|---|---|---|
| A | H99 | MATα | 43 |
| H99-ura5 | MATα ura5 | This study | |
| M001 | MATα ade2 | 44 | |
| AAC1 | MATα ade2 gpa1::ADE2 | 3 | |
| AAC3 | MATα ade2 gpa1::ADE2 GPA1-hph | 3 | |
| AAC17 | MATα ade2 gpa1::ADE2 CAC1-hph | This study | |
| RPC3 | MATα ura5 cac1::URA5 | This study | |
| RPC7 | MATα ura5 cac1::URA5 CAC1-hph | This study | |
| LCC22 | MATα ade2 cac1::ADE2 | This study | |
| LCC23 | MATα ade2 cac1::ADE2 | This study | |
| LCC22-1 | MATα ade2 cac1::ADE2 CAC1-hph | This study | |
| D | JEC20 | MATa | 31 |
Standard yeast media were used for most experiments as described previously (51). Niger seed agar (30), Dulbecco’s modified Eagle’s medium (DMEM) with 22 mM NaHCO3 (20), and V8 mating medium (30) were prepared as previously described. When needed, cAMP was added at 2.5 to 5 mM.
PCR.
All PCRs were performed by use of a Perkin-Elmer GeneAmp 9600 thermocycler with 50 ng of template DNA, 100 ng of each oligonucleotide primer, and standard reagents from a TaKaRa kit (Takara Shuzo Co.). For initial identification of the adenylyl cyclase gene in C. neoformans, primers were designed based on conserved regions of fungal adenylyl cyclases: primer DF2, 5′-AGTIAAGACIGARGGIGAYATG, and primer DF5, 5′-AYTGICCICCRTCIGC (I, inosine; R, purine; Y, pyrimidine). Genomic DNA from strain H99 was used as a template for the PCRs. The PCR conditions were 35 cycles at 94°C for 30 s, 35°C for 30 s, and 72°C for 30 s. The resulting 316-bp PCR fragment was TA cloned (Invitrogen) and sequenced.
Southern hybridization and cloning of the CAC1 gene.
Genomic DNA was isolated from strain H99 as described previously (45). Restriction digestion, gel electrophoresis, DNA transfer, prehybridization, hybridization, and autoradiography were performed as described previously (49) with the initial 316-bp CAC1 PCR fragment as the probe. The probe was labeled by using a Random Primed DNA labeling kit (Boehringer Mannheim) and 32P-dCTP (Amersham).
Based on the Southern hybridization data, subgenomic libraries of NheI-digested genomic fragments were cloned into pBluescript, and the clones containing the CAC1 gene fragments were identified by colony hybridization.
Identification of 5′ and 3′ regions of the CAC1 gene.
Strain H99 was incubated at 30°C for 18 h in yeast extract, peptone, dextrose (YPD) medium. Aliquots were subcultured for 4 h at 30 and 37°C in YPD medium and in DMEM–22 mM NaHCO3. The cells were pelleted, and total RNA was isolated by using an RNeasy Mini kit (Qiagen). The RNA samples were pooled, and the polyadenylated RNA fraction was purified by using an Oligotex mRNA Midi kit (Qiagen). This sample was used as the template for cDNA production and subsequent PCR amplification of the 5′ and 3′ ends of the CAC1 gene message by using a Marathon cDNA amplification kit (Clontech).
Disruption of the CAC1 gene.
To create a cac1::URA5 disruption construct, we subcloned a 6.7-kb SacI/XhoI fragment of the CAC1 locus (extending from 523 bp before the start codon to 6,234 bp after the start codon) into plasmid pUC18. The URA5 gene was inserted as a selectable marker into the BamHI-digested CAC1 fragment, resulting in the loss of 1,652 nucleotides internal to the CAC1 open reading frame (ORF). The cac1Δ::URA5 linear fragment was precipitated onto 0.6 μg of gold microcarrier beads (Bio-Rad) and biolistically transformed into strain H99-ura5 as previously described (44). Stable transformants were selected on synthetic medium lacking uracil and containing 1 M sorbitol.
To screen for cac1 mutant strains, genomic DNA from each transformant was isolated and used as the template for PCR amplification with the CAC1-specific primer 3273 (5′-CCAACATCTCTCAACGTGACG) and the URA5-specific primer 5151 (5′-CCTCTTCTTCATCTAGTCGG). Because the recognition sequence of primer 3273 lies outside the disruption construct, only strains in which the cac1::URA5 disruption allele was integrated at the endogenous CAC1 locus amplified a 2-kb PCR band in this reaction. One strain (RPC3) of 120 transformants screened in this manner was found by PCR to have a cac1Δ::URA5 mutation. Southern hybridization was performed by using genomic DNA digested with PstI and the 3.7-kb XbaI/KpnI fragment of the CAC1 gene (corresponding to nucleotides 1153 to 4900 of the CAC1 GenBank sequence) as the probe. We observed that the wild-type bands at 2.2 and 1.7 kb were missing and that only the expected band at 4.1 kb was present in the samples of strain RPC3, confirming that the CAC1 locus was replaced by the cac1Δ::URA5 allele, with no ectopic integrations.
Two independent cac1 mutant strains were made by using the ADE2 gene as the selectable marker for gene disruption. Both of these strains were made by using the serotype A ade2 strain M001 as the recipient for transformation (44, 53). Strain LCC22 was created by using a cac1Δ::ADE2 disruption construct with the ADE2 gene cloned into a StuI site at nucleotide position 5176 of the published CAC1 sequence. The majority of the CAC1 open reading frame was replaced by the ADE2 gene in strain LCC23 by using a cac1Δ::ADE2 construct with the ADE2 gene inserted into the XbaI-digested CAC1 gene, resulting in the loss of 6,590 nucleotides of CAC1 sequence.
The cac1 CAC1- and gpa1 CAC1-reconstituted strains were created by biolistically transforming the wild-type CAC1 gene, linked to the hph gene conferring resistance to hygromycin B, into the cac1 mutant strains RPC3 and LCC22 and the gpa1 mutant strain AAC1 as previously described (3, 13).
Northern analysis.
Strains were incubated in YPD medium at 30°C for 18 h. Cells were pelleted and divided equally for incubation for 4 h in synthetic complete medium with 2% glucose, synthetic complete medium with 0% glucose, or synthetic low-ammonium–dextrose (SLAD) medium (18). Cells were pelleted at 4°C and frozen on dry ice, and total RNA was isolated as previously described (3). Fifteen micrograms of RNA was analyzed for each sample. Gel electrophoresis, RNA transfer, hybridization, and autoradiography were performed as described previously (49).
Mating assays.
All strains to be tested for mating were initially grown in YPD medium for 48 h at 30°C. Mating reactions were performed by coincubating cells of opposite mating types on V8 mating medium in the dark at room temperature for 1 to 2 weeks. Mating mixtures were analyzed for filamentation, and photomicroscopy was performed on representative sectors of the mating mixtures.
Capsule assessment by packed-cell-volume measurement.
Packed-cell volume was assessed as previously described with modifications (20). Strains were incubated in DMEM–22 mM NaHCO3 at 30°C for 24 h, treated with 10% formalin, and normalized to 109 cells/ml. The normalized samples were added to heparinized Microhematocrit capillary tubes (Fisher 02-668-66) and spun for 5 min in a model MB Microhematocrit centrifuge (International Equipment Co.). Packed-cell volume, or cryptocrit, was measured as the length of the packed-cell phase divided by the length of the total suspension within the capillary tube.
Determination of the intracellular cAMP concentration.
Cells were preincubated at 30°C for 18 h in YPD medium. The overnight culture was inoculated into fresh YPD medium to an optical density at 600 nm of 0.05 and grown under the same conditions for 20 h. Cells were collected by centrifugation and washed twice with water and once with buffer (10 mM morpholineethanesulfonic acid [MES] [pH 6.0], 0.1 mM EDTA). Cells were resuspended in buffer and incubated at 30°C with shaking so that they would be subjected to glucose starvation. After 2 h, glucose was added to a final concentration of 2%. At various time points, 0.5 ml of cell suspension was transferred to a tube containing an equal volume of ice-cold 10% trichloroacetic acid and 0.3 ml of glass beads and was immediately frozen in liquid nitrogen. Crude cell extracts were prepared by homogenization with a bead beater at 4°C and were lyophilized. cAMP assays were performed by using a cAMP enzyme immunoassay kit (Amersham) as previously described (38).
Virulence experiments.
In the murine inhalation model of systemic cryptococcosis, A/Jcr mice were intranasally inoculated with 5 × 105 cells as previously described (12). Groups of 10 mice were infected with each strain, and animals were observed twice daily. Symptoms due to the experimental infection included lethargy, ruffled fur, and inability to maintain daily care. In this model, mice develop meningitis and resulting hydrocephalus due to C. neoformans, mimicking the natural history of infection in humans. Moribund mice were sacrificed prior to death. The Kruskal-Wallis algorithm was used to determine the statistical significance of differences in survival.
In the rabbit model, New Zealand White rabbits were sedated with ketamine (Fort Dodge) and xylazine (Vedco) and intrathecally inoculated with the CAC1 wild-type strain (H99), the gpa1 mutant strain (AAC1), and the cac1 mutant strain (LCC22) as previously described (3). Three rabbits were infected with each strain and were treated daily with 1.2 mg of betamethasone sodium-betamethasone acetate (Schering). Cerebrospinal fluid (CSF) was obtained by cisternal puncture after sedation on experimental days 4, 7, and 11 after infection, and the total CFU per milliliter of CSF was determined by quantitative culturing on YPD medium. All virulence studies were performed in compliance with institutional guidelines for animal experimentation.
Nucleotide sequence accession number.
The CAC1 has been assigned GenBank accession no. AF290191.
RESULTS
Identification and disruption of the C. neoformans adenylyl cyclase gene.
In previous studies, it was found that mutants lacking the Gα protein Gpa1 were viable and exhibited cAMP-remediable phenotypes (3; Allen et al., unpublished). Here we tested the hypothesis that Gpa1 regulates cAMP production by adenylyl cyclase. Because adenylyl cyclase is essential in S. cerevisiae, we wished to distinguish between two alternative models. In the first model, adenylyl cyclase is essential and Gpa1 is not essential because redundant upstream factors regulate adenylyl cyclase in gpa1 mutants. In the second model, neither Gpa1 nor adenylyl cyclase is essential for viability in C. neoformans.
We identified a fragment of the gene encoding C. neoformans adenylyl cyclase, CAC1 (Cryptococcus adenylyl cyclase), from serotype A strain H99 by using low-stringency PCR and degenerate primers based on conserved regions of other fungal adenylyl cyclase genes. Southern hybridization, under high or low stringency, revealed a single copy of the CAC1 gene. Two adjacent NheI restriction fragments, spanning the entire CAC1 open reading frame, were isolated from the genomic DNA of strain H99 by colony hybridization of size-selected libraries and then sequenced (GenBank accession number AF290191). The 5′ and 3′ regions of the CAC1 gene were determined by rapid amplification of cDNA ends, and the intron-exon boundaries were identified by comparing CAC1 cDNA fragments and the genomic sequence.
The CAC1 gene consists of 7,188 nucleotides from the start to the termination codons, contains 7 introns, and encodes a predicted protein of 2,271 amino acids. The C. neoformans adenylyl cyclase shares 62% amino acid sequence similarity and 45% identity from residues 838 to 2228 with the analogous region of its closest homolog, U. maydis adenylyl cyclase Uac1. Like other fungal adenylyl cyclase proteins (25, 63, 64), the C. neoformans Cac1 enzyme lacks the hydrophobic transmembrane domains characteristic of the mammalian enzymes (28). It does, however, share the tandemly repeated leucine-rich motifs found in the S. cerevisiae and S. pombe adenylyl cyclase proteins. These regions are predicted to serve regulatory or cell localization functions, since they are not required for catalytic activity (25).
To disrupt the C. neoformans CAC1 gene, an internal portion of the gene was replaced with the URA5 gene. The resulting cac1Δ::URA5 disruption construct was introduced into the serotype A ura5 strain H99-ura5 by biolistic transformation. In 1 isolate (RPC3) from among 120 Ura+ transformants, the CAC1 gene was replaced by integration of the cac1Δ::URA5 mutant allele, with no ectopic integrations (see Materials and Methods). Two independent cac1 mutant strains (LCC22 and LCC23) were also isolated by using the ADE2 gene as a selectable marker in ade2 mutant strain M001 (44, 53). The in vitro phenotypes of the three independent cac1 mutants were identical. It is important that all three mutants exhibited a budding morphology like that of wild-type cells. This observation is in contrast to the situation for another basidiomycete pathogen, U. maydis, in which disruption of the gene for adenylyl cyclase results in constitutive filamentation and the loss of budding growth (19).
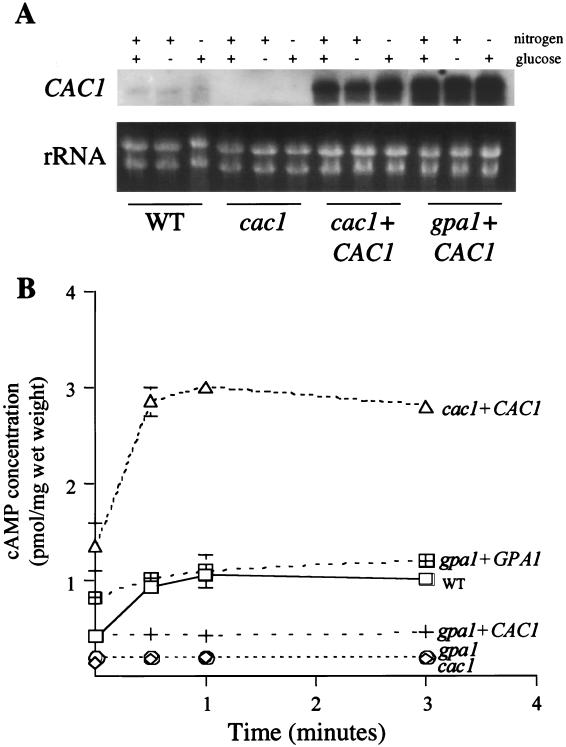
To ensure that the phenotypes observed were attributable to the cac1 adenylyl cyclase mutation, the wild-type CAC1 gene was reintroduced into the cac1 mutant background. The wild-type CAC1 gene was linked to the hph gene, encoding resistance to hygromycin B, and ectopically integrated into the genome of the cac1 mutant strains (RPC3 and LCC22) by biolistic transformation to generate cac1 CAC1-reconstituted strains. Three reconstituted strains in the RPC3 strain background and nine reconstituted strains in the LCC22 strain background demonstrated identical phenotypes in vitro. Therefore, one cac1 CAC1 strain from each mutant background was selected for a more detailed evaluation (RPC7 and LCC22-1). This transformation technique has been used to integrate C. neoformans genes and typically results in the integration of multiple tandem copies of the linked hph gene, which are apparently necessary for sufficient expression to confer hygromycin B resistance (3). We note that as a consequence, the reintroduced linked CAC1 gene is also present in multiple copies, often resulting in functional overexpression, which can be used for epistasis analysis. We have used this approach to achieve overexpression, since the serotype D GAL7-regulatable promoter is not tightly regulated or highly expressed in serotype A strains (14). Additionally, the pCnTel1 (16) and pPM8 (41) episomal plasmids, which have been used for overexpression studies with serotype D strains, are often unstable and integrated in serotype A strains (unpublished observations). To observe the effects of CAC1 overexpression in the gpa1 mutant background, the CAC1 gene was similarly introduced into a gpa1 mutant strain (AAC1). The in vitro phenotypes of four such gpa1 CAC1 strains were identical, and one (AAC17) was chosen for further studies. Northern analysis confirmed that the CAC1 gene was not expressed in the cac1 mutant strain but was markedly overexpressed in the gpa1 CAC1 and cac1 CAC1 strains, albeit to different extents, independent of medium conditions (Fig. 1A). In contrast to the important role of nutrient deprivation in the transcriptional regulation of the GPA1 gene (59), expression of the CAC1 gene was not induced by either glucose or nitrogen limitation (Fig. 1A).
FIG. 1.
Disruption of the adenylyl cyclase gene CAC1 abolishes cAMP production. (A) CAC1 wild-type (WT) (H99), cac1 mutant (RPC3), cac1 CAC1-reconstituted (RPC7), and gpa1 CAC1 (AAC17) strains were incubated in YPD medium for 18 h. The cells were divided for incubation for 4 h in one of three different media: synthetic complete medium with 2% glucose (nitrogen +, glucose +), synthetic complete medium with 0% glucose (nitrogen +, glucose −), or SLAD (nitrogen −, glucose +). Total RNA from each sample was assessed by Northern analysis with the CAC1 gene as a probe. The rRNA bands of the ethidium bromide-stained gel (rRNA) are shown to demonstrate RNA loading. (B) CAC1 wild-type (H99) (squares), cac1 mutant (LCC22) (diamonds), cac1 CAC1-reconstituted (LCC22-1) (triangles), gpa1 mutant (AAC1) (circles), gpa1 GPA1-reconstituted (AAC3) (crossed squares), and gpa1 CAC1 (AAC17) (crosses) strains were starved for glucose for 2 h. At the indicated time after a glucose pulse, aliquots of the cell suspensions were frozen, and intracellular cAMP concentrations were determined. Data points represent the mean and standard deviation for duplicate samples in two identical experiments (four samples for each data point).
The Gα protein Gpa1 and adenylyl cyclase control intracellular cAMP production.
The hypothesis that mutation of the CAC1 gene affects cAMP production in C. neoformans was tested by assaying intracellular cAMP levels (Fig. 1B). The CAC1 wild-type (H99), cac1Δ::ADE2 mutant (LCC22), cac1 CAC1-reconstituted (LCC22-1), gpa1 mutant (AAC1), gpa1 GPA1-reconstituted (AAC3), and gpa1 CAC1 (AAC17) strains were grown overnight in YPD medium and then starved in glucose-free buffer for 2 h. Following the readdition of glucose, cells were collected and frozen, and cAMP levels were determined by an enzyme immunoassay. The cAMP concentration was modestly increased in wild-type cells in response to glucose readdition, as in previous studies with S. cerevisiae (56) (Fig. 1B). No cAMP was detectable in total cell extracts from either the cac1 or the gpa1 mutant strains under any conditions (Fig. 1B). When the wild-type GPA1 gene was reintroduced into the gpa1 mutant strain, the basal cAMP level was modestly increased compared to that in wild-type cells, a result which may be attributable to partial activation of the pathway by the increased level of the Gpa1 protein. cAMP production in response to glucose readdition was also restored in the gpa1 GPA1 strain (Fig. 1B). In comparison, both the basal cAMP level and the glucose-induced cAMP increase were significantly enhanced in the cac1 CAC1 strain, in which adenylyl cyclase is overexpressed. Importantly, overexpression of adenylyl cyclase in the gpa1 mutant strain restored basal cAMP production but not glucose-stimulated cAMP synthesis. These findings support a model in which a receptor coupled to Gpa1 detects extracellular glucose and activated Gpa1 then stimulates cAMP production by adenylyl cyclase.
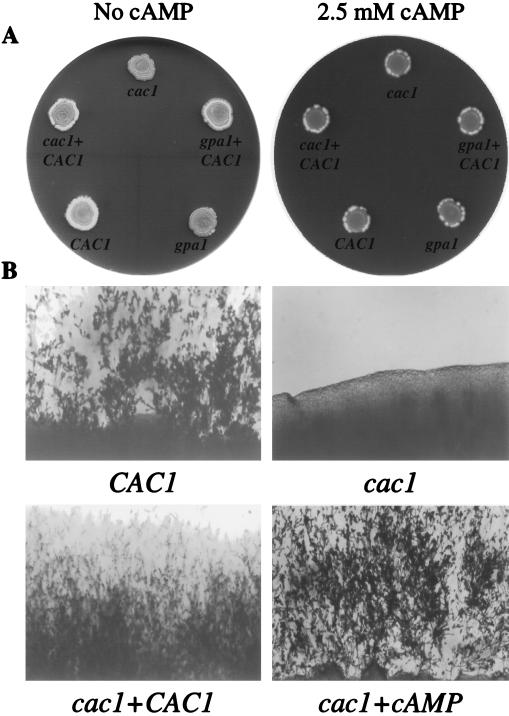
C. neoformans adenylyl cyclase is required for efficient mating.
The cac1 mutants lacking adenylyl cyclase showed significantly reduced mating compared to isogenic wild-type strains. The CAC1 wild-type (H99), cac1 mutant (RPC3), cac1 CAC1-reconstituted (RPC7), gpa1 mutant (AAC1), and gpa1 CAC1 (AAC17) strains were incubated in mating reactions with the serotype D MATa strain JEC20 on V8 mating medium. The mating reaction mixtures containing the wild-type and cac1 CAC1-reconstituted strains produced extensive mating hyphae after 7 days of incubation. In contrast, the cac1 and gpa1 mutant strains produced no mating hyphae after 7 days (Fig. 2). As previously described, isolated foci of mating hyphae were observed with the gpa1 mutant strain following prolonged incubation (≥14 days); however, no significant mating was observed with the cac1 mutant strain. The addition of 2.5 mM cAMP to V8 mating medium restored the mating of both gpa1 and cac1 mutant strains (Fig. 2). Similarly, overexpression of adenylyl cyclase restored the mating of the gpa1 mutant strain. These findings further support a model in which the Gα protein Gpa1 regulates cAMP production by the Cac1 fungal adenylyl cyclase.
FIG. 2.
Adenylyl cyclase is required for mating in C. neoformans. (A) CAC1 wild-type, cac1 mutant, cac1 CAC1-reconstituted, gpa1 mutant, and gpa1 CAC1 strains were coincubated with the MATa strain JEC20 on V8 mating medium in the dark for 14 days at 30°C with and without cAMP (2.5 mM). (B) The edges of the mating mixtures were examined microscopically each day for mating hyphae and photographed after 7 days (×61).
cAMP restores melanin production in adenylyl cyclase mutant strains.
We next tested whether adenylyl cyclase is required for melanin production. The CAC1 wild-type (H99), cac1 mutant (RPC3), cac1 CAC1-reconstituted (PRC7), gpa1 mutant (AAC1), and gpa1 CAC1 (AAC17) strains were incubated on Niger seed medium with and without cAMP. The wild-type and cac1 CAC1-reconstituted strains produced similar amounts of melanin after 3 to 4 days of incubation at 37°C, whereas the cac1 and gpa1 mutant strains made little or no visible melanin, even after 7 days of incubation (Fig. 3). Exogenous cAMP restored melanin production by both gpa1 and cac1 mutant strains. Interestingly, overexpression of adenylyl cyclase in the gpa1 CAC1 strain partially restored melanin production after 72 h of incubation on Niger seed medium, and exogenous cAMP further enhanced melanin production by this strain. These observations demonstrate that adenylyl cyclase is necessary for melanin production and provide additional evidence that Gpa1 normally functions to regulate cAMP production by adenylyl cyclase.
FIG. 3.
Adenylyl cyclase mutants have defects in melanin production. CAC1 wild-type (WT), cac1 mutant, cac1 CAC1-reconstituted, gpa1 mutant, and gpa1 CAC1 strains were grown on Niger seed medium with (+) and without (−) cAMP (2.5 mM) at 37°C and photographed after 4 days. Strains that produce melanin are brown (gray on figure), whereas strains that produce less or no melanin are white.
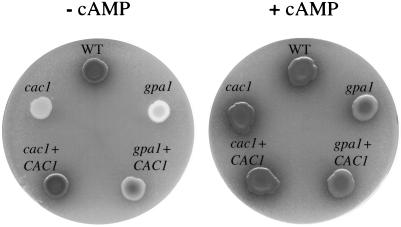
cAMP suppresses the capsule defect of adenylyl cyclase mutant strains.
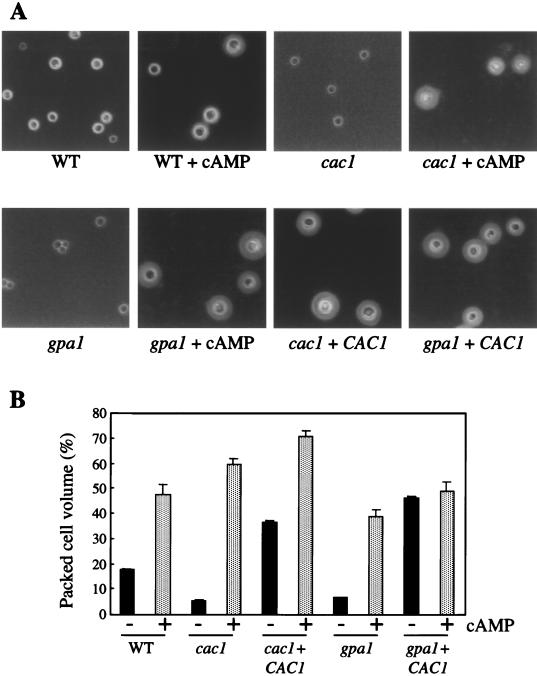
The polysaccharide capsule of C. neoformans is induced by conditions that mimic environmental signals encountered by this pathogenic yeast in the infected host, including iron deprivation and physiological CO2/HCO3− levels (20). To assess the role of adenylyl cyclase in C. neoformans capsule production, the CAC1 wild-type (H99), cac1 mutant (RPC3), cac1 CAC1-reconstituted (RPC7), gpa1 mutant (AAC1), and gpa1 CAC1 (AAC17) strains were incubated in DMEM–22 mM NaHCO3 for 24 h. India ink analysis revealed a marked decrease in the size of the capsule in the cac1 and gpa1 mutant strains compared with the wild-type strain (Fig. 4A). When the CAC1 gene was introduced into either the cac1 or the gpa1 mutant strains, capsule induction was restored (Fig. 4A). Exogenous cAMP also restored capsule production by the cac1 and gpa1 mutant strains and increased capsule size in all of the strains tested (Fig. 4A).
FIG. 4.
Adenylyl cyclase mutants fail to induce capsule. (A) CAC1 wild-type (WT), cac1 mutant, cac1 CAC1-reconstituted, gpa1 mutant, and gpa1 CAC1 strains were incubated for 2 days under capsule-inducing conditions (DMEM–22 mM NaHCO3) with and without cAMP (20 mM). Capsule induction was qualitatively assessed with a standard India ink preparation and photographed (×61). (B) Capsule size was quantified by determining the packed-cell volume of normalized cell suspensions (109 cells/ml) for each sample. Data points represent the mean and standard error for triplicate samples.
To quantify the differences in capsule production of these strains, the packed-cell volume (3, 20), which is related to capsule size, was determined for normalized suspensions of strains incubated in capsule-inducing medium in the absence or presence of cAMP. The quantitative findings indicated a significant decrease in capsule size in the cac1 and gpa1 mutant strains as well as complete restoration of encapsulation in these mutant strains by exogenous cAMP or the CAC1 adenylyl cyclase gene (Fig. 4B).
Adenylyl cyclase is required for C. neoformans virulence.
The roles of melanin and the polysaccharide capsule in the pathogenesis of cryptococcal infections are well established. Strains deficient in either melanin or capsule production are dramatically attenuated for virulence in animal models of cryptococcosis (7–9, 32, 48). Moreover, C. neoformans gpa1 mutant strains are significantly less virulent than wild-type parent strains in both rabbit and murine models of cryptococcal meningitis (3, 15). Based on models in which Gpa1 regulates adenylyl cyclase, we hypothesized that disruption of the CAC1 gene would similarly impair virulence.
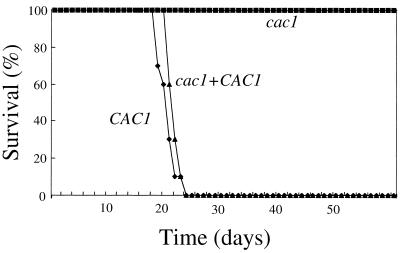
Ten A/Jcr mice were infected by inhalation with the CAC1 wild-type (H99), cac1 mutant (RPC3), and cac1 CAC1-reconstituted (RPC7) strains. In this model, inhaled cells initially infect the lung and then disseminate hematogenously to infect the brain, resulting in meningoencephalitis in all infected animals. Survival was monitored over the course of a 60-day infection. Mice infected with the CAC1 wild-type strain survived a median of 20 days (Fig. 5). In contrast, the cac1 mutant strain was avirulent, and no lethal infection was observed in any infected animal (P < 0.01) (Fig. 5). Importantly, reintroduction of the wild-type CAC1 gene restored the virulence of the cac1 mutant strain to the wild-type level (median survival, 21 days; P = 0.076 in a comparison with the wild-type strain).
FIG. 5.
Adenylyl cyclase is required for virulence of C. neoformans. Ten A/Jcr mice were intranasally inoculated with the CAC1 wild-type strain (diamonds), the cac1 mutant strain (squares), or the cac1 CAC1-reconstituted strain (triangles). The three groups of mice (30 total) were monitored for survival over 60 days.
Two independent cac1Δ::ADE2 mutant strains were also found to be avirulent. In the murine inhalation model of systemic cryptococcosis, the cac1Δ::ADE2 mutant strains (LCC22 and LCC23) produced no lethal infections after 120 days of observation. Isogenic CAC1 wild-type strains induced lethal infections in all animals by day 23 in the same experiment. Additionally, in the rabbit model of cryptococcal meningitis, cac1 mutant strain LCC22 showed a significant reduction in the ability to survive in the host compared to the wild-type strain. The number of viable cells recovered from the CSF of animals infected with cac1 mutant strain LCC22 was reduced approximately 10,000-fold compared to the results obtained with the wild-type strain after 10 days of infection (1.5 × 105 CFU/ml of CSF for the wild-type strain; 60 CFU/ml of CSF for the cac1 mutant strain). In summary, in three separate experiments with three independent cac1 mutant strains, adenylyl cyclase was found to be required for C. neoformans virulence.
DISCUSSION
All cells must sense and respond to changes in the extracellular environment. In pathogenic microorganisms, dramatic cellular adaptation occurs as these organisms infect their host. An understanding of the mechanisms by which these organisms adapt to and infect their host underlies the basis for the molecular dissection of microbial pathogenesis.
Here we demonstrate that the enzyme adenylyl cyclase and its second messenger product, cAMP, play a central role in the differentiation and virulence of the opportunistic fungal pathogen C. neoformans. First, we identified a single, nonessential gene encoding adenylyl cyclase in this fungus. Second, we showed that the adenylyl cyclase Cac1 plays a central role in the induction of two virulence factors, capsule and melanin. Third, we demonstrated that adenylyl cyclase is required for virulence. Mutant phenotypes conferred by the adenylyl cyclase mutation were completely remediated by cAMP in vitro, indicating that the enzyme is catalytic and that it is not required for scaffolding of other signaling components.
Previously it was found that the differentiation and virulence of C. neoformans are controlled by the Gα protein Gpa1 (3; Allen et al., unpublished). Because the gpa1 mutant phenotypes were suppressed by exogenous cAMP, it was proposed that Gpa1 might function by regulating cAMP production. Here we isolated the CAC1 gene encoding adenylyl cyclase and, by molecular genetic approaches, provide compelling support for this model. Strains lacking the adenylyl cyclase gene were sterile, similar to gpa1 mutants. Additionally, both the gpa1 and the cac1 mutant strains failed to induce the expression of the major virulence determinants capsule and melanin. All mutant phenotypes were suppressed by exogenous cAMP or by overexpression of adenylyl cyclase. These findings support a model in which the Gpa1 and Cac1 proteins function in a linear pathway controlling cAMP production. Our studies and recent findings obtained with budding and fission yeasts underscore how this pathway is conserved between microorganisms and humans.
Determination of intracellular cAMP levels provides additional evidence that the C. neoformans Gα protein Gpa1 and adenylyl cyclase functionally interact to regulate cAMP production. The cAMP level increased when glucose was added to glucose-starved C. neoformans cells, directly implicating cAMP as a central element in C. neoformans nutrient-sensing pathways. Similar responses to glucose have been observed for S. cerevisiae (56). In contrast, no cAMP was detectable in the cac1 mutant strain. That no cAMP was produced in the gpa1 mutant strain further supports a central role for the Gα protein Gpa1 in cAMP signaling. Interestingly, the basal cAMP level was restored in the gpa1 mutant strain overexpressing adenylyl cyclase, but glucose failed to stimulate cAMP production. In the same strain background, reintroduction of a wild-type GPA1 gene complemented the mutant cAMP defect to wild-type levels. Although the interpretation of these results may be limited by the number of strains tested, together these findings suggest that the Gpa1 protein is required to link a glucose-sensing G-protein-coupled receptor to adenylyl cyclase. This receptor has not yet been identified for C. neoformans, but the corresponding receptors in budding yeast (Gpr1) and fission yeast (git3) are known (61, 62).
Other fungal G protein subunits in this conserved signal transduction pathway have been identified. The fission yeast Gβ subunit git5 and the Gγ subunit git11 interact with the Gα protein gpa2 and are required for adenylyl cyclase activation by glucose (34). Thus far, only one Gβ subunit (Gpb1) and no Gγ subunits have been identified for C. neoformans (60). The Gpb1 protein clearly functions in pheromone sensing and mating and is not coupled to the Gpa1-cAMP cascade (60). Its role is analogous to those of the βγ subunits Ste4 and Ste18 in pheromone sensing in S. cerevisiae but is quite distinct from the role of the βγ subunits git5 and git11 that function with gpa2 in nutrient sensing in S. pombe. Therefore, in budding yeast and in C. neoformans, the nutrient-sensing Gα subunits function in the absence of other G protein subunits or with novel subunits that remain to be identified (reviewed in reference 35).
Mating in the fission yeast S. pombe requires a nutrient-poor medium, and the S. pombe gpa2-adenylyl cyclase pathway plays a central role in signaling nutrient-rich conditions. cAMP levels are regulated in response to carbon source, although nitrogen source may also play a role (52). Mutation of the S. pombe cyr1 adenylyl cyclase gene leads to starvation-independent mating on nutrient-rich medium (39). Similarly, gpa2 mutant cells mate and sporulate in rich medium and fail to produce cAMP in response to glucose (23). Thus, the cAMP pathway in S. pombe functions to signal the presence of abundant nutrients, either carbon source or nitrogen source, and mutations in this pathway result in starvation-independent mating (52).
Like fission yeast mating, C. neoformans mating can occur on a nutrient-poor medium limiting for nitrogen source but containing abundant fermentable carbon source, such as SLAD medium. However, disruption of cAMP signaling in C. neoformans impairs mating. These observations could suggest that the C. neoformans Gpa1-Cac1-cAMP pathway is activated by nutrient deprivation signals rather than the presence of abundant nutrients. Alternatively, the Gpa1-Cac1-cAMP pathway might function to sense abundant fermentable carbon sources and thereby stimulate the mating of C. neoformans. In this model, the pathway inhibits mating in S. pombe and stimulates mating in C. neoformans. Nitrogen limitation stimulates mating and meiosis in S. pombe, pseudohyphal differentiation in S. cerevisiae, and haploid fruiting and mating in C. neoformans. These events are also regulated by fermentable carbon sources that are sensed by the cAMP signaling cascade. cAMP inhibits mating in S. pombe, possibly to restrict mating and sporulation until both nitrogen and fermentable carbon sources have been exhausted. In contrast, pseudohyphal growth in S. cerevisiae and filamentous differentiation and mating in C. neoformans are stimulated by fermentable carbon sources and activated by cAMP. Thus, mutations in the Gα protein and adenylyl cyclase lead to a loss of carbon source sensing and defects in development in budding yeast and C. neoformans on minimal medium and yet to precocious mating of S. pombe on rich medium. Although the precise roles of components of the cAMP pathway differ between organisms, the basic functions of Gα proteins and cAMP in nutrient sensing and mating are conserved among divergent fungal species.
The mating processes of other fungi are also dependent on nutrient-sensing Gα protein-cAMP signal transduction pathways. In U. maydis, a basidiomycete plant pathogen, a Gα protein-cAMP pathway regulates mating. Strains with mutations in the ubc1 gene, encoding a PKA regulatory subunit, are mating defective (19). Additionally, the Gα protein Gpa3, homologous to C. neoformans Gpa1, regulates cAMP signaling, mating, and virulence in this organism (27). The cAMP pathway also plays a major role in morphogenesis in U. maydis because mutants defective in adenylyl cyclase display a constitutively filamentous phenotype (19). As we have demonstrated here, disruption of the CAC1 gene does not trigger filamentous growth in C. neoformans.
G protein activation of adenylyl cyclases has been intensively investigated in other systems, including the slime mold Dictyostelium discoideum. Cell surface receptors sense extracellular cAMP to monitor cell density signals (22, 26, 50). The cAMP receptors in turn control intracellular adenylyl cyclase and PKA activities through G protein activation. The molecular dissociation and reassociation of heterotrimeric G protein subunits in response to receptor activation by the cAMP ligand were recently demonstrated by fluorescence resonance energy transfer (24). At present, there is no evidence that fungi express extracellular cAMP receptors. However, the basic signaling machinery by which G proteins regulate adenylyl cyclases in response to extracellular signals to control developmental processes is remarkably conserved.
In addition to regulating mating and differentiation, the C. neoformans cAMP pathway controls two major virulence factors: capsule and melanin. There was no difference in the growth rates of the isogenic wild-type and cac1 mutant strains at 30 or 37°C, arguing that the phenotypes of the mutant are not simply attributable to defects in growth. In animal experiments, the cac1 adenylyl cyclase mutant strain had defects in capsule and melanin that conferred a severe disadvantage in the host. This strain was completely avirulent in two animal models with different modes of infection. Therefore, a functioning cAMP pathway is necessary for the expression of virulence irrespective of animal host or site of infection. Furthermore, in comparison to C. neoformans mutants lacking Gpa1 (15), phospholipase B (11), or urease (12), which are attenuated but not avirulent, the cac1 mutant strain was more severely compromised in the host. This mutational block may result in an in vivo fungicidal response, which could have therapeutic implications in the design of fungicidal drugs that target this enzyme.
While the phenotypes of adenylyl cyclase mutant strains are strikingly similar to those of strains lacking the Gα protein Gpa1, the cac1 mutation confers more severe phenotypes. Some degree of mating by gpa1 mutants is observed after prolonged incubation, whereas cac1 mutants are completely sterile. Higher concentrations of cAMP are required to suppress the capsule defect of cac1 mutant strains compared to gpa1 mutant strains (3). In the murine inhalation model of cryptococcosis, gpa1 mutant strains are attenuated for virulence but are still capable of causing lethal infections (15). In contrast, no lethal infections are observed with cac1 mutant strains. These observations could be explained by a basal level of adenylyl cyclase activity present in gpa1 mutants but not in cac1 mutants, possibly implicating other signaling elements that also act on adenylyl cyclase function. For example, the C. neoformans RAS1 protein may play a dual role in the regulation of both cAMP and pheromone-responsive pathways (1). We note that the cAMP measurements (Fig. 1B) showing an undetectable level of cAMP in the gpa1 and cac1 mutants argue that the quantitative phenotypic differences between the gpa1 and cac1 mutant strains may not simply be attributable to differences in cAMP levels in total extracts prepared from bulk cultures. We offer three possible explanations. First, since the cAMP levels were below the limits of detection of this assay, gpa1 mutant cells may have a higher level of cAMP than cac1 mutant cells that we cannot detect. Second, there may be a difference in cAMP levels in some cells of the population such that some gpa1 mutant cells have more cAMP than cac1 mutant cells. Third, there may be differences in localized cAMP levels that are not detected in total cell extracts. The identification of other signaling elements in this conserved pathway should further illuminate the molecular regulation of microbial pathogenicity and eukaryotic cellular differentiation.
Acknowledgments
We thank Cameron DeCesare for technical assistance and Tina J. Wilkins for assistance with manuscript preparation.
This work was supported in part by NIAID K08 award AI01556 (to J.A.A.) and NIAID R01 grants AI39115, AI41937, and AI42159 (to J.H.) and AI28388 (to J.R.P.). This work was also supported by P01 award AI44975 from the NIAID to the Duke University Mycology Research Unit and by a grant from the Canadian Institutes of Health Research (to J.W.K.). J. Andrew Alspaugh is an IDSA Merck Young Investigator in Medical Mycology. Joseph Heitman and James W. Kronstad are Burroughs Wellcome Scholars in Molecular Pathogenic Mycology. Gary M. Cox and J. Andrew Alspaugh are Burroughs Wellcome New Investigators in Molecular Pathogenic Mycology. Joseph Heitman is an associate investigator of the Howard Hughes Medical Institute.
REFERENCES
- 1.Alspaugh, J. A., L. M. Cavallo, J. R. Perfect, and J. Heitman. 2000. RAS1 regulates filamentation, mating and growth at high temperature of Cryptococcus neoformans. Mol. Microbiol. 36: 352–365. [DOI] [PubMed] [Google Scholar]
- 2.Alspaugh, J. A., R. C. Davidson, and J. Heitman. 2000. Morphogenesis of Cryptococcus neoformans. Contrib. Microbiol. 5: 217–238. [DOI] [PubMed] [Google Scholar]
- 3.Alspaugh, J. A., J. R. Perfect, and J. Heitman. 1997. Cryptococcus neoformans mating and virulence are regulated by the G-protein α subunit GPA1 and cAMP. Genes Dev. 11: 3206–3217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Alspaugh, J. A., J. R. Perfect, and J. Heitman. 1998. Signal transduction pathways regulating differentiation and pathogenicity of Cryptococcus neoformans. Fungal Genet. Biol. 25: 1–14. [DOI] [PubMed] [Google Scholar]
- 5.Borges-Walmsley, M. I., and A. R. Walmsley. 2000. cAMP signalling in pathogenic fungi: control of dimorphic switching and pathogenicity. Trends Microbiol. 8: 133–141. [DOI] [PubMed] [Google Scholar]
- 6.Broach, J. R., and R. J. Deschenes. 1990. The functions of RAS genes in Saccharomyces cerevisiae. Adv. Cancer Res. 54: 79–138. [DOI] [PubMed] [Google Scholar]
- 7.Chang, Y. C., and K. J. Kwon-Chung. 1994. Complementation of a capsule-deficient mutation of Cryptococcus neoformans restores its virulence. Mol. Cell. Biol. 14: 4912–4919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chang, Y. C., and K. J. Kwon-Chung. 1998. Isolation of the third capsule-associated gene, CAP60, required for virulence in Cryptococcus neoformans. Infect. Immun. 66: 2230–2236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chang, Y. C., L. A. Penoyer, and K. J. Kwon-Chung. 1996. The second capsule gene of Cryptococcus neoformans, CAP64, is essential for virulence. Infect. Immun. 64: 1977–1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Colombo, S., P. Ma, L. Cauwenberg, J. Winderickx, M. Crauwels, A. Teunissen, D. Nauwelaers, J. H. de Winde, M. Gorwa, D. Colavizza, and J. M. Thevelein. 1998. Involvement of distinct G-proteins, Gpa2 and Ras, in glucose- and intracellular acidification-induced cAMP signalling in the yeast Saccharomyces cerevisiae. EMBO J. 17: 3326–3341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cox, G. M., H. C. McDade, S. C. A. Chen, S. C. Tucker, M. Gottredsson, L. C. Wright, T. C. Sorrell, S. D. Leidich, A. Casadevall, M. A. Ghannoum, and J. R. Perfect. 2001. Extracellular phospholipase activity is a virulence factor for Cryptococcus neoformans. Mol. Microbiol. 39: 166–175. [DOI] [PubMed] [Google Scholar]
- 12.Cox, G. M., J. Mukherjee, G. T. Cole, A. Casadevall, and J. R. Perfect. 2000. Urease as a virulence factor in experimental cryptococcosis. Infect. Immun. 68: 443–448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cox, G. M., D. L. Toffaletti, and J. R. Perfect. 1996. Dominant selection system for use in Cryptococcus neoformans. J. Med. Vet. Mycol. 34: 385–391. [PubMed] [Google Scholar]
- 14.Del Poeta, M., D. L. Toffaletti, T. H. Rude, C. C. Dykstra, J. Heitman, and J. R. Perfect. 1999. Topoisomerase I is essential in Cryptococcus neoformans: role in pathobiology and as an antifungal target. Genetics 152: 167–178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.D’Souza, C. A., J. A. Alspaugh, C. Yue, T. Harashima, G. M. Cox, J. R. Perfect, and J. Heitman. 2001. Cyclic AMP-dependent protein kinase controls virulence of the fungal pathogen Cryptococcus neoformans. Mol. Cell. Biol. 21: 3179–3191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Edman, J. C. 1992. Isolation of telomerelike sequences from Cryptococcus neoformans and their use in high-efficiency transformation. Mol. Cell. Biol. 12: 2777–2783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Fukui, Y., T. Kozasa, Y. Kaziro, T. Takeda, and M. Yamamoto. 1986. Role of ras homolog in the life cycle of Schizosaccharomyces pombe. Cell 44: 329–336. [DOI] [PubMed] [Google Scholar]
- 18.Gimeno, C. J., P. O. Ljungdahl, C. A. Styles, and G. R. Fink. 1992. Unipolar cell divisions in the yeast S. cerevisiae lead to filamentous growth: regulation by starvation and RAS. Cell 68: 1077–1090. [DOI] [PubMed] [Google Scholar]
- 19.Gold, S., G. Duncan, K. Barrett, and J. Kronstad. 1994. cAMP regulates morphogenesis in the fungal pathogen Ustilago maydis. Genes Dev. 8: 2805–2816. [DOI] [PubMed] [Google Scholar]
- 20.Granger, D. L., J. R. Perfect, and D. T. Durack. 1985. Virulence of Cryptococcus neoformans: regulation of capsule synthesis by carbon dioxide. J. Clin. Investig. 76: 508–516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hartmann, H. A., J. Krüger, F. Lottspeich, and R. Kahmann. 1999. Environmental signals controlling sexual development of the corn smut fungus Ustilago maydis through the transcriptional regulator Prfl. Plant Cell 11: 1293–1305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Insall, R. H., R. D. M. Soede, P. Schaap, and P. N. Devreotes. 1994. Two cAMP receptors activate common signaling pathways in Dictyostelium. Mol. Biol. Cell 5: 703–711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Isshiki, T., N. Mochizuki, T. Maeda, and M. Yamamoto. 1992. Characterization of a fission yeast gene, gpa2, that encodes a Gα subunit involved in the monitoring of nutrition. Genes Dev. 6: 2455–2462. [DOI] [PubMed] [Google Scholar]
- 24.Janetopoulos, C., T. Jin, and P. Devreotes. 2001. Receptor-mediated activation of heterotrimeric G-proteins in living cells. Science 291: 2408–2411. [DOI] [PubMed] [Google Scholar]
- 25.Kataoka, T., D. Broek, and M. Wigler. 1985. DNA sequence and characterization of the S. cerevisiae gene encoding adenylate cyclase. Cell 43: 493–505. [DOI] [PubMed] [Google Scholar]
- 26.Klein, P. S., T. J. Sun, I. C. L. Saxe, A. R. Kimmel, R. L. Johnson, and P. N. Devreotes. 1988. A chemoattractant receptor controls development in Dictyostelium discoideum. Science 241: 1467–1472. [DOI] [PubMed] [Google Scholar]
- 27.Krüger, J., G. Loubradou, E. Regenfelder, A. Hartmann, and R. Kahmann. 1998. Crosstalk between cAMP and pheromone signalling pathways in Ustilago maydis. Mol. Gen. Genet. 260: 193–198. [DOI] [PubMed] [Google Scholar]
- 28.Krupinski, J., F. Coussen, H. A. Bakalyar, W. J. Tang, P. G. Feinstein, K. Orth, C. Slaughter, R. R. Reed, and A. G. Gilman. 1989. Adenylyl cyclase amino acid sequence: possible channel- or transporter-like structure. Science 244: 1558–1564. [DOI] [PubMed] [Google Scholar]
- 29.Kübler, E., H. U. Mösch, S. Rupp, and M. P. Lisanti. 1997. Gpa2p, a G-protein alpha-subunit, regulates growth and pseudohyphal development in Saccharomyces cerevisiae via a cAMP-dependent mechanism. J. Biol. Chem. 272: 20321–20323. [DOI] [PubMed] [Google Scholar]
- 30.Kwon-Chung, K. J., and J. E. Bennett. 1991. Medical mycology, p. 397–446. Lea and Febinger, Malvern, Pa.
- 31.Kwon-Chung, K. J., J. C. Edman, and B. L. Wickes. 1992. Genetic association of mating types and virulence in Cryptococcus neoformans. Infect. Immun. 60: 602–605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kwon-Chung, K. J., and J. C. Rhodes. 1986. Encapsulation and melanin formation as indicators of virulence in Cryptococcus neoformans. Infect. Immun. 51: 218–223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kwon-Chung, K. J., A. Varma, J. C. Edman, and J. E. Bennett. 1992. Selection of ura5 and ura3 mutants from the two varieties of Cryptococcus neoformans on 5-fluoroorotic acid medium. J. Med. Vet. Mycol. 30: 61–69. [PubMed] [Google Scholar]
- 34.Landry, S., M. T. Pettit, E. Apolinario, and C. S. Hoffman. 2000. The fission yeast git5 gene encodes a Gβ subunit required for glucose-triggered adenylate cyclase activation. Genetics 154: 1463–1471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lengeler, K. B., R. C. Davidson, C. D’Souza, T. Harashima, W.-C. Shen, P. Wang, X. Pan, M. Waugh, and J. Heitman. 2000. Signal transduction cascades regulating fungal development and virulence. Microbiol. Mol. Biol. Rev. 64: 746–785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lengeler, K. B., P. Wang, G. M. Cox, J. R. Perfect, and J. Heitman. 2000. Identification of the MATa mating type locus of Cryptococcus neoformans reveals a serotype A MATa strain thought to have been extinct. Proc. Natl. Acad. Sci. USA 97: 14455–14460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lorenz, M. C., and J. Heitman. 1997. Yeast pseudohyphal growth is regulated by GPA2, a G protein α homolog. EMBO J. 16: 7008–7018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lorenz, M. C., X. Pan, T. Harashima, M. E. Cardenas, Y. Xue, J. P. Hirsch, and J. Heitman. 2000. The G protein-coupled receptor GPR1 is a nutrient sensor that regulates pseudohyphal differentiation in Saccharomyces cerevisiae. Genetics 154: 609–622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Maeda, T., N. Mochizuki, and M. Yamamoto. 1990. Adenylyl cyclase is dispensable for vegetative cell growth in the fission yeast Schizosaccharomyces pombe. Proc. Natl. Acad. Sci. USA 87: 7814–7818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Matsumoto, K., I. Uno, Y. Oshima, and T. Ishikawa. 1982. Isolation and characterization of yeast mutants deficient in adenylate cyclase and cAMP-dependent protein kinase. Proc. Natl. Acad. Sci. USA 79: 2355–2359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Mondon, P., Y. C. Chang, A. Varma, and K. J. Kwon-Chung. 2000. A novel episomal shuttle vector for transformation of Cryptococcus neoformans with the ccdB gene as a positive selection marker in bacteria. FEMS Microbiol. Lett. 187: 41–45. [DOI] [PubMed] [Google Scholar]
- 42.Pan, X., and J. Heitman. 1999. Cyclic AMP-dependent protein kinase regulates pseudohyphal differentiation in Saccharomyces cerevisiae. Mol. Cell. Biol. 19: 4874–4887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Perfect, J. R., S. D. R. Lang, and D. T. Durack. 1980. Chronic cryptococcal meningitis: a new experimental model in rabbits. Am. J. Pathol. 101: 177–194. [PMC free article] [PubMed] [Google Scholar]
- 44.Perfect, J. R., D. L. Toffaletti, and T. H. Rude. 1993. The gene encoding phosphoribosylaminoimidazole carboxylase (ADE2) is essential for growth of Cryptococcus neoformans in cerebrospinal fluid. Infect. Immun. 61: 4446–4451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Pitkin, J. W., D. G. Panaccione, and J. D. Walton. 1996. A putative cyclic peptide efflux pump encoded by the TOXA gene of the plant-pathogenic fungus Cochliobolus carbonum. Microbiology 142: 1557–1565. [DOI] [PubMed] [Google Scholar]
- 46.Regenfelder, E., T. Spellig, A. Hartmann, S. Lauenstein, M. Bölker, and R. Kahmann. 1997. G proteins in Ustilago maydis: transmission of multiple signals? EMBO J. 16: 1934–1942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Robertson, L. S., and G. R. Fink. 1998. The three yeast A kinases have specific signaling functions in pseudohyphal growth. Proc. Natl. Acad. Sci. USA 95: 13783–13787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Salas, S. D., J. E. Bennett, K. J. Kwon-Chung, J. R. Perfect, and P. R. Williamson. 1996. Effect of the laccase gene, CNLAC1, on virulence of Cryptococcus neoformans. J. Exp. Med. 184: 377–386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 50.Saxe, I. C. L., G. T. Ginsburg, J. M. Louis, R. Johnson, P. N. Devreotes, and A. R. Kimmel. 1993. CAR2, a prestalk cAMP receptor required for normal tip formation and late development of Dictyostelium discoideum. Genes Dev. 7: 262–272. [DOI] [PubMed] [Google Scholar]
- 51.Sherman, F. 1991. Getting started with yeast. Methods Enzymol. 194: 3–21. [DOI] [PubMed] [Google Scholar]
- 52.Stettler, S., E. Warbrick, S. Prochnik, S. Mackie, and P. Fantes. 1996. The wis1 signal transduction pathway is required for expression of cAMP-repressed genes in fission yeast. J. Cell Sci. 109: 1927–1935. [DOI] [PubMed] [Google Scholar]
- 53.Sudarshan, S., R. C. Davidson, J. Heitman, and J. A. Alspaugh. 1999. Molecular analysis of the Cryptococcus neoformans ADE2 gene, a selectable marker for transformation and gene disruption. Fungal Genet. Biol. 27: 36–48. [DOI] [PubMed] [Google Scholar]
- 54.Taussig, R., and A. G. Gilman. 1995. Mammalian membrane-bound adenylyl cyclases. J. Biol. Chem. 270: 1–4. [DOI] [PubMed] [Google Scholar]
- 55.Tesmer, J. J., R. K. Sunahara, A. G. Gilman, and S. R. Sprang. 1997. Crystal structure of the catalytic domains of adenylyl cyclase in a complex with Gsα.GTPγS. Science 278: 1907–1916. [DOI] [PubMed] [Google Scholar]
- 56.Thevelein, J. M., and J. H. de Winde. 1999. Novel sensing mechanisms and targets for the cAMP-protein kinase A pathway in the yeast Saccharomyces cerevisiae. Mol. Microbiol. 33: 904–918. [DOI] [PubMed] [Google Scholar]
- 57.Toda, T., S. Cameron, P. Sass, and M. Wigler. 1988. SCH9, a gene of Saccharomyces cerevisiae that encodes a protein distinct from, but functionally and structurally related to, cAMP-dependent protein kinase catalytic subunits. Genes Dev. 2: 517–527. [DOI] [PubMed] [Google Scholar]
- 58.Toda, T., I. Uno, T. Ishikawa, S. Powers, T. Kataoka, D. Broek, S. Cameron, J. Broach, K. Matsumoto, and M. Wigler. 1985. In yeast, RAS proteins are controlling elements of adenylate cyclase. Cell 40: 27–36. [DOI] [PubMed] [Google Scholar]
- 59.Tolkacheva, T., P. McNamara, E. Piekarz, and W. Courchesne. 1994. Cloning of a Cryptococcus neoformans gene, GPA1, encoding a G-protein α-subunit homolog. Infect. Immun. 62: 2849–2856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Wang, P., J. R. Perfect, and J. Heitman. 2000. The G-protein β-subunit GPB1 is required for mating and haploid fruiting in Cryptococcus neoformans. Mol. Cell. Biol. 20: 352–362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Welton, R. M., and C. S. Hoffman. 2000. Glucose monitoring in fission yeast via the gpa2 Gα, the git5 Gβ and the git3 putative glucose receptor. Genetics 156: 513–521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Xue, Y., M. Batlle, and J. P. Hirsch. 1998. GPR1 encodes a putative G protein-coupled receptor that associates with the Gpa2p Gα subunit and functions in a Ras-independent pathway. EMBO J. 17: 1996–2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Yamawaki-Kataoka, Y., T. Tamaoki, H. R. Choe, H. Tanaka, and T. Kataoka. 1989. Adenylate cyclases in yeast: a comparison of the genes from Schizosaccharomyces pombe and Saccharomyces cerevisiae. Proc. Natl. Acad. Sci. USA 86: 5693–5697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Young, D., M. Riggs, J. Field, A. Vojtek, D. Broek, and M. Wigler. 1989. The adenylyl cyclase gene from Schizosaccharomyces pombe. Proc. Natl. Acad. Sci. USA 86: 7989–7993. [DOI] [PMC free article] [PubMed] [Google Scholar]