Abstract
Purpose
Head and neck squamous cell carcinoma (HNSCC) is a common cancer with high mortality and poor prognosis partially owing to lack of application of predictive markers. Increasing evidence has suggested that metabolic dysregulation plays an important part in tumorigenesis. We aim to identify a prognostic metabolic pathway (MP) signature in HNSCC.
Methods
Single sample gene-set enrichment analysis (ssGSEA) was used in metabolic gene sets to develop a metabolism-based prognostic risk score (MPRS) for HNSCC using Cox regression analysis (univariate, LASSO, and stepwise multiple cox analysis), which was then validated in different subgroups, and association with clinical and mutational features was analyzed.
Results
Seventy-two dysregulated metabolic pathways were identified, and a six-MP signature (6MPS) was constructed which can effectively distinguish between the high- and low-risk patients in both training and testing sets, accompanied with high sensitivity and specificity (AUC = 0.7) in prognosis prediction. 6MPS was also applicable to patients of different subgroups. Furthermore, 6MPS is not only an independent prognostic predictor but also associated with clinicopathological and mutational features. Higher tumor stage and tumor mutation burden (TMB) have a higher MPRS.
Conclusion
6MPS functions not only as a promising predictor of prognosis and survival but also as potential marker for therapeutic schedule monitoring.
Electronic supplementary material
The online version of this article (10.1007/s00432-020-03155-4) contains supplementary material, which is available to authorized users.
Keywords: Biomarker, Prognosis, Survival, Head and neck squamous cell carcinoma, Metabolic pathway
Introduction
Head and neck cancers consist of tumors occurring in oral cavity, throat, and pharynx, and the predominant histology is squamous cell carcinoma (Cai et al. 2017; Moy et al. 2017). More than 600,000 cases of head and neck squamous cell carcinoma (HNSCC) are diagnosed each year, making it the sixth most common malignancy around the world (Moskovitz et al. 2017). The high mortality and poor prognosis of HNSCC, partially blaming the lack of suitable predictors for diagnosis and prognosis, calls for an urgent need for applicable biomarkers in this field (Budach and Tinhofer 2019). Despite advances in the therapeutic methods of HNSCC, its survival rate remained lower than 50% (Siegel et al. 2017). Stratification of HNSCC patients into high- and low-risk group upon multiple markers could be a new approach for improvement of the prognosis of HNSCC patients. Patients with lower risk could receive conventional treatment, while higher risk patients will be going through a stricter monitoring and can be assigned to novel clinical trials. This could in turn elevate the benefits of clinical trials (Guo et al. 2018), and result in a more personalized treatment which could improve the overall clinical outcome of HNSCC patients (Quon et al. 2017). Therefore, identifying efficient, sensitive, and independent biomarkers to predict prognosis of HNSCC patients is essential.
Compared to normal tissue, cancer cells usually exhibit abnormal metabolic profiles, which is also referred to as “cancer metabolome” (Buck et al. 2017). These aberrant metabolites can be considered to be related with oncogenesis as well as the body’s response to cancer (Aboud and Weiss 2013). There are six known metabolic hallmarks of cancer: (1) deregulated uptake of glucose and amino acids, (2) use of opportunistic modes of nutrient acquisition, (3) use of glycolysis/TCA cycle intermediates for biosynthesis and NADPH production, (4) increased demand for nitrogen, (5) alterations in metabolite-driven gene regulation, and (6) metabolic interactions (Pavlova and Thompson 2016). Currently, cancer metabolome has been widely used in the discovery of diagnostic as well as prognostic biomarkers, as well as anti-tumor drug targets (Zhang et al. 2017). For example, Li et al. identified ten metabolic biomarkers by analyzing serum lipid metabolites in colorectal cancer patients (Li et al. 2013). Another study found four salivary biomarkers for early diagnosis of oral squamous cell carcinoma (Wang et al. 2014). However, most of these studies only focused on analyzing metabolites extracted from tissue or discharge from a relatively small cohort, and most of these metabolites identified lacks clinical significance. Thus, a metabolic signature in transcriptional level is urgently needed to explore their impact on overall survival (OS), as well as locating potential therapeutic targets of cancer metabolism.
Recently, the fast-paced development in bioinformatics as well as the increasing availability of large-scale RNA-sequencing (RNA-seq) transcriptome data of multiple cancers combined with clinical follow-up information makes exploring biomarkers for diagnosis and prognosis much more efficient and accurate, providing better approaches for the classification of patients and the implementation of personalized therapy, as well as uncovering the underlying mechanism of tumorigenesis (He et al. 2018; Xing et al. 2019c; Zhang et al. 2018a, b). However, most of these studies only focused on the diagnostic/prognostic signature based on single marker, such as DNA methylation, lncRNAs, pseudogenes, snoRNA, mRNA, miRNAs, and alternative splicing(Guo et al. 2018; He et al. 2018; Liu et al. 2019, 2018; Xing et al. 2019b, 2020; Zhang et al. 2018c). Using a gene set to construct prognostic signature is increasing, in which a more complicated biological processes can be considered (Shen et al. 2019). Thus, we aim to construct a metabolism-based prognostic risk score (MPRS) using single sample gene-set enrichment (ssGSEA) analysis on transcriptome data and a series of machine learning analysis, which could provide new insights into predicting and evaluating the clinical outcome of HNSCC.
Materials and methods
HNSCC cohort acquisition
HNSCC RNA-seq expression profile combined with clinical follow-up are coming from the public database UCSC Xena (https://xenabrowser.net/datapages/). We included patients who has complete follow-up information and survival status combined with RNA-seq data. Following this criteria, 494 HNSCC cases were involved in this study.
Metabolism-related gene definition
The metabolism-related gene sets were obtained from KEGG pathway database (https://www.genome.jp/kegg/pathway.html) which included 12 categories of metabolic pathways (MP), such as carbohydrate metabolism, energy metabolism, and lipid metabolism, and there are 90 human MPs in total. Under the organism menu, choose “homo sapiens”, the genes involved in each of the 90 MPs were in the KO/Gene/Compound list [for example, the gene set of Glycolysis (Embden–Meyerhof pathway) were in https://www.genome.jp/kegg-bin/show_module?hsa_M00001]. The gene list of the 90 MPs is in Table S1.
Development of the metabolism-based prognostic signature for HNSCC
We randomly divided the whole data set into training set and testing set (training: testing = 7:3). The signature was developed through a five-step strategy in training set, and was validated in testing set and whole set to verify the robustness of the signature. First, using ssGSEA, an enrichment score for each MP was evaluated which represents the level of absolute enrichment of each MP in each sample (Foroutan et al. 2018; Shen et al. 2019). For each MP, the enrichment scores were normalized. Second, dysregulated MPs were identified between normal and tumor samples by cut-off FDR < 0.05. Third, dysregulated MPs were passed on to the univariate cox regression analysis from which survival-related MPs (p value < 0.1) could be identified. Then, LASSO regression was performed on survival-related MPs and MPs with non-zero coefficients were screened for further analysis. Finally, stepwise multiple cox analysis was performed to construct the MP signature. MPs consisted of cox model with least AIC, largest C-index, and least variants were determined to construct the final signature. The risk score for the 6MPS of each patient was calculated using the formula: MPRS = where Coefi is the coefficient and xi is the ssGSEA score for each MP. All of these processes were conducted using R software (version 3.5.1).
Statistical analysis
Using the median MPRS in training group, we can divide the patients into high-risk and low-risk groups; for these groups, Kaplan–Meier (K–M) survival analysis was performed, and OS differences were determined using two-sided log-rank test. To further evaluate the precision of the predictive model in predicting the 1-, 3-, and 5-year survival, we performed receiver-operating characteristic (ROC) analysis and the area under curve (AUC) was obtained. To evaluate the agreement between the predicted outcomes with the actual ones, calibration curve was plotted. Using Chi-square tests, the clinical feature distribution (grade, age, gender, stage, subdivision, and margin status) and mutation status (TMB and top ten mutational genes) were compared between the two risk groups. T test or one-way ANOVA was used to compare the MPRS level in different subgroups. Univariate and multivariate Cox regression analyses were carried out to determine the prognostic value of the 6MPS and various clinical characteristics. All of these processes above were conducted using R software (version 3.5.1).
Results
Clinical profile of the HNSCC patients
The HNSCC cohort includes RNA-sequencing data from a total of 546 patients. After excluding samples without complete clinical follow-up information, we obtained RNA-seq data from 494 tumor samples and 44 normal samples. Clinicopathological characteristics of the included patients are presented in table S2.
Identification of dysregulated metabolic pathways
To determine the dysregulated metabolic pathways, 90 MPs were further performed t test of the metabolic pathway enrichment score between tumor and normal samples from which 79 were significantly dysregulated (FDR < 0.05).
Construction of metabolic signature for HNSCC
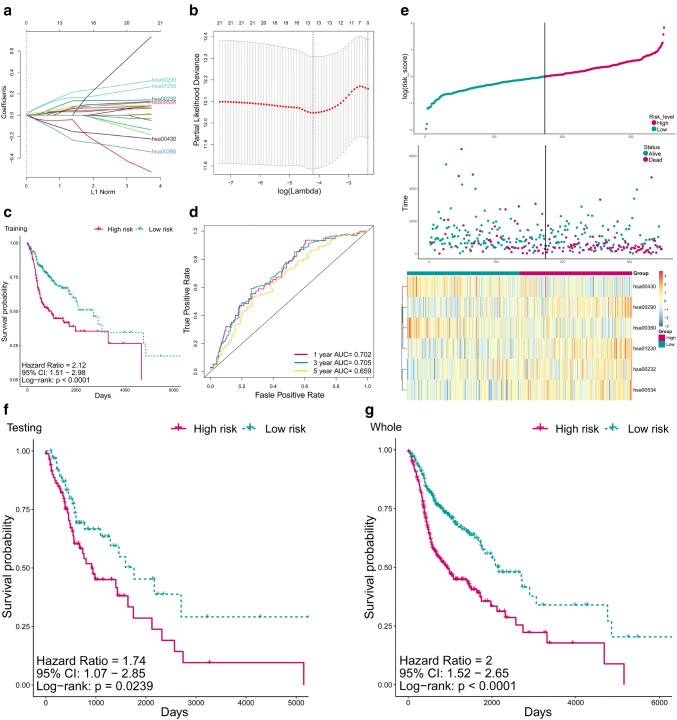
To explore the prognostic prediction potential of the dysregulated metabolic pathways in HNSCC, we employed univariate regression, LASSO regression analysis, and stepwise cox regression analysis on the dysregulated MPs in the training set (Fig. 1a, b), and a 6MPS was constructed (hsa00290: Valine, leucine and isoleucine biosynthesis, hsa01230: Biosynthesis of amino acids, hsa00430: Taurine and hypotaurine metabolism, hsa00380: Tryptophan metabolism, hsa00232: Caffeine metabolism, hsa00534: Glycosaminoglycan biosynthesis—heparan sulfate/heparin). The coefficients of the 6 MPs are as follows: “hsa00232: 0.20369, hsa00290: 0.23850, hsa00380: − 0.28431, hsa00430: − 0.19614, hsa00534: 0.13115, hsa01230: 0.32494”.
Fig. 1.
Construction of 6MPS and its prognostic value. a LASSO coefficient profiles of the 21 survival-related MPs. A coefficient profile plot was produced against the log lambda sequence. b Tuning parameter (lambda) selection in the LASSO model used tenfold cross-validation via minimum criteria. A lambda value of 0.01485 was chosen (minimum criteria). Dotted vertical lines were drawn at the optimal values using the minimum criteria and the 1 standard error of the minimum criteria (the 1-SE criteria). c The Kaplan–Meier estimates of the OS for high-risk and low-risk patient cohorts grouping by the 6MPS in the training set (N = 346). The OS differences between the two groups were determined by the two-sided log-rank test. It can be concluded that higher MPRS are significantly associated with worse OS (p < 0.0001). d ROC analysis of sensitivity and specificity for the 6MPS in predicting the OS of patients for 1, 3, and 5 years in training set. e The distribution of MPRS, patients’ survival status and five MP enrichment scores in the training set were presented. As the risk score rising, the patients had a shorter survival time, more dead events, and a higher MP enrichment score. f, g The Kaplan–Meier estimates of the OS for high-risk and low-risk patients grouping by the 6MPS in the testing set (N = 148) and whole set (N = 494). The OS differences between the two groups were determined by the two-sided log-rank test. It can be concluded that higher MPRS are significantly associated with worse OS (p < 0.05). MP: metabolic pathway, MPRS: metabolism-based prognostic risk score, 6MPS: 6-MP signature, hsa00290: valine, leucine, and isoleucine biosynthesis, hsa01230: biosynthesis of amino acids, hsa00430: taurine and hypotaurine metabolism, hsa00380: tryptophan metabolism, hsa00232: caffeine metabolism, hsa00534: glycosaminoglycan biosynthesis—heparan sulfate/heparin
Prognostic value of 6MPS
To assess the prognostic value of the 6MPS, Kaplan–Meier survival analysis was performed on the training set, and high-risk group patients have a significantly worse overall survival (p < 0.0001, Fig. 1c). Furthermore, ROC analysis was carried out to evaluate the sensitivity and specificity of the 1-, 3-, and 5-year survival of the signature, and the respective areas under the ROC curve (AUCs) were 0.702, 0.705, and 0.659 (Fig. 1d, Fig S1). In addition, higher risk score was related with shorter survival time, more death events, and higher MP enrichment score of the five MPs (Fig. 1e). To further validate the 6MPS, we applied the signature to the testing set and the entire set, and it can be concluded that the signature performed well to distinguish patients in overall survival (both p < 0.05, Fig. 1f, g). Figure S2 presents the calibration curve estimated the predictive value of the metabolic pathway signature for predicting the probability of OS at 1, 3, and 5 years.
MPRS showed significant association with clinical features
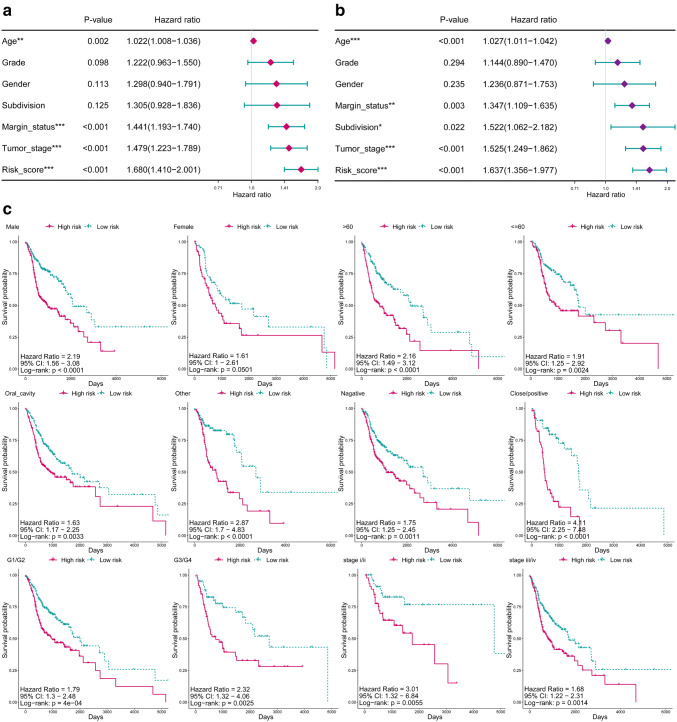
A heatmap was plotted to reveal the potential associations between the MPRS and some clinical features in the high- and low-risk groups (Fig. 2a). Tumor stage (p < 0.05), gender (p < 0.05), and status (p < 0.01) are observed to be significantly associated with the low-risk or high-risk group. Next, we compared the MPRS distribution between different clinical subgroups to explore the possible association between clinical features and MPRS (Fig. 2b). Significant difference in the distribution of MPRS can be seen between male and female (p < 0.05), grade of G1 and G2 (p < 0.05), stage i and ii (p < 0.05), stage i and iii (p < 0.05), stage i and iv (p < 0.001), as well as stage ii and stage iv (p < 0.05).
Fig. 2.
The clinical association of MPRS. a The heatmap shows the MPRS level of the five metabolic pathways in low- and high-risk HNSCC. The distribution of clinical features was compared between the low- and high-risk groups. b Distribution of MPRS in different subgroups was compared, which stratified by gender, age, anatomic subdivision, grade, stage, and margin status. MP: metabolic pathway, MPRS: metabolism-based prognostic risk score, 6MPS: 6-MP signature, hsa00290: valine, leucine, and isoleucine biosynthesis, hsa01230: biosynthesis of amino acids, hsa00430: taurine and hypotaurine metabolism, hsa00380: tryptophan metabolism, hsa00232: caffeine metabolism, hsa00534: glycosaminoglycan biosynthesis—heparan sulfate / heparin. *p < 0.05, **p < 0.01
6MPS is an independent prognostic factor
To evaluate the independence of 6MPS for OS, we performed Cox univariate and multivariate regression analyses on the 6MPS and some clinical features. Univariate regression analysis showed that age, grade, gender, subdivision, margin status, tumor stage, and risk score are significantly related to OS (Fig. 3a). While including the same factors into multivariate regression analysis, it can be concluded that the 6MPS is still significantly related to OS (p < 0.001, Fig. 3b). Taken together, the MPRS derived from the metabolic pathway signature is an independent factor for predicting the prognosis of HNSCC patients. Furthermore, we performed Kaplan–Meier analysis on clinical subgroups including gender, age, subdivision, margin status, grade, and tumor stage to evaluate the prediction ability of the signature in different clinical subgroups (Fig. 3c). The signature performed well in most of the clinical subgroups. However, the signature cannot extinguish patients of high and low risks in subgroups of female.
Fig. 3.
6MPS is an independent prognostic factor. a Univariate and b multivariate Cox regression analyses of the association between clinical factors (including the MPRS) and overall survival of patients in the HNSCC. The 6MPS is an independent prognostic factor (p value < 0.001). c Kaplan–Meier analyses of patients with HNSCC in different clinical subgroups, grouping based on their gender, age, anatomic subdivision, grade, stage, and margin status. Kaplan–Meier analysis with two-sided log-rank test was performed to estimate the differences in OS between the low-risk and high-risk patients. MPRS: metabolism-based prognostic risk score, 6MPS: 6-MP signature, *p < 0.05, **p < 0.001, ***p < 0.0001
Associations between MPRS and mutation
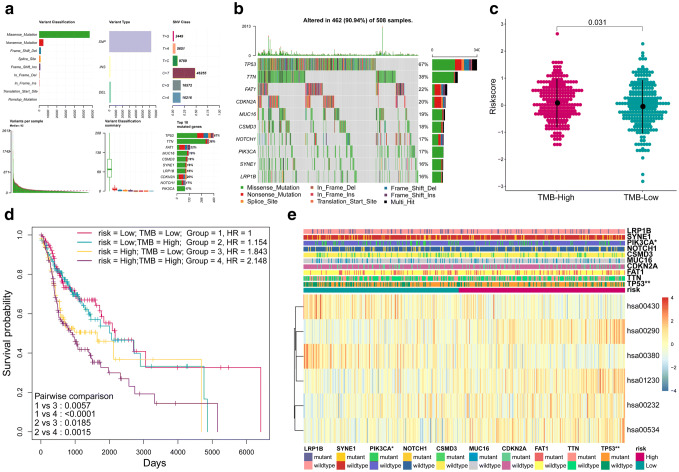
To explore the relationship between the 6MPS and mutation status of HNSCC (TMB and top ten mutational genes, Fig. 4a, b), a heatmap was plotted to compare the differences between high- and low-risk groups (Fig. 4e). PIK3CA (p < 0.05) and TP53 (p < 0.01) are observed to be significantly associated with the MPRS. Next, we compared the MPRS distribution between high-TMB and low-TMB groups to explore the possible association between TMB and MPRS (Fig. 4c). Significant difference in the distribution of MPS can be seen between high-TMB and low-TMB groups (p < 0.05). And the K–M plot revealed in high-risk group the patients with high-TMB had a worse prognosis and short survival (Fig. 4d).
Fig. 4.
Associations between MPRS and mutation. a The overall summary of mutation information in HNSCC cohort by mutect2. b The oncoplot shows the top ten mutated genes in HNSCC cohort and their mutation information. c Distribution of MPRS in high-TMB and low-TMB groups was compared, The MPRS is higher in high-TMB group than low-TMB groups. d The Kaplan–Meier estimates of the OS for groups using combinations of MRPS and TMB. The OS differences between the four groups were determined by the two-sided log-rank test. TMB: Tumor mutation burden. e The heatmap shows the MP enrichment score of the five metabolic pathways in low- and high-risk HNSCC. The distributions of top ten mutation genes status were compared between the low- and high-risk groups. MP metabolic pathway, MPRS metabolism-based prognostic risk score, 6MPS 6-MP signature, hsa00290: valine, leucine, and isoleucine biosynthesis, hsa01230: biosynthesis of amino acids, hsa00430: taurine and hypotaurine metabolism, hsa00380: tryptophan metabolism, hsa00232: caffeine metabolism, hsa00534: glycosaminoglycan biosynthesis—heparan sulfate/heparin. **p < 0.01, *p < 0.05
Discussion
Patients with HNSCC frequently have poor prognosis and a low survival rate. Despite great improvements in diagnostic and therapeutic methods, the survival rate of HNSCC still remains unsatisfying (Troiano et al. 2018). However, prognostic as well as diagnostic signature might be able to shed some light on the probability of a certain disease as well as its relative progression, recurrence, and death (Ballman 2015), which could contribute in patient classification, treatment management, and monitoring disease status in clinical practice, for example, offering personalized therapeutic schedules to HNSCC patients who would benefit enormously (Nonaka and Wong 2018). In the past few years, multiple molecular biomarkers have proved themselves to be useful in the prediction of the prognosis of HNSCC such as mRNA (Shen et al. 2017a), miRNA (Wong et al. 2016), lncRNA (Liu et al. 2018; Xing et al. 2019a), and methylation (Shen et al. 2017b). In our study, we did not adapt the usual methods in which single gene is the center of attention to explore prognostic markers, while a novel method named ssGSEA was employed in which a gene-set is the focus of signature construction (Shen et al. 2019). To obtain the metabolic pathway status in each sample, we performed ssGSEA analysis in which a metabolic gene matrix has been transformed to an MP enrichment score matrix (Foroutan et al. 2018). This gave the chance to analyze the cancer metabolism and metabolic signature in transcriptional level. Therefore, we performed a systematic analysis of MPs in HNSCC, and have explored the potential role of MPs as prognostic marker for HNSCC, from which the survival-related MPs were the first to be discovered. During the course of exploration, several important discoveries were made. First, we identified 72 dysregulated MPs which their enrichment score are statistically different (FDR < 0.001) between normal and tumor samples. Second, we identified 14 survival-related MPs using univariate Cox regression. Third, a 6MPS was identified and a scoring system was established which was significantly related to the OS of HNSCC patients. The 6MPS demonstrated great efficiency in stratifying patients into low- and high-risk groups and predicting the OS. The signature was first applied in the training set and was then validated in the testing set and whole set, suggesting that it was reliable. Third, to testify the universality in different patients and to verify its application in different clinicopathological subgroups, Kaplan–Meier survival analysis was performed in various subgroups. We found that the 6MPS was independent of other potential predictors, including age, sex, anatomical subdivision, margin status, stage, and grade, and its performances were of satisfaction. Fourth, to clarify the potential associations between the MPRS and clinical features, heatmap was plotted and the distributions of the MPRS in different clinical subgroups was compared. We found that the 6MPS was associated with stage and anatomical subdivision. Finally, we explored the associations between the 6MPS and gene mutation. Our results demonstrated that the 6MPS is associated with PIC3KA and TP53 mutation. High TMB is accompanied with high MPRS and patients in high-risk group with high-TMB have a worse prognosis than low-TMB. These results indicate the 6MPS is an efficient tool for the prediction of the prognosis of HNSCC patients and revealed that the up- or down-regulation of the five metabolic pathways is linked to some clinical features as well as gene mutation.
Still limitations in this study cannot be overlooked. First, the study was mainly based on methodology, such as machine learning and bioinformatic analysis, and experimental validation will be further required. Second, one must take it into consideration of application when constructing a signature. Since the model in our study was mainly based on RNA-seq data, the procedure of detection, quantification, and determination of transcriptional activity of RNAs must be standardized (Guglas et al. 2017). We also need to stress the limitation of how far transcriptome data can be transferred into metabolic dysregulation, which occurs to mostly at the post-transcriptional and post-translational levels. Thus, the interpretation of the result obtained from such analysis should be followed by proteomic and metabolomic data, to thoroughly investigate the metabolic dysregulation for therapeutic and prognostic exploitation (Rosario et al. 2018).
Conclusions
In conclusion, this study highlighted the prognostic value of metabolic pathways in transcriptional level and explored their relationship between mutation statuses. Some prognosis-related metabolic pathways have been revealed, and the survival of HNSCC patients could be predicted by a risk model based on these pathways, which could serve as prognostic markers in clinical practice. These results may provide new potential prognostic and therapeutic implications for HNSCC patient management.
Electronic supplementary material
Below is the link to the electronic supplementary material.
The metabolism-related gene sets (XLSX 165 kb)
Clinicopathological characteristics of the included HNSCC patients (XLSX 44 kb)
Acknowledgements
We are grateful for the kind help of Dr. Xinhua Liu from Yantai Yuhuangding Hospital; Dr. Di Liu from School of Stomatology, Shandong University; and Professor Minqi Li from Department of bone metabolism of School of Stomatology, Shandong University.
Author contributions
Conceptualization, FL; data curation, FL; formal analysis, LX and MG; methodology, LX and MG; project administration, FL; software, XZ and XZ; validation, XZ and FL; visualization, XZ; writing—original draft, LX and XZ; writing—review and editing, FL.
Funding
This research received no external funding.
Data availability statement
The TCGA-HNSCC data set that supports the findings of this study is available in [UCSC Xena] at [https://xenabrowser.net/datapages/] and [KEGG] at [https://www.genome.jp/kegg/pathway.html].
Compliance with ethical standards
Conflict of interest
The authors declare that they have no conflict of interest.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Lu Xing and Mingzhu Guo have contributed equally to this work and should be considered to be co-first authors.
References
- Aboud OA, Weiss RH (2013) New opportunities from the cancer metabolome. Clin Chem 59:138–146. 10.1373/clinchem.2012.184598 [DOI] [PubMed] [Google Scholar]
- Ballman KV (2015) Biomarker: predictive or prognostic? J Clin Oncol 33:3968–3971. 10.1200/JCO.2015.63.3651 [DOI] [PubMed] [Google Scholar]
- Buck A, Aichler M, Huber K, Walch A (2017) Situ metabolomics in cancer by mass spectrometry imaging. Adv Cancer Res 134:117–132. 10.1016/bs.acr.2016.11.004 [DOI] [PubMed] [Google Scholar]
- Budach V, Tinhofer I (2019) Novel prognostic clinical factors and biomarkers for outcome prediction in head and neck cancer: a systematic review. Lancet Oncol 20:e313–e326. 10.1016/S1470-2045(19)30177-9 [DOI] [PubMed] [Google Scholar]
- Cai Y, Dodhia S, Su GH (2017) Dysregulations in the PI3K pathway and targeted therapies for head and neck squamous cell carcinoma. Oncotarget 8: 22203–22217. 10.18632/oncotarget.14729. [DOI] [PMC free article] [PubMed]
- Foroutan M, Bhuva DD, Lyu R, Horan K, Cursons J, Davis MJ (2018) Single sample scoring of molecular phenotypes. BMC Bioinform 19:404. 10.1186/s12859-018-2435-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guglas K et al (2017) lncRNA in HNSCC: challenges and potential. Contemp Oncol (Pozn) 21:259–266. 10.5114/wo.2017.72382 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo W, Zhu L, Yu M, Zhu R, Chen Q, Wang Q (2018) A five-DNA methylation signature act as a novel prognostic biomarker in patients with ovarian serous cystadenocarcinoma. Clin Epigenet 10:142. 10.1186/s13148-018-0574-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- He Y, Ma J, Wang A, Wang W, Luo S, Liu Y, Ye X (2018) A support vector machine and a random forest classifier indicates a 15-miRNA set related to osteosarcoma recurrence. Onco Targets Ther 11:253–269. 10.2147/OTT.S148394 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li F et al (2013) Lipid profiling for early diagnosis and progression of colorectal cancer using direct-infusion electrospray ionization Fourier transform ion cyclotron resonance mass spectrometry. Rapid Commun Mass Spectrom 27:24–34. 10.1002/rcm.6420 [DOI] [PubMed] [Google Scholar]
- Liu G et al (2018) A prognostic 5-lncRNA expression signature for head and neck squamous cell carcinoma. Sci Rep 8:15250. 10.1038/s41598-018-33642-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu F, Xing L, Zhang X, Zhang X (2019) A Four-pseudogene classifier identified by machine learning serves as a novel prognostic marker for survival of osteosarcoma. Genes (Basel). 10.3390/genes10060414 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moskovitz JM, Moy J, Seiwert TY, Ferris RL (2017) Immunotherapy for head and neck squamous cell carcinoma: a review of current and emerging therapeutic options. Oncologist 22:680–693. 10.1634/theoncologist.2016-0318 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moy JD, Moskovitz JM, Ferris RL (2017) Biological mechanisms of immune escape and implications for immunotherapy in head and neck squamous cell carcinoma. Eur J Cancer 76:152–166. 10.1016/j.ejca.2016.12.035 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nonaka T, Wong DTW (2018) Liquid biopsy in head and neck cancer: promises and challenges. J Dent Res 97:701–708. 10.1177/0022034518762071 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pavlova NN, Thompson CB (2016) The emerging hallmarks of cancer metabolism. Cell Metab 23:27–47. 10.1016/j.cmet.2015.12.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quon H et al (2017) Quantitative evaluation of head and neck cancer treatment-related dysphagia in the development of a personalized treatment deintensification paradigm. Int J Radiat Oncol Biol Phys 99:1271–1278. 10.1016/j.ijrobp.2017.08.004 [DOI] [PubMed] [Google Scholar]
- Rosario SR, Long MD, Affronti HC, Rowsam AM, Eng KH, Smiraglia DJ (2018) Pan-cancer analysis of transcriptional metabolic dysregulation using The Cancer Genome Atlas. Nat Commun 9:5330. 10.1038/s41467-018-07232-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen S et al (2017a) A seven-gene prognostic signature for rapid determination of head and neck squamous cell carcinoma survival. Oncol Rep 38:3403–3411. 10.3892/or.2017.6057 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen S et al (2017b) Seven-CpG-based prognostic signature coupled with gene expression predicts survival of oral squamous cell carcinoma. Clin Epigenet 9:88. 10.1186/s13148-017-0392-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen S, Wang G, Zhang R, Zhao Y, Yu H, Wei Y, Chen F (2019) Development and validation of an immune gene-set based Prognostic signature in ovarian cancer. EBioMedicine 40:318–326. 10.1016/j.ebiom.2018.12.054 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siegel RL, Miller KD, Jemal A (2017) Cancer statistics. CA Cancer J Clin 67:7–30. 10.3322/caac.21387 [DOI] [PubMed] [Google Scholar]
- Troiano G, Mastrangelo F, Caponio VCA, Laino L, Cirillo N, Lo Muzio L (2018) Predictive prognostic value of tissue-based microrna expression in oral squamous cell carcinoma: a systematic review and meta-analysis. J Dent Res 97:759–766. 10.1177/0022034518762090 [DOI] [PubMed] [Google Scholar]
- Wang Q, Gao P, Wang X, Duan Y (2014) Investigation and identification of potential biomarkers in human saliva for the early diagnosis of oral squamous cell carcinoma. Clin Chim Acta 427:79–85. 10.1016/j.cca.2013.10.004 [DOI] [PubMed] [Google Scholar]
- Wong N et al (2016) Prognostic microRNA signatures derived from The Cancer Genome Atlas for head and neck squamous cell carcinomas. Cancer Med 5:1619–1628. 10.1002/cam4.718 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xing L, Zhang X, Chen A (2019a) Prognostic 4-lncRNA-based risk model predicts survival time of patients with head and neck squamous cell carcinoma. Oncol Lett 18:3304–3316. 10.3892/ol.2019.10670 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xing L, Zhang X, Tong D (2019b) Systematic profile analysis of prognostic alternative messenger RNA splicing signatures and splicing factors in head and neck squamous cell carcinoma. DNA Cell Biol. 10.1089/dna.2019.4644 [DOI] [PubMed] [Google Scholar]
- Xing L, Zhang X, Zhang X, Tong D (2020) Expression scoring of a small-nucleolar-RNA signature identified by machine learning serves as a prognostic predictor for head and neck cancer. J Cell Physiol. 10.1002/jcp.29462 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang F et al (2017) Metabolomics for biomarker discovery in the diagnosis, prognosis, survival and recurrence of colorectal cancer: a systematic review. Oncotarget 8: 35460–35472. 10.18632/oncotarget.16727 [DOI] [PMC free article] [PubMed]
- Zhang X, Feng H, Li D, Liu S, Amizuka N, Li M (2018a) Identification of differentially expressed genes induced by aberrant methylation in oral squamous cell carcinomas using integrated bioinformatic analysis. Int J Mol Sci. 10.3390/ijms19061698 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X, Feng H, Li Z, Guo J, Li M (2018b) Aspirin is involved in the cell cycle arrest, apoptosis, cell migration, and invasion of oral squamous cell carcinoma. Int J Mol Sci. 10.3390/ijms19072029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X, Feng H, Li Z, Li D, Liu S, Huang H, Li M (2018c) Application of weighted gene co-expression network analysis to identify key modules and hub genes in oral squamous cell carcinoma tumorigenesis. Onco Targets Ther 11:6001–6021. 10.2147/OTT.S171791 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
The metabolism-related gene sets (XLSX 165 kb)
Clinicopathological characteristics of the included HNSCC patients (XLSX 44 kb)
Data Availability Statement
The TCGA-HNSCC data set that supports the findings of this study is available in [UCSC Xena] at [https://xenabrowser.net/datapages/] and [KEGG] at [https://www.genome.jp/kegg/pathway.html].