Abstract
Purpose
Metastasis is an unavoidable event happened among almost all small cell lung cancer (SCLC) patients. However, the molecular driven factors have not been elucidated. Recently, a novel hydrolase called cell migration inducing hyaluronidase (CEMIP) triggered both migration and invasion in many tumors but not SCLC. Therefore, in this study, we verified that CEMIP promoted migration and invasion in SCLC and applied proteomics analysis to screen out potential target profiles and the signaling pathway related to CEMIP regulation.
Method
Immunofluorescence was conducted to exam the expression of CEMIP on SCLC and paired adjacent normal tissues among enrollment. RT-qPCR and Western blot (WB) assays were conducted to valuate cellular protein and mRNA expression of CEMIP and EMT markers. Lentivirus-CEMIP-shRNAs and CEMIP plasmid were used for expression manipulating. Changes of cellular migration and invasion were tested through transwell assays. Tandem Mass Tag (TMT) peptide labeling coupled with LC–MS/MS was used for quantifying proteins affected by reducing expression of CEMIP on H446 cells.
Results
The expression of CEMIP showed 1.64 ± 0.16-fold higher in SCLC tissues than their normal counterpart. Decreasing the expression of CEMIP on SCLC cells H446 regressed both cellular migration and invasion ability, whereas the promoting cellular migration and invasion was investigated through over-expressing CEMIP on H1688. Proteomic and bioinformatics analysis revealed that total 215 differentially expressed proteins (DEPs) that either their increasing or decreasing relative expression met threshold of 1.2-fold changes with p value ≤ 0.05. The dramatic up-regulated DEPs included an unidentified peptide sequence (encoded by cDNA FLJ52096) SPICE1 and CRYAB, while the expression of S100A6 was largely down-regulated. DEPs mainly enriched on caveolae of cellular component, calcium ion binding of biological process and epithelial cell migration of molecular function. KEGG enrichment indicated that DEPs mainly exerted their function on TGF-β, GABAergic synapse and MAPK signaling pathway.
Conclusion
It is the first report illustrating that CEMIP might be one of the metastatic triggers in SCLC. And also, it provided possible molecular mechanism cue and potential downstream target on CEMIP-induced cellular migration and invasion on SCLC.
Electronic supplementary material
The online version of this article (10.1007/s00432-020-03308-5) contains supplementary material, which is available to authorized users.
Keywords: SCLC, CEMIP, Proteome analysis, Metastasis, Signaling pathway
Introduction
Since the GLOBOCAN project was held in 1980s, global incidence and mortality of lung cancer was recorded with fluctuated rate around 12% and 18% of total cancer, respectively, during past 3 decades (Jemal et al. 2011; Siegel and Miller 2020; Torre et al. 2015). Among all types of cancer, lung cancer has long been remained the top health-killer among the whole world wherein the fatality rate reached 90% (Chen et al. 2017). Small cell lung cancer is regarded as a special subtype of lung carcinoma due to its origin of pulmonary neuroendocrine cells (Semenova et al. 2015; van Meerbeeck et al. 2011). Although annual incidence of SCLC remains around 10% of total incidence of lung cancer, the high mortality since the metastasis often occurred at the time of diagnosis (van Meerbeeck et al. 2011). Moreover, almost all advanced SCLC patients were challenged by tumor metastasis mainly due to the therapeutic resistance (Powell et al. 2014). So far, the effective treatment based on platinum-reagents and topoisomerase inhibitors could only extend the life span of patients around 1–2 years (Amarasena et al. 2015; van Meerbeeck et al. 2011). However, there still was no predominate genetic driven factor was found that mainly facilitated on metastasis of SCLC.
CEMIP, encoded by KIAA1199, is a novel discovered protein that promotes tumor migration and invasion, owning enzyme ability that degraded hyaluronan (Li et al. 2017b). Apart its enzyme function, CEMIP was found over-expressed in many cancers and high CEMIP expression stimulated the migration and invasion of tumor cells. Analysis of tissue slice indicated the cytoplasmic CEMIP remained higher expression in tumor than adjacent normal tissues in colorectal (Fink et al. 2015), gastric (Oneyama et al. 2019; Wang et al. 2019), pancreatic cancer (Koga et al. 2017; Kohi et al. 2017) and NSCLC (Tang et al. 2019) which also predicted the high possibility of metastasis and bad outcomes. In colorectal cancer, CEMIP accelerated EMT process, enhancing mesenchymal transformation to shuttle into circulation (Duong et al. 2018). Also, CEMIP could delivery cytoplasmic β-catenin into nuclei for activating transcription of down-sites of gene (Birkenkamp-Demtroder et al. 2011). The translocated β-catenin then triggered TCF region to generate Wnt over-expression that could launch metastasis (Birkenkamp-Demtroder et al. 2011). Tang Z. et al. reported that CEMIP triggered EMT of tumor cell through PI3K/AKT pathway (Tang et al. 2019). Besides, CEMIP could facilitate inflammation secretion such as IL-6 and IL-8 that keeping chronic inflammatory infiltration around tumor microenvironment and induced precancerosis (Kohi et al. 2017; Suh et al. 2016). Mechanical movement of tumor and other cells was also affected by CEMIP. Colorectal tumor cell rearranged their microtubule associated by CEMIP (Zhao et al. 2019). And CEMIP also arouse cytoskeleton rearrangement signal passed by β-catenin that lead to pseudopodia hyperplasia in osteoblast (Chen et al. 2019). Based on these evidences, we assumed CEMIP might function in tumor migration in SCLC, but the precise intelligence of mechanism in how it was triggered and what signal pathways participated in cell movement remained largely blank.
In this study, we first enrolled a small scale of clinical samples to discover the high expression of CEMIP in SCLC, and set up in vitro model according to results of histopathological CEMIP staining. Then, declined migration and invasion were confirmed when the expression of CEMIP was decreased in SCLC cell H446, while the inverse phenomenon appeared on over-expressing CEMIP in SCLC cell H1688. Since the cellular characteristic of SCLC had blurry boundary between classic epithelium and neuroendocrine cell, the common signaling pathway such as EMT transformation and TGF-β might not interpret its molecular mechanism might in some subtype. Therefore, we conducted the proteomics analysis which demonstrated that cellular signaling pathway affected the most in TGF-β, GABAergic synapse and MAPK when CEMIP was inhibited in SCLC.
Methods
Collection of SCLC pathological sample and clinical data
We retrospectively reviewed medical charts of patient diagnosed with SCLC from Aug 2013 to Jan 2019 in Affiliated Tumor Hospital of Guangxi Medical University. Enrollment should meet the criteria: (1) patients with complete medical chart; (2) ESCO score less than 2; (3) pathological diagnosis confirmed SCLC; (4) patients did not receive either chemotherapy or radiotherapy ahead of surgery; (5) patients that could be reviewed for prognosis. This study was agreed by Ethic Committee of Guangxi Medical University. All patients were signed informed consent. Total of nine patients fulfilled all criteria and provided collectable paraffin block.
Immunofluorescence
Each paraffin tissue samples were sliced into 5 μm thick. Oven was preheated to 72 ℃ and then placed all slides for 5 h followed by the deparaffinating and rehydration. Slides then were restored by high-pressure method using cirate acid/sodium cirate buffer (pH 6) and blocked with 5% BSA. Experimental groups were incubated with rabbit anti human CEMIP (1:100) primary antibody whereas the blank group was incubated with PBS overnight at 4 ℃ then recovered at 37 ℃ for 1 h. Fluorescence secondary antibody (Alexa 555-goat anti rabbit 1:100, BBI) was then used for 1 h at room temperature and nuclei were dyed with 1 μg/ml DAPI for 5 min at room temperature. All slides were washed by PBS for five times and displayed into oven which was preheated to 50 ℃. Staining of samples was sealed by neutral gum after drying. Confocal laser scanning microscope (Leica Microsystems CMS GmbH/Model DMi8) was used for determination the staining area and density of each slides, fluorescence density was analyzed according to the appendant software.
Cell culture
Normal human lung bronchial epithelial cell lines BEAS-2B were purchased from Cell bank of Chinese Academy of Sciences Kunming branch. Human small cell lung cancer cell lines H446 (RRID:CVCL_1562), H1688 (RRID:CVCL_1487) and H209 (RRID:CVCL_1525) were purchased from American Type Culture Collection (ACTT, Manassas, VA, USA). BEAS-2B (RRID:CVCL_0168) was incubated in specific BEGM (Lonza co. USA) which contained basal BEBM with addition of 0.4% BPE, 0.1% hydrocortisone, 0.1% hEGF, 0.1% epinephrine, 0.1% insulin, 0.1% transferrin, 0.1% triiodothyronine, 0.1% retinoic acid, 0.1% GA. H446 and H1688 were cultivated in complete RIPM-1640 medium containing 1% mixture of 100U/mL penicillin and 100 μg/mL streptomycin (Invitrogen, Thermo Fischer Scientific co. USA) and 10% Fetal Bovine Serum (Gibco, Thermo Fischer Scientific co. USA). H209 was cultivated in complete RIPM-1640 medium with supplement of 15% Fetal Bovine Serum.
Lentivirus and plasmid transfection
Three small interference RNA targets siRNA-CEMIP#1 (5′-GAGCCGGAACATCATAGTGAT-3′), siRNA-CEMIP#2 (5′-GAGGTTATGACCCACCCACAT-3′), siRNA-CEMIP#3 (5′-gcGAATGAAGATCATCAAGAA-3′), and one off-target siRNA-NC (5′-TTCTCCGAACGTGTCACGT-3′) were parceled into lentiviral vector GV248 (hU6-MCS-Ubiquitin-EGFP-IRES-puromycin) to achieve shRNA-expressing lentiviral. The packages of lentiviral were designed and synthesized by Genechem, Shanghai. The expression of CEMIP was interfered in H446 by transducing #1, #2, #3 as listed above, parallelly one off-targeted counterpart was transduced as negative control. Transfection efficiency was detected by fluorescence microscope (BioTek, USA) after 72 h cultivation. Due to the puromycin resistance transcription was written in vector, 2.5 μg/mL puromycin (Sorabol, China) was used for purifying untransfected cells. H446 cells stably silencing CEMIP was enlarged propagation for further assay when transfection efficiency reached up to 95%. H1688 cells were harvested and plated into 6-well plates. When cells confluence reach above 90%, complex liposome encapsulated either 2.5 μg CEMIP plasmid or equal amount of negative control plasmid by Lipo3000 (Invitrogen, Thermo Fischer Scientific co. USA) was diluted into 2 mL complete culture medium followed by replacement of complete 1640 medium daily. Thereafter, H1688 that transiently over-expressed CEMIP was used for further assays between 48 and 96 h.
RNA extraction and RT-qPCR
Total RNA of tumor cells was extracted by RNAiso Plus (TAKARA, Japan) according to manufacturer instructions. Then, PrimeScript™ RT reagent Kit with gDNA Eraser (TAKARA, Japan) was used for reverse-transcription of 1 μg of total RNA to synthesize cDNA solution. Gene expression was analyzed by SYBR method (TB Green® Premix Ex Taq™ II (Tli RNaseH Plus, TAKARA, Japan) and following primers: GADPH (Forward: 5′-CAAATTCCATGGCACCGTCA-3′, Reverse:5′-GACTCCACGACGTACTCAGC-3′) CEMIP (Forward: 5′-TCCTCTCGGATGTTCACAATC-3′, Reverse: 5′-ATGGTCCTTTGTTTTCCTGAGT-3′). RT-qPCR analysis then was performed on Applied Biosystems 7500 Real-time PCR System.
Western blot
Cells were planted into six-well plate 24 h ahead before harvestation When confluence reached 90–100%, 100 μL RIPA lysis buffer (Sorabol, China) mixed with 1% PMSF (Sorabol, China) and 2.5% phosphatase inhibitor (Sorabol, China) was added into each wells to get cell lysis. After centrifuging cell lysis, protein supernatants were then quantified through BCA reagent kit (Beyotime Biotechnology, Shanghai). Then 10% SDS-PAGE gel was prepared and loaded with 10 μg protein sample in each lane for electrophoresis separation followed by the 150 min electrotransformation to blot into PVDF membrane (Millipore, Billerica, MA). Blotting membrane then was soaked into 5% non-fatty milk, and specific primary antibody was prepared as followed: rabbit anti human KIAA1199 (1:800, Proteintech, China), and loading control rabbit anti human GADPH (1:2000, BBI, Canada). For detecting EMT-markers, loading amount of protein sample was raised up to 80 μg, and the primary antibody was diluted as instruction: E-cadherin (1:1000, CST, US), Cytokeratin 18 (CK18 1:1000, Abcam, UK), N-cadherin (1:1000, Proteintech, China), Vimentin (1:1000, CST, US), Twist1 (1:1000, Proteintech, China), and Snail1 (1:1000, CST, US). Each band was soaked into diluted antibody solution for 4 ℃ overnight. All membrane bands were then washed three times by PBST, and incubated with secondary antibody, anti-rabbit IgG (DyLight™ 800 4X PEG Conjugate, CST, US) for 1 h at room temperature, followed by washing three times. Protein bands were then imaged using infrared fluorescence imaging system Odyssey (LI-COR, USA). Intensity of each band was valuated using ImagJ Plus.
Transwell assay
Migration: Cells were grown in six-well plate until confluence reached 80–90%, then culture medium was changed to basal 1640 to starve 12 h before assay. Cell suspension was prepared using basal 1640 with concentration of 106 cell/mL, each upper chamber was added with 100 μL cell suspension while the lower chamber was filled with 600 μL complete 1640 supplemented with 15% FBS and incubated at 5% CO2, 37 ℃ for 12 h. Culture medium of upper chamber then was discarded and was gently wiped by wet swab. Cells attached on the lower chamber then were fixed by treating with 4% paraformaldehyde for 30 min and later was stained with 0.1% crystal violet for 10 min. Cell count was presented in a rule of average count of 5 high power fields.
Invasion: Matrigel was pre-thaw and diluted with basal 1640 (1:8 v/v), 100 μL liquid diluted Matrigel was added into upper chamber and incubated in 37℃ for 4 h until it turned to solid gel. The following procedure was the same as migration part.
Protein digestion and tandem mass tag (TMT) labeling
Total of 2 × 107 H446 shNC and H446 shCEMIP was collected and digested by SDT lysis buffer (4%(w/v) SDS, 100 mM Tris/HCl pH7.6, 0.1 M DTT). Total protein quantity was determined using BCA method, and trypsinized using the Filter aided proteome preparation (FASP) method. Peptide fragment was quantized by ultraviolet spectrophotometry with 280 nm wavelength (Wisniewski et al. 2009). Then, 100 μg protein of each sample was labeled by isotope kit TMT (Thermo Fischer Scientific co. US) according to the instruction. Each group was repeated above procedure in triplicates. The labeled peptides of each group were mixed in equal amounts and fractionated using the High pH Reversed Phase Peptide Fractionation Kit. The column was equilibrated with acetonitrile and 0.1% trifluoroacetic acid (TFA), and then was loaded the mixed labeled peptide sample within column. Addition of pure water in column and low-speed centrifugation was for desalination. The column was eluted by elution with gradient increased pH acetonitrile to separate exclusive peptides. Each eluted peptide sample was vacuumed and reconstituted with 12 μL 0.1% FA, and the peptide concentration was determined by ultraviolet spectrophotometry with 280 nm wavelength.
Proteomic analysis based on LC–MS/MS system
Each fractionated sample was analyzed with HPLC liquid system Easy nLC and Q Exactive mass spectrometer. Sample was loaded on the pretreatment C-18 reverse column (Thermo Scientific Acclaim PepMap100, 100 μm × 2 cm, nanoViper C18), and was separated through analytical column (Thermo scientific EASY column, 10 cm, ID75 μm, 3 μm, C18-A2). Eluent A was 0.1% formic acid aqueous solution. The diluted eluent A with 5% water was used for balancing LC system, eluent B, 0.1% formic acetonitrile aqueous solution (84% acetonitrile) was used for fragment separation. Elution flow rate was 300 nL/min. MS data detection method applied to positive ion, the scanning range of the precursor ion was 300–1800 m/z, the resolution of the primary mass spectrum was 70,000 at 200 m/z, the automatic gain control (AGC) target was 1e6, the maximum IT was 50 ms, and the dynamic exclusion time was 60.0 s. The mass-to-charge ratios of peptides and peptide fragments were collected according to the following method: 20 fragment spectra were acquired after each full scan. MS2 Activation Type was HCD. Isolation window was 2 m/z, second-order mass spectrometry resolution 17,500 at 200 m/z (TMT 6 plex). Normalized Collision Energy was 30 eV, with underfill ratio of 0.1%.
MS spectra data were identified through making score by Mascot2.2 and searching protein library by Proteome Discoverer 1.4 among Uniprot. Related parameters were listed: Enzyme: trypsin; Missed cleavage: 2; Fixed modification: Carbamidomethyl (C), TMT 6/10plex (N-term), TMT6/10 plex (K), Variable modification: oxidation (M), TMT 6/10plex (Y), peptide mass tolerance: ± 20 ppm; fragment mass tolerance: 0.1 Da; database pattern: decoy; false discovery rate (FDR) ≤ 0.01. To screen out DEPs, the protein ratios are calculated as the median of only unique peptides of the protein based on integrating average ion intensities report of biological triplicates. Avoiding experimental bias, normalization of all peptide ratios were measured by the median protein ratio. The median protein ratio should be 1 after the normalization. DEPs should meet criteria: matching of at least one unique peptide, and the quantitative analysis revealed protein fold changes ≥ 1.2 or ≤ 0.83 and p value < 0.05.
Bioinformatic analysis
Bioinformatic software for clustering biological function of DEPs involved Blast2GO, KAAS, Complexheatmap R package (R Version 3.4), STRING (https://stringdb.org/org/) and CytoScape software (version number: 3.2.1).
Statistics analysis
Statistical significance was calculated by SPSS19.0 using non-pair Student’s t test. Statistical result of each figures was indicated as mean value with ± SD. GO and KEGG enrichment was analyzed by the Fisher’s Exact Test. Every experiments were processed at least three times both biological replicate and technical replicate to avoid accidental error among inter- and intra-groups. Statistics significance was illustrated in each figure as *< 0.05, **< 0.01, ***< 0.001 when necessary.
Result
Clinical sample illustrated elevated expression of CEMIP on SCLC
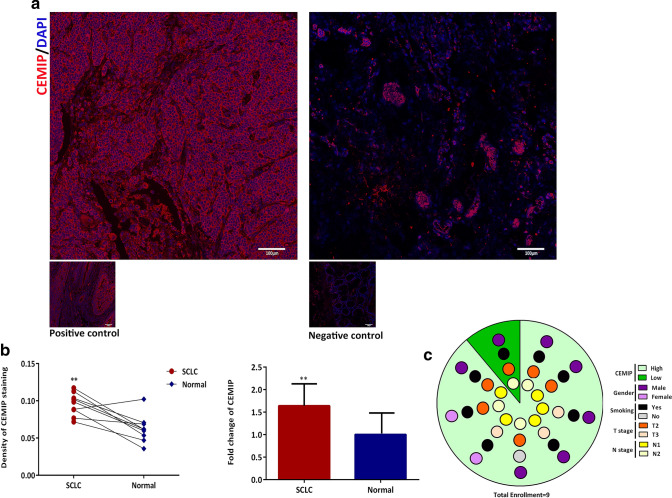
Due to our previous background survey of CEMIP (Li et al. 2017b), its protein expression was up-regulated among many cancers, promoting cell migration and invasion. According to the latest relevant report, CEMIP was up-regulated and associated EMT process of NSCLC to trigger metastasis (Tang et al. 2019). When considering the certain similarity and homology between NSCLC and SCLC, we hypothesized that the expression of CEMIP might be elevated in SCLC and it also might be a crucial promoter of metastasis. We retrospectively searched SCLC cases among Aug 2013–Jan 2019 and found out nine cases met all criteria. Among nine enrollment, eight of them stayed higher CEMIP expression in SCLC tissue than adjacent normal lung, the fold change was 1.64 ± 0.16, p < 0.01, as shown in Fig. 1a, b. CEMIP mostly located in cytoplasm of tumor cells and sporadically spread on tissues stroma (Fig. 1a). As for the basic characteristic of enrolled individuals, the average age was 61.89 ± 12.25 years, the average smoking amount was 42.22 ± 30.73 packs per year, and the average tumor size was 23.234 ± 40.09 cm3. Specific information was listed and summarized in Table 1 and Fig. 1c.
Fig. 1.
Expression of CEMIP on SCLC patients. a Left confocal image showed protein expression of CEMIP on SCLC tissue slice which was stained in red, and the nuclei was stained by DAPI as in blue color, whereas the right picture showed its pair normal counterpart. The positive and negative control of CEMIP staining, correspond to colon tumor tissue and its adjacent normal tissue, was placed in the down left of each pictures respectively. b Fluorescence density of CEMIP staining for SCLC and paired adjacent normal tissue was matched after statistic analysis wherein expression of CEMIP on SCLC with 1.64 ± 0.64 fold higher than that on paired normal, shown as in bar chart. c Demographic characteristics of enrolled patients. Background of the pie chart presented the expression level of CEMIP, each rounds with different color in each parallel cycles indicated the gender, smoking statue, T stage and N stage, respectively. p value ≤ 0.05 remains statistic significance, ***p value ≤ 0.001, **p value ≤ 0.01
Table 1.
Demographic of enrolled SCLC patients
| Items | Total | High expression of CEMIP | Low expression of CEMIP |
|---|---|---|---|
| No. of patients | 9 | 8 | 1 |
| Gender | |||
| Male | 7 | 6 | 1 |
| Female | 2 | 2 | 0 |
| Smoking | 42.22 ± 30.73 | 43.75 ± 32.48 | 30 |
| Age | 61.89 ± 12.25 | 64 ± 11.21 | 45 |
| Stage I–IV | |||
| II | 2 | 2 | 0 |
| III | 7 | 6 | 1 |
| Tumor size | 23.234 ± 40.09 | 10.1 ± 1.39 | 128.3 |
| T stage | |||
| T2 | 3 | 5 | 1 |
| T3 | 6 | 3 | 0 |
| N stage | |||
| N1 | 5 | 5 | 0 |
| N2 | 4 | 3 | 1 |
| M stage | |||
| M0 | 7 | 7 | 0 |
| M1 | 1 | 1 | 0 |
Establishment of CEMIP silence and over-repression in vitro model on SCLC
To build up the suitable in vitro model, we first detected both mRNA and protein expression of CEMIP in three SCLC cell lines and one cell line of normal lung bronchial epithelial cells. As shown in Fig. 2a, b, the mRNA expression of three SCLC cell lines, H446, H209 and H1688 was increased compared with that of BEAS-2B. The high-expressed CEMIP cell line H446 and low-expressed CEMIP cell lines H1688 was selected for setting up either expression-silencing or over-expression model, respectively. Both mRNA and protein expression of CEMIP was successfully declined by transduction LV-shCEMIP (Fig. 2c, d), wherein mRNA was silenced to 0.21 ± 0.12, 0.26 ± 0.07 and 0.58 ± 0.18 fold compared with their NC reference respectively. Besides, these three stable strains attained 0.36 ± 0.07, 0.49 ± 0.16 and 0.67 ± 0.05 reduced fold of CEMIP expression at protein level in correspond to the strain #1, #2 and #3, respectively. After transfecting CEMIP plasmid into H1688 cell, both mRNA expression of protein expression achieved 8.89 ± 3.4-fold and 1.90 ± 0.11-fold increase, respectively, when compared with the correspond expression of NC counterpart (Fig. 2e, f).
Fig. 2.
Selection of cell model and movement ability test. Screening results of mRNA and the protein expression of CEMIP among SCLC cell lines H446, H1688 and H209 and normal lung cell lines BEAS-2B by qRT-PCR and Western blot was presented in a and b respectively. p value ≤ 0.05 remains statistic significance. Efficiency of silencing and over-expressing CEMIP achieved by each LV-shRNA targets or plasmid transduction was examined both on mRNA (c and e) and protein level (d and f). In part g and h, transwell assays were carried out for comparing both migrated and invasive ability between H446 shCEMIP, H1688 OE and their negative control and parental cell lines. p value ≤ 0.05 remains statistic significance, ***p value ≤ 0.001, **p value ≤ 0.01, *p value ≤ 0.05
CEMIP enhanced cellular migration and invasion in SCLC
According to changing the expression of CEMIP might alter cell migration and invasion, in vitro model was prepared for verifying our assumption that whether CEMIP might exert as migratory and invasive promoter on SCLC. The migration and invasion ability was determined by transwell assays. Decreased protein expression of H446 leaded to significant drop of both cell migration and invasion as data were indicated in Fig. 2g. On the other hand, over-expressing the expression of CEMIP on H1688 resulted in increase of both migrated and invaded cells when compared to the parental and negative control groups (Fig. 2h).
Protein identification and differential protein expression
Based on the proteomic analysis, total amount of 51,611 peptides was screened out wherein 42,853 fragments were unique peptides. After blasting peptides from protein database, there were 7125 protein was identified. Quality control data including mass error distribution, peptide ionscore distribution and protein ratio distribution (Supplemental Fig. S1a, b) confirmed the stability and reliability of profiling procedure during data collection, all the distribution of peptides ranged among 10 ppm, and 60% of MS diagram illustrated the MASCOT score was larger than 20, with median score of 24.37. Protein ratio distribution of two groups concentrated on 1 (Supplemental Fig. S1c). All identified protein then was characterized in aspect of molecule weight, isoelectric point, protein length, peptides count and protein sequence coverage distribution to confirm the profiling results (Supplemental Fig. S2–3).
Among all identified proteins, the number of DEPs between H446 shCEMIP and its counterpart counted 215 proteins of which 109 was significantly up-regulated and 106 was significantly down-regulated (fold change > 1.20 vs < 0.83, p value < 0.05, FDR ≤ 0.01).
The classification of total 215 DEPs was described as Fig. 3a, and integrated the specific expression changes into each biological sample by clustering individual similar function, presenting in Fig. 3b. In the list of up-regulated proteins, the most expressed protein with around sevenfold changes was a novel protein which could not match any existed one in the databases. The expression of spindle and centriole-associated protein 1, SPICE1 (3.96), and Alpha-crystallin B chain CRYAB (1.99) ranked second and third up-regulation in the list. Details of top 20 up-regulated proteins were listed in Table 2. Table 3 otherwise summarized the most down-regulated proteins profiles ranging among 0.41–0.72-fold reduction. The most decreased protein was protein S100A6, whose expression reduced 0.41-fold referent to control group. The secondary and tertiary decreased DEPs would be hydroxyacid oxidase 1, HAO1 and AN1-type zinc finger protein 1.
Fig. 3.
a Distribution of changes of DEPs. Volcano plot showed the distribution of DEPs after silencing CEMIP on H446 cells, DEPs located away from two vertical dash lines indicated their expressed fold change were either higher or lower the cut-off value among total expressed change of DEPs. Among them, the pink pots beyond the horizontal dash line presented that their changes also met statistical threshold where p value ≤ 0.05. Bar chart located below this volcano plot briefly summarized the rule of distributed DEPs due to expression changes. b The identified DEPs was hierarchical clustered DEPs whose change fold over 1.2 with statistic significance. The result was displayed as Heat-map. Red cubes indicated up-regulated one while blue cube meant down-regulated one
Table 2.
Increased DEPs of H446 shCEMIP to control in TOP 20 list
| Accesion | Gene name | Protein name | Ratio | p value |
|---|---|---|---|---|
| B4DK97 | cDNA FLJ52096 | 6.968207 | 0.000849 | |
| Q8N0Z3 | SPICE1 | Spindle and centriole-associated protein 1 | 3.95636 | 1.51E−05 |
| E9PR44 | CRYAB | Alpha-crystallin B chain | 1.990323 | 4.28E−05 |
| A0A096LP69 | CD99 | CD99 antigen | 1.938855 | 0.030121 |
| H6VRF8 | KRT1 | Keratin 1 | 1.932469 | 0.015112 |
| P13645 | KRT10 | Keratin, type I cytoskeletal 10 | 1.840443 | 0.001536 |
| E0CX15 | UNC5C | Netrin receptor UNC5C | 1.795837 | 0.00085 |
| E7EW28 | SLC4A10 | Anion exchange protein | 1.724796 | 0.005225 |
| Q14894 | CRYM | Ketimine reductase mu-crystallin | 1.705285 | 0.001199 |
| Q01469 | FABP5 | Fatty acid-binding protein 5 | 1.691514 | 0.000297 |
| P35908 | KRT2 | Keratin, type II cytoskeletal 2 epidermal | 1.682743 | 0.006394 |
| Q49AL4 | NMNAT3 | NMNAT3 protein | 1.654057 | 0.004019 |
| H0YNS1 | PSTPIP1 | Proline-serine-threonine phosphatase-interacting protein 1 | 1.646509 | 0.00227 |
| Q96N83 | PODXL | Podocalyxin | 1.579259 | 0.000214 |
| F8VWZ8 | ARHGAP29 | Rho GTPase-activating protein 29 | 1.555096 | 0.001379 |
| P04083 | ANXA1 | Annexin A1 | 1.554867 | 1.95E-05 |
| P08493 | MGP | Matrix Gla protein | 1.514115 | 0.006992 |
| E9PCT3 | CAV2 | Caveolin | 1.472081 | 0.002359 |
| Q53TK7 | ACYP2 | Acylphosphatase | 1.472049 | 0.002279 |
| C9JW52 | IGFBP2 | Insulin-like growth factor-binding protein 2 | 1.450286 | 0.00169 |
Table 3.
Decreased DEPs of H446 shCEMIP to control in DOWN 20 list
| Accesion | Gene name | Protein name | Ratio | p value |
|---|---|---|---|---|
| P21359 | NF1 | Neurofibromin | 0.725099 | 0.000914 |
| C9JE82 | CACNA2D2 | Voltage-dependent calcium channel subunit alpha-2/delta-2 | 0.720526 | 0.002618 |
| P16402 | HIST1H1D | Histone H1.3 | 0.708203 | 1.34E−05 |
| Q59GS6 | DGK | Diacylglycerol kinase | 0.706853 | 1.5E−05 |
| P37837 | TALDO1 | Transaldolase | 0.704169 | 1.25E−06 |
| M0QZH0 | RCN3 | Reticulocalbin-3 | 0.700083 | 0.004536 |
| Q96DE7 | SMURF2 | SMURF2 protein | 0.684759 | 0.013039 |
| A0A087X0E5 | GGNBP2 | Gametogenetin-binding protein 2 | 0.68207 | 0.007184 |
| J3QL69 | LIMD2 | LIM domain-containing protein 2 | 0.675713 | 0.000813 |
| B2RCC2 | YBX2 | Y box binding protein 2 | 0.659611 | 0.00257 |
| B2R7H9 | cDNA, FLJ93450 | 0.655346 | 0.000113 | |
| H7C269 | TNRC6A | Trinucleotide repeat-containing gene 6A protein | 0.648618 | 0.000944 |
| P29762 | CRABP1 | Cellular retinoic acid-binding protein 1 | 0.64462 | 0.000551 |
| F4MHI6 | UTY | Ubiquitously transcribed tetratricopeptide repeat protein Y-linked transcript variant 283 | 0.619642 | 0.001545 |
| Q12771 | p37 AUF1 | 0.618075 | 0.019008 | |
| O14682 | ENC1 | Ectoderm-neural cortex protein 1 | 0.566633 | 0.006615 |
| A0A0A0MT55 | SARDH | Sarcosine dehydrogenase, mitochondrial | 0.540431 | 0.042912 |
| Q05BD2 | ZFAND1 | AN1-type zinc finger protein 1 | 0.480825 | 0.048354 |
| Q9UJM8 | HAO1 | Hydroxyacid oxidase 1 | 0.45578 | 0.025734 |
| R4GN98 | S100A6 | Protein S100A6 | 0.413797 | 0.003698 |
GO analysis of DEPs
Next, GO analysis (Fig. 4a) by online tool BLAST2 indicated that these DEPs facilitated cellular component (Fig. 4b) of protein belonged to caveolae, cornified envelope, and intermediate filament. These cellular component were annotated under the GO term "cell" in which most of DEPs were annotated. They participated in biological process (Fig. 4c) category including epithelial cell migration (GO:0010631), epithelium migration (GO:0090132), and tissue migration (GO:0090130) (Fig. 4a) whose general function attributed to "cellular process" term. As for molecule function (Fig. 4d), they enriched in secondary active transmembrane transporter activity, calcium ion binding, and inorganic anion exchanger activity, all of which either belonged to "binding" or "catalytic activity" term. Since we intended to discover the downstream targets of movement changes, we then concentrated on the enrichment term with gradually expanding their belonging layer under cellular component, biological process and molecular function which contained the most various proteins involved (Fig. 4e). The top two enrichment of cellular component level caveolae and cornified envelop belonged to intracellular organelle membrane. Additional classification of epithelium cell migration, it was originated from a brunch of protein controlled the cellular locomotion process, as well as 4 regulator protein with contrary direction was included in this term. In terms of molecular function, most of DEPs related to calcium ion channel, which work with binding tunnel protein to regulate ion alternation.
Fig. 4.
GO analysis of DEPs on H446 silencing CEMIP. Integrated classification of DEPs enriched towards cellular component, molecular function and biological process respectively (a). Bar length presented the participated number of protein, bar color illustrated the degree of p value, and the label number at the end of each illustrated the rich factor in specific activity. Number and proportion of involved DEPs were classified into these three terms individually in b–d. Part e displayed the specific terms listed in a to its originally belonging in more general layers. Protein name in black was up-regulated DEPs while the down-regulated showed in red
Biological annotation of DEPs and pathway enrichment
All the protein then was analyzed with their pathway enrichment KEGG database, overall distribution and protein amount of involved pathway were described in Fig. 5a, b and Table 4. DEPs enriched in each pathway were re-classified among all protein, representing as rich-factor. There were three clustering pathways affected the most from amount of DEPs and statistics analysis ranking in order of TGF-β, GABAergic synapse and MAPK signaling pathway, wherein the specific DEPs of each pathway were listed in Table 5. Most of identified protein assembled on MAPK pathway which was the main and powerful signaling hub connecting proliferation, organelle stress, apoptosis and metastasis. According to the enrichment analysis, 8 DEPs belonged to any site of MAPK pathway among total 73 identified ones, Fig. 6 displayed the position of these proteins and how their biological action worked. The common TGF-β pathway involved 3 DEPs among total 17 proteins which were NEO1, RBX1 and SMURF2. Concomitant decline of SMURF2 and rise of RBX1 contributed to suppression of Smad site. However, an unreported pathway of lung cancer, GABAergic synapse included 3 affected DEPs which were GS, GLS and SLC1A5 of total 14 proteins. GS and GLS took over the turnover of glutamic acid (Glu) and glutamine (Gln) which showed up-regulation while SLC1A5 located on membrane controlling releasing of Gln was down-regulated. Figure 5c depicted the interaction of DEPs using STRING tool wherein the SRC bridged most signal transduction to other DEPs, since it was reported of major upstream site for initiating many signaling such as EGFR, FGFR, and MAPK. PPI diagram displayed the connection of DEPs due to parts of current evidence, and the protein involvers were clustered within individual cycle that presented their major function.
Fig. 5.
KEGG enrichment and PPI network of DEPs on H446 silencing CEMIP. Top 10 enriched signaling pathway listed in a part. Bar length presented the participated number of protein, bar color illustrated the degree of p value, and the label number at the end of each illustrated the rich factor in specific activity. Number of DEPs enriched in top 20 signaling pathway was summarized in b. Part c demonstrated the interaction among part of DEPs with change over 1.25 fold. Thickness connecting line indicated the evidence level. Circles with different color presented the specific biological activities according to the database records
Table 4.
DEPs involved in specific biological function
| Function Type | GO term | Protein |
|---|---|---|
| Biological Process | Epithelial cell migration | EFNB2, GLUL, SRC, PFN2, HSPB1, NF1, SCG2, KRT2, ANXA1 |
| Epithelium migration | ||
| Tissue migration | ||
| Molecule Function | Secondary active transmembrane transporter activity | SLC6A15, SLC4A7, SLC4A7, SLC4A10, SLC24A2, SLC1A5 |
| Calcium ion binding | ANXA1, ACTN4, ANXA4, PLS3, VSNL1, HPCAL1, MGP, RCN3, SCGN, DGK, S100A2, FSRP, PLCB2, AIF1L, S100A6 | |
| Inorganic anion exchanger activity | SLC4A7, SLC4A7, SLC4A10 | |
| Cellular Component | Caveola | CAV1, SRC, PTRF, NOS1AP, CAV2 |
| Cornified envelope | ANXA1, KRT10, KRT2 | |
| Intermediate filament | KRT1, KRT2, KRT10 |
Table 5.
DEPs involved in specific signaling pathway
| Term | Protein |
|---|---|
| MAPK signaling | HSPB1, MAPT, CACNA2D1, CACNA2D2, IL1RAP, MAP3K20, NF1, DUSP7 |
| TGF-β signaling | NEO1, RBX1, SMURF2 |
| GABAergic synapse | GS, GLUL, SRC |
Fig. 6.
Hypothesis of CEMIP regulative way among top three signaling was drawn in flow chart based on the current researches. Green circle indicated the down-regulated protein whereas the Red one presented up-regulated protein. Plain line with arrow indicated the transmit direction of activated signals whereas the plain line with notch indicated the station of inhibition signals. Dash line with arrow directed the release way of involved items
Discussion
SCLC originated from lung neuroendocrine cell, retaining some feature of epithelial cell (Gazdar et al. 2017; van Meerbeeck et al. 2011). Normally, the diagnosis accompanied with either unresectable large fusion of lesion or distant metastasis resulted in very limited therapy choice for those patients. The promising therapy is still combination of platinum compounds and other chemo-reagent. Although a large scale of single nucleotide polymorphism (SNP) analysis revealed that mutation p53 and Rb1 was carcinogenic trigger for SCLC, these two major oncogene remained less specific (Gazdar et al. 2017; Peifer et al. 2012; van Meerbeeck et al. 2011). Therefore, we searched new protein target CEMIP which mainly functioned in promoting tumor migration, hoping to explore some part of molecule mechanism of SCLC metastasis. Our data first confirmed decreasing CEMIP expression attenuated both invasion and migration of SCLC. Similar results were found that CEMIP not only promoted metastasis by tumor itself, but also modified tumor microenvironment to assist tumor migration and invasion. CEMIP was previously reported as promoter of EMT transformation in other cancer types. Typical epithelial-derived tumor might arouse CEMIP up-regulation to cut down cyto-adherence and trigger EMT process, involving cross-talk between Wnt/β-catenin and EGFR pathway in cancer (Wang et al. 2019; Xu et al. 2019). Over-expression of CEMIP was also found in NSCLC that bridged PI3K/AKT on EMT process (Tang et al. 2019) and assisted colorectal cancer cell in EMT progression through sponge mechanism of lncRNA TUG1 and its ceRNA miR-600 (Sun et al. 2018). Similarly, our data showed that the expression of four mesenchymal markers (N-cadherin, Vimentin, Snail1 and Twist1) were significantly down-regulated, while the expression of epithelial marker cytokeratin 18 (CK18) was up-regulated, but E-cadherin could not be detected in H446 shCEMIP cells compared to those in H446 shNC cells (Supplementary Fig. S4a, b). Consistently, the H446 shNC cells exhibited scattered, elongated, and mesenchymal-like morphology, while the H446 shCEMIP cells showed rounded shape, epithelial cobblestone appearance (Supplementary Fig. S4c). Taken together, our results suggest a potential role of CEMIP in the acquisition of EMT in SCLC cells. However, further studies are still needed to clarify the underlying mechanism.
The other way CEMIP induced tumor progression would be through induction of inflammation, which ensured the metastatic environment. CEMIP over-expression enhanced the COX2 activity in secreting IL-8 and IL-1β in pancreatic cancer (Kohi et al. 2017). CEMIP could stimulate expression and secretion of IL-6 to sustain chronic filtration (Soroosh et al. 2016). Metastatic breast tumor cells secreted exosome highly expressed CEMIP around brain tissues to create inflammation that allowed remodeling vessel net for lesion colonization (Rodrigues et al. 2019). CEMIP also facilitated the HPV-infection to cause cervical carcinogenesis through continuously phosphorylated MEK/ERK axis (Shostak et al. 2014). Worthy to be noted that our results showed one of inflammatory receptor associated protein IL1RAP was down-regulated by silencing CEMIP expression. Down-regulation of IL1RAP could deprive the inflammation respond of SCLC cells since the receptor complex lessened combination to inflammatory factors. Since we aimed to discover the targets under the CEMIP controls, the down-regulated DEPs might receive its directly positive regulation as the down-regulated DEPs would be inhibited by CEMIP, when the protein expression of CEMIP on H446 was descended. Based on these evidence, we searched out current information of the most down-regulated protein S100A6 whether it was suggested being related to inflammations. Li C et al. found out S100A6 was up-regulated in inflammation-associated corneal neovascularization (Li et al. 2010), and the high expression of S100A6 also related to asthma-related airway inflammation (Calvo et al. 2009). Recent report explored that S100A6 was launched by ZEB1 which stimulated translation of IL-6 and IL-11 associating invasion of pancreatic ductal adenocarcinoma (Al-Ismaeel et al. 2019). Besides, Kikuchi et al. reported that ZEB1 located at upstream sites of CEMIP (Kikuchi et al. 2016), therefore, we suspected that S100A6 might be a vital downstream of CEMIP with its positive regulation in cellular migration and invasion. Since few researches focused on CEMIP and S100A6, how CEMIP activated S100A6 and in what pathway it played still nevertheless demand to be urgently solved.
Besides, the enrichment gene ontology term then was traced back to its general functional category. Step-by-steps introduction were assembled in Fig. 4e, first we found out that the cellular location of DEPs in two of the high score enriched cellular component were consisted of intracellular organelle membrane. This evidence implied declining CEMIP expression could interrupt the movement of membrane system. As all of the biological process centralized on migration, its bi-direction regulator therefore was included, wherein the SRC, a big ‘signal transfer-station’ and positive regulator of cell migration exhibited down-regulation. This analysis was accordance with the results of our transwell assays. Since our next stage was building up on whether CEMIP bound other candidates to play role of signals transmitter, we then turned to figure what molecular function contained calcium ion binding wherein the largest amount of DEPs and the most down-regulated S100A6 participated.
Aiming at interpret what signaling might act on tumorous function when CEMIP expression was deficiency, hypothesis of CEMIP-induced metastatic pathway was redrew in Fig. 6 including GABAergic synapse, TGF-β and MAPK. Combined our data and the current researches of MAPK, three isoforms directed each branch to survive, stress respond, apoptosis and cross-talked with migration pathway (Wagner and Nebreda 2009). When expression of CEMIP was descended, inhibition of ERK1/2 and p38 branches lead by NF1 was relieved. Besides, the dephosphorylation enzyme MKP was up-regulated, thereby attenuating degradation of p-ERK1/2 and p-p38. Evidence proved the S100A6 indirectly interacted with p38/MAPK in colon cancer (Duan et al. 2014), nasopharyngeal carcinoma (Li et al. 2017a) and ectopic endometrial stromal cells (Peng et al. 2018). Surprisingly, silence of CEMIP suppressed expression of its binding protein Src, a vital tyrosine kinase, thereby inactivating phosphorylation of many signaling proteins including MEK/ERK, FAK and EGFR (Parkin et al. 2019; Shostak et al. 2014). Another pathway TGF-β signaling was reported for controlling cell migration on many tumors (Jakowlew, 2006). The signals from MAPK repressed the downstream of Smad2/3 could inhibit migration (Li et al. 2019). In the results of DEPs, a key protein SUMRF2 assisted ubiquitin smad2/3 to activate signaling transduction (Izzi and Attisano, 2004). Reduction of SUMRF2 suppressed the inhibition of TGF-β signals, cell migration in pancreatic cancer cells (Wu et al. 2016). Jin et al. also found the abrogated SMURF2 expression resulted in attenuation of migration and invasion by reorganizing skeleton protein (Jin et al. 2009). Concordance to KEGG enrichment result, declined SUMRF2 and raising RBX1 affected by silencing CEMIP might be another promising line in SCLC migration and invasion. Another metastatic inhibitor K10 was increased by silencing CEMIP, its expression negatively correlated to cell invasion and malignancy through TGF-β signaling in oral squamous cell carcinoma (Huntley et al. 2004).
Moreover, MAPK signaling also cross-talks to GABAergic synapse signaling. CEMIP positively regulated Gln-releasing channel SLC1A5 through c-Myc suppression from ERK activation. Although GABAergic synapse pathway was barely studied in tumor diseases except aging impairment of nervous system and brain cancer, none of reports found this pathway dominated in lung cancer (Rozycka and Liguz-Lecznar 2017). However, ascending GABA content and expression of its receptor might be biomarker for paraneoplastic limbic encephalitis which might be the specific sign of pulmonary symptoms in SCLC (Hoftberger et al. 2013). Due to the complicated tumourigenesis of SCLC, CEMIP controlled signaling pathways as cross-linked network rather than action alone. In total, we pursued the speculation of this study that CEMIP promoted the migration and invasion of SCLC cells in detail: CEMIP deprived the binding ability of regulator of ion channel protein such as S100A6, disturbing cellular ion flow, thereby triggering the MAPK signaling which owning potential to cross-talk TGF-β pathway.
Unavoidable limitation also appeared in our study. As surgery remained less suggestive intervention for SCLC, clinical samples were extremely hard to collect, thereby only a small amount of SCLC tissues was provided. Thus, our further research focused on animal model in aspect of recurrence of SCLC metastatic phenomenon and tracking the precisely locus among ‘soil’. Besides, the reports of some researchers who have validated high expression of CEMIP was involved in generating chemo-resistance when administrating with targeted agents such as sorafenib (Xu et al. 2019), selumetinib (Duong et al. 2018) and EGFR-tyrosine kinase inhibitors (Rizzolio et al. 2018). However, whether and how CEMIP facilitates cells to resist to chemotherapeutic agents need to be further studied. Nevertheless, our results do shall light on the research gap of CEMIP in SCLC.
Conclusion
In this study, we found that CEMIP showed similar biological function in promoting cellular migration and invasion in SCLC cells. After proteomic analysis based on TMT coupled LC–MS systems, there were 251 DEPs related to downstream of CEMIP enriching in cellular migration. DEPs affected by CEMIP might predominately participate in MAPK and TGF-β signaling to trigger metastasis possibly through reducing S100A6 expression and cross-talked to other involved signaling proteins. S100A6, the most down-regulated target which was reported in MAPK mediated inflammation and migration in other tumor researches might be the key connection of CEMIP to intracellular homeostasis in aspect of SCLC metastasis. Although there is still limitation of current data since disease rarity and long-duration collection of large scale of clinical samples, further study of in vivo verification and precise signaling mechanism in how CEMIP promoted metastasis associated drug resistance in SCLC has been conducted by our research team. So far, these data remained the first report of CEMIP in promoting SCLC metastasis, and block of CEMIP might be the potential site delayed the occurrence of metastasis.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Acknowledgements
We express our gratitude to the research team of Prof. Huaxin Hou and Prof. Danrong Li, who provided countless assistance during experiment process.
Abbreviations
- BLAST
Basic local alignment search tool
- CEMIP
Cell migration inducing hyaluronidase
- CRYAB
Alpha-crystallin B chain
- DEPs
Differentially expressed proteins
- GABA
γ-Aminobutyric acid
- GO
Gene ontology
- KEGG
Kyoto Encyclopedia of Genes and Genomes
- LC–MS
Liquid chromatography–tandem mass
- MAPK
Mitogen-activated protein kinase
- PVDF
Polyvinylidene fluoride
- RT-qPCR
Real-time quantitative polymerase chain reaction
- S100A6
Protein S100-A6
- SCLC
Small cell lung cancer
- SDS-PAGE
Sodium dodecyl sulfate polyacrylamide gel electrophoresis
- SMURF2
E3 ubiquitin-protein ligase SMURF2
- SPICE1
Spindle and centriole-associated protein 1
- TGF-β
Transforming growth factor-β
- TMT
Tandem mass tag
- WB
Western blot
Author contributions
Experiments procedure and paper writing were conducted by LL, JY and LL designed study proposal, JY provided technology support, GH, CT and YP provided assistance in some experiments. XM collected the clinical sample and XM, ML, TW and JS analyzed data. KL and NM provided clinical consultation. All authors read and reached agreement on final manuscript.
Funding
This study is funded by the National Natural Science Foundation of China (No. 81860649), Natural Science Foundation Program of Guangxi (No. 2018GXNSFAA050053) and Innovation Project of Guangxi Graduate Education (No. YCBZ2020054). This research was also funded by the first batch of cultivating talents of young and middle-aged backbone teachers in Guangxi universities and Gunagxi First-class Discipline Project for Pharmaceutical Sciences (No. GXFCDP-PS-2018) and the Project of Innovation, Entrepreneurship, and Joint Training Base for Pharmaceutical Postgraduates.
Availability of data and materials
All data generated or analyzed during this study were included in this published article and its supplementary information files.
Compliance with ethical standards
Conflict of interest
All authors declare no conflict of interest.
Ethics approval
Study protocol obeyed the Declaration of Helsinki for all human or animal experimental investigations and was approved and filed by Ethic committee board of Guangxi Medical University. Patients signed the written informed consent and the study was approved by Ethic committee board of Guangxi Medical University, China.
Informed consent
All enrolled cases were signed informed consents.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Li Li, Xiaoxiang Mo, Yingxing Pan are co-authors and equally contributed their work in this study.
References
- Al-Ismaeel Q, Neal CP, Al-Mahmoodi H, Almutairi Z, Al-Shamarti I, Straatman K, Jaunbocus N, Irvine A, Issa E, Moreman C et al (2019) ZEB1 and IL-6/11-STAT3 signalling cooperate to define invasive potential of pancreatic cancer cells via differential regulation of the expression of S100 proteins. Br J Cancer 121(1):65–75 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amarasena IU, Chatterjee S, Walters JA, Wood-Baker R, Fong KM (2015) Platinum versus non-platinum chemotherapy regimens for small cell lung cancer. Cochrane Database Syst Rev 8:CD006849 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Birkenkamp-Demtroder K, Maghnouj A, Mansilla F, Thorsen K, Andersen CL, Oster B, Hahn S, Orntoft TF (2011) Repression of KIAA1199 attenuates Wnt-signalling and decreases the proliferation of colon cancer cells. Br J Cancer 105(4):552–561 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calvo FQ, Fillet M, de Seny D, Meuwis MA, Maree R, Crahay C, Paulissen G, Rocks N, Gueders M, Wehenkel L et al (2009) Biomarker discovery in asthma-related inflammation and remodeling. Proteomics 9(8):2163–2170 [DOI] [PubMed] [Google Scholar]
- Chen L, Shi K, Andersen TL, Qiu W, Kassem M (2019) KIAA1199 is a secreted molecule that enhances osteoblastic stem cell migration and recruitment. Cell Death Dis 10(2):126 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen W, Zheng R, Zhang S, Zeng H, Zuo T, Xia C, Yang Z, He J (2017) Cancer incidence and mortality in China in 2013: an analysis based on urbanization level. Chin J Cancer Res 29(1):1–10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duan L, Wu R, Zou Z, Wang H, Ye L, Li H, Yuan S, Li X, Zha H, Sun H et al (2014) S100A6 stimulates proliferation and migration of colorectal carcinoma cells through activation of the MAPK pathways. Int J Oncol 44(3):781–790 [DOI] [PubMed] [Google Scholar]
- Duong HQ, Nemazanyy I, Rambow F, Tang SC, Delaunay S, Tharun L, Florin A, Buttner R, Vandaele D, Close P (2018) The endosomal protein CEMIP links WNT signaling to MEK1-ERK1/2 activation in selumetinib-resistant intestinal organoids. Cancer Res 78(16):4533–4548 [DOI] [PubMed] [Google Scholar]
- Fink SP, Myeroff LL, Kariv R, Platzer P, Xin B, Mikkola D, Lawrence E, Morris N, Nosrati A, Willson JK et al (2015) Induction of KIAA1199/CEMIP is associated with colon cancer phenotype and poor patient survival. Oncotarget 6(31):30500–30515 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gazdar AF, Bunn PA, Minna JD (2017) Small-cell lung cancer: what we know, what we need to know and the path forward. Nat Rev Cancer 17(12):725–737 [DOI] [PubMed] [Google Scholar]
- Hoftberger R, Titulaer MJ, Sabater L, Dome B, Rozsas A, Hegedus B, Hoda MA, Laszlo AHJ, Harms L et al (2013) Encephalitis and GABAB receptor antibodies: novel findings in a new case series of 20 patients. Neurology 81(17):1500–1506 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huntley SP, Davies M, Matthews JB, Thomas G, Marshall J, Robinson CM, Eveson JW, Paterson IC, Prime SS (2004) Attenuated type II TGF-beta receptor signalling in human malignant oral keratinocytes induces a less differentiated and more aggressive phenotype that is associated with metastatic dissemination. Int J Cancer 110(2):170–176 [DOI] [PubMed] [Google Scholar]
- Izzi L, Attisano L (2004) Regulation of the TGFbeta signalling pathway by ubiquitin-mediated degradation. Oncogene 23(11):2071–2078 [DOI] [PubMed] [Google Scholar]
- Jakowlew SB (2006) Transforming growth factor-beta in cancer and metastasis. Cancer Metastasis Rev 25(3):435–457 [DOI] [PubMed] [Google Scholar]
- Jemal A, Bray F, Center MM, Ferlay J, Ward E, Forman D (2011) Global cancer statistics. CA Cancer J Clin 6(2):69–90 [DOI] [PubMed] [Google Scholar]
- Jin C, Yang YA, Anver MR, Morris N, Wang X, Zhang YE (2009) Smad ubiquitination regulatory factor 2 promotes metastasis of breast cancer cells by enhancing migration and invasiveness. Cancer Res 69(3):735–740 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kikuchi M, Yamashita K, Waraya M, Minatani N, Ushiku H, Kojo K, Ema A, Kosaka Y, Katoh H, Sengoku N et al (2016) Epigenetic regulation of ZEB1-RAB25/ESRP1 axis plays a critical role in phenylbutyrate treatment-resistant breast cancer. Oncotarget 7(2):1741–1753 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koga A, Sato N, Kohi S, Yabuki K, Cheng XB, Hisaoka M, Hirata K (2017) KIAA1199/CEMIP/HYBID overexpression predicts poor prognosis in pancreatic ductal adenocarcinoma. Pancreatology 17(11):115–122 [DOI] [PubMed] [Google Scholar]
- Kohi S, Sato N, Koga A, Matayoshi N, Hirata K (2017) KIAA1199 is induced by inflammation and enhances malignant phenotype in pancreatic cancer. Oncotarget 8(10):17156–17163 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li A, Shi D, Xu B, Wang J, Tang YL, Xiao W, Shen G, Deng W, Zhao C (2017a) S100A6 promotes cell proliferation in human nasopharyngeal carcinoma via the p38/MAPK signaling pathway. Mol Carcinog 56(3):972–984 [DOI] [PubMed] [Google Scholar]
- Li C, Zhang F, Wang Y (2010) S100A proteins in the pathogenesis of experimental corneal neovascularization. Mol Vis 16:2225–2235 [PMC free article] [PubMed] [Google Scholar]
- Li H, Zhang Z, Chen L, Sun X, Zhao Y, Guo Q, Zhu S, Li P, Min L, Zhang S (2019) Cytoplasmic Asporin promotes cell migration by regulating TGF-beta/Smad2/3 pathway and indicates a poor prognosis in colorectal cancer. Cell Death Dis 10(2):109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li L, Yan LH, Manoj S, Li Y, Lu L (2017b) Central role of CEMIP in tumorigenesis and its potential as therapeutic target. J Cancer 8(12):2238–2246 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oneyama M, Sakamoto N, Oue N, Kimura Y, Hiroshima Y, Hashimoto I, Hara K, Maezawa Y, Kano K, Aoyama T et al (2019) Clinical significance of KIAA1199 as a novel target for gastric cancer drug therapy. Anticancer Res 39(12):6567–6573 [DOI] [PubMed] [Google Scholar]
- Parkin A, Man J, Timpson P, Pajic M (2019) Targeting the complexity of Src signalling in the tumour microenvironment of pancreatic cancer: from mechanism to therapy. FBES J 286(18):3510–3539 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peifer M, Fernandez-Cuesta L, Sos ML, George J, Seidel D, Kasper LH, Plenker D, Leenders F, Sun R, Zander T et al (2012) Integrative genome analyses identify key somatic driver mutations of small-cell lung cancer. Nat Genet 44(10):1104–1110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng Y, Lin J, Ma J, Lin K, Xu K, Lin J (2018) Upregulation of S100A6 in patients with endometriosis and its role in ectopic endometrial stromal cells. Gynecol Endocrinol 34(9):815–820 [DOI] [PubMed] [Google Scholar]
- Powell HA, Tata LJ, Baldwin DR, Potter VA, Stanley RA, Khakwani A, Hubbard RB (2014) Treatment decisions and survival for people with small-cell lung cancer. Br J Cancer 110(4):908–915 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rizzolio S, Battistini C, Cagnoni G, Apicella M, Vella V, Giordano S, Tamagnone L (2018) Downregulating neuropilin-2 triggers a novel mechanism enabling EGFR-dependent resistance to oncogene-targeted therapies. Cancer Res 78(4):1058–1068 [DOI] [PubMed] [Google Scholar]
- Rodrigues G, Hoshino A, Kenific CM, Matei IR, Steiner L, Freitas D (2019) Tumour exosomal CEMIP protein promotes cancer cell colonization in brain metastasis. Nat Cell Biol 21(11):1403–1412 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rozycka A, Liguz-Lecznar M (2017) The space where aging acts: focus on the GABAergic synapse. Aging Cell 16(4):634–643 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Semenova EA, Nagel R, Berns A (2015) Origins, genetic landscape, and emerging therapies of small cell lung cancer. Genes Dev 29(14):1447–1462 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shostak K, Zhang X, Hubert P, Goktuna SI, Jiang Z, Klevernic I, Hildebrand J, Roncarati P, Hennuy B, Ladang A et al (2014) NF-kappaB-induced KIAA1199 promotes survival through EGFR signalling. Nat Commun 5:5232 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siegel RL, Miller KD (2020) Cancer statistics, 2020. CA Cancer J Clin 70(1):7–30 [DOI] [PubMed] [Google Scholar]
- Soroosh A, Albeiroti S, West GA, Willard B, Fiocchi C, de la Motte CA (2016) Crohn's disease fibroblasts overproduce the novel protein KIAA1199 to create proinflammatory hyaluronan fragments. Cell Mol Gastroenterol Hepatol 2(3):358–368.e354 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suh HN, Jun S, Oh AY, Srivastava M, Lee S, Taniguchi CM, Zhang S, Lee WS, Chen J, Park BJ et al (2016) Identification of KIAA1199 as a biomarker for pancreatic intraepithelial neoplasia. Sci Rep 6:38273 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun JF, Hu JY, Wang GJ, Yang Z, Zhao CL, Zhang XF, Wang JX (2018) LncRNA TUG1 promoted KIAA1199 expression via miR-600 to accelerate cell metastasis and epithelial-mesenchymal transition in colorectal cancer. J Exp Clin Cancer Res 37(1):106 [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Tang Z, Ding Y, Shen Q, Zhang C, Li J, Nazar M, Wang Y, Zhou X, Huang J (2019) KIAA1199 promotes invasion and migration in non-small-cell lung cancer (NSCLC) via PI3K-Akt mediated EMT. J Mol Med (Berl) 97(1):127–140 [DOI] [PubMed] [Google Scholar]
- Torre LA, Bray F, Siegel RL, Ferlay J, Lortet-Tieulent J, Jemal A (2015) Global cancer statistics, 2012. CA Cancer J Clin 65(2015):87–108 [DOI] [PubMed] [Google Scholar]
- van Meerbeeck JP, Fennell DA, De Ruysscher DK (2011) Small-cell lung cancer. Lancet 378:1741–1755 [DOI] [PubMed] [Google Scholar]
- Wagner EF, Nebreda AR (2009) Signal integration by JNK and p38 MAPK pathways in cancer development. Nat Rev Cancer 9(8):537–549 [DOI] [PubMed] [Google Scholar]
- Wang L, Yu T, Li W, Li M, Zuo Q, Zou Q, Xiao B (2019) The miR-29c-KIAA1199 axis regulates gastric cancer migration by binding with WBP11 and PTP4A3. Oncogene 38(17):334–3150 [DOI] [PubMed] [Google Scholar]
- Wisniewski JR, Zougman A, Nagaraj N, Mann M (2009) Universal sample preparation method for proteome analysis. Nat Methods 6(5):359–362 [DOI] [PubMed] [Google Scholar]
- Wu B, Guo B, Kang J, Deng X, Fan Y, Zhang X, Ai K (2016) Downregulation of Smurf2 ubiquitin ligase in pancreatic cancer cells reversed TGF-beta-induced tumor formation. Tumour Biol. 10.1007/s13277-016-5432-0 [DOI] [PubMed] [Google Scholar]
- Xu Y, Xu H, Li M, Wu H, Guo Y, Chen J, Shan J, Chen X, Shen J, Ma Q et al (2019) KIAA1199 promotes sorafenib tolerance and the metastasis of hepatocellular carcinoma by activating the EGF/EGFR-dependent epithelial-mesenchymal transition program. Cancer Lett 454:78–89 [DOI] [PubMed] [Google Scholar]
- Zhao L, Zhang D, Shen Q, Jin M, Lin Z, Ma H, Huang S, Zhou P, Wu G, Zhang T (2019) KIAA1199 promotes metastasis of colorectal cancer cells via microtubule destabilization regulated by a PP2A/stathmin pathway. Oncogene 38(7):935–949 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data generated or analyzed during this study were included in this published article and its supplementary information files.