Abstract
Chronic wounds are increasing in numbers and biofilm-producing bacteria are highly prevalent in these wounds and often create resilient polymicrobial infections. Moreover, estimates suggest that up to 23 % of wounds contain fungi, particularly Candida albicans. Currently, inter-kingdom chronic wound models are scarce; thus, this study presents one of the few in vitro models that incorporate both bacterial and fungal species in a wound-relevant environment, addressing a critical gap in current biofilm research. The newly developed model contained the commonly isolated wound bacteria Pseudomonas aeruginosa and Staphylococcus aureus, and the fungus Candida albicans. Inter-species interactions were investigated through selective plate counting and pH and oxygen measurements, as well as confocal microscopy. Investigations were carried out before and after exposure to commonly used clinical antimicrobial treatments, including silver-infused bandages. When grown in a tri-species consortium, P. aeruginosa and S. aureus exhibited a higher tolerance towards silver-infused bandages than when they were grown individually. This suggests that C. albicans plays a protective role for the bacteria. In addition, the treatment also caused a shift in species ratios, moving from a P. aeruginosa-dominated consortium to a S. aureus-dominated consortium. Moreover, confocal microscopy revealed a change in biofilm architecture when comparing single-species models to tri-species models. Finally, we observed that silver-infused bandages increased the pH in the tri-species model as well as partially restoring the oxygenation within the wound model. In conclusion, our novel model exemplifies how inter-kingdom interactions in fungal-bacterial infections can complicate both the microenvironment and treatment efficacy.
Keywords: In vitro models, Biofilms, Multispecies, Wound models, Inter-kingdom interactions, Silver-infused bandages
Highlights
-
•
A novel tri-species wound model was created containing bacteria and fungi.
-
•
Silver-infused bandages were tested in both single-species and tri-species models.
-
•
Bandages had a reduced effect in tri-species models suggesting a protective effect.
-
•
Different biofilm architecture might play a role in protection.
1. Introduction
Fungal biofilm infections pose a significant burden on hospitals due to several predisposing factors in patients, such as immunosuppression, catheters, disruption of mucosal barriers, chemotherapy and the use of broad-spectrum antibiotics [1]. Fungi are responsible for at least 13 million infections globally each year, and the prevalence is steadily increasing [2]. Candida albicans, a commensal fungus living on mucosal surfaces, is one of the fungi most frequently associated with biofilm formation [1]. Candida species are increasingly attracting attention due to their role in the pathophysiological processes within the wound microenvironment and interactions across biological kingdoms. Biofilm production enables fungi and bacteria to adapt to and resist environmental changes – including antimicrobial treatments [3]. The increased resistance to conventional antibiotic treatments raises significant concerns in relation to chronic wounds, as the microenvironment provides an ideal habitat for biofilm-forming microbes [4]. Moreover, a recent rise in general fungal resistance to antifungal agents has been observed [5]. This includes resistance to a single class of drugs as well as multidrug resistance towards several classes [6], both of which have been observed in wounds.
It is estimated, that up to 80 % of chronic wounds contain biofilm, although it is often speculated that all chronic wounds may contain some amount of biofilm [7]. Pseudomonas aeruginosa and Staphylococcus aureus are two of the most frequently isolated bacteria from chronic wounds, and both organisms can adapt to co-exist [[8], [9], [10], [11]]. This co-existence creates a polymicrobial infection, where the two organisms often display intricate interactions that complicate treatment [8]. The fungal involvement in wounds is often overlooked, although recently, the presence of fungi in wounds has received more interest. Some studies estimate that up to 23 % of chronic wounds contain fungal species [5,12] However, a study on diabetic foot ulcers, using molecular techniques, found fungal species in 80 % of the assayed wounds, with Candida being one of the most predominant genera [13,14]. Furthermore, it is believed that fungi contribute to more than 50 % of the microbial quantity burden in most wounds [12].
Interactions between fungal and bacterial species are thought to complicate treatment further. Although they are only sparsely studied, initial reports suggest that co-infections may exacerbate the infection. Studies have found that in vitro co-cultures of S. aureus and C. albicans display increased virulence factors, upregulated drug resistance genes, and heightened toxin production [15,16]. Animal studies conducted in mice showed that single-species intra-abdominal infections of either S. aureus or C. albicans were non-lethal, but co-infections with the same doses caused 100 % mortality [17,18].
The human chronic wound environment is a complex milieu, containing several distinct features. The microenvironment is alkaline with pH levels ranging from 7.5 to 8.9 [19]. Wounds with a very alkaline pH have been reported to heal significantly slower than those with neutral pH [20]. Hence, pH levels play a significant role in healing and are also found to affect microbial growth, as some microbes favour alkaline conditions [21]. The chronic wound environment is believed to be highly anoxic due to the large oxygen consumption exhibited by microbes as well as human skin and immune cells [22]. Gradients of oxygen within wounds are believed to create microniches allowing for a larger variety of phenotypically distinct populations [23,24]. Thus, the biogeography and the heterogeneity of the species found within a wound may depend on gradients of oxygen and nutrients, although this topic is not yet fully understood.
Inter-kingdom in vitro models that mimic the wound environment are scarce, yet highly valuable. Initial reports investigating the effects of treatment on such tri-species consortia suggest that protective interactions may occur [25]. In this study, we aimed to establish a robust tri-species inter-kingdom chronic wound model consisting of C. albicans, P. aeruginosa, and S. aureus. The model was based on a previously published version which only included P. aeruginosa and S. aureus [26]. In this newly established model, we tested the effect of a commonly used clinical treatment, namely silver-infused bandages. Additionally, we treated the models with a combination of silver bandages and the antifungal compound caspofungin. Caspofungin was chosen because many clinical guidelines suggests using this echinocandin as first-line therapy for candidal infections and its effectiveness against Candida biofilms has been found to be more potent compared to other antifungal agents [27]. The treatment effects were compared between tri-species models and single-species models. These effects were evaluated using selective agar plate counts, oxygen and pH microsensor measurements, and confocal laser scanning microscopy (CLSM).
2. Materials and methods
2.1. Strains
GFP-tagged S. aureus (strain RN4220) [28] and mCherry-tagged P. aeruginosa (strain PAO1) [29] were grown as overnight (ON) cultures in peptone (Oxoid). C. albicans (strain ATCC 18804) were grown in peptone supplemented with glucose (1 g/L). ON cultures were grown at 37 °C with shaking (160 rpm).
2.2. The collagen-based layered chronic wound biofilm model
The models were made as previously described [26]. In short, a subcutaneous fat-like layer composed of peptone (Oxoid), cattle serum (Sigma-Aldrich), laked horse blood (SSI Diagnostica), highly concentrated rat tail tendon collagen type 1 (Corning Inc.) at a final concentration of 2 mg/ml, alpha-amylase (A6255, Sigma–Aldrich), pig fat, 1 N NaOH, 10x PBS, and distilled water was cast in ThinCert cell culture inserts (0.4 μm pore size, Greiner Bio-One GmbH). On top of this, a dermis-like layer was cast with the same components at slightly different concentrations, but with the pig fat omitted. See Fig. 1 for a schematic of the model.
Fig. 1.
Schematic of the model.
For tri-species models, P. aeruginosa was added on top of the dermis-like layer, followed by a short incubation period, after which S. aureus and C. albicans were added together. An estimate of 25–100 colony-forming units (CFU) of P. aeruginosa, 75–300 CFU of S. aureus, and 200–500 CFU of C. albicans were added. For single-species models, only one species was added on top. For dual-species models, the two chosen species were added in the same manner as in tri-species models; P. aeruginosa was always added first, followed by S. aureus and C. albicans together.
Once the models were made, the inserts were placed in a 12-well plate containing wound simulating medium (WSM), composed of 50 % bovine serum mixed with 50 % saline-buffered peptone at a 0.1 % concentration. This medium has previously been used in similar studies to simulate wound fluid [[30], [31], [32]] as the major components of wound fluid are damaged host tissue and plasma [33]. The models were incubated at 33 °C on a shaker set to 60 rpm. The WSM was replaced every 24 h.
2.3. Microsensor measurements
Microsensor measurements of oxygen and pH were performed using electrochemical sensors (Unisense A/S). The oxygen microelectrode had a sensor tip diameter of 25 μm and both the pH microelectrode and reference electrode had sensor tip diameters of 100 μm. The software Sensortrace Suite (Unisense A/S) was used to control the sensors, which were connected via a Microsensor Multimeter. The microsensors were angled vertically and positioned directly above the center of each model before measurements were taken. Readings were obtained every 100 μm within the model until a depth of 1 mm was reached.
2.4. Confocal laser scanning microscopy
Before the models could be visualised by confocal laser scanning microscopy (CLSM), they were wrapped in foil, snap-frozen in dry ice for 5 min, and then thin vertical slices were cut using a razor blade. The slices were subsequently placed on 0.17 mm glass slides. When baseline models were visualised, a drop of Calcofluor white (Sigma-Aldrich) was added to stain the fungi and Syto™62 (Invitrogen) was added as well. When comparing treated to untreated models, a drop of Sytox™Orange Dead Cell Stain (Invitrogen) or Propidium Iodide (PI) (Sigma-Aldrich) was added to visualise dead microbes. A Zeiss LSM 880 inverted confocal laser scanning microscope (Carl Zeiss GmbH) was used to visualise the models using a 10x, 20x, or 63x oil objective. The final images were processed using Imaris 10.0.1 software (Bitplane AG). Overview images were taken using a tile-scan approach with a 10 % overlap and z-stacks with step sizes of 50 μm covering the entirety of the sample.
2.5. Treatments and caspofungin MIC determination
All treatments were applied to 48-h-old models. Silver-infused bandages (Acticoat Flex 3, Smith & Nephew) were cut into 1 × 1 cm pieces and gently placed on top of the models. When testing three times the normal amount, three layers of bandages were placed on top of the models. Minimum inhibitory concentration (MIC) values of caspofungin (Sigma-Aldrich) were determined by performing broth microdilution following the protocol of Wiegand et al. [34]. In short, the turbidity of C. albicans was adjusted to the OD600 equivalent of a 0.5 McFarland standard. A dilution series of caspofungin was made, ranging from 0.03 μg/ml to 64 μg/ml. C. albicans were inoculated in a 96-well plate with the different antibiotic concentrations in duplicates at 37 °C for 20 h before the results were read by visual examination. When models were treated, they were treated with 1000x MIC, as biofilms are often described to be drastically more tolerant towards antimicrobials compared to planktonic cells [35].
2.6. CFU enumeration
When CFU enumeration of the models was performed, the total content of each insert was transferred to a bead-beating tube containing 6 ceramic beads (2.8 mm) (Precellys) and 1 ml of Dey-Engley neutralising broth (Sigma-Aldrich). The broth was found to effectively neutralize the effect of the silver bandages, as previously described [36], hence all models were harvested using this broth. The tubes were bead-beaten on a MagNA Lyser (Roche) for 2 × 10 s at 6000 rpm. Subsequently, the tubes underwent 5 min of degassing, followed by 15 min of sonication in a water bath. Dilution series were made in 0.9 % saline, and spots from each dilution were plated on selective media: Azide agar (Sigma-Aldrich) was used to enumerate S. aureus, Cetrimide agar (Sigma-Aldrich) was used to enumerate P. aeruginosa, and Sabouraud Glucose agar with chloramphenicol (Sigma-Aldrich), supplemented with ciprofloxacin (10 μg/ml), was used to enumerate C. albicans.
2.7. Statistical analysis
All statistical analyses were performed in GraphPad Prism version 10 (GraphPad Software Inc.). For comparing several groups, ANOVA tests were used if the data had a Gaussian distribution, and Kruskal-Wallis were used if not. For comparing only two groups, a t-test was used for data with a Gaussian distribution, and a Mann-Whitney U test was used if not. The lower limit of detection (LOD) was set at 100 CFU/mL; hence, if no colonies were observed, the value was plotted as ‘99’ for all data visualisation and statistical tests. To denote statistical significance, the following symbols were used: ns = not significant (p > 0.05), ∗p < 0.05, ∗∗p < 0.01 and ∗∗∗p < 0.001.
3. Results
3.1. Establishment of a tri-species model
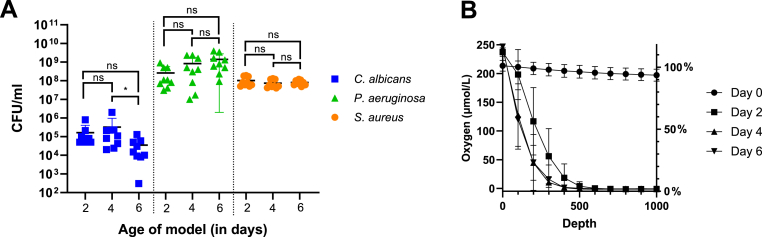
A stable consortium of P. aeruginosa, S. aureus, and C. albicans was established within the chronic wound model. The CFUs remained stable over the tested duration of 6 days, with a slight decrease in C. albicans observed on day 6 (Fig. 2A). Oxygen measurements were taken at 100 μm intervals in the top 1 mm of the model on days 0, 2, 4, and 6 (Fig. 2B). A percentage drop in oxygen at 1 mm was calculated for each day. The model was almost fully oxygenated at day 0, with a modest decrease to 93 % at 1 mm, but a sharp decrease in oxygen availability was observed on day 2, with even steeper gradients on days 4 and 6. pH measurements were performed in the same manner as the oxygen measurements, but since the pH levels remained constant throughout the model's depth (Suppl Figure 1), the pH values are presented as averages instead (Table 1). On day 0, the models were neutral, tending towards alkaline, with an average pH of 7.99. On day 2, the pH level had increased by almost one unit to 8.88. The pH levels remained alkaline during all 6 days.
Fig. 2.
A) Baseline CFUs of the tri-species model at days 2, 4, and 6. CFU numbers for each day were compared within each species using a Kruskal-Wallis test. Lines and bars represent mean ± SD, n = 9 and LOD = 100 CFUs. B) Oxygen levels measured within the top 1 mm layer of the tri-species model on days 0, 2, 4, and 6. Error bars represent SD, n = 9.
Table 1.
pH levels within the top 1 mm depth of the tri-species model on days 0, 2, 4, and 6. All 10 values from 0 μm to 1000 μm were averaged, and then the mean was calculated from n=9 replicates. An ANOVA was used to compare the value of day 0 to the other days.
| Baseline pH of tri-species models (mean ± SD)(statistical significance) | |||
|---|---|---|---|
| Day 0 | Day 2 | Day 4 | Day 6 |
| 7.99 ± 0.008 | 8.88 ± 0.019(∗∗∗) | 8.54 ± 0.041(∗∗∗) | 8.70 ± 0.054(∗∗∗) |
3.2. Models treated with silver-infused bandages
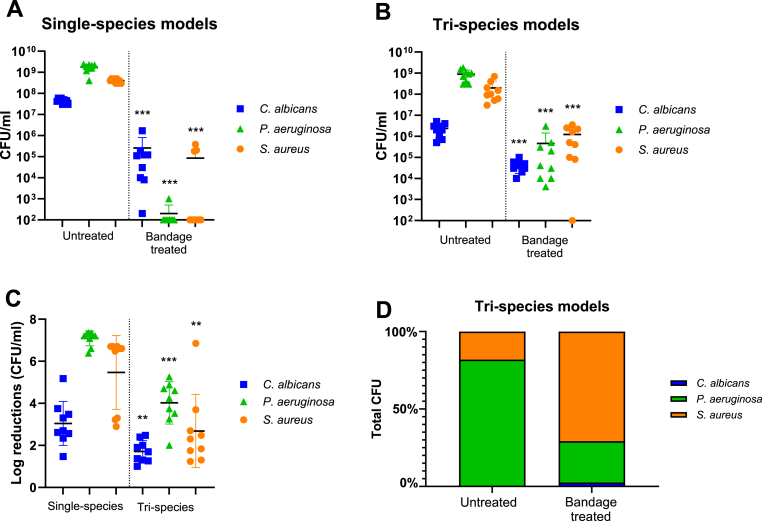
Tri-species models and single-species models were treated with silver-infused bandages for 24 h. When single-species C. albicans models were treated with silver-infused bandages, a slight reduction in CFUs was observed compared to untreated models (Fig. 3A). When P. aeruginosa single-species models were treated with silver-infused bandages, it caused an almost complete reduction in bacterial numbers, with growth observed in only one out of nine replicates (Fig. 3A). When single-species S. aureus models were treated with silver-infused bandages, it caused a complete reduction in six out of nine replicas, while some growth was still observed in the remaining three replicas (Fig. 3A).
Fig. 3.
A) CFUs of single-species models, both untreated and treated. The treated models have been compared with their respective untreated models for statistical testing. Lines and bars represent mean ± SD, n = 9, and LOD = 100 CFUs. B) CFUs of tri-species models, both untreated and treated. The treated models have been compared with their respective untreated models for statistical testing. Lines and bars represent mean ± SD, n = 9, and LOD = 100 CFUs. C) Log reductions were calculated from single-species and tri-species data in A) and B) and tested for statistical difference using Mann-Whitney U tests for each species. Lines and bars represent mean ± SD, n = 9, and LOD = 100 CFUs. D) The total CFU counts of both untreated and bandage-treated tri-species models were converted to 100 %, and the proportion of each species was subsequently calculated and plotted.
When tri-species models were treated with silver-infused bandages, a curious trend was observed. Both P. aeruginosa and S. aureus showed significantly higher survival in the tri-species models (Fig. 3B). Viable P. aeruginosa was found in all models, and viable S. aureus was present in all but one model, showing a large difference between the CFU numbers of single-species models and tri-species models treated with bandages. To determine if this difference was significant, despite slightly higher CFU numbers in the untreated single-species models compared to tri-species models, log reductions were calculated for both single-species and tri-species models after bandage treatment (Fig. 3C). A statistically significant difference between the CFU numbers of the model types was observed, with lower log reductions in the tri-species model. This suggested better survival in the tri-species models than in the single-species models following bandage treatment. Moreover, the CFU of each species was calculated as a percentage of the total CFU within the tri-species models for both untreated and bandage-treated conditions (Fig. 3D). A shift in species dominance was observed; for untreated models, P. aeruginosa comprised over 80 % of the total CFU, while in bandage-treated models, S. aureus became the dominant species, accounting for over 70 % of the total CFU.
Since all untreated single-species models showed slightly higher CFU counts than their untreated tri-species counterparts (Suppl Fig. 2A), a direct comparison of the CFUs in treated models was also performed. This test aimed to determine if there was a significant difference between the single-species and tri-species models treated with bandages, regardless of their untreated counterparts. For both P. aeruginosa and S. aureus, this was the case; their survival in the tri-species models was significantly higher (Suppl Figure 2B).
3.3. Dual-species models
To investigate whether the reduced effect of silver bandages observed in the tri-species models would also occur when only two species were present, dual-species models were created (see Suppl Figure 3). Most notably, it was observed that when treated with silver bandages, P. aeruginosa did not grow, whether it was grown with C. albicans or S. aureus. S. aureus, on the other hand, remained viable at all times when grown with either C. albicans or P. aeruginosa.
3.4. Oxygen measurements
Oxygen measurements were obtained in both single-species and tri-species models. The addition of bandages had a restorative effect on the oxygen levels in all models, causing the levels to rise after only 24 h of treatment (Fig. 4). All models had dropped to 0 % oxygen at a 1 mm depth before treatments were added, except single-species C. albicans models, which were at 2 %. The day after, all untreated models remained at 0 % oxygen, except for the single-species C. albicans models, which averaged 3 % oxygen. When treated with bandages, single-species C. albicans models reached 81 % oxygen at a 1 mm depth. Single-species P. aeruginosa models reached 90 % oxygen after bandage treatments, and single-species S. aureus models reached 61 %. Tri-species models treated with bandages were at 69 % oxygen at 1 mm depth, indicating that although several microorganisms survived the bandage treatment in the tri-species models, oxygenation within the models was still partially restored.
Fig. 4.
Oxygen measurements were taken at every 100 μm within the models to a depth of 1 mm. Measurements were performed before treatment and after 24 h of bandage treatment, or in models left untreated. Each symbol represents mean ± SD, n=9. The measurements were conducted for A) tri-species models, b) single-species C. albicans models, C) single-species P. aeruginosa models, and D) single-species S. aureus models.
3.5. pH measurements
pH measurements were performed in all models before and after treatments (Table 2). Before treatment, the pH level was 8.67 in the tri-species models, however, after 24 h of silver-infused bandage treatment, the pH level had slightly increased to 8.84. while the pH in the untreated models was virtually unchanged. All single-species models had similar starting pH values, ranging from 8.84 in C. albicans models to 8.49 in S. aureus models. After bandage treatments, the pH levels in C. albicans and P. aeruginosa models increased to above 9, measuring 9.29 for C. albicans and 9.06 for P. aeruginosa. The pH in S. aureus models barely increased, with a post-treatment pH of 8.53.
Table 2.
pH levels within the top 1 mm depth of the tri-species model and single-species model before treatments, after bandage application, and for untreated models. All 10 values from 0 μm to 1000 μm were averaged, and the mean was calculated from n = 9 replicates. ANOVA tests were conducted to compare “Before treatment”-values to “After bandages” and “Untreated”-values.
| pH of models before and after treatments (mean ± SD)(statistical significance) | |||
|---|---|---|---|
| Type of model | Before treatment | After bandages | Untreated |
| Tri-species | 8.67 ± 0.031 | 8.84 ± 0.023(∗∗∗) | 8.69 ± 0.025(ns) |
| Single-species C. albicans | 8.84 ± 0.018 | 9.29 ± 0.030(∗∗∗) | 8.91 ± 0.016(∗∗∗) |
| Single-species P. aeruginosa | 8.74 ± 0.020 | 9.06 ± 0.016(∗∗∗) | 8.89 ± 0.031(∗∗∗) |
| Single-species S. aureus | 8.49 ± 0.011 | 8.53 ± 0.031(∗∗) | 8.50 ± 0.030(ns) |
3.6. Models treated with silver-infused bandages and caspofungin
We tested the antifungal compound caspofungin in the hope that a combination of silver-infused bandages and caspofungin would successfully kill all microbes in the tri-species model. We hypothesised that the caspofungin would reduce the numbers of C. albicans which would in turn reduce its protective effect. Using a broth microdilution method, a caspofungin MIC value of 0.125 μg/ml was determined for C. albicans. For the treatments, a concentration of 1000 times the MIC was used. In the tri-species models, the combination of silver-bandages and caspofungin did not affect the C. albicans nor the S. aureus CFU. The numbers of P. aeruginosa increased slightly after the combination treatment (Suppl Figure 4A). The oxygen level, however, did appear to decrease slightly less when caspofungin was added (Suppl Figure 4C). Caspofungin was also tested in single-species models of C. albicans but was found to be less effective than the silver-infused bandages (Suppl Figure 4B). Nonetheless, caspofungin did affect the oxygen levels within the model, resulting in higher levels than in untreated models (Suppl Figure 4D). The tri-species model was also treated with three times the normal amount of silver bandages. This was done to ensure that the amount of silver particles was not a limiting factor in treating the tri-species models and it was performed by placing three layers of bandages on top of each other on the models. The average untreated tri-species model contained around 1.1x109 organisms in total. While models containing S. aureus or C. albicans alone had fewer organisms (4.0x108 and 4.5x107, respectively), models with P. aeruginosa alone actually contained more organisms (1.7x109). Nonetheless, we wanted to ensure that the tri-species models were saturated with the silver treatment. However, there was no difference in CFU between models treated with one layer and models treated with three layers of silver-infused bandages (Suppl Figure 4A).
3.7. Biogeography of the different species in the models
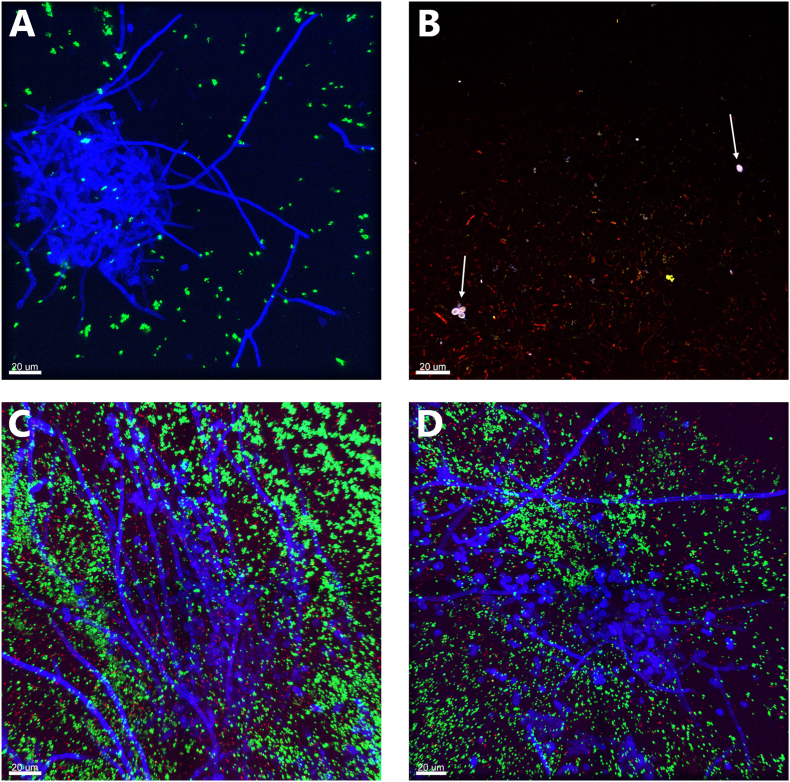
To establish the baseline, tri-species models were visualised by confocal microscopy after 2, 4, and 6 days (Fig. 5). At all time points, P. aeruginosa was observed to form a thick biofilm layer near the top of the models. Smaller biofilms, microcolonies, and single cells of P. aeruginosa could also be seen, primarily in the top parts of the models. S. aureus and C. albicans were distributed throughout the model, in both top and bottom regions. S. aureus appeared as both single cells and larger biofilms but was primarily seen as microcolonies. C. albicans formed both yeast-biofilms and hyphal networks, as well as single cells. The hyphal networks were often found in close contact with both S. aureus and P. aeruginosa. Single-species and multi-species biofilms, in various combinations, were observed within the tri-species models. Additionally, special forms of C. albicans were also observed, including forms suspected to be clumps of blastospores and chlamydospores. Occasionally, the bacteria dropped their plasmid containing their fluorescence, hence Syto62 was added to the models. Small S. aureus microcolonies that were not fluorescently green were occasionally detected but were only visible when using Syto62 stain. Interestingly, the blastospores and chlamydospores of C. albicans exhibited strong fluorescence with Syto62, suggesting high DNA content.
Fig. 5.
CLSM images of the tri-species models containing P. aeruginosa (red), S. aureus (green), C. albicans (blue), and DNA (purple). A) An overview image of the model on day 2. B) A hyphal network in the middle of a P. aeruginosa biofilm on day 2, with microcolonies of S. aureus visible. C) Microcolonies of all species present on day 2. The white arrows point to two special C. albicans forms suspected to be clumped blastospores, and the red arrow points to a possible chlamydospore. D) Microcolonies, single cells, and mixed biofilms of S. aureus and P. aeruginosa on day 4. E) A hyphal network surrounding an S. aureus biofilm on day 6. (For interpretation of the references to colour in this figure legend, the reader is referred to the Web version of this article.)
Treated and untreated tri-species models were visualised as well (see Fig. 6). In the bandage-treated models, the top layers were almost empty, but some growth was observed deeper within the models. Hyphal networks and yeast biofilms were still present, although yeast biofilms appeared to be confined to the deeper layers of the model. Large biofilms of S. aureus and P. aeruginosa were no longer observed. Instead, only smaller colonies or single cells were seen, both near and far from the C. albicans biofilms. Cells with clear SytoxOrange staining were observed as well for all three species. The untreated models resembled the baseline models, with large mixed biofilms observed throughout the models.
Fig. 6.
CLSM images of treated (A and B) and untreated (C and D) tri-species models containing P. aeruginosa (red), S. aureus (green), C. albicans (blue), and dead cells (orange). A) A hyphal network and smaller microcolonies of S. aureus in a bandage-treated model. B) Dead and alive P. aeruginosa stained in orange and red. The white arrows point to dead C. albicans yeast, which appear both orange and blue. C- D) Untreated models showing large hyphal networks surrounded by S. aureus and P. aeruginosa. (For interpretation of the references to colour in this figure legend, the reader is referred to the Web version of this article.)
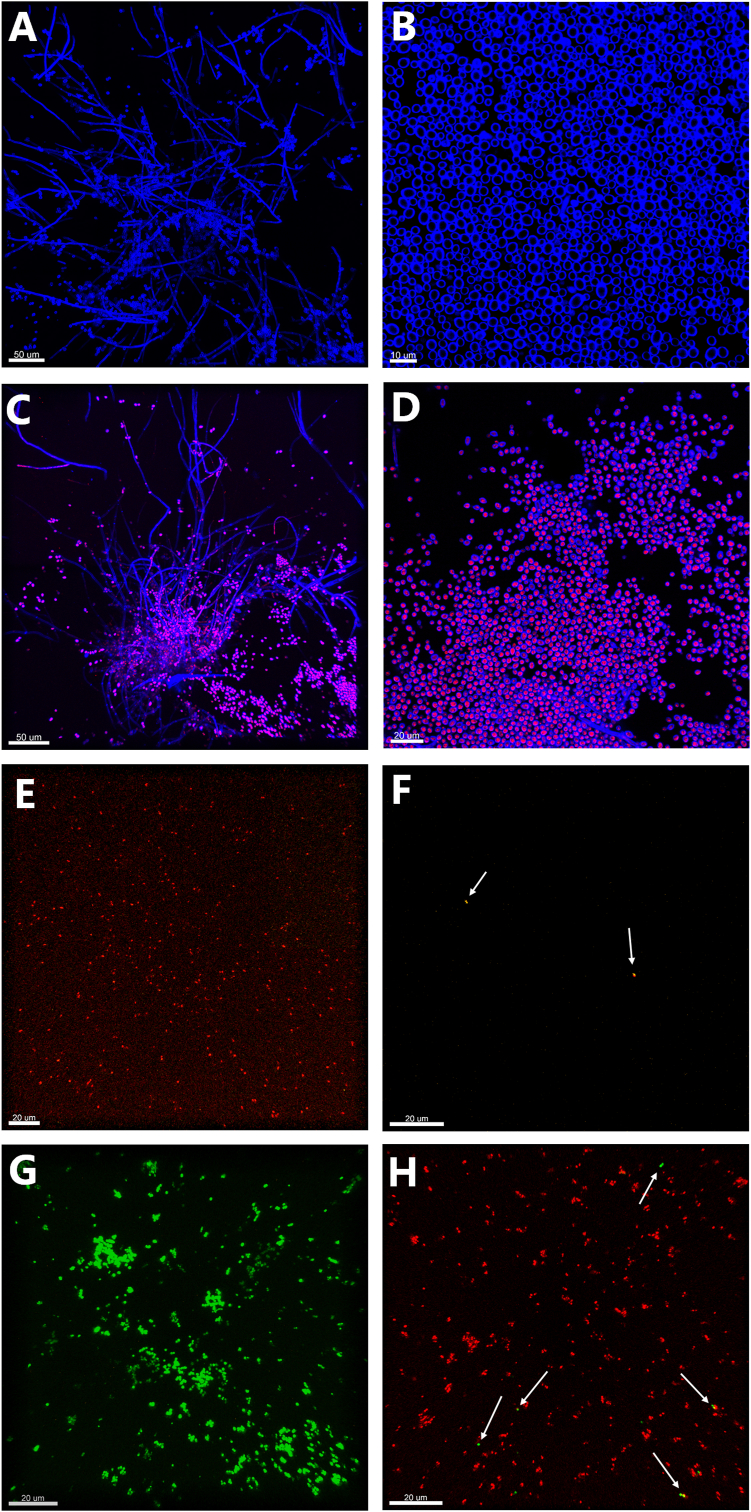
Single-species models of each microbe were visualised as well. Single-species C. albicans models contained large biofilms throughout the models (Fig. 7). When treated with silver bandages, the biofilms remained, but several of the cells, particularly the yeast forms, were stained with PI, indicating permeable membranes (Fig. 7C and D). Strikingly, untreated single-species models of P. aeruginosa and S. aureus did not contain any large biofilms (Fig. 7E and G). Instead, they mostly contained microcolonies, thin layers of biofilms, or single cells spread throughout the model. When treated with silver bandages, there was barely any P. aeruginosa left to observe in the models, except for a few cells stained with SytoxOrange (Fig. 7F). The single-species S. aureus models treated with bandages still contained numerous microcolonies, but they all stained with PI, except for a few sporadic cells (Fig. 7H).
Fig. 7.
CLSM images of single-species models of C. albicans in A-D), P. aeruginosa in E-F) and S. aureus in G-H). A-B) Untreated C. albicans (blue) models displaying hyphal networks and yeast biofilms. C-D) Bandage-treated models showing PI-stained cells (red) indicating permeable cell walls. E) Untreated P. aeruginosa models showing several live cells (red). F) Bandage-treated P. aeruginosa models. The two white arrows point to SytoxOrange-stained P. aeruginosa cells (orange), indicating cell permeability. G) Single-species model of S. aureus (green) left untreated displaying both single cells and microcolonies. H) Bandage-treated S. aureus models, in which most of the cells are stained with PI (red), but the white arrows point to a few that remain alive. (For interpretation of the references to colour in this figure legend, the reader is referred to the Web version of this article.)
4. Discussion and conclusion
In this paper, we have developed a tri-species inter-kingdom model simulating a realistic wound environment. Very few such models currently exist that incorporate both bacterial and fungal species while also providing a suitable, wound-relevant 3D environment.
When establishing the wound model presented in this paper, the key to maintaining stable CFU numbers over time appeared to be the initial inoculum size. Initial failed attempts to create a stable consortium were performed with much lower numbers of C. albicans. However, as previous studies have found that the initial inoculum plays a significant role in cultures growing together [8], we adjusted the starting inoculum to the current level, which resulted in a stable C. albicans presence within the model for at least 6 days. When evaluating the stability of the model, we also examined the presence of C. albicans hyphae, interpreting their establishment as visual confirmation that the fungus had established itself within the model. Hyphal growth compensates for the lack of motility in fungi and serves as a nutrient-scavenging mechanism [37]. Moreover, hyphae are key drivers of fungal pathogenesis and strong indicators of virulence [37].
In addition to hyphae, alternative forms of C. albicans were observed using CLSM. We suspect these forms to be tightly clumped blastospores and chlamydospores. Chlamydospores are often described as occurring under sub-optimal growth conditions as a survival mechanism for C. albicans during stressful conditions [38]. The formation of chlamydospores could be a result of the stress C. albicans might endure when growing with two bacterial species, as these structures were not observed in single-species C. albicans models.
After establishing a stable presence of all three species, we investigated the antimicrobial effect of silver-infused bandages in both single-species models and the tri-species model. We observed a protective effect when the three species were grown together. Particularly the survival of P. aeruginosa and S. aureus was markedly higher when grown in the tri-species model compared to single-species models. However, we are not the first to report this protective phenomenon. Townsend et al. [39] created an in vitro model that included the same species investigated here. The substrate in their models was composed of a cellulose matrix and a polymerised hydrogel made of 10 % 3-sulfopropyl acrylate potassium salt, 0.95 % v v–1 poly(ethylene glycol) diacrylate, 0.01 % v v–1 1-hydroxycyclohexyl phenyl ketone, to which 50 % heat-inactivated horse serum was added. Using this model, Townsend et al. [25] found that S. aureus in their tri-species model was more tolerant to ciprofloxacin than the single-species S. aureus counterpart. Kean et al. [40] grew a biofilm-defective strain of S. aureus together with C. albicans on the same model scaffold as Townsend et al. [25], and observed significantly higher survival of S. aureus against miconazole in dual-species models than in single-species models. Using CLSM and scanning electron microscopy, Kean et al. observed that S. aureus grew in close association with C. albicans hyphae. Their co-culture also demonstrated increased secretion of eDNA, and the tolerance to miconazole appeared to correlate with eDNA levels. Additionally, a metabolomic investigation suggested that S. aureus may scavenge components of the fungal cell wall, promoting its growth. Hence, the relationship between S. aureus and C. albicans seems to have many facets. In our bandage-treated tri-species models, we did not observe a consistent close contact between C. albicans biofilms and either of the bacterial species. Therefore, we speculate that the protective effect was caused by factors beyond close physical contact, such as secreted molecules, alterations in the microenvironment or gene expression changes.
Several in vitro studies have investigated the co-culture of P. aeruginosa with S. aureus. Among other benefits, these studies have found that P. aeruginosa exhibits increased tolerance to antibiotics, altered metabolism, and modified biofilm architecture, which can provide increased protection [8,41,42]. Using CLSM, we observed that neither of the single species models of P. aeruginosa nor S. aureus developed large biofilms after 48 h of growth. However, when grown together in a dual-species model (as seen in a previous version of the model in [26]) or in a tri-species model as shown in this paper, large biofilms were observed for both species. We hypothesize that the presence of other microorganisms may drive the microbes to produce a denser biofilm more rapidly, contributing to the persistent species stability within the model and, in turn, leading to increased tolerance to silver-infused bandages. Curiously, when P. aeruginosa was grown in dual-species models and treated with bandages, it did not survive, suggesting that the presence of both S. aureus and C. albicans was necessary for its survival, and that the altered biofilm architecture alone may not provide full protective effects.
S. aureus, similar to P. aeruginosa, may also experience several benefits from being co-cultured. When grown in co-cultures, S. aureus has previously been observed to convert to the phenotype known as small colony variant (SCV). SCV is often associated with a decreased metabolic rate and increased resistance to various stresses, including antibiotics [43,44]. We speculate that co-cultures containing S. aureus might drive its conversion to the SCV phenotype, enhancing its tolerance to stress, including silver-infused bandages. It is possible that this conversion to SCV may even occur occasionally when S. aureus is grown alone and challenged by stress, which would explain why some single-species models of S. aureus survived the bandage treatment.
Using microsensor measurements, we observed that although the silver bandages did not eradicate the infection in the tri-species model, they still caused a marked increase in oxygen levels. We suspect this to be due to two main factors: the lower microbial burden in the model and possibly halted metabolic state of the microorganisms, leading to slower oxygen consumption. There is substantial evidence supporting the benefits of using silver-infused bandages in clinical settings, and we postulate that this may be partly due to the reduction in microbial burden, as well as a slower metabolism, which leaves more oxygen available for human skin and immune cells.
Bandage treatments appeared to cause alkalinisation in all models, both tri-species and single-species, as all treated models had a higher pH than their untreated counterparts. The pH of the bandage itself was not investigated in this study, but has been reported previously in Parsons et al. [45]. After soaking the Acticoat™ dressing in deionized water for 3 h, the authors found the pH to be 9.5, but after 24 h, it had decreased to 7.7. We suspect that the silver-infused bandages, in concert with microbial metabolism, contributed to the elevated pH in the models. The effect of pH on silver alginate wound dressings has previously been investigated, showing that the silver dressings were more effective at lower pH levels [46]. However, it is uncertain if this is also applies to the silver-infused dressings used in this study. The alkaline environment of infected chronic wounds is often considered a significant hindrance to wound healing, hence if a treatment increases the pH in a wound even further, caution should be advised.
The models in this study were only treated with a single layer of silver-infused dressings. Therefore, to ensure that the presence of bacteria observed in the tri-species model treated with silver bandages was not due to a limited amount of the treatment, i.e. silver particles, we also treated some tri-species models with thrice the amount of silver bandages. However, there was no difference between three layers or one layer of treatment, suggesting that the increased survival observed in the tri-species model was not due to a lack of silver particles.
In conclusion, we believe we have successfully created a reliable inter-kingdom consortium in a realistic chronic wound model environment. The model closely mimics the infectious environment found in patients suffering from chronic wounds, hence we believe results obtained in the model to have high translatability. Our model maintains stable CFU numbers for at least 6 days, exhibits high oxygen consumption, and shows alkalinisation of the microenvironment. Biofilms, microcolonies, single cells, as well as fungal hyphae, are present within the model at all time points. Finally, we have demonstrated that within the model, a protective effect of the bacteria appears to occur, as treatment with silver-infused bandages does not result in the same antimicrobial impairment observed in single-species models. We believe this model platform is highly useful for future studies related to bacterial-fungal interactions and for investigating the efficacy of various wound treatments. It provides a platform for testing novel antimicrobial strategies that target both bacterial and fungal components of biofilms, potentially leading to more effective treatments for chronic wounds and other inter-kingdom infections. Future work on the model could explore the molecular mechanisms underlying the observed inter-species interactions and investigate the efficacy of combination therapies targeting both bacterial and fungal components.
CRediT authorship contribution statement
Stine Sørensen: Writing – original draft, Methodology, Investigation. Lasse Kvich: Writing – review & editing, Methodology, Conceptualization. Yijuan Xu: Writing – review & editing, Methodology. Trine R. Thomsen: Writing – review & editing, Methodology. Thomas Bjarnsholt: Writing – review & editing, Supervision, Conceptualization. Ida Thaarup: Writing – original draft, Supervision, Methodology, Investigation, Formal analysis, Conceptualization.
Declaration of competing interest
The authors declare the following financial interests/personal relationships which may be considered as potential competing interests: Ida Thaarup reports financial support was provided by Magle Chemoswed AB. Stine Sorensen reports a relationship with Novo Nordisk that includes: employment. Yijuan Xu reports a relationship with SEGES Innovation that includes: employment. If there are other authors, they declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgements
We would like to thank Gordon Ramage for interesting C. albicans discussions, Bethan Roberts for laboratory assistance, Blaine Fritz for statistical help and Claus Lex for proof reading.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.bioflm.2025.100256.
Appendix A. Supplementary data
The following are the Supplementary data to this article.
Data availability
Data will be made available on request.
References
- 1.Ramage G., Rajendran R., Sherry L., Williams C. Fungal biofilm resistance. International Journal of Microbiology. 2012;2012 doi: 10.1155/2012/528521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Rayens E., Norris K.A. Prevalence and healthcare burden of fungal infections in the United States, 2018. Open Forum Infect Dis. 2022;9(1) doi: 10.1093/ofid/ofab593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bahamondez-Canas T.F., Heersema L.A., Smyth H.D.C. Current status of in vitro models and assays for susceptibility testing for wound biofilm infections. Biomedicines. 2019;7(2):34. doi: 10.3390/biomedicines7020034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Parvin F., Vickery K., Deva A.K., Hu H. Efficacy of surgical/wound washes against bacteria: effect of different in vitro models. Materials. 2022;15(10) doi: 10.3390/ma15103630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ge Y., Wang Q. Current research on fungi in chronic wounds. Front Mol Biosci. 2023;9(January):1–10. doi: 10.3389/fmolb.2022.1057766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Sanglard D. Emerging threats in antifungal-resistant fungal pathogens. Front Med. 2016;3(MAR):1–10. doi: 10.3389/fmed.2016.00011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Malone M., Bjarnsholt T., McBain A.J., James G.A., Stoodley P., Leaper D., Tachi M., Schultz G., Swanson T., Wolcott R.D. The prevalence of biofilms in chronic wounds: a systematic review and meta-analysis of published data. J Wound Care. 2017;26(1):20–25. doi: 10.12968/jowc.2017.26.1.20. [DOI] [PubMed] [Google Scholar]
- 8.DeLeon S., Clinton A., Fowler H., Everett J., Horswill A.R., Rumbaugh K.P. Synergistic interactions of Pseudomonas aeruginosa and Staphylococcus aureus in an in vitro wound model. Infect Immun. 2014;82(11):4718–4728. doi: 10.1128/IAI.02198-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kvich L., Crone S., Christensen M.H., Lima R., Alhede M., Alhede M., Staerk D., Bjarnsholt T. Investigation of the mechanism and chemistry underlying Staphylococcus aureus ’ ability to inhibit Pseudomonas aeruginosa growth in vitro. J Bacteriol. 2022;204(11):1–16. doi: 10.1128/jb.00174-22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Mitchell G., Séguin D.L., Asselin A.E., Déziel E., Cantin A.M., Frost E.H., Michaud S., Malouin F. Staphylococcus aureus sigma B-dependent emergence of small-colony variants and biofilm production following exposure to Pseudomonas aeruginosa 4-hydroxy-2-heptylquinoline-N-oxide. BMC Microbiol. 2010;10:1–15. doi: 10.1186/1471-2180-10-33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Pastar I., Nusbaum A.G., Gil J., Patel S.B., Chen J., Valdes J., Stojadinovic O., Plano L.R., Tomic-Canic M., Davis S.C. Interactions of methicillin resistant Staphylococcus aureus USA300 and Pseudomonas aeruginosa in polymicrobial wound infection. PLoS One. 2013;8(2):1–11. doi: 10.1371/journal.pone.0056846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Harriott M.M., Noverr M.C. Importance of Candida-bacterial polymicrobial biofilms in disease. Trends Microbiol. 2011;19(11):557–563. doi: 10.1016/j.tim.2011.07.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kalan L., Loesche M., Hodkinson B.P., Heilmann K., Ruthel G., Gardner S.E., Grice E.A. Redefining the chronic-wound microbiome: fungal communities are prevalent, dynamic, and associated with delayed healing. mBio. 2016;7(5):1–12. doi: 10.1128/mBio.01058-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kalan L., Grice E.A. Fungi in the wound microbiome. Adv Wound Care. 2018;7(7):247–255. doi: 10.1089/wound.2017.0756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hu Y., Niu Y., Ye X., Zhu C., Tong T., Zhou Y., Zhou X., Cheng L., Ren B. Staphylococcus aureus synergized with Candida albicans to increase the pathogenesis and drug resistance in cutaneous abscess and peritonitis murine models. Pathogens. 2021;10(8) doi: 10.3390/pathogens10081036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Paul S., Todd O.A., Eichelberger K.R., Tkaczyk C., Sellman B.R., Noverr M.C., Cassat J.E., Fidel P.L., Peters B.M. A fungal metabolic regulator underlies infectious synergism during Candida albicans-Staphylococcusaureus intra-abdominal co-infection. Nat Commun. 2024;15(1) doi: 10.1038/s41467-024-50058-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Carlson E. Synergistic effect of Candida albicans and Staphylococcus aureus on mouse mortality. Infect Immun. 1982;38(3):921–924. doi: 10.1128/iai.38.3.921-924.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Todd O.A., Fidel P.L., Harro J.M., Hilliard J.J., Tkaczyk C., Sellman B.R., Noverr M.C., Peters B.M. Candida albicans augments staphylococcus aureus virulence by engaging the staphylococcal agr quorum sensing system. mBio. 2019;10(3) doi: 10.1128/mBio.00910-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Schneider L.A., Korber A., Grabbe S., Dissemond J. Influence of pH on wound-healing: a new perspective for wound-therapy? Arch Dermatol Res. 2007;298(9):413–420. doi: 10.1007/s00403-006-0713-x. [DOI] [PubMed] [Google Scholar]
- 20.Nagoba B.S., Suryawanshi N.M., Wadher B., Selkar S. Acidic environment and wound healing: a review. Wounds. 2015;27(1):5–11. [Google Scholar]
- 21.Cendra M. del M., Blanco-Cabra N., Pedraz L., Torrents E. Optimal environmental and culture conditions allow the in vitro coexistence of Pseudomonas aeruginosa and Staphylococcus aureus in stable biofilms. Sci Rep. 2019;9(1):1–17. doi: 10.1038/s41598-019-52726-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lichtenberg M., Jakobsen T.H., Kühl M., Kolpen M., Jensen P.Ø., Bjarnsholt T. The structure-function relationship of Pseudomonas aeruginosa in infections and its influence on the microenvironment. FEMS (Fed Eur Microbiol Soc) Microbiol Rev. 2022:1–13. doi: 10.1093/femsre/fuac018. April. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Pabst B., Pitts B., Lauchnor E., Stewart P.S. Gel-entrapped Staphylococcus aureus bacteria as models of biofilm infection exhibit growth in dense aggregates, oxygen limitation, antibiotic tolerance, and heterogeneous gene expression. Antimicrob Agents Chemother. 2016;60(10):6294–6301. doi: 10.1128/aac.01336-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Thaarup Ida C., Bjarnsholt T. Current in vitro biofilm-infected chronic wound models for developing new treatment possibilities. Adv Wound Care. 2021;10(2):91–102. doi: 10.1089/wound.2020.1176. [DOI] [PubMed] [Google Scholar]
- 25.Townsend E.M., Sherry L., Kean R., Hansom D., Mackay W.G., Williams C., Butcher J., Ramage G. Implications of antimicrobial combinations in complex wound biofilms containing fungi. Antimicrob Agents Chemother. 2017;61(9):1–5. doi: 10.1128/aac.00672-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Thaarup Ida C., Lichtenberg M., Nørgaard K.T.H., Xu Y., Lorenzen J., Thomsen T.R., Bjarnsholt T. A collagen-based layered chronic wound biofilm model for testing antimicrobial wound products. Wound Repair Regen. 2023:1–16. doi: 10.1111/wrr.13087. June 2022. [DOI] [PubMed] [Google Scholar]
- 27.Pappas P.G., Kauffman C.A., Andes D.R., Clancy C.J., Marr K.A., Ostrosky-Zeichner L., Reboli A.C., Schuster M.G., Vazquez J.A., Walsh T.J., Zaoutis T.E., Sobel J.D. Clinical practice guideline for the management of candidiasis: 2016 update by the infectious diseases society of America. Clin Infect Dis. 2015;62(4):e1–e50. doi: 10.1093/cid/civ933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Malone C.L., Boles B.R., Lauderdale K.J., Thoendel M., Kavanaugh J.S., Horswill A.R. Fluorescent reporters for Staphylococcus aureus. J Microbiol Methods. 2009;77(3):251–260. doi: 10.1016/j.mimet.2009.02.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Irie Y., Borlee B.R., O’Connor J.R., Hill P.J., Harwood C.S., Wozniak D.J., Parsek M.R. Self-produced exopolysaccharide is a signal that stimulates biofilm formation in Pseudomonas aeruginosa. Proc Natl Acad Sci USA. 2012;109(50):20632–20636. doi: 10.1073/pnas.1217993109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hakonen B., Lönnberg L.K., Larkö E., Blom K. A novel qualitative and quantitative biofilm assay based on 3D soft tissue. International Journal of Biomaterials. 2014:1–5. doi: 10.1155/2014/768136. 2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Price B.L., Lovering A.M., Bowling F.L., Dobson C.B. Development of a novel collagen wound model to simulate the activity and distribution of antimicrobials in soft tissue during diabetic foot infection. Antimicrob Agents Chemother. 2016;60(11):6880–6889. doi: 10.1128/aac.01064-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Werthén M., Henriksson L., Jensen P.Ø., Sternberg C., Givskov M., Bjarnsholt T. An in vitro model of bacterial infections in wounds and other soft tissues. Apmis. 2010;118(2):156–164. doi: 10.1111/j.1600-0463.2009.02580.x. [DOI] [PubMed] [Google Scholar]
- 33.Trivedi U., Madsen J.S., Rumbaugh K.P., Wolcott R.D., Burmølle M., Sørensen S.J. A post-planktonic era of in vitro infectious models: issues and changes addressed by a clinically relevant wound like media. Crit Rev Microbiol. 2017;43(4):453–465. doi: 10.1080/1040841X.2016.1252312. [DOI] [PubMed] [Google Scholar]
- 34.Wiegand I., Hilpert K., Hancock R.E.W. Agar and broth dilution methods to determine the minimal inhibitory concentration (MIC) of antimicrobial substances. Nat Protoc. 2008;3(2):163–175. doi: 10.1038/nprot.2007.521. [DOI] [PubMed] [Google Scholar]
- 35.Yadav M.K., Song J.J., Singh B.P., Vidal J.E. New and future developments in microbial biotechnology and bioengineering: microbial biofilms current research and future trends in microbial biofilms. Elsevier B.V; 2019. Microbial biofilms and human disease: a concise review. (vol. 1) [DOI] [Google Scholar]
- 36.Bourdillon K.A., Delury C.P., Cullen B.M. Biofilms and delayed healing – an in vitro evaluation of silver- and iodine-containing dressings and their effect on bacterial and human cells. Int Wound J. 2017;14(6):1066–1075. doi: 10.1111/iwj.12761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Desai J.V. Candida albicans hyphae: from growth initiation to invasion. Journal of Fungi. 2018;4(1) doi: 10.3390/jof4010010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Whiteway M., Bachewich C. Morphogenesis in Candida albicans. Annu Rev Microbiol. 2007;61:529–553. doi: 10.1146/annurev.micro.61.080706.093341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Townsend E.M., Sherry L., Rajendran R., Hansom D., Butcher J., Mackay W.G., Williams C., Ramage G. Development and characterisation of a novel three-dimensional inter-kingdom wound biofilm model. Biofouling. 2016;32(10):1259–1270. doi: 10.1080/08927014.2016.1252337. [DOI] [PubMed] [Google Scholar]
- 40.Kean R., Rajendran R., Haggarty J., Townsend E.M., Short B., Burgess K.E., Lang S., Millington O., Mackay W.G., Williams C., Ramage G. Candida albicans mycofilms support Staphylococcus aureus colonization and enhances miconazole resistance in dual-species interactions. Front Microbiol. 2017;8(FEB):1–11. doi: 10.3389/fmicb.2017.00258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Beaudoin T., Yau Y.C.W., Stapleton P.J., Gong Y., Wang P.W., Guttman D.S., Waters V. Staphylococcus aureus interaction with Pseudomonas aeruginosa biofilm enhances tobramycin resistance. Npj Biofilms and Microbiomes. 2017;3(1):1–8. doi: 10.1038/s41522-017-0035-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Yung D.B.Y., Sircombe K.J., Pletzer D. Friends or enemies? The complicated relationship between Pseudomonas aeruginosa and Staphylococcus aureus. Mol Microbiol. 2021;116(1):1–15. doi: 10.1111/mmi.14699. [DOI] [PubMed] [Google Scholar]
- 43.Hoffman L.R., Deziel E., D’Argenio D.A., Lepine F., Emerson J., McNamara S., Gibson R.L., Ramsey B.W., Miller S.I. Selection for Staphylococcus aureus small-colony variants due to growth in the presence of Pseudomonas aeruginosa. Proc Natl Acad Sci USA. 2006;103(52):19890–19895. doi: 10.1073/pnas.0606756104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Woods P.W., Haynes Z.M., Mina E.G., Marques C.N.H. Maintenance of S. aureus in Co-culture with P. aeruginosa while growing as biofilms. Front Microbiol. 2019;10(JAN):1–9. doi: 10.3389/fmicb.2018.03291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Parsons D., Bowler P.G., Myles V., Jones S. Silver antimicrobial dressings in wound management: a comparison of antibacterial, physical, and chemical characteristics. Wounds. 2005;17(8):222–232. [Google Scholar]
- 46.Slone W., Linton S., Okel T., Corum L., Thomas J.G., Percival S.L. The effect of pH on the antimicrobial efficiency of silver alginate on chronic wound isolates. Journal of the American College of Certified Wound Specialists. 2010;2(4):86–90. doi: 10.1016/j.jcws.2011.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Data will be made available on request.