Abstract
To explore the structural basis for the essential role of calmodulin (CaM) in Aspergillus nidulans, we have compared the biochemical and in vivo properties of A. nidulans CaM (AnCaM) with those of heterologous CaMs. Neither Saccharomyces cerevisiae CaM (ScCaM) nor a Ca2+ binding mutant of A. nidulans CaM (1234) interacts appreciably with A. nidulans CaM binding proteins by an overlay assay or activates two essential CaMKs, CMKA and CMKB. In contrast, although vertebrate CaM (VCaM) binds a spectrum of proteins similar to that for AnCaM, it is unable to fully activate CMKA and CMKB, displaying a higher KCaM and reduced Vmax for both enzymes. In correlation with the biochemical analysis, neither ScCaM nor 1234 can support A. nidulans growth in the absence of the endogenous protein, whereas VCaM only partially complements the absence of wild-type CaM. Analysis of VCaM and AnCaM chimeras demonstrates that amino acid variations in both N- and C-terminal domains contribute to the inability of VCaM to activate CMKB, but differences in the N terminus are largely responsible for the reduced activity towards CMKA. In vivo, the chimeric molecules support growth equivalently, but only to levels intermediate between those of VCaM and AnCaM, suggesting that the reduced ability to activate the CaMKs is not solely responsible for the inability of VCaM to complement the absence of the wild-type protein. Thus, not only is Ca2+ binding required for CaM function in A. nidulans, but the essential in vivo functions of A. nidulans CaM are uniquely sensitive to the subtle amino acid variations present in vertebrate CaM.
Calmodulin (CaM), an EF-hand Ca2+ binding protein, is highly conserved throughout evolution, with the vertebrate CaMs being identical at the amino acid level (2). In Saccharomyces cerevisiae (6), Schizosaccharomyces pombe (44), Aspergillus nidulans (40), and Drosophila melanogaster (14), the unique CaM gene is essential. CaM regulates its binding partners in both Ca2+-dependent and Ca2+-independent manners (2, 17). Genetic studies suggest that Ca2+/CaM-dependent pathways are essential for the growth of fission yeast but not of budding yeast (27). In S. pombe, CaM requires at least one functional Ca2+ binding site to support growth (30). However, the S. cerevisiae CaM protein contains only three functional EF-hand motifs rather than the four Ca2+ binding sites found in all other CaMs and, in contrast to results of complementation studies with S. pombe (30), does not require Ca2+ binding to support S. cerevisiae growth (13). However, in both budding and fission yeast, vertebrate CaM complements the yeast null strains, suggesting that vertebrate CaM can perform both Ca2+-dependent and Ca2+-independent essential functions in vivo (5, 30, 34).
For S. cerevisiae, all identified CaM binding proteins that are essential for growth under normal circumstances do not require Ca2+ binding, consistent with the importance of the Ca2+-independent roles of CaM in this organism. However, several critical functions for Ca2+-dependent CaM targets, calcineurin and CaM kinases, have been identified for S. cerevisiae, including survival of pheromone-induced growth arrest (3, 4, 29, 45), salt tolerance (1, 12, 15, 28, 31), and acquired thermotolerance (18). In S. pombe, no Ca2+-independent targets of CaM essential for growth have been identified. Whereas calcineurin is not essential for growth (47), other Ca2+/CaM-dependent enzymes, such as the serine/threonine kinase CMK, have been identified but not evaluated in the context of gene deletion studies. Thus, neither S. pombe nor S. cerevisiae provides a system in which Ca2+-regulated CaM structure-function relationships can be studied in vitro and in vivo in the context of known essential Ca2+/CaM-dependent targets.
In contrast to the yeasts, other fungi such as A. nidulans and Cryptococcus neoformans do contain genes encoding Ca2+/CaM binding proteins that are essential for growth. In C. neoformans, the fungal pathogen that causes meningitis in immunocompromised patients, both calcineurin A and B genes are required for growth at 37°C and for virulence (11, 33). The filamentous fungus A. nidulans possesses both Ca2+-dependent and Ca2+-independent targets of CaM that are essential for growth. In A. nidulans, four Ca2+/CaM-dependent enzymes, consisting of three serine/threonine protein kinases and one phosphatase (calcineurin), are homologous to known mammalian enzymes, and three of the four are essential. The protein kinases CMKA/CMPK and CMKB are required for the G2-to-M transition and for the reentry of spores into the proliferative cycle, respectively(7, 20), whereas calcineurin is required in G1 (39, 32). On the other hand, A. nidulans also possesses Ca2+-independent targets of CaM, such as the class I myosin MyoA, that are essential for growth (24).
We have examined structural requirements of A. nidulans CaM both in vitro and in vivo. First, we demonstrate that Ca2+ binding is essential for the activation of CMKA and CMKB and is required to support growth in A. nidulans. Although vertebrate CaM appears to bind most CaM binding proteins in A. nidulans extracts, it is unable to fully activate CMKA and CMKB and only partially supports growth in the absence of endogenous CaM. Analysis of chimeras between A. nidulans and vertebrate CaM demonstrates that the N-terminal domain of VCaM harbors variations resulting in a severe defect in the activation of CMKA. However, neither chimeric CaM fully complements the loss of endogenous CaM in A. nidulans. These results demonstrate that while the CaM amino acid sequence is highly conserved, the protein does possess subtle sequence and structural variations that can dramatically alter the essential functions of the molecule in an organism-specific manner.
MATERIALS AND METHODS
Medium and growth conditions.
A. nidulans conidia were grown in minimal medium containing 50 mM glycerol (MMG), 50 mM glycerol plus 100 mM Thr (MMGT), or 50 mM glucose (MMD) as the sole carbon source as described by Lu and Means (22). For strains with the pyrG89 or pyroA4 phenotype, the medium was supplemented with 5 mM uridine and 10 mM uracil or 2 μg of pyridoxine hydrochloride/liter, respectively. All strains were propagated at 37°C.
Generation of CaM expression vectors.
A multistep process was used to generate a CaM expression plasmid containing the gpdA promoter, the pyroA nutritional marker, and the trpC terminator sequence. First, a 1.42-kb XbaI/NcoI fragment of pAN 7-1 was ligated into a 4.75-kb XbaI/NcoI fragment of pAN 8-1 (both pAN 7-1 and 8-1 were gifts of A. M. J. J. Van Den Hondel [36]) to generate pAN 8-7. Second, pAN 8-7 was digested with XbaI, blunt ended, and digested with EcoRI. The resulting 3.22-kb fragment, containing the gpdA promoter and trpC terminator, was subcloned into the EcoRI and blunted PstI sites of pSX, pUC18 containing the 2.5-kb pyroA nutritional marker, thus generating pSXG. All CaMs were expressed as hemagglutinin (HA)-tagged fusion proteins to facilitate detection. The HA epitope was incorporated into the A. nidulans CaM cDNA by PCR, and the PCR product was cloned into NcoI/BamHI-digested pSXG as an AflIII/BamHI fragment generating pSXG-AnCaM. The S. cerevisiae (gift of T. Davis) cDNA and all A. nidulans mutant cDNAs were subcloned into the NcoI/BamHI-digested pSXG-AnCaM. The chicken CaM cDNA was subcloned into the NcoI and blunted BamHI sites as an NcoI/blunted XbaI fragment from pCaMpl (37). All point mutations and chimeric CaM cDNAs were generated using the megaprimer PCR-based mutagenesis technique (42). All CaM cDNAs were sequenced prior to use.
Ectopic CaM expression in A. nidulans.
A. nidulans strains used in this study are listed in Table 1. The AlcCaM strain of A. nidulans was transformed with the CaM expression vectors by either electroporation (41) or conventional polyethylene glycol-mediated transformation (22). The control strains were generated by transforming the GR5 strain with pAL5 and pSXG vectors and the AlcCaM strain with pSXG. Positive transformants were screened for ectopic CaM protein expression. Extracts were generated from spores germinated for 15 h in MMG. Following filtration, the mycelia were frozen in liquid N2. The frozen mycelia were ground in a cold mortar and pestle in protein extraction buffer (50 mM Tris [pH 7.5], 5 mM EDTA, 10 mM benzamidine, 1 μg of pepstatin A/ml, 1 μg of leupeptin/ml and 1 μg of aprotinin/ml). The protein extracts were clarified by centrifugation at 16,000 × g for 10 min at 4°C. Western analysis was performed using 100 μg of crude protein extract, the 12CA5 anti-HA antibody (Boehringer-Mannheim, Indianapolis, Ind.), goat anti-mouse horseradish peroxidase-conjugated immunoglobulin G (Jackson Laboratory, Bar Harbor, Maine), and the Enhanced Chemiluminescence kit (Amersham, Piscataway, N.J.).
TABLE 1.
A. nidulans strains used in this study
| Strain | Genotype | Source |
|---|---|---|
| GR5 | A773; pyr G89; w A2; pyro A4 | Rasmussen et al. (40) |
| JJ10 | A773; w A2 | This study |
| AlcCaM | A773; alc:cam; w A2; pyro A4 | Lu et al. (23) |
| JJ12 | A773; alc:cam; w A2 | This study |
| JJ13 | A773; alc:cam; gpd:cam; w A2 | This study |
| JJ14 | A773; alc:cam; gpd:cmd 1; w A2 | This study |
| JJ15 | A773; alc:cam; gpd:vCaM; w A2 | This study |
| JJ16 | A773; alc:cam; gpd:cam 1234; w A2 | This study |
| JJ17 | A773; alc:cam; gpd:cam ANVC; w A2 | This study |
| JJ18 | A773; alc:cam; gpd:cam VNAC; w A2 | This study |
CaM quantification.
The concentrations of endogenous, ectopically expressed A. nidulans CaM and the four-Ca2+-binding-site mutant CaM were determined by radioimmunoassay (RIA), as described by Rasmussen et al. (40). Semiquantitative analysis of S. cerevisiae and chicken CaM expression was performed by comparative Western blot analysis, comparing crude extracts from strains to amino acid analyzed standards using a polyclonal anti-cmd1 antibody (gift of T. Davis) and a monoclonal anti-CaM antibody (UBI, Lake Placid, N.Y.), respectively. The relative expression levels of the chimeric CaMs were determined by direct comparison to the AnCaM expressing strain by Western analysis using a monoclonal 12CA5 anti-HA antibody.
A. nidulans growth.
The ability of the ectopically expressed CaMs to functionally complement the absence of wild-type CaM was determined by a colony diameter assay as described by McGoldrick et al. (24). Briefly, the spores were diluted and plated on either MMG or MMD agar plates at a concentration of approximately four spores per plate. The average colony diameter was measured every 24 h for 5 days.
Calmodulin overlay.
CaM overlay was performed using protein A-CaM fusion proteins as described by Stirling et al. (43). The protein A-CaM fusion proteins were generated in a multistep process. First, pALP1 (gift of M. Stark) was digested with EcoRI, blunted, and digested with BamHI. A HA-tagged A. nidulans CaM cDNA PCR product was subcloned as a blunted AflIII/BamHI fragment, generating pALP-AnCaM. To place the chicken CaM and the four-Ca2+-binding-site mutant CaM into the pALP1 vector as HA-tagged protein A CaM fusion proteins, the cDNAs were first shuttled into the blunted EcoRI/BamHI sites of the original pALP1 plasmid as PvuII/BamHI fragments from the pSXG-based expression plasmids described above. Finally, the cDNAs were interchanged with the AnCaM cDNA in pALP-AnCaM as NcoI/BamHI fragments, thereby creating pALP-VCaM and pALP-1234. The S. cerevisiae CaM-protein A fusion protein was expressed using pDS100 that was kindly provided by M. Stark (43). The fusion proteins were expressed constitutively in BL-21 bacteria and purified on phenyl-Sepharose resin (38). The concentration of the CaM fusion protein used in the overlay medium was 175 nM (6 μg/ml).
CaM kinase purification and assays.
The CaM concentrations required for half-maximal kinase activation (KCaM) of both CMKA and CMKB were determined using bacterially expressed hexa-histidine-tagged protein kinases and bacterially expressed CaM. Both CMKA and CMKB were subcloned into pTrcHisB (Invitrogen, Carlsbad, Calif.) independently, CMKA as a SacI/EcoRI fragment and CMKB as a BamHI/EcoRI fragment. The kinases were expressed and purified as described by Joseph and Means (20). Kinase assays were performed in 30-μl reaction mixtures in a buffer containing 50 mM Tris (pH 7.5), 10 mM MgCl2, 1 mM CaCl2, 1 mM dithiothreitol, 0.1% Tween 20, 200 μM ATP, 0.2 μl of [γ-32P]ATP/reaction, and a 200 μM concentration of the peptide substrate ADR1G (LKKLTRRASFSGQ) (8). CaM was added to reactions at various concentrations up to 1 μM, and the reactions were initiated with the addition of either 200 or 400 ng of CMKB and incubated at 30°C for either 5 or 10 min for CMKA and CMKB, respectively. The reactions were terminated by transferring 20 μl of the reaction mixture onto p81 phosphocellulose filters (Millipore, Bedford, Mass.), followed by extensive washing in 75 mM phosphoric acid (9). The specific activity was determined following liquid scintillation counting of the dried filters.
RESULTS
Vertebrate CaM is unable to fully activate CMKA and CMKB.
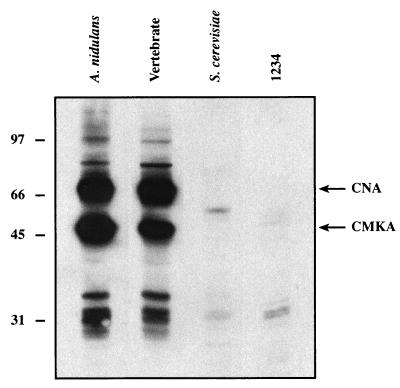
We first compared the abilities of A. nidulans CaM (AnCaM), S. cerevisiae CaM (ScCaM), vertebrate CaM (VCaM), and a Ca2+ binding mutant of A. nidulans CaM (1234) to interact with A. nidulans CaM binding proteins in an overlay assay. AnCaM is highly homologous with VCaM and ScCaM, sharing 84 and 60% amino acid identity, respectively (Fig. 1). Although the sequence is highly conserved, the abilities of these CaMs to bind proteins from crude A. nidulans extracts vary remarkably. AnCaM and VCaM bind similar spectra of proteins by CaM overlay, including two abundant and essential Ca2+/CaM-dependent enzymes, CNA and CMKA (Fig. 2). In contrast, ScCaM binds few, if any, proteins and displays a pattern similar to that of the Ca2+ binding mutant of AnCaM (1234), in which the Glu residue at position 12 of each EF hand is mutated to Ala (Fig. 1). Ye and Bretscher (46) used a similar overlay assay to show that S. cerevisiae CaM also fails to bind the CaM binding proteins present in bovine brain.
FIG. 1.
Alignment of A. nidulans CaM with vertebrate and S. cerevisiae CaMs. The A. nidulans (40) (J05545), vertebrate (M36167), and S. cerevisiae (6) (P06787) CaM sequences were aligned using FASTA and shaded by BOXSHADE. Asterisks indicate the Glu residues mutated to Ala to generate the Ca2+-binding mutant CaM. The junction of the A. nidulans/vertebrate chimeric CaMs lies between the amino acids marked with pluses.
FIG. 2.
Identification of A. nidulans CaM binding proteins by overlay assay. A. nidulans crude protein extract was separated by polyacrylamide gel electrophoresis, transferred to an Immobilon-P membrane, and probed with either A. nidulans CaM, vertebrate CaM , S. cerevisiae CaM, or the Ca2+ binding mutant CaM-protein A fusion protein. The blots are identical exposures of 100 μg of crude protein extract probed in the presence of Ca2+ as described in Materials and Methods.
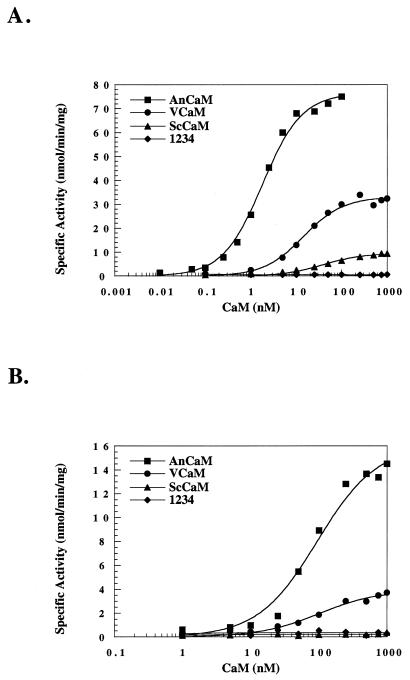
To further investigate the differential functions of these heterologous CaM proteins, we tested their ability to activate two essential CaMKs present in A. nidulans, CMKA and CMKB (Fig. 3 and Table 2). Consistent with the CaM overlay, neither ScCaM nor 1234 is able to appreciably activate these kinases under the assay conditions. However, VCaM is unable to fully activate these kinases despite its similarly to AnCaM in the CaM overlay assay. The most profound difference between VCaM and AnCaM is that VCaM exhibits an eightfold increase in KCaM for CMKA. Additionally, the Vmax of VCaM is reduced to about one-half for CMKA and one-third for CMKB. Therefore, the qualitative results of the CaM overlay are not predictive of the ability of VCaM to activate CMKA and CMKB.
FIG. 3.
Activation of CMKA and CMKB by A. nidulans and heterologous CaMs. The specific activity of hexa-histidine-tagged bacterially expressed and purified CMKA (A) and CMKB (B) was measured at different concentrations of purified AnCaM (closed square), VCaM (closed circle), ScCaM (closed triangle), or 1234 (closed diamond) CaM as described in Materials and Methods. The graphs are representative of three independent experiments.
TABLE 2.
CaM expression in medium containing glycerol or glucose
| Strain | Amt of endogenous CaMa (ng/mg)
|
Amt of heterologous CaMb (ng/mg)
|
|
|---|---|---|---|
| Glycerol | Glucose | Glucose | |
| GR5 | 256 | 193 | NAc |
| AlcCaM | 40 | <2 | NA |
| AlcCaM/VCaM | NDd | ND | 233 |
| AlcCaM/ScCaM | 24 | <2 | 158 |
| AlcCaM/1234 | ND | ND | 1,418 |
| AlcCaM/ANVC | ND | ND | 276 |
| AlcCaM/VNAC | ND | ND | 489 |
CaM concentration determined by RIA.
CaM concentration determined by quantitative Western analysis.
NA, not applicable.
ND, not determined due to antibody cross-reactivity.
Activation of A. nidulans CaMKs correlates with the ability of heterologous CaMs to support fungal growth.
To evaluate the abilities of heterologous CaMs or the AnCaM 1234 mutant to complement the loss of endogenous CaM in A. nidulans, we ectopically expressed different CaM proteins under the control of the constitutive gpdA promoter in the background of the AlcCaM strain. In the AlcCaM strain, the expression of the essential CaM gene is regulated by carbon source. In glucose, expression is repressed and the fungus fails to grow. In glycerol, expression is low and the fungus is able to maintain growth (23). Multiple strains expressing the heterologous CaMs were generated, and the expression levels were examined by quantitative Western analysis or radioimmunoassay (Table 3). In glucose, the VCaM and ScCaM expression is similar to the levels of endogenous CaM in the parental GR5 strain, and all these values are similar to previously reported CaM concentrations (40, 23), while the Ca2+ binding mutant (1234) is expressed at a concentration about seven times higher than that of endogenous CaM.
TABLE 3.
Activation of A. nidulans CaM kinases
| CaM | CMKA
|
CMKB
|
||
|---|---|---|---|---|
| KCaM (nM) | Vmax (nmol/min/mg) | KCaM (nM) | Vmax (nmol/min/mg) | |
| AnCaM | 1.9 ± 0.9 | 77.0 ± 1.6 | 88.6 ± 24.7 | 14.1 ± 2.2 |
| VCaM | 15.9 ± 7.1 | 33.2 ± 3.2 | 133.1 ± 34.7 | 4.3 ± 0.4 |
| ScCaM | 37.8 ± 8.5 | 9.4 ± 2.8 | ND | ND |
| 1234 | NDa | ND | ND | ND |
| ANVC | 2.1 ± 0.4 | 50.5 ± 9.5 | 94.3 ± 18.1 | 11.6 ± 3.2 |
| VNAC | 7.7 ± 0.6 | 56.1 ± 3.8 | 102.9 ± 16.3 | 10.3 ± 2.6 |
ND, no detectable activity
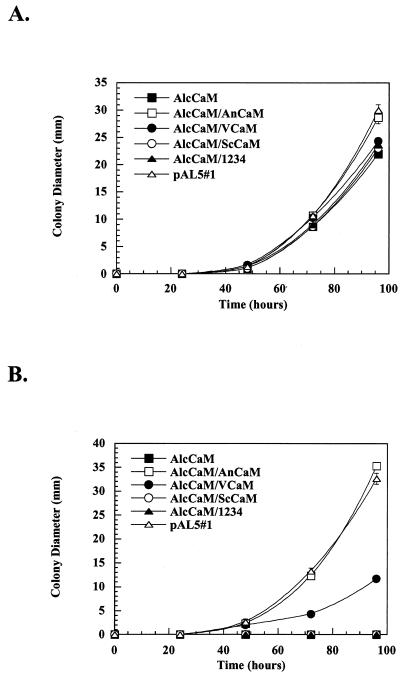
Next, we wanted to determine how well the ability of CaM to activate CMKA and CMKB correlated with its ability to support growth in the absence of endogenous CaM. As seen in Fig. 4A, all strains grew at a rate similar to that of the control when germinated on glycerol medium (allowing wild-type CaM expression). When wild-type CaM expression was repressed by the presence of glucose, as demonstrated in Fig. 4B, the AnCaM/AlcCaM strain and the control strain grew at comparable rates, while the nutritionally complemented AlcCaM strain without any heterologous CaM failed to grow. Importantly, neither AlcCaM/ScCaM nor AlcCaM/1234 grows in the absence of A. nidulans CaM. Thus, ScCaM and 1234 are essentially inactive in biochemical assays in vitro (CaM overlay and CaMK activation), and those results correlate with their inability to support growth in vivo .
FIG. 4.
Complementation of A. nidulans by heterologous CaMs. AlcCaM strains expressing the ectopically expressed AnCaM (open squares), VCaM (closed circles), ScCaM (open circles), and 1234 (closed triangles) were inoculated onto either MMG (A) or MMD (B) agar plates, and their colony diameters were measured over a period of 5 days. A nutritionally complemented pAL5#1 strain (open triangles) was used to represent wild-type A. nidulans (positive control), and the nutritionally complemented AlcCaM strain (closed squares) was used as a negative control.
We also evaluated whether VCaM could functionally complement the absence of the endogenous A. nidulans CaM. In glucose, AlcCaM/VCaM grows at a rate approximately one-third that of both AlcCaM/AnCaM and the nutritionally complemented control. Thus, VCaM partially performs the essential functions of wild-type CaM in vivo, a result that is consistent with its ability to partially activate the A. nidulans CaMKs in vitro.
The N terminus of CaM is critical for proper activation of CMKA and CMKB.
Since VCaM cannot fully complement A. nidulans CaM despite sharing 84% amino acid identity, we questioned whether the amino acid differences responsible could be localized to the N- or C-terminal lobe of the protein. The C terminus is more divergent between these species, so we predicted that it would be most important in determining functional differences in vitro and in vivo. We created two chimeric CaM cDNAs in which the N- and C-terminal globular domains of the A. nidulans and vertebrate CaM are interchanged. The junction of the proteins (Fig. 1) is following Asn 61, so the chimeras generated are the Aspergillus N terminus-vertebrate C terminus (ANVC) or the Aspergillus C terminus-vertebrate N terminus (VNAC) proteins. First, we tested the activation of CMKA and CMKB by ANVC and VNAC (Table 3). Notably, the KCaM for CMKA using ANVC is similar to that obtained with AnCaM rather than to that obtained with VCaM. In contrast, the KCaM using VNAC falls between the values obtained with AnCaM and VCaM. Both chimeric CaMs demonstrate intermediate effects on Vmax for CMKA. Interestingly, the chimeras display only subtle differences in ability to activate CMKB and are most similar to AnCaM. These biochemical assays support two conclusions: first, the activation of CMKA is more sensitive to differences in the CaM sequence than that of CMKB, and second, amino acid variations in the N terminus, not the C terminus, of VCaM are the most important in determining the KCaM for activation of CMKA.
Both the N and C termini of AnCaM are required for normal growth.
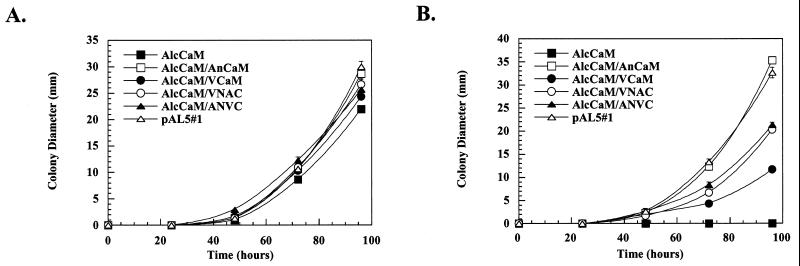
If the activation parameters of CMKA in vitro correlate with the ability to support growth in vivo, then ANVC would be better able to support the growth of the fungus than VNAC in the absence of endogenous CaM. To test this, the cDNAs were expressed in the AlcCaM strain, and their ability to support growth was compared to that of AnCaM and VCaM. When germinated in glycerol, all of the strains grew at approximately the same rate (Fig. 5A). However, when germinated in glucose, which represses expression of the endogenous CaM, the strains containing the chimeric proteins at two to four times the level of endogenous CaM (Table 2) grew to an intermediate diameter two-thirds that of the wild type but significantly greater than that of AlcCaM/VCaM (Fig. 5B). Thus, even though ANVC and VNAC demonstrate differences in biochemical properties in vitro, these two chimeric proteins behave similarly in vivo and neither can completely complement the absence of endogenous CaM.
FIG. 5.
Complementation of A. nidulans by A. nidulans/vertebrate chimeric CaMs. The AlcCaM strains expressing AnCaM (open squares), VCaM (closed circles), VNAC (open circles) and ANVC (closed triangles) were grown on either MMG (A) or MMD (B) agar plates, and growth was quantified by measuring colony diameter over a period of 5 days. The nutritionally complemented pAL5#1 strain (open triangles) was used to represent the wild-type A. nidulans (positive control), and the nutritionally complemented AlcCaM strain (closed squares) was used as a negative control.
DISCUSSION
In this study, we investigated the ability of the Ca2+ binding mutant of AnCaM (1234) and heterologous CaMs from yeast and vertebrate systems to complement the functions of AnCaM biochemically and genetically. As predicted based on the identification of multiple and essential Ca2+/CaM-dependent enzymes in A. nidulans, 1234 is unable to activate CMKA or CMKB and cannot support growth of the fungus. Therefore, as with S. pombe (30) and in contrast to S. cerevisiae (13), CaM of A. nidulans requires high-affinity Ca2+ binding to support growth. Similar to 1234, ScCaM was unable to activate CMKA and CMKB or functionally complement the absence of the endogenous protein. This inability of ScCaM to complement AnCaM may be due to the significant amino acid alterations that occur throughout the two proteins, the inability of the fourth EF hand to bind Ca2+, or a combination of both types of differences.
Despite sharing 84% amino acid identity with AnCaM, VCaM is unable to fully substitute for AnCaM in vitro or in vivo. Although the CaM overlays showed that AnCaM and VCaM interact with similar spectra of proteins present in A. nidulans crude extracts, VCaM cannot maximally activate either of A. nidulans' essential Ca2+/CaM-dependent protein kinases, CMKA and CMKB. For both enzymes, the Vmax is reduced relative to AnCaM, but the most striking defect is an eightfold increase in KCaM for CMKA. In correlation with these biochemical results, VCaM only partially complements the loss of AnCaM in vivo. This result is in stark contrast to results of studies with both budding and fission yeast, where VCaM and the endogenous CaM are functionally interchangeable in vivo (5, 30, 34).
Between AnCaM and VCaM, there are only 23 amino acid differences, 13 of which are conservative. Examination of the contacts between vertebrate Ca2+/CaM- and CaM binding peptides based on available three-dimensional X-ray crystal and nuclear magnetic resonance structures reveals that only five of the variant residues interact with CaM binding peptides (10, 19, 25, 26, 35), whereas the other 19 residues lie outside of the CaM/CaM binding peptide interfaces. Although the residues involved in peptide binding may be implicated in the inability of VCaM to complement AnCaM, results of prior biochemical and genetic studies argue that the solvent-exposed residues of CaM are also critical for enzyme activation. Biochemically, both AnCaM/VCaM chimeras are able to rescue the ability of VCaM to activate CMKB. However, the results with CMKA are markedly different. For CMKA the amino acid variation in the N-terminal globular domain is largely responsible for the inability of VCaM to fully activate the kinase. The KCaM for ANVC is similar to that of AnCaM, while the KCaM for VNAC is much greater than that of AnCaM. Within the N-terminal domain of AnCaM and VCaM there are only nine amino acid differences. Intriguingly, amino acids 9 to 12, just prior to the first EF hand, differ in three out of four residues. For AnCaM these residues are VSEY, but for VCaM (as well as for ScCaM and the CaMs of several other lower eukaryotes) these residues are IAEF. Both Ala10 and Phe12 interact with CaM binding peptides in crystallographic studies, and mutation of Phe12 to Ala reduces the ability of ScCaM to activate both S. cerevisiae calcineurin and Cmk1p (16). Based on these results, it seems reasonable to suggest that this four-amino-acid cluster may be functionally important. Another difference of potential significance lies at the fourth residue of the second EF hand, Asn in AnCaM and Gly in most other CaMs. Genetic studies of Paramecium identified a mutation at this position (Gly59 to Ser) that abrogates the function of Ca2+-dependent Na+ channels in vivo (21). Interestingly, the Gly-to-Asn variation is also found in the fourth position of the third EF hand in AnCaM. Although these amino acid differences can be speculated to result in biochemical and/or biological consequences, the actual residue or residues responsible for the functional differences cannot be conclusively identified based solely on sequence or structural analysis, since variation in residues within the hydrophobic pockets, on the outer surface of the CaM molecule, or in internally located residues that contribute to stabilizing the structure of the molecule are all potential culprits.
Amino acid substitutions in both globular domains of CaM contribute to the inability of VCaM to fully support growth. Surprisingly, both ANVC and VNAC support A. nidulans growth to roughly the same degree, better than VCaM but not as well as AnCaM, even though the biochemical deficits of VCaM can be largely attributed to the N-terminal globular domain. Although the activation of CMKA and CMKB correlates with the ability of ScCaM, VCaM, and 1234 to support growth in vivo, the analysis of the AnCaM/VCaM chimeras indicates that the in vivo phenotype cannot be simply attributed to the efficacy of these proteins to activate CaMKs. Indeed, several studies suggest that reductions in CaMK activity may not severely impact the in vivo functions of the enzyme. Previous work in our laboratory has demonstrated that for A. nidulans, a discernible growth phenotype is observed only when CMKA protein expression (and presumably activity) is repressed to undetectable levels (7). Similarly, in S. cerevisiae, biochemical and in vivo analyses of CaM Phe-to-Ala mutants demonstrate that mutations that subtly attenuate calcineurin and CaMK activation in vitro do not appreciably alter the ability of yeast to survive an α-factor-induced growth arrest (16).
Perhaps it should come as no surprise that while the in vitro activation of CMKA and CMKB may correlate with the ability of the heterologous proteins to support growth, the in vivo target(s) involved in causing the phenotypes may be considerably more complex. For example, two essential CaM binding proteins in addition to CMKA and CMKB have been identified in A. nidulans. The Ca2+/CaM-dependent phosphatase, calcineurin, and MyoA, a class I myosin, are each required for fungal growth (39, 24). Additionally, a recently identified homologue of S. cerevisiae Spc110p and S. pombe Pcp1p would be predicted to perform an essential mitotic role in A. nidulans based on the null phenotype in yeast (10a). Therefore, alterations in CaM binding or activation of at least five CaM binding proteins may be contributing to the inability of the heterologous proteins to functionally replace the endogenous A. nidulans CaM. It will be an interesting challenge to dissect the multiple, essential functions of Ca2+/CaM-dependent and -independent pathways in the growth of this fungus.
Acknowledgments
We thank Christina Kahl, Katharine Winkler, Ethan Corcoran, and David Chin for valuable discussions and critical reading of the manuscript. We also thank Michael Stark for the protein A-CaM expression vectors, Trisha Davis for the S. cerevisiae CaM cDNA and antibody, and A. M. J. J. Van Den Hondel for the A. nidulans vectors.
National Institutes of Health grant GM-33976 awarded to A.R.M. funded this study.
REFERENCES
- 1.Breuder, T., C. S. Hemenway, N. R. Movva, M. E. Cardenas, and J. Heitman. 1994. Calcineurin is essential in cyclosporin A- and FK506-sensitive yeast strains. Proc. Natl. Acad. Sci. USA 91:5372-5376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Chin, D., and A. R. Means. 2000. Calmodulin: a prototypical calcium sensor. Trends Cell Biol. 10:322-328. [DOI] [PubMed] [Google Scholar]
- 3.Cyert, M. S., R. Kunisawa, and J. Thorner. 1991. Yeast has homologs (CNA1 and CNA2) of mammalian calcineurin, a calmodulin-regulated phosphoprotein phosphatase. Proc. Natl. Acad. Sci. USA 88:7376-7380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cyert, M. S., and J. Thorner. 1992. Regulatory subunit (CNB1 gene product) of yeast Ca2+/calmodulin-dependent phosphoprotein phosphatases is required for adaptation to pheromone. Mol. Cell. Biol. 12:3460-3469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Davis, T. N., and J. Thorner. 1989. Vertebrate and yeast calmodulin, despite significant sequence divergence, are functionally interchangeable. Proc. Natl. Acad. Sci. USA 86:7909-7913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Davis, T. N., M. S. Urdea, F. R. Masiarz, and J. Thorner. 1986. Isolation of the yeast calmodulin gene: calmodulin is an essential protein. Cell 47:423-431. [DOI] [PubMed] [Google Scholar]
- 7.Dayton, J. S., and A. R. Means. 1996. Ca(2+)/calmodulin-dependent kinase is essential for both growth and nuclear division in Aspergillus nidulans. Mol. Biol. Cell 7:1511-1519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Denis, C. L., S. C. Fontaine, D. Chase, B. E. Kemp, and L. T. Bemis. 1992. ADR1c mutations enhance the ability of ADR1 to activate transcription by a mechanism that is independent of effects on cyclic AMP-dependent protein kinase phosphorylation of Ser-230. Mol. Cell. Biol. 12:1507-1514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.DeRemer, M. F., R. J. Saeli, D. L. Brautigan, and A. M. Edelman. 1992. Ca(2+)-calmodulin-dependent protein kinases Ia and Ib from rat brain. II. Enzymatic characteristics and regulation of activities by phosphorylation and dephosphorylation. J. Biol. Chem. 267:13466-13471. [PubMed] [Google Scholar]
- 10.Elshorst, B., M. Hennig, H. Forsterling, A. Diener, M. Maurer, P. Schulte, H. Schwalbe, C. Griesinger, J. Krebs, H. Schmid, T. Vorherr, and E. Carafoli. 1999. NMR solution structure of a complex calmodulin with a binding peptide of the Ca2+ pump. Biochemistry 38:12320-12332. [DOI] [PubMed] [Google Scholar]
- 10a.Flory, M. R., M. Morphew, J. D. Joseph, A. R. Means, and T. N. Davis. Pcp1p, an Spc110p-related calmodulin target at the centrosome of the fission yeast Schizosaccharomyces pombe. Cell Growth Differ., in press. [PubMed]
- 11.Fox, D. S., M. C. Cruz, R. A. Sia, H. Ke, G. M. Cox, M. E. Cardenas, and J. Heitman. 2001. Calcineurin regulatory subunit is essential for virulence and mediates interactions with FKBP12-FK506 in Cryptococcus neoformans. Mol. Microbiol. 39:835-849. [DOI] [PubMed] [Google Scholar]
- 12.Garrett-Engele, P., B. Moilanen, and M. S. Cyert. 1995. Calcineurin, the Ca2+/calmodulin-dependent protein phosphatase, is essential in yeast mutants with cell integrity defects and in mutants that lack a functional vacuolar H(+)-ATPase. Mol. Cell. Biol. 15:4103-4114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Geiser, J. R., D. van Tuinen, S. E. Brockerhoff, M. M. Neff, and T. N. Davis. 1991. Can calmodulin function without binding calcium? Cell 65:949-959. [DOI] [PubMed] [Google Scholar]
- 14.Heiman, R. G., R. C. Atkinson, B. F. Andruss, C. Bolduc, G. E. Kovalick, and K. Beckingham. 1996. Spontaneous avoidance behavior in Drosophilia null for calmodulin expression. Proc. Natl. Acad. Sci. USA 93:2420-2425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hemenway, C. S., K. Dolinski, M. E. Cardenas, M. A. Hiller, E. W. Jones, and J. Heitman. 1995. vph6 mutants of Saccharomyces cerevisiae require calcineurin for growth and are defective in vacuolar H(+)-ATPase assembly. Genetics 141:833-844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hiroyuki, O., M. S. Cyert, and Y. Ohya. 1998. Importance of phenylalanine residues of yeast calmodulin for target binding and activation. J. Biol. Chem. 273:26375-26382. [DOI] [PubMed] [Google Scholar]
- 17.Hook, S. S., and A. R. Means. 2001. Ca(2+)/CaM-dependent kinases: from activation to function. Annu. Rev. Pharmacol. Toxicol. 41:471-505. [DOI] [PubMed] [Google Scholar]
- 18.Iida, H., Y. Ohya, and Y. Anraku. 1995. Calmodulin-dependent protein kinase II and calmodulin are required for induced thermotolerance in Saccharomyces cerevisiae. Curr. Genet. 27:190-193. [DOI] [PubMed] [Google Scholar]
- 19.Ikura, M., M. Clore, A. M. Gronenborn, G. Zhu, C. B. Klee, and A. Bax. 1992. Solution structure of a calmodulin-target peptide complex by multidimensional NMR. Science 256:632-638. [DOI] [PubMed] [Google Scholar]
- 20.Joseph, J. D., and A. R. Means. 2000. Identification and characterization of two Ca2+/CaM-dependent protein kinases required for normal nuclear division in Aspergillus nidulans. J. Biol. Chem. 275:38230-38238. [DOI] [PubMed] [Google Scholar]
- 21.Ling, K., M. E. Maley, R. R. Preston, Y. Saimi, and C. Kung. 1994. New non-lethal calmodulin mutations in Paramecium. Eur. J. Biochem. 222:433-439. [DOI] [PubMed] [Google Scholar]
- 22.Lu, K. P., and A. R. Means. 1993. Conditional mutants for studying functions of calmodulin in Aspergillus nidulans. Methods Mol. Genet. 2:255-275. [Google Scholar]
- 23.Lu, K. P., C. D. Rasmussen, G. S. May, and A. R. Means. 1992. Cooperative regulation of cell proliferation by calcium and calmodulin in Aspergillus nidulans. Mol. Endocrinol. 6:365-374. [DOI] [PubMed] [Google Scholar]
- 24.McGoldrick, C. S., C. Gruver, and G. S. May. 1995. myoA of Aspergillus nidulans encodes an essential myosin I required for secretion and polarized growth. J. Cell Biol. 128:577-587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Meador, W. E., A. R. Means, and F. A. Quiocho. 1993. Modulation of calmodulin plasticity in molecular recognition on the basis of X-ray structures. Science 262:1718-1721. [DOI] [PubMed] [Google Scholar]
- 26.Meador, W. E., A. R. Means, and F. A. Quiocho. 1992. Target enzyme recognition by calmodulin: 2.4 A structure of a calmodulin-peptide complex. Science 257:1251-1255. [DOI] [PubMed] [Google Scholar]
- 27.Means, A. R., C. R. Kahl, D. G. Crenshaw, and J. S. Dayton. 1999. Traversing the cell cycle: the calcium/calmodulin connection, p. 512-528. In E. Carafoli and C. Klee (ed.), Calcium as a cellular regulator. Oxford University Press, New York, N.Y.
- 28.Mendoza, I., F. Rubio, A. Rodriguez-Navarro, and J. M. Pardo. 1994. The protein phosphatase calcineurin is essential for NaCl tolerance of Saccharomyces cerevisiae. J. Biol. Chem. 269:8792-8796. [PubMed] [Google Scholar]
- 29.Moser, M. J., J. R. Geiser, and T. N. Davis. 1996. Ca2+-calmodulin promotes survival of pheromone-induced growth arrest by activation of calcineurin and Ca2+-calmodulin-dependent protein kinase. Mol. Cell. Biol. 16:4824-4831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Moser, M. J., S. Y. Lee, R. E. Klevit, and T. N. Davis. 1995. Ca2+ binding to calmodulin and its role in Schizosaccharomyces pombe as revealed by mutagenesis and NMR spectroscopy. J. Biol. Chem. 270:20643-20652. [DOI] [PubMed] [Google Scholar]
- 31.Nakamura, T., Y. Liu, D. Hirata, H. Namba, S. Harada, T. Hirokawa, and T. Miyakawa. 1993. Protein phosphatase type 2B (calcineurin)-mediated, FK506-sensitive regulation of intracellular ions in yeast is an important determinant for adaptation to high salt stress conditions. EMBO J. 11:4063-4071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Nanthakumar, N. N., J. S. Dayton, and A. R. Means. 1996. Role of Ca++/calmodulin binding proteins in Aspergillus nidulans cell cycle regulation. Prog. Cell Cycle Res. 2:217-228. [DOI] [PubMed] [Google Scholar]
- 33.Odom, A., S. Muir, E. Lim, D. L. Toffaletti, J. Perfect, and J. Heitman. 1997. Calcineurin is required for virulence of Cryptococcus neoformans. EMBO J. 16:2576-2589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ohya, Y., and Y. Anraku. 1989. Functional expression of chicken calmodulin in yeast. Biochem. Biophys. Res. Commun. 158:541-547. [DOI] [PubMed] [Google Scholar]
- 35.Osawa, M., H. Tokumitsu, M. B. Swindells, H. Kurihara, M. Orita, T. Shibanuma, T. Furuya, and M. Ikura. 1999. A novel target recognition revealed by calmodulin in complex with Ca2+-calmodulin-dependent kinase kinase. Nat. Struct. Biol. 6:819-824. [DOI] [PubMed] [Google Scholar]
- 36.Punt, P. J., and A. M. J. J. Van Den Hondel. 1992. Transformation of filamentous fungi based on hygromycin B and phleomycin resistance markers. Methods Enzymol. 216:447-457. [DOI] [PubMed] [Google Scholar]
- 37.Putkey, J. A., T. Ono, M. F. A. VanBerkum, and A. R. Means. 1988. Functional significance of the central helix in calmodulin. J. Biol. Chem. 263:11242-11249. [PubMed] [Google Scholar]
- 38.Putkey, J. A., G. R. Slaughter, and A. R. Means. 1988. Genetically engineered calmodulins differentially activate target enzymes. J. Biol. Chem. 260:9896-9903. [PubMed] [Google Scholar]
- 39.Rasmussen, C., C. Garen, S. Brining, R. L. Kincaid, R. L. Means, and A. R. Means. 1994. The calmodulin-dependent protein phosphatase catalytic subunit (calcineurin A) is an essential gene in Aspergillus nidulans. EMBO J. 13:3917-3924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Rasmussen, C. D., R. L. Means, K. P. Lu, G. S. May, and A. R. Means. 1990. Characterization and expression of the unique calmodulin gene of Aspergillus nidulans. J. Biol. Chem. 265:13767-13775. [PubMed] [Google Scholar]
- 41.Sanchez, O., and J. Aguirre. 1996. Efficient transformation of Aspergillus nidulans by electroporation of germinated conidia. Fungal Genet. Newsl. 43:48-51. [Google Scholar]
- 42.Sarkar, G., and S. S. Sommer. 1990. The "megaprimer' method of site-directed mutagenesis. BioTechniques 8:404-407. [PubMed] [Google Scholar]
- 43.Stirling, D. A., A. Petrie, D. J. Pulford, D. T. Paterson, and M. J. Stark. 1992. Protein A-calmodulin fusions: a novel approach for investigating calmodulin function in yeast. Mol. Microbiol. 6:703-713. [DOI] [PubMed] [Google Scholar]
- 44.Takeda, T., and M. Yamamoto. 1987. Analysis and in vivo disruption of the gene encoding calmodulin in Schizosaccharomyces pombe. Proc. Natl. Acad. Sci. USA 84:3580-3584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Withee, J. L., J. Mulholland, R. Jeng, and M. S. Cyert. 1997. An essential role of the yeast pheromone-induced Ca2+ signal is to activate calcineurin. Mol. Biol. Cell 8:263-277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Ye, R. R., and A. Bretscher. 1992. Identification and molecular characterization of the calmodulin-binding subunit gene (CMP1) of protein phosphatase 2B from Saccharomyces cerevisiae. An alpha-factor inducible gene. Eur. J. Biochem. 204:713-723. [DOI] [PubMed] [Google Scholar]
- 47.Yoshida, T., T. Toda, and M. Yanagida. 1994. A calcineurin-like gene ppb1+ in fission yeast: mutant defects in cytokinesis, cell polarity, mating and spindle pole body positioning. J. Cell Sci. 107:1725-1735. [DOI] [PubMed] [Google Scholar]