Abstract
Alzheimer disease (AD) is a complex disorder characterized by a wide range, within and between families, of ages at onset of symptoms. Consideration of age at onset as a covariate in genetic-linkage studies may reduce genetic heterogeneity and increase statistical power. Ordered-subsets analysis includes continuous covariates in linkage analysis by rank ordering families by a covariate and summing LOD scores to find a subset giving a significantly increased LOD score relative to the overall sample. We have analyzed data from 336 markers in 437 multiplex (⩾2 sampled individuals with AD) families included in a recent genomic screen for AD loci. To identify genetic heterogeneity by age at onset, families were ordered by increasing and decreasing mean and minimum ages at onset. Chromosomewide significance of increases in the LOD score in subsets relative to the overall sample was assessed by permutation. A statistically significant increase in the nonparametric multipoint LOD score was observed on chromosome 2q34, with a peak LOD score of 3.2 at D2S2944 (P=.008) in 31 families with a minimum age at onset between 50 and 60 years. The LOD score in the chromosome 9p region previously linked to AD increased to 4.6 at D9S741 (P=.01) in 334 families with minimum age at onset between 60 and 75 years. LOD scores were also significantly increased on chromosome 15q22: a peak LOD score of 2.8 (P=.0004) was detected at D15S1507 (60 cM) in 38 families with minimum age at onset ⩾79 years, and a peak LOD score of 3.1 (P=.0006) was obtained at D15S153 (62 cM) in 43 families with mean age at onset >80 years. Thirty-one families were contained in both 15q22 subsets, indicating that these results are likely detecting the same locus. There is little overlap in these subsets, underscoring the utility of age at onset as a marker of genetic heterogeneity. These results indicate that linkage to chromosome 9p is strongest in late-onset AD and that regions on chromosome 2q34 and 15q22 are linked to early-onset AD and very-late-onset AD, respectively.
Introduction
Alzheimer disease (AD [MIM 104300]) is a neurodegenerative disorder that is the most common cause of dementia in older adults. The etiology of AD is complex, comprising both genetic and environmental factors. Four genes are known to be associated with AD. Mutations in three of these genes (amyloid precursor protein [APP], presenilin 1 [PS1], and presenilin 2 [PS2]) cause autosomal dominant, early-onset familial AD (Goate et al. 1991; Levy-Lahad et al. 1995; Sherrington et al. 1995). Together, these genes account for <2% of all cases of AD. The fourth gene, apolipoprotein E (APOE), increases risk of the more common late-onset familial and sporadic forms of AD. The APOE-4 allele increases risk and reduces age at onset of AD in a dose-dependent manner (Corder et al. 1993, 1994; Saunders et al. 1993; Strittmatter et al. 1993). These genes were identified by studying sets of families chosen by age at onset and inheritance pattern. Age at onset is clearly an important covariate for the identification of susceptibility genes for AD.
In addition to the relationship of these known genes with age at onset of AD, several lines of evidence suggest that age at onset may have other genetic determinants. Segregation analysis of large pedigrees with AD suggested that multiple loci associated with age at onset exist (Daw et al. 2000). Quantitative trait linkage analysis, with age at onset as the trait of interest, identified several genomic regions that might contain genes modulating age at onset of AD symptoms (Li et al. 2002).
Although the identification of APP, PS1, PS2, and APOE has significantly advanced the understanding of the etiology of AD, together these loci explain, at most, 50% of the genetic effect in the disease (Farrer 1997). Several large families with early-onset familial AD have been reported that do not have mutations in APP, PS1, or PS2, indicating that at least one additional early-onset AD gene exists (Janssen et al. 2003). Also, familial aggregation of AD has been described in populations with low frequency of the APOE-4 allele, indicating that additional late-onset AD genes exist (Pericak-Vance et al. 1996). Therefore, significant effort has been invested in identifying additional susceptibility loci, particularly for late-onset (age at onset >60 years) AD.
To this end, many genomic screens have been performed in families with multiple individuals diagnosed with late-onset AD (Pericak-Vance et al. 1997, 2000; Kehoe et al. 1999; Hiltunen et al. 2001; Myers et al. 2002; Blacker et al. 2003; Farrer et al. 2003). Although these studies have detected suggestive evidence for linkage on many chromosomes, the strongest and most consistent linkages across these studies were to regions of chromosomes 9, 10, and 12. However, aside from considering late-onset AD separately (variously defined as ⩾60 and ⩾65 years old), these studies have not thoroughly considered linkage heterogeneity by age at onset. Two recent studies (Olson et al. 2001, 2002) describe analyses including age as a covariate in linkage analysis of affected relative pairs. These studies report significant differences in linkage on chromosomes 20 and 21 when current age was used as the covariate, which was interpreted as meaning that these loci influenced duration of AD rather than age at onset.
Since the effects of all known AD loci have been shown to be age dependent, the limited consideration of age at onset may have low power to detect linkage to loci with age-dependent effects on risk of AD. Therefore, examination of genomic-screen data with consideration of age at onset as a covariate may refine estimates of linkage.
Several methods of incorporating covariates into linkage analysis have been developed. The simplest to apply is stratification of families into defined subsets according to age at onset (Pericak-Vance et al. 1991). Blacker and colleagues (2003) recently employed this approach, classifying families as “late onset” if all affected individuals had onset of AD at age ⩾65 years and “early/mixed” if at least one individual was affected at onset age <65 years. One disadvantage of this method is that a predetermined cut point must be used for stratification and may not result in the most homogeneous subsets. For example, the APOE gene has its maximum effect on risk of AD between the ages of 60 and 70 years; therefore, a cut point of age 65 years might decrease the power of the analysis to detect the APOE gene. An alternative to stratification is ordered-subsets analysis (OSA) (Hauser et al. 1998, in press; Ghosh et al. 2000; Shao et al. 2003). OSA orders families by a continuous covariate (such as mean age at onset) and then finds the subset with maximum evidence for linkage to a particular map of markers. The statistical significance of the increased evidence for linkage relative to evidence for linkage in the entire sample is assessed via permutation. This approach identifies a set of families in which the LOD score in a particular region is higher than in the overall data set. Thus, the primary goal of OSA is to identify regions of increased linkage in a subset of families, even though genetic heterogeneity significantly reduces the evidence for linkage in the overall data set. Subsets identified by OSA may then be used for candidate-gene analysis and fine mapping in that region of interest. OSA limits the possibility of missing such a covariate-dependent linkage result because it adaptively chooses the cut point for a stratified analysis on the basis of the observed linkage evidence. We have applied OSA to data from a recently completed genomic screen in late-onset AD (Pericak-Vance et al. 2000) to identify loci with increased evidence for linkage (relative to the overall sample) in subsets of families defined by mean and minimum ages at onset.
Subjects and Methods
Subjects
Families with two or more individuals with a diagnosis of probable or definite AD were ascertained by the Collaborative Alzheimer Project (CAP [Duke University Medical Center, Vanderbilt University Medical Center, and University of California at Los Angeles]), the National Institute of Mental Health (NIMH) AD Genetics Initiative (Massachusetts General Hospital, Johns Hopkins University, and the University of Alabama at Birmingham), and the Indiana University (IU) AD Cell Repository. Informed consent was obtained for each participant, under protocols approved by the institutional review boards at the participating sites. The current study was conducted under protocols approved by the Duke University Medical Center institutional review board. A subset of 437 white families with complete age-at-onset information was selected from the original 455 families included in the genomic screen (Pericak-Vance et al. 2000). Two Latino and three African American families were removed from the original genomic screen data set to control for potential linkage heterogeneity by ethnicity; linkage results in regions of interest became more significant when these families were removed (data not shown). These 437 families contained 1,252 sampled individuals (1,014 affected individuals), 543 affected sibling pairs, and 62 other affected relative pairs.
The diagnosis of AD was consistent with National Institute of Neurological and Communicative Disorders and Stroke–Alzheimer's Disease and Related Disorders Association consensus diagnostic criteria (McKhann et al. 1984). Autopsy confirmation of the AD diagnosis, by use of consensus criteria (Mirra et al. 1991; Hyman and Trojanowski 1997), was obtained for at least one individual with AD in 227 of the 437 (52%) families. Age at onset of AD was recorded as that age at which the first symptoms were noted by the participant or a family member. The mean age at onset for individuals with AD was 72.7±6.7 years (range 49–93 years). Two covariates were used to divide the families into subsets, mean age at onset and minimum age at onset. To examine the notion that early onset, even in only one member of the family, may be a marker for increased genetic susceptibility in a given region of the genome, the minimum age at onset was determined by the youngest age at onset in each family. Use of minimum age at onset as a covariate may therefore increase the power to detect a locus that causes earlier-onset AD with wide intrafamilial variation in age at onset, as is seen in families with PS2 mutations (Sherrington et al. 1996).
Laboratory Analysis
Genomic DNA was extracted from whole blood by use of the Puregene system (Gentra Systems) (for CAP samples) or was obtained directly from repositories (for NIMH and IU samples). Microsatellite-marker genotyping was performed for 336 markers by use of the FASST method (Vance and Ben Othmane 1998). Markers were selected on the basis of heterozygosity, ease of genotyping, and location, to generate a marker map of ∼10 cM density.
Systematic genotyping errors were minimized by use of a system of quality control (QC) checks with duplicated samples (Rimmler et al. 1998). On each 96-well PCR plate, two standard samples from CEPH families were included, and six additional samples were duplicates of samples either on that plate or on another plate in the screen. Laboratory technicians were blinded to the location of these QC samples to avoid bias in interpretation of results. Automated computer scripts checked each set of genotypes submitted by the technician for mismatches between the duplicated samples; mismatches were indicative of potential genotype reading errors, misloading of samples, and sample misallocations. Mismatched reads were returned to the laboratory technician with a set of surrounding samples for rereading. As an additional QC measure, potential pedigree errors were checked with the program RELPAIR (Boehnke and Cox 1997), which infers likely relationships between pairs of relatives, using identity-by-descent sharing estimates from a set of microsatellite markers.
Statistical Analysis
The affected relative pair–allele-sharing method implemented in the ASM module of GENEHUNTER-PLUS (Kong and Cox 1997) was used to calculate multipoint nonparametric LOD scores with the Spairs statistic and a linear model. Intermarker distances were obtained from the genetic linkage maps developed by Marshfield Medical Research Foundation (Broman et al. 1998).
The OSA program (Hauser et al. 1998, in press) was used to identify significant increases in multipoint nonparametric LOD score curves in subsets of families defined by mean and minimum ages at onset. The OSA method proceeds as follows:
-
1.
Order families by a family-specific covariate value.
-
2.
Assign ranks to each family in ascending and descending order. Families with the same covariate value are given the same rank.
-
3.
Beginning with the first-ranked families, calculate the multipoint LOD scores for the subset across all map positions for a chromosome. Store the maximum LOD score and map position.
-
4.
Add the family (or families) next in order after the previous subset (e.g., 1+2). Calculate the multipoint LOD scores. Store the maximum LOD score and map position. Consider successively larger subsets by repeating this step (e.g., 1+2+3, 1+2+3+4,…), adding the remaining families in order until all families are included in the final subset (i.e., 1+…+N).
-
5.
Select the subset with the highest maximum LOD score.
-
6.
Perform permutation tests of the chromosomewide significance of the increase in this maximum LOD score over baseline:
-
A.
Randomly order families with the same number of ranks as in step 2.
-
B.
Repeat steps 3–5.
-
C.
Determine if the subset generates a multipoint LOD score anywhere on the chromosome at least as high as the maximum subset LOD score found in step 5.
-
D.
Repeat 10,000 times.
-
E.
Calculate an empirical P value on the basis of the proportion of 10,000 random orderings of families that gave a maximum subset LOD score greater than that seen in step 5.
-
A.
-
7.
Repeat the procedure for each covariate and ranking order (ascending and descending).
In addition, the “optimal slice” option was used to define the subset (of any size) of adjacent families from anywhere in the covariate distribution that maximized the LOD score. This option follows the procedure outlined above but repeats the process beginning with successively higher ranks (e.g., start with the second rank, then the third rank, etc.). The optimal-slice analysis considers a much greater number of potential subsets than the other two rank orders. Therefore, if the same subset of families is identified in the ascending or descending orders and the optimal-slice option, the optimal-slice subset may have a larger P value than the other analyses.
For each chromosome, six maximum nonparametric LOD scores were obtained. To adjust for the use of two covariates (minimum and mean ages at onset) and two independent ranking orders (ascending and descending), a nominal significance level of 0.0125 (0.05/4) was used. As OSA results for the optimal-slice option are highly correlated with those for ascending and descending rankings, no correction was made for the optimal-slice analysis. Since analysis was performed on 23 chromosomes, results with P values <.0005 (.0125/23) may be considered as having genomewide significance. Clinical features for each subset with a statistically significant increase in nonparametric LOD score were analyzed using SAS version 8 (SAS Institute).
To assess the potential modification of linkage by APOE genotype in OSA-defined subsets, the ASM module of GENEHUNTER-PLUS (Kong and Cox 1997) was used to generate nonparametric LOD scores in OSA subsets weighted by APOE-4 carrier status. Increases in LOD scores when weighting by the proportion carrying the APOE-4 allele indicates epistasis, whereas increases when weighting by the proportion not carrying an APOE-4 allele indicates linkage heterogeneity.
Results
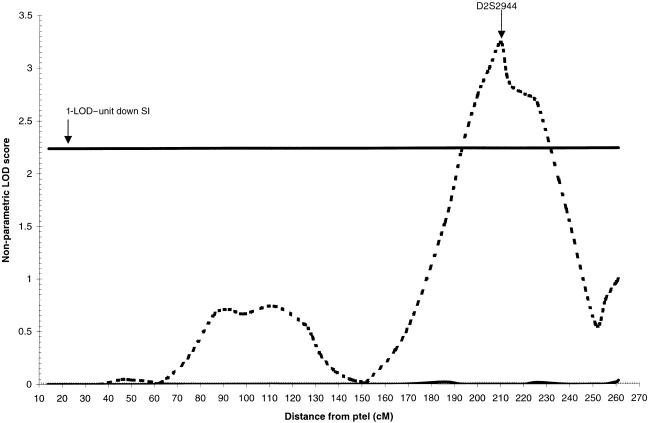
Nominally significant increases in LOD scores were observed in three regions of the genome, on chromosomes 2q34, 9p22, and 15q22 (table 1). When ordering families by ascending minimum age at onset, a maximum LOD score of 3.0 was obtained at D2S2944 (210 cM) in the 32 families with minimum age at onset ⩽60 years. This was a significant increase from the baseline LOD score of 0 (P=.002). The optimal slice identified for this region was 31 families with minimum age at onset of 50–60 years; the maximum LOD score was 3.2 at D2S2944 (P=.008). The 1-LOD–unit down support interval (SI) for this peak is 46 cM wide, from 193 cM to 239 cM (fig. 1).
Table 1.
Significant Increases in LOD Score Associated with Age at Onset, Detected by OSA in the Overall Data Set
| Chromosomeand Peak Marker | cM | MaxLOD | Changefrom Baseline | P | No. of Familiesin Subseta | Variableb | Order | Age(years) |
| 2q34: | ||||||||
| D2S2944 | 210 | 3.0 | 3.0 | .002 | 32 | Minimum | Ascending | ⩽60 |
| D2S2944 | 210 | 3.2 | 3.2 | .008 | 31 | Minimum | Optimal slice | 50–60 |
| 9p22: | ||||||||
| D9S741 | 42 | 4.6 | 1.6 | .01 | 334 | Minimum | Optimal slice | 60–75 |
| 15q22: | ||||||||
| D15S1507 | 60 | 2.8 | 2.8 | .0004 | 38 | Minimum | Descending | ⩾79 |
| D15S1507 | 60 | 2.8 | 2.8 | .004 | 38 | Minimum | Optimal slice | ⩾79 |
| D15S153 | 62 | 3.1 | 3.1 | .0006 | 43 | Mean | Descending | ⩾80 |
| D15S153 | 62 | 3.1 | 3.1 | .01 | 43 | Mean | Optimal slice | ⩾80 |
n=437.
Age at onset.
Figure 1.
Nonparametric LOD score plot for chromosome 2. LOD scores for the overall sample (n=437 [solid line]) and the subset with minimum age at onset with a range of 50–60 years (n=31 [dashed line]) are presented. The 1-LOD–unit down SI (192–239 cM) is marked by the bold horizontal line.
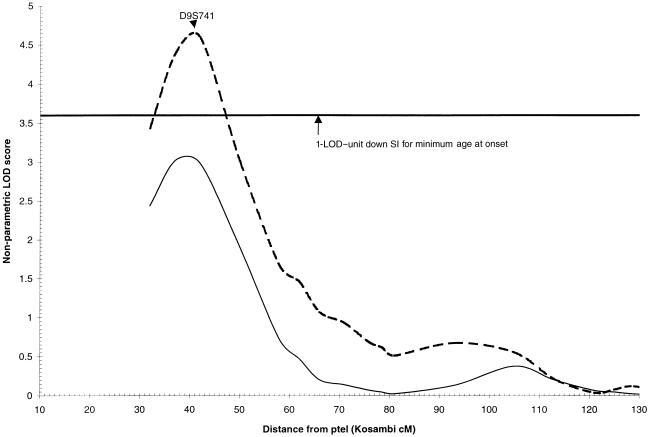
Consistent with the original genomic screen analysis (Pericak-Vance et al. 2000), the region on 9p22 generated strong evidence for linkage in the overall data set (LOD=3.0). Although neither the ascending nor descending orders generated a significant increase in the LOD score, the optimal-slice ranking by minimum age at onset generated a maximum LOD score of 4.6 at D9S741 (P=.01) in the 334 families with minimum age at onset between 60 and 75 years. The 1-LOD–unit down SI for this peak is 15 cM wide, from 33 cM to 48 cM (fig. 2).
Figure 2.
Nonparametric LOD score plot for chromosome 9. LOD scores for the overall sample (n=437 [solid line]) and the subset with minimum age at onset with a range of 60–75 years (n=32 [dashed line]) are presented. The 1-LOD–unit down SI (33–48 cM) is marked by the bold horizontal line.
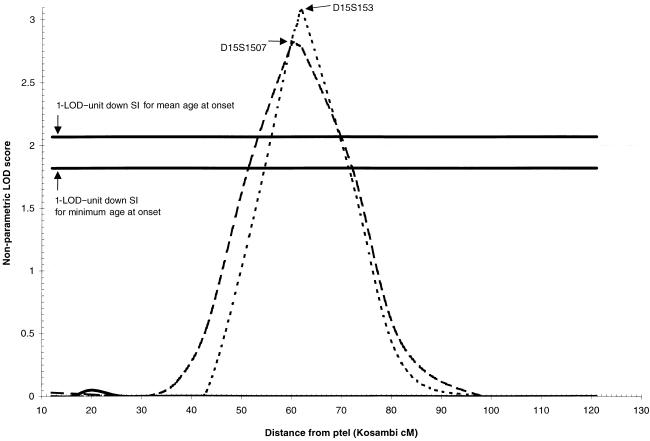
When descending minimum age at onset was considered as the covariate, a maximum LOD score of 2.8 was obtained at D15S1507 (60 cM) in the 38 families with minimum age at onset ⩾79 years. This was also a significant increase from the baseline LOD score of 0 (P=.0004). The same 38 families were identified as the optimal slice for this covariate. The 1-LOD–unit down SI for this peak was 21 cM wide, from 51 cM to 72 cM (fig. 3).
Figure 3.
Nonparametric LOD score plot for chromosome 15. LOD scores for the overall sample (n=437 [solid line]), the subset with minimum age at onset >79 years (n=38 [dashed line]), and the subset with mean age at onset >80 years (n=43 [dotted line]) are presented. The 1-LOD–unit down SIs for the minimum–age-at-onset curve (51–72 cM) and the mean age at onset curve (56–70 cM) are marked by the bold horizontal lines.
Similarly, when considering family mean age at onset as the covariate, a maximum LOD score of 3.1 was obtained at D15S153 (62 cM) in the 43 families with mean age at onset ⩾80 years (P=.0006). The optimal slice consisted of the same 43 families; however, the P value for this subset was much less significant (P=.01), owing to the greater number of subsets considered in the optimal-slice analysis. The 1-LOD–unit down SI for this region is 14 cM wide, from 56 cM to 70 cM (fig. 3).
The two chromosome 15 regions overlap significantly. Both subsets contain 31 of the same families, and the SI for the descending mean age-at-onset peak is completely contained within the SI for the descending minimum–age at onset peak. The peak markers (D15S1507 and D15S153) are only 2 cM apart. Thus, these two results point to the same very-late-onset AD locus on 15q22 in a subset of 50 families.
A description of these subsets by family size, origin of data set (CAP, IU, and NIMH), age at onset, and APOE-4 carrier status is provided in table 2. There is some overlap between the subsets of families linked to the three regions: the chromosome 2q and 15q subsets do not overlap at all, three families are in both the chromosome 9p and 15q subsets, and 13 families are in both the 2q and 9p subsets. A greater percentage (19%) of the 31 families in the 2q34 subset were “extended families” (defined as “those families with sampled affected relative pairs other than sibling pairs”), compared with the overall data set (6%). In addition, this subset contained a higher proportion of families from the CAP (18%) and IU (12%) data sets, compared with the NIMH data set (3%). This is perhaps because the publicly distributed NIMH data set is composed primarily of families containing affected sibling pairs, with few other affected-relative pairs available. It is not surprising that this subset had an earlier mean age at onset (66.1 years) than did the overall data set (72.7 years), but the range of age at onset was not substantially different. The mean within-family difference was greater in this subset than overall (14.8 years vs. 6.7 years), indicating that age at onset is more variable within families in this subset. The proportion of families in which all affected individuals had at least one copy of the APOE-4 allele was higher in this subset (71%) than overall (61%).
Table 2.
Clinical Features of Subsets Identified by OSA
|
No. (%) of Families in Sample |
||||||||||
| Region | Total | CAP | IU | NIMH | % Extended | Mean Age at Onset | Range | Mean within-Family Difference in Age at Onset | Range | Families Positive for APOE-4a(%) |
| 2q34 | 31 (7) | 10 (18) | 13 (12) | 8 (3) | 19 | 66.1 | 50–85 | 14.8 | 6–35 | 71 |
| 9p22 | 334 (76) | 45 (79) | 77 (73) | 212 (77) | 6 | 71.5 | 60–88 | 7.0 | 0–22 | 65 |
| 15q22 | 50 (11) | 5 (9) | 6 (6) | 39 (14) | 2 | 82.2 | 74–93 | 4.5 | 0–13 | 34 |
| Total | 437 (100) | 57 (100) | 105 (100) | 275 (100) | 6 | 72.7 | 49–93 | 6.7 | 0–35 | 61 |
Families in which all affected individuals have at least one copy of the APOE-4 allele.
In contrast, the families in the 15q22 subset contained, almost exclusively, affected sibling pairs (2% extended families), had substantially later age at onset than the overall data set (and had a smaller within-family difference and range of age at onset), and included a much lower proportion of families in which all affected individuals carried the APOE-4 allele (34% vs. 61%). This subset contained a slightly greater percentage of families from the NIMH data set (14%), compared with the CAP (9%) and IU (6%) data sets. The 9p22 subset contained similar proportions of families from all three data sets and was not substantially different from the overall sample, with respect to percentage of extended families, APOE genotype, or within-family differences in age at onset.
To test if linkage in these three regions is modified by the APOE genotype, conditional linkage analysis was performed using GENEHUNTER-PLUS. Weighted nonparametric LOD scores were calculated for each OSA subset by weighting family-specific LOD scores by the proportion of affected members carrying an APOE-4 allele (to model epistasis) and by the proportion of affected members not carrying an APOE-4 allele (to model heterogeneity). No significant increases in the LOD score were seen for any subset, indicating that evidence for linkage to these regions is not strongly modified by the APOE genotype.
Discussion
We have identified three regions with age-dependent evidence for linkage to AD: a locus on chromosome 2q34 associated with earlier onset, a locus on 9p associated with late onset, and a locus on 15q22 associated with very late onset. To our knowledge, the 2q34 locus has not been reported elsewhere. The 9p locus was detected in the original genomic screen in these families (Pericak-Vance et al. 2000). The region on 15q22 is ∼20 cM from D15S659, a marker at which a Finnish case-control study detected linkage disequilibrium with AD (Hiltunen et al. 2001). Neither of the genomic screens that considered age at onset, in some fashion, as a covariate (Olson et al. 2002; Blacker et al. 2003) detected linkage to any of these regions. This is surprising, since 39 of 50 families in the 15q subset were from the NIMH family data set common to this study and to the studies by Olson et al. (2002) and Blacker et al. (2003). The current study did not detect significant improvement in linkage results in families with “early/mixed” onset AD (families with a minimum age at onset <65 years) in any of the six “early/mixed” regions (1p31, 3p26, 10p14, 14q22, 15q26, or 19q13) reported by Blacker and colleagues (2003). Similarly, this study did not identify linkage of very-late-onset AD to chromosomes 20 and 21, as others have reported elsewhere (Olson et al. 2001, 2002). Therefore, either the composition of the current data set or the specific manner in which OSA incorporates covariates into linkage analysis has resulted in the novel results.
To explore this issue further, we used OSA to reanalyze the data from chromosomes 20 and 21 in the subset of 275 families in this study collected by the NIMH AD Genetics Initiative. No significant differences in LOD score were detected on chromosome 21 by minimum or mean age at onset. The variation in results on chromosome 21 may be due to differences in methodology, since OSA utilizes family-specific covariates, and the likelihood-ratio methods used by Olson et al. (2001) utilize relative pair–specific covariates. In contrast, a significant increase in LOD score at 12 cM (LOD=2.6; P=.002) was detected on chromosome 20 in the 61 families with mean age at onset ⩾78 years. This result is 12 cM from the peak LOD score reported by Olson et al. (2002) and suggests that variation in chromosome 20 linkage results in very-late-onset AD may be due to differences in the composition of the family data sets used rather than differences in statistical methodologies. However, as expected from our overall results, the non-NIMH families showed no increase in LOD scores in this region.
These results illustrate the utility of OSA for incorporating continuous covariates into linkage analysis of complex traits. OSA avoids arbitrary cut points (such as early/mixed versus late onset, defined as “age at onset ⩾60 or 65 years”) by considering the entire distribution of the covariate. This approach identifies loci that may be strongly linked to disease in a small portion of the covariate distribution. Controlling for the covariate on the family level rather than the relative-pair level focuses on identifying between-family linkage heterogeneity due to the covariate and identifies a subset of families to use in fine-mapping or candidate-gene studies.
OSA analysis detected a significant increase in LOD score (3.2) on chromosome 2q34 in 31 families with minimum age at onset between 50 and 60 years. To our knowledge, no previous study has reported significant evidence for linkage within 50 cM of this peak. These families (mean age at onset 66 years; range 50–85 years) all contained at least one individual with early-onset AD (age at onset ⩽60 years) and at least one individual with late-onset AD (age at onset >60 years). The within-family difference in age at onset was larger than that in the overall sample (14.8 years vs. 6.7 years), suggesting that there was more variability in age at onset in this subset. Thus, this region may contain a locus, like PS2, that causes AD in families with both early- and late-onset cases of AD and is marked by wide variability in the age at onset of symptoms (Sherrington et al. 1996). Furthermore, a greater proportion of families (19% vs. 6%) contained an affected relative pair other than a sibling pair, indicating that this locus may be associated with a family history of AD in multiple generations. Whereas all individuals with AD carried the APOE-4 allele in a greater proportion of families in this subset (72%) versus the overall data set (61%), there was no modification of the linkage results when conditioning on the APOE genotype.
A search of the Ensembl database identified 454 known genes in the 1-LOD–unit down SI (192–239 cM) for this locus. Two genes are of particular interest: microtubule-associated protein 2 (MAP2) and ATP-binding cassette A12 (ABCA12). MAP2 is located within 4 Mb of the peak marker, D2S2944. MAP2 is a dendrite-specific microtubule-binding protein from the same protein family as microtubule-associated protein τ (MAPT), which is found in the characteristic neurofibrillary tangles seen in the brains of patients with AD (Al-Bassam et al. 2002). The ABCA12 gene, located <1 Mb from the peak marker, is a member of the ATP–binding cassette (ABC) transporter superfamily, the members of which move substrates across membranes. Other members of the ABCA gene family have been implicated in disorders of cholesterol transport (ABCA1) and early-onset forms of macular degeneration (ABCA4) (Dean et al. 2001). Since AD is a neurodegenerative condition and the risk of AD is influenced by other genes involved in cholesterol transport (APOE), ABCA12 is a plausible candidate gene. Farther away from the peak marker, but still in the SI, are the genes for caspases 8 and 10 (involved in apoptosis) and the GTPase regulator alsin, mutations of which have been recently reported in juvenile amyotropic lateral sclerosis (Hadano et al. 2001; Yang et al. 2001).
OSA analysis also increased the LOD score in the region of chromosome 9p that provided the strongest evidence for linkage in the original genomic screen (Pericak-Vance et al. 2000) and subsequent studies in independent samples (Farrer et al. 2003). A subset of 334 families with minimum age at onset between 60 and 75 years was identified using the optimal-slice option, and the LOD score increased from a baseline of 3.0 to 4.6. Mean age at onset, proportion of extended families, and proportion of families with APOE-4 were all similar to those in the overall sample. These results indicate that this region influences the development of AD in a broad subset of families with late-onset AD, and they narrow the linkage to a 15-cM 1-LOD–unit down SI. According to Ensembl, this interval contains 44 genes, including several biologically plausible candidate genes for AD in this data set—such as MTAP, p15, and p16—that are currently being evaluated for association (Xu et al. 2002).
The other significant increase in LOD score, when controlling for age at onset, was on chromosome 15q22. The greatest evidence for linkage (LOD=3.1) was detected in 43 families with mean age at onset ⩾80 years, and the P value for this increase met our criteria for genomewide significance (P<.0005). A similar increase was detected when minimum age at onset was used as the covariate. These results suggest that the effect of this locus is greatest in families with the oldest age at onset. These subsets contained families consisting mostly of affected sibling pairs, suggesting that this locus is involved in causing AD in smaller family aggregates. Although a larger proportion of families had at least one individual with AD who did not carry the APOE-4 allele, relative to the overall sample, there was no statistically significant modification of linkage results by the APOE genotype. These observations regarding family size and APOE genotype are consistent with the average age at onset in this subset: multiple family members need to survive past age 80 years to develop very-late-onset AD, and APOE-4 has its maximum impact on risk of AD between ages 60 and 70 years. Only one previous study has reported evidence for linkage to this region. Hiltunen et al. (2001) detected evidence for linkage disequilibrium between late-onset AD and a marker ∼20 cM from the peak markers in the 15q22 region reported here. The influence of age at onset and APOE genotype on the linkage disequilibrium in the Finnish sample was not reported; therefore, it is not possible to determine if the previous association was stronger in very-late-onset AD.
The 1-LOD–unit down SI for the mean age at onset peak on 15q22 was completely contained in the SI for the minimum age-at-onset peak. This 21-Mb SI contains 183 known genes, according to the Ensembl database. Interesting candidate genes in this interval include: MAP2K1 and MAP2K5, which encode protein kinases that may influence apoptosis; RAB11A, which encodes an RAS-related GTPase; KIF23, which enclodes a kinesinlike microtubule motor protein; and CYP1A2 and CYP1A1, members of the cytochrome P450 gene family.
No other nominally significant (P<.0125) increases in LOD score, when controlling for age at onset, were detected in this data set. Genomic screens, reported elsewhere, of families included in this data set detected evidence for linkage to chromosomes 19 (Pericak-Vance et al. 1991), 12 (Pericak-Vance et al. 1997), and 10 (Pericak-Vance et al. 2000); however, evidence for linkage to these regions did not significantly increase when age at onset was considered. This could be because OSA has reduced power to detect small increases in regions with strong baseline LOD scores, since such strong overall results may reflect an effect spread across much of the age-at-onset distribution. Also, the analysis performed here focused on the effect of one continuous covariate (age at onset) on linkage; other categorical covariates—such as APOE genotype, autopsy confirmation, and source of data (CAP, IU, or NIMH)—could not be easily incorporated in the analysis. On chromosome 10, the maximum subset LOD score was 1.8 (from 0.6 overall), and, on chromosome 12, the maximum subset LOD score was 1.0 (from 0.22). It is important to note that this OSA analysis did not consider the known effect of the APOE genotype on linkage to chromosome 12 (where linkage is strongest in APOE-4–negative families). However, the LOD score on chromosome 19q13 (near APOE) increased to 3.4 (from 1.8) in 103 families with mean age at onset <70 years. These results, although not statistically significant relative to the linkage results in the entire data set, indicate that the strongest linkage in this region is in families with age at onset <70 years of age; this is consistent with the known age-related effect of the APOE-4 allele.
These results suggest that OSA is most powerful for identifying genetic effects that are restricted to a small subset of families with extreme covariate values; such effects are likely to be lost in the overall sample because of the substantial heterogeneity of the LOD scores. In contrast, linkage signals that may be detected in the overall sample may be derived from a much larger set of families, and a “ceiling effect” may exist. Therefore, the improvement in LOD score when considering a covariate may not be statistically significant in regions with strong baseline linkage results.
These results also differ significantly from our study of age at onset as a quantitative trait locus (QTL) in this same data set (Li et al. 2002). The study of Li et al. (2002) reported strong evidence for linkage (LOD>2) of age at onset to regions of chromosomes 4q, 8q, and 10q. None of these regions was detected by our analysis using AD as the trait and age at onset as a covariate. Similarly, quantitative linkage analysis of age at onset as the trait did not detect evidence for linkage to 2q or 15q (Li et al. 2002). This disparity in results reflects the differences in the statistical methodology used in the two studies. Analysis of age at onset as a QTL aims to identify genetic loci that explain within- and between-family variation in age at onset across the entire age-at-onset distribution. In contrast, OSA–LOD score analysis aims to identify genetic loci that have stronger effects in subsets of families defined by a continuous covariate. Although the two methods have common goals (identifying genes related to risk of AD), the approaches may detect loci with different mechanisms of action.
The results of this study underscore the utility of using covariate analyses to identify potential linkage heterogeneity in genomic screens of complex diseases. In AD, age at onset is clearly an important covariate, because each of the four known genes for AD has a distinctive age-at-onset distribution. Although the effect of the locus on 9p was broad enough to be detectable in the overall data set, the LOD score improves when considering a large subset of families with onset of AD between ages 60 and 75 years. The two novel regions identified in this study generate strong evidence for linkage only in a small subset of families with extreme values of age at onset. If candidate-gene analysis in these regions is focused on families and individuals with extreme age at onset, it is possible to maximize the chances of identifyings the genes responsible for these linkages. Although the percentage of cases of AD attributable to these loci might be relatively small, the discovery of additional genes that influence risk of AD would greatly advance the understanding of the pathophysiology of the disease.
Acknowledgments
We thank the families for their participation and support of AD research. We thank the research staffs of the Duke Center for Human Genetics and Vanderbilt Program in Human Genetics for their efforts on this study. This study was supported by research grants NS31153, MH59528, AG05128, AG11268, AG09029, MH52453, AG10123, RR00856, and AG019726 from the National Institutes of Health; grants II-RG94101, RG2-96044, II-RG00-05, and TLL-97-012 from the Alzheimer's Association; an Alzheimer's Association Zenith award; and a grant from the Neurosciences Education and Research Foundation. Data and biomaterials were collected in three projects that participated in the NIMH Alzheimer Disease Genetics Initiative. From 1991 to 1998, the principal investigators and coinvestigators were: Marilyn S. Albert, Ph.D., and Deborah Blacker, M.D., Sc.D., Massachusetts General Hospital, Boston (NIMH grant U01 MH46281); Susan S. Bassett, Ph.D., Gary A. Chase, Ph.D., and Marshal F. Folstein, M.D., Johns Hopkins University, Baltimore (NIMH grant U01 MH46290); and Rodney C. P. Go, Ph.D., and Lindy E. Harrell, M.D., University of Alabama, Birmingham (NIMH grant U01 MH46373).
Electronic-Database Information
URLs for data presented herein are as follows:
- Center for Human Genetics, http://www.chg.duke.edu/software/osa.html (for OSA software)
- Ensembl, http://www.ensembl.org/
- Marshfield Medical Research Foundation, Center for Medical Genetics, http://www.marshfieldclinic.org/research/genetics/
- Online Mendelian Inheritance in Man (OMIM), http://www.ncbi.nlm.nih.gov/Omim/ (for AD) [PubMed]
References
- Al-Bassam J, Ozer RS, Safer D, Halpain S, Milligan RA (2002) MAP2 and tau bind longitudinally along the outer ridges of microtubule protofilaments. J Cell Biol 157:1187–1196 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blacker D, Bertram L, Saunders AJ, Moscarillo TJ, Albert MS, Wiener H, Perry RT, Collins JS, Harrell LE, Go RC, Mahoney A, Beaty T, Fallin MD, Avramopoulos D, Chase GA, Folstein MF, McInnis MG, Bassett SS, Doheny KJ, Pugh EW, Tanzi RE (2003) Results of a high-resolution genome screen of 437 Alzheimer’s disease families. Hum Mol Genet 12:23–32 [DOI] [PubMed] [Google Scholar]
- Boehnke M, Cox NJ (1997) Accurate inference of relationships in sib-pair linkage studies. Am J Hum Genet 61:423–429 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Broman KW, Murray JC, Sheffield VC, White RL, Weber JL (1998) Comprehensive human genetic maps: individuals and sex-specific variation in recombination. Am J Hum Genet 63:861–869 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corder EH, Saunders AM, Risch NJ, Strittmatter WJ, Schmechel DE, Gaskell PC Jr, Rimmler JB, Locke PA, Conneally PM, Schmader KE, Small GW, Roses AD, Haines JL, Pericak-Vance MA (1994) Protective effect of apolipoprotein E type 2 allele for late onset Alzheimer disease. Nat Genet 7:180–184 [DOI] [PubMed] [Google Scholar]
- Corder EH, Saunders AM, Strittmatter WJ, Schmechel DE, Gaskell PC, Small GW, Roses AD, Haines JL, Pericak-Vance MA (1993) Gene dose of apolipoprotein E type 4 allele and the risk of Alzheimer’s disease in late onset families. Science 261:921–923 [DOI] [PubMed] [Google Scholar]
- Daw EW, Payami H, Nemens EJ, Nochlin D, Bird TD, Schellenberg GD, Wijsman EM (2000) The number of trait loci in late-onset Alzheimer disease. Am J Hum Genet 66:196–204 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dean M, Rzhetsky A, Allikmets R (2001) The human ATP-binding cassette (ABC) transporter superfamily. Genome Res 11:1156–1166 [DOI] [PubMed] [Google Scholar]
- Farrer LA (1997) Genetics and the dementia patient. Neurolog 3:13–30 [Google Scholar]
- Farrer LA, Bowirrat A, Friedl RP, Waraska K, Korczyn AD, Baldwin CT (2003) Identification of multiple loci for Alzheimer disease in a consanguineous Israeli-Arab community. Hum Mol Genet 12:415–422 [DOI] [PubMed] [Google Scholar]
- Ghosh S, Watanabe RM, Valle TT, Hauser ER, Magnuson VL, Langefeld CD, Ally DS, et al (2000) The Finland–United States investigation of non–insulin-dependent diabetes mellitus genetics (FUSION) study. I. An autosomal genome scan for genes that predispose to type 2 diabetes. Am J Hum Genet 67:1174–1185 [PMC free article] [PubMed] [Google Scholar]
- Goate A, Chartier-Harlin MC, Mullan M, Brown J, Crawford F, Fidani L, Giuffra L, Haynes A, Irving N, James L, Mant R, Newton P, Rooke K, Roques P, Talbot C, Pericak-Vance, MA, Roses A, Williamson R, Rossor M, Owen M, Hardy J (1991) Segregation of a missense mutation in the amyloid precursor protein gene with familial Alzheimer’s disease. Nature 349:704–706 [DOI] [PubMed] [Google Scholar]
- Hadano S, Hand CK, Osuga H, Yanagisawa Y, Otomo A, Devon RS, Miyamoto N, Showguchi-Miyata J, Okada Y, Singaraja R, Figlewicz DA, Kwiatkowski T, Hosler BA, Sagie T, Skaug J, Nasir J, Brown RH Jr, Scherer SW, Rouleau GA, Hayden MR, Ikeda JE (2001) A gene encoding a putative GTPase regulator is mutated in familial amyotrophic lateral sclerosis 2. Nat Genet 29:166–173 [DOI] [PubMed] [Google Scholar]
- Hauser ER, Watanabe RM, Duren WL, Bass MP, Langefeld CD, Boehnke M. Ordered subset analysis in genetic linkage mapping of complex traits. Genet Epidemiol (in press) [DOI] [PubMed] [Google Scholar]
- Hauser ER, Watanabe RM, Duren WL, Boehnke M, the Fusion Study Group (1998) Stratified linkage analysis of complex genetic traits using related covariates. Am J Hum Genet Suppl 63:A45 [Google Scholar]
- Hiltunen M, Mannermaa A, Thompson D, Easton D, Pirskanen M, Helisalmi S, Koivisto AM, Lehtovirta M, Ryynanen M, Soininen H (2001) Genome-wide linkage disequilibrium mapping of late-onset Alzheimer’s disease in Finland. Neurology 57:1663–1668 [DOI] [PubMed] [Google Scholar]
- Hyman BT, Trojanowski JQ (1997) Consensus recommendations for the postmortem diagnosis of Alzheimer disease from the National Institute on Aging and the Reagan Institute working group on diagnostic criteria for the neuropathological assessment of Alzheimer disease. J Neuropathol Exp Neurol 56:1095–1097 [DOI] [PubMed] [Google Scholar]
- Janssen JC, Beck JA, Campbell TA, Dickinson A, Fox NC, Harvey RJ, Houlden H, Rossor MN, Collinge J (2003) Early onset familial Alzheimer’s disease: mutation frequency in 31 families. Neurology 60:235–239 [DOI] [PubMed] [Google Scholar]
- Kehoe P, Wavrant-De Vrieze F, Crook R, Wu WS, Holmans P, Fenton I, Spurlock G, Norton N, Williams H, Williams N, Lovestone S, Perez-Tur J, Hutton M, Chartier-Harlin MC, Shears S, Roehl K, Booth J, Van Voorst W, Ramic D, Williams J, Goate A, Hardy J, Owen MJ (1999) A full genome scan for late onset Alzheimer’s disease. Hum Mol Genet 8:237–245 [DOI] [PubMed] [Google Scholar]
- Kong A, Cox NJ (1997) Allele-sharing models: LOD scores and accurate linkage tests. Am J Hum Genet 61:1179–1188 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levy-Lahad, E, Wasco W, Poorkaj P, Romano DM, Oshima J, Pettingell WH, Yu CE, Jondro PD, Schmidt SD, Wang K, Crowley AC, Fu YH, Guenette SY, Galas D, Nemens E, Wijsman EM, Bird TD, Schellenberg GD, Tanzi RE (1995) Candidate gene for the chromosome 1 familial Alzheimer’s disease locus. Science 269:973–977 [DOI] [PubMed] [Google Scholar]
- Li Y-J, Scott WK, Hedges DJ, Zhang F, Gaskell PC, Nance MA, Watts RL, et al (2002) Age at onset in two common neurodegenerative diseases is genetically controlled. Am J Hum Genet 70:985–993 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKhann G, Drachman D, Folstein M (1984) Clinical diagnosis of Alzheimer’s disease: report of the NINCDS-ADRDA Work Group under the auspices of the Department of Health and Human Services Task Force on Alzheimer’s Disease. Neurology 34:939–944 [DOI] [PubMed] [Google Scholar]
- Mirra SS, Heyman A, McKeel D, Sumi SM, Crain BJ, Brownlee LM, Vogel FS, Hughes JP, Van Belle G, Berg L (1991) The consortium to establish a registry for Alzheimer’s disease (CERAD). II. Standardization of the neuropathologic assessment of Alzheimer’s disease. Neurology 41:479–486 [DOI] [PubMed] [Google Scholar]
- Myers A, De Vrieze FW, Holmans P, Hamshere M, Crook R, Compton D, Marshall H, et al (2002) Full genome screen for Alzheimer disease: stage II analysis. Am J Med Genet 114:235–244 [DOI] [PubMed] [Google Scholar]
- Olson JM, Goddard KAB, Dudek DM (2001) The amyloid precursor protein locus and very-late-onset Alzheimer disease. Am J Hum Genet 69:895–899 [DOI] [PMC free article] [PubMed] [Google Scholar]
- ——— (2002) A second locus for very-late-onset Alzheimer disease: a genome scan reveals linkage to 20p and epistasis between 20p and the amyloid precursor protein region. Am J Hum Genet 71:154–161 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pericak-Vance MA, Bass MP, Yamaoka LH, Gaskell PC, Scott WK, Terwedow HA, Menold MM, Conneally PM, Small GW, Vance JM, Saunders AM, Roses AD, Haines JL (1997) Complete genomic screen in late-onset familial Alzheimer disease: evidence for a new locus on chromosome 12. JAMA 278:1237–1241 [PubMed] [Google Scholar]
- Pericak-Vance MA, Bebout JL, Gaskell PC Jr, Yamaoka LH, Hung W-Y, Alberts MJ, Walker AP, Bartlett RJ, Haynes CA, Welsh KA, Earl NL, Heyman A, Clark CM, Roses AD (1991) Linkage studies in familial Alzheimer disease: evidence for chromosome 19 linkage. Am J Hum Genet 48:1034–1050 [PMC free article] [PubMed] [Google Scholar]
- Pericak-Vance MA, Grubber J, Bailey LR, Hedges D, West S, Kemmerer B, Hall JL, Saunders AM, Roses AD, Small GW, Scott WK, Conneally PM, Vance JM, Haines JL (2000) Identification of novel genes in late-onset Alzheimer disease. Exp Gerontol 35:1343–1352 [DOI] [PubMed] [Google Scholar]
- Pericak-Vance MA, Johnson CC, Rimmler JB, Saunders AM, Robinson LC, D’Hondt EG, Jackson CE, Haines JL (1996) Alzheimer’s disease and apolipoprotein E-4 allele in an Amish population. Ann Neurol 39:700–704 [DOI] [PubMed] [Google Scholar]
- Rimmler J, McDowell JG, Slotterback BD, Haynes CS, Menold MM, Rogala A, Speer MC, Gilbert JR, Hauser ER, Vance JM, Pericak-Vance MA (1998) Development of a data coordinating center (DCC): data quality control for complex disease studies. Am J Hum Genet Suppl 63:A240 [Google Scholar]
- Saunders AM, Strittmatter WJ, Schmechel D, George-Hyslop PH, Pericak-Vance MA, Joo SH, Rosi BL, Gusella JF, Crapper-MacLachlan DR, Alberts MJ (1993) Association of apolipoprotein E allele epsilon 4 with late-onset familial and sporadic Alzheimer’s disease. Neurology 43:1467–1472 [DOI] [PubMed] [Google Scholar]
- Shao Y, Cuccaro ML, Hauser ER, Raiford KL, Menold MM, Wolpert CM, Ravan SA, Elston L, Decena K, Donnelly SL, Abramson RK, Wright HH, DeLong GR, Gilbert JR, Pericak-Vance MA (2003) Fine mapping of autistic disorder to chromosome 15q11-q13 by use of phenotypic subtypes. Am J Hum Genet 72:539–548 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sherrington R, Froelich S, Sorbi S, Campion D, Chi H, Rogaeva EA, Levesque G, Rogaev EI, Lin C, Liang Y, Ikeda M, Mar L, Brice A, Agid Y, Percy ME, Clerget-Darpoux F, Piacentini S, Marcon G, Nacmias B, Amaducci L, Frebourg T, Lannfelt L, Rommens JM, St George-Hyslop PH (1996) Alzheimer’s disease associated with mutations in presenilin 2 is rare and variably penetrant. Hum Mol Genet 5:985–988 [DOI] [PubMed] [Google Scholar]
- Sherrington R, Rogaev EI, Liang Y, Rogaeva EA, Levesque G, Ikeda M, Chi H, et al (1995) Cloning of a gene bearing missense mutations in early-onset familial Alzheimer’s disease. Nature 375:754–760 [DOI] [PubMed] [Google Scholar]
- Strittmatter WJ, Saunders AM, Schmechel D, Pericak-Vance M, Enghild J, Salvesen GS, Roses AD (1993) Apolipoprotein E: high-avidity binding to β-amyloid and increased frequency of type 4 allele in late-onset familial Alzheimer disease. Proc Natl Acad Sci USA 90:1977–1981 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vance JM, Ben Othmane K (1998) Methods of genotyping. In: Haines JL, Pericak-Vance MA (eds) Approaches to gene mapping in complex human diseases. Wiley-Liss, New York, pp 213–228 [Google Scholar]
- Xu P-T, Gilbert JR, Walters SN, Browning C, Desombre KA, Nicodemus K, Scott WK, Haines JL, Pericak-Vance MA (2002) Characterization and analyses of AD candidate genes P16 and MTAP on chromosome 9p21. Am J Hum Genet Suppl 71:A490 [Google Scholar]
- Yang Y, Hentati A, Deng HX, Dabbagh O, Sasaki T, Hirano M, Hung WY, Ouahchi K, Yan J, Azim AC, Cole N, Gascon G, Yagmour A, Ben-Hamida M, Pericak-Vance M, Hentati F, Siddique T (2001) The gene encoding alsin, a protein with three guanine-nucleotide exchange factor domains, is mutated in a form of recessive amyotrophic lateral sclerosis. Nat Genet 29:160–165 [DOI] [PubMed] [Google Scholar]