Abstract
Supernumerary marker chromosomes (SMCs) of chromosome 15, designated “SMC(15)s,” are the most common SMC in humans, accounting for as much as 60% of all those observed. We report the characterization of 46 large SMC(15)s, using both fluorescence in situ hybridization and polymerase chain reaction analysis within and distal to the Prader-Willi/Angelman syndrome critical region (PWACR). Our aim was to establish detailed information on origin, content, and breakpoints, to address the formation of SMC(15)s, and to facilitate genotype-phenotype correlations. For all patients in whom we were able to establish the parental origin, the SMC(15)s were maternally derived. Two patients were observed who had familial SMC(15)s, both inherited from the mother; however, in all remaining patients for whom parental samples were available, the SMC(15)s were shown to have arisen de novo. With one exception, all the SMC(15)s were shown to include the entire PWACR. Detailed investigations of the distal breakpoints categorized the SMC(15)s into two groups. Group A, representing approximately two-thirds of the SMC(15)s, had a breakpoint beyond the standard distal PWS/AS deletion breakpoint BP3, at a position close to the microsatellite marker D15S1010 and the bacterial artificial chromosome 10I10. The group B SMC(15)s were shorter, with more variable breakpoints located around BP3. The majority of the SMC(15)s were shown to have asymmetrical breakpoints, with the two inverted arms of the SMC being unequal in length. Our study revealed an unexpected level of complexity and heterogeneity among SMC(15)s that is not seen in other chromosome 15 rearrangements, such as deletions and duplications. This suggests that multiple mechanisms are involved in the formation of large SMC(15)s.
Introduction
Supernumerary marker chromosomes (SMCs) occur at a frequency of 0.3 per 1,000 live births (Buckton et al. 1985), and ∼60% of all SMCs are derived from chromosome 15 and are designated “SMC(15)s” (Blennow et al. 1994). SMC(15)s were first described by Van Dyke et al. (1977) and consist of two inverted copies of the short arm, centromere(s), and proximal long arm of chromosome 15. The majority of SMC(15)s are dicentric, with one centromere inactivated, and are also referred to as “pseudodicentric chromosome 15” or “inv dup(15).” By conventional cytogenetics, they can be classified into two main groups: (1) small SMC(15)s, which are metacentric chromosomes without euchromatic material; and (2) large SMC(15)s, which are acrocentric chromosomes containing two copies of the 15q11-q13 region. Small SMC(15)s can be familial or de novo and are not directly associated with an abnormal phenotype. In contrast, large SMC(15)s are almost always de novo in origin and maternally derived and are associated with an abnormal phenotype that includes severe mental retardation, developmental delay, behavioral problems, and seizures (Leana-Cox et al. 1994; Crolla et al. 1995).
In addition to SMC(15)s, the 15q11-q13 region is prone to a variety of other structural rearrangements, including deletions, interstitial duplications, and triplications. Deletions on the paternally inherited chromosome cause Prader-Willi syndrome (PWS [MIM 176270]), and maternally derived deletions result in Angelman syndrome (AS [MIM 105830]). The majority of deletions and interstitial duplications are uniform in size, covering the ∼4-Mb Prader-Willi/Angelman syndrome critical region (PWACR), and have tightly clustered breakpoints (Knoll et al. 1989; Repetto et al. 1998; Roberts et al. 2002). There are two common proximal breakpoints, BP1 and BP2, defining class I and class II rearrangements, and a single common distal breakpoint, BP3 (Knoll et al. 1989; Kuwano et al. 1992; Christian et al. 1995). The common deletion breakpoints, BP1 and BP2, are also found in small SMC(15)s (Huang et al. 1997).
In contrast, previous studies of large SMC(15)s identified multiple distal breakpoints that frequently extended beyond BP3. The study by Wandstrat et al. (1998) demonstrated two types of large inv dup(15), one with a distal breakpoint equivalent to the common breakpoint BP3, and a second type with a more distal breakpoint close to D15S1010. Mignon et al. (1996) and Webb et al. (1998) also described large SMC(15)s with distal breakpoints located at BP3, as well as at other sites distal to the PWACR. Although large SMC(15)s are defined by inclusion of the PWACR, the location of the distal breakpoint appears to be much more variable than that associated with other structural rearrangements involving the PWACR. However, since the studies described above used different probes and PCR loci, it is difficult to achieve a clear consensus of where the distal breakpoints lie.
We report the detailed molecular and FISH analysis of 46 large SMC(15)s, including inv dup(15)s, supernumerary ring chromosomes 15 and SMC(15)s with more-complex characteristics and composition. Therefore, the present study is, to our knowledge, the largest and most comprehensive of its kind to date. Our principal aim was to characterize the distal breakpoints of the SMC(15)s as precisely as possible and to address their mechanism of formation. Individuals with SMC(15)s containing the PWACR exhibit highly variable clinical phenotypes, and detailed characterization of these large SMC(15)s may help in determining genotype/phenotype correlations. The results of detailed psychometric and clinical genetic evaluations of the majority of the patients presented here are currently being analyzed and will be reported separately.
Subjects and Methods
Subjects
Information about all the subjects is summarized in table 1. A total of 46 patients with SMC(15) were ascertained either directly through the Wessex Regional Genetics Laboratory or by referral via a number of collaborators and a patient support group called “Unique.” The study of these patients was approved by the South West Multicentre Research Ethics Committee, United Kingdom. Both DNA and FISH analyses were possible for 27 patients (A1–A18, B1–B3, and B5–B10) from whom peripheral blood samples were received. Buccal smear samples were received from a further 18 patients (A19–A32 and B11–B14), restricting analysis to PCR. A cytogenetic cell suspension for FISH analysis was available for only one patient (B4). In total, PCR analysis was possible for 45 patients, and results of FISH were available for 28. Six patients, A2 (case 3), A6 (case 4), A28 (case 1), B6 (case 5), B2 (case 2) (Crolla et al. 1995), and case A17 (Maggouta et al. 2003) have been reported elsewhere, and only additional molecular studies relating to these patients are reported here. DNA samples were available from both parents of 32 patients, from only the mother of 11 patients, and from neither parent of 3 patients.
Table 1.
Genetic Information about Subjects with SMC(15)
| Subject | Conventional Karyotype | S/AS/Ringa | ParentalOriginb | Inheritanceb | Mosaic |
| A1 | 47,XY,+idic(15) (pter→q13::q13→pter) | S | M | de novo | N |
| A2 | 47,XY,+idic(15)(q12::q12)[49]/46,XY[11] | S | U | U | Y |
| A3 | 47,XX,+psu dic(15)(pter→q15::?q11→p11:) | AS | M | de novo | N |
| A4 | 47,XY,+idic(15)(pter→q11.2::q11.2→pter) | S | M | de novo | N |
| A5 | 47,XX,+idic(15)(pter→q13::q13→pter) | S | M | de novoc | N |
| A6 | 47,XX,+idic(15)(pter→q13::q13→pter) | S | M | de novo | N |
| A7 | 46,XY/47,XY+dic(15) | U | M | de novoc | N |
| A8 | 47,XY,+idic(15)(pter→q12::q12→pter) | S | M | de novo | N |
| A9 | 47 XX,+idic(15) (pter-q12::q12-pter) | S | U | de novo | N |
| A10 | Conventional karyotype not available | U | M | de novoc | N |
| A11 | 47,XY,+idic(15)(pter→q13::q13→pter) | S | Md | U | N |
| A12 | 47,XY,+idic(15)(pter→q12::q12→pter) | S | M | de novo | N |
| A13 | 47,XX,+idic(15)(pter→q13::q13→pter) | S | M | de novoc | N |
| A14, Proband | 47,XY,+psu dic(15)(pter→q13::q11.2→pter) | AS | Md | Familial | N |
| A14, Mother | 47,XX,+psu dic(15)(pter→q13::q11.2→pter) | AS | Md | U | N |
| A14, Uncle | 47,XY,+psu dic(15)(pter→q13::q11.2→pter) | AS | Md | U | N |
| A15 | 47,XX,+mar.ish r(15)[12]/46,XX[18] | R | M | de novo | Y |
| A16 | 47,XX,+mar.ish r(15)[20]/46,XX[10] | R | U | de novo | Y |
| A17 | 47 XY,+der(15)[21]/46 XY[9] | S | M | de novo | Y |
| A18 | 47,XY,+idic(15)(pter→q13::q13→pter) | S | U | de novo | N |
| A19 | 47,XY,+idic(15)(pter→q13::q13→pter)[21]/46,XY[9] | S | Md | de novo | N |
| A20 | 47,XY,+idic(15)(pter→q13::q13→pter) | S | M | de novoc | N |
| A21 | 47,XY,+idic(15)(pter→q13::q13→pter) | S | M | de novoc | N |
| A22 | 47,XX,+idic(15)(pter→q13::q13→pter) | S | M | de novoc | N |
| A23 | 47,XY,+psu dic(15)(pter→q11::q13→pter) | AS | M | de novoc | N |
| A24 | 47,XY,+idic(15)(pter→q12::q12→pter) | S | U | U | N |
| A25 | 47,XY,+idic(15)(pter→q12::q12→pter) | S | M | de novoc | N |
| A26 | 47,XX,+idic(15)(pter→q13::q13→pter) | S | M | de novo | N |
| A27 | 47,XX,+idic(15)(pter→q12::q12→pter) | S | U | de novo | N |
| A28 | 47,XY,+idic(15)(pter→q11::q11→pter)[20]/48,XY,+idic(15),+idic(15)[18] | S | M | de novo | Y |
| A29 | 46,XX,inv(4)(p16q12)[10]/47,XX,inv(4)(p16q12)+idic(15)(q12q12)[20] | S | Md | U | Y |
| A30 | 47,XX,+idic(15)(pter→q13::q13→pter) | S | Md | U | N |
| A31 | 47,XX,+idic(15)(pter→q13::q13→pter) | S | M | U | N |
| A32 | 47,XY,+idic(15)(pter→q12::q12→pter) | S | M | de novoc | N |
| B1 | 47,XX,+idic(15)(pter→q12::q12→pter) | S | U | U | N |
| B2 | 47 XY,+psu dic(15)(pter→q11::q13→pter) | AS | M | de novo | N |
| B3 | 47,XY,+idic(15)(pter→q11::q11→pter)[49]/46,XY[11] | S | Md | de novo | Y |
| B4 | 47,XY,+idic(15)(pter→q11::q11→pter) | S | U | de novo | N |
| B5 | Conventional karyotype not available | U | M | de novoc | N |
| B6 | 47 XX,+psu dic(15)(pter→q11::q13→pter) | AS | M | de novo | N |
| B7 | 47,XY,+idic(15)(pter→q13::q13→pter) | S | M | de novoc | N |
| B8 | 47,XY,+idic(15)(pter→q13::q13→pter) | S | M | de novoc | N |
| B9 | 48 XY. SMC(15) x2 inv dup(15) (pter-q12::q12-pter) | S | M | de novoc | N |
| B10, Proband | 47,XX,+idic(15)(pter→q12::q12→pter) | S | M | Familial | N |
| B10, Mother | 47,XX,+idic(15)(pter→q12::q12→pter)[23]/46,XX[7] | S | Md | de novo | Y |
| B11 | 47,XX,+idic(15)(pter→q13::q13→pter) | S | M | de novoc | N |
| B12 | 47,XX,+idic(15)(pter→q12::q12→pter) | S | M | de novoc | N |
| B13 | 47 XX,+psu dic(15)(pter→q11::q12→pter) | AS | U | U | N |
| B14 | 47XY, +psu dic(15)(pter-q12::q12-q11.1::q11.2-pter) | AS | M | de novo | N |
AS = asymmetrical from given cytogenetic breakpoints; R = ring(15); S = symmetrical from given cytogenetic breakpoints.
M = maternal; U = unknown.
No evidence available that parents have been examined cytogenetically. Inheritance established from PCR results only.
Parental origin established from methylation-specific PCR only.
Cytogenetic Techniques
Cytogenetic analysis was performed on 17 samples at the Wessex Regional Genetics Laboratory. Karyotypes were determined by analysis of G-banded metaphase chromosomes harvested from peripheral blood lymphocytes. The karyotypes of the remaining patients were obtained from collaborating centers. In three patients known to carry an SMC(15), we were unable to trace the original laboratory report.
FISH Techniques
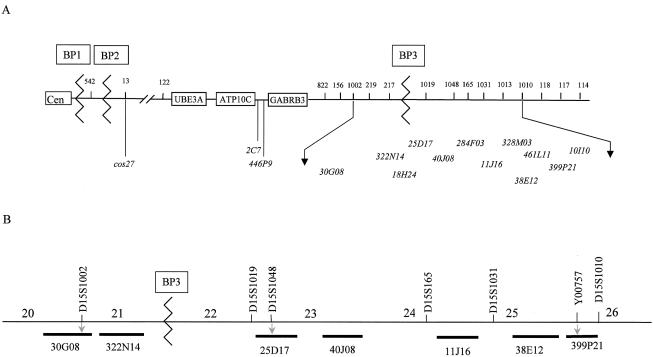
Chromosome preparations were made from peripheral blood cultures, using standard methods. FISH was performed according to the method described by Pinkel et al. (1988), with slight modifications. Nick-translated biotin or digoxigenin-labeled BAC and cosmid probes used in this study are shown in figure 1. FISH slides were analyzed using a Zeiss Axioskop microscope with a cooled charged-coupled-device camera (Photometrics) and Applied Imaging MacProbe software. FISH was performed on metaphase chromosomes of 28 patients who had large SMC(15)s. BACs were chosen to span the regions immediately proximal and distal to breakpoint BP3, using information in the Whitehead database (fig. 1). However, the exact relative order of the BACs is not known, and the order given is taken directly from the Ensembl maps (release 11.31.1). Additional FISH probes used in the present study are pTRA25 (centromere 15–specific) (Choo et al. 1990) and cos27 (D15S13). Probe cos27 was used to confirm the presence of the PWACR in the SMCs.
Figure 1.
BAC and microsatellite positions. A, Schematic map of chromosome 15q, showing the relative positions of the BAC probes (shown beneath the line) and microsatellite markers used in the FISH and PCR analysis. The numbers shown above the line are the microsatellite markers, which, for brevity, are listed without the preceding “D15S.” The exact orders of both the BACs and microsatellite markers are not known, because information is unavailable for several of those used. Genes are boxed (UBE3A [MIM 601623] and GABRB3 [MIM 137192]). B, More detailed BAC and microsatellite marker positions, as given in Ensembl release 11.31.1 (February 27, 2003). The distance (expressed as Mb from pter) is shown at the top. The BACs shown at the bottom (dark lines) are those that have exact positions defined in Ensembl. Electronic information on the position of 399P21 is not available, because this BAC has become “buried,” but it contains the STS Y00757. BACs that are PCR positive for the respective microsatellite markers are indicated (gray downward arrows).
Molecular Techniques
PCR
DNA was extracted from peripheral blood and buccal cells by a salt precipitation technique (Miller et al.1988). PCR was performed under standard conditions with primers spanning proximal 15q (see fig. 1). All primers and conditions are available from the Genome Database. A total of 45 large SMC(15)s were characterized using microsatellite markers at intervals throughout 15q11-q14. The relative order of the molecular markers was based on the Ensembl (version 11.31.1, February 27, 2003) and Entrez maps (Genethon, 1996, and DeCode, July 2002) (see fig. 1). The positions of the microsatellite markers with respect to the BACs is not precisely defined, but figure 1B shows all the currently available information gained both from electronic resources and from our own PCR analysis using the microsatellite markers and BACs. A quantitative PCR technique (Roberts and Thomas 2003) was also used in one patient to establish the increased copy number of the PWACR. When parental DNA was unavailable, the origin of the SMC(15)s was determined using methylation-specific PCR (Zeschnigk et al. 1997).
Methylation-Specific PCR Assay
Alleles were scored on the basis of the number of distinct peaks seen and on the basis of their relative dosage as measured by the peak heights. A breakpoint is defined by a change in copy number between adjacent loci (i.e., four copies to three or two copies, or three copies to two copies). Breakpoints could not always be precisely defined, because of shared alleles in both parents or a lack of parental DNA.
Results
Cytogenetics
The cytogenetic results, if known, are shown in table 1. There is a high degree of variability in the use of cytogenetic nomenclature used to describe SMC(15)s. The majority are described with apparently symmetrical breakpoints (e.g., pter→q12::q12→pter or pter→q13::q13→pter) although others are described with asymmetrical breakpoints (e.g., pter→q12::q13→pter). This variability reflects the difficulty of accurately defining the content of these SMC(15)s using cytogenetic techniques alone. For ease of description, in table 1 the SMC(15)s are described as either symmetrical or asymmetrical, and the breakpoints are given if they are known. Eight cases were reported to be mosaic (see table 1).
Origins of the SMC(15)s
Of the 46 probands with SMC(15), 27 were analyzed using both PCR and FISH, 18 by PCR only, and 1 by FISH only. Two patients (A14 and B10) were seen with familial large SMC(15)s, both maternally transmitted. In one case (A14) the original mutation could not be traced, but in the other case (B10) the SMC(15) was shown to have arisen de novo in the proband’s maternal grandmother. When cytogenetic and PCR results were combined, a total of 37 of the SMC(15)s (including that seen in the mother of patient B10) were shown to have arisen de novo (see table 1). In the remaining eight patients, the inheritance could not be established because of a lack of parental or proband DNA.
Analysis of the allele inheritance patterns and/or methylation-specific PCR and parental cytogenetic analyses showed that, in 37 patients, the SMC(15) had been derived maternally (see table 1). The origin in the remaining nine patients could not be determined because of unavailability of parental DNA and/or insufficient proband DNA.
Distal Breakpoint Mapping: FISH Studies
FISH with the use of the probe cos27 (D15S13) (figure 1) confirmed the presence of the PWACR in the SMC(15)s. Twenty-four SMC(15)s contained two copies of cos27; however, only a single copy was found in three SMC(15)s, and four copies were seen in one patient (tables 2 and 3). FISH analysis with the use of the BAC probes divided the SMC(15)s into two groups, according to the location of their most-distal breakpoint (table 2 [group A] and table 3 [group B]). Eighteen SMC(15)s (A1–A18) (group A) were positive (i.e., gave at least one signal) for the panel of FISH probes 30G08 through to 399P21. No FISH signal was found in this group of SMC(15)s with the use of probe 10I10, indicating that these 18 SMC(15)s share a common breakpoint distal to the PWACR between BAC 399P21 and BAC 10I10.
Table 2.
Results of FISH in Patients A1–A10 with an SMC(15)[Note]
| Probea | A1 | A2 | A3 | A4 | A5 | A6 | A7 | A8 | A9 | A10b | A11 | A12 | A13c | A14 | A15b | A16b | A17b | A18b |
| cos27 | ++ | ++ | ++ | ++ | ++ | ++ | ++ | ++ | ++ | + | ++ | ++ | ++ | + | ++ | ++ | ++++ | ++ |
| 30G08 | ++ | ++ | ++ | ++ | ++ | ++ | ++ | ++ | ++ | ++ | ++ | ++ | ++ | + | ++ | ++ | ++++ | ++ |
| 322N14 | ++ | ++ | ++ | ++ | ++ | ++ | ++ | ++ | ++ | ++ | ++ | ++ | ++ | + | ++ | ++ | ++++ | ++ |
| 18H24 | + | + | + | + | + | ++ | ++ | ++ | ++ | ++ | ++ | ++ | ++ | + | + | ++ | ++++ | ++ |
| 25D17 | + | + | + | + | NT | + | + | ++ | ++ | ++ | ++ | ++ | ++ | + | + | ++ | ++ | ++ |
| 40J08 | + | + | + | + | NT | + | + | + | + | + | + | + | + | + | + | + | NT | + |
| 284F03 | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | NT | + |
| 11J16 | + | + | + | + | NT | + | + | + | + | + | + | + | ++ | + | + | + | NT | + |
| 328M03 | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | ++ | + |
| 38E12 | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | ++ | + |
| 461L11 | + | + | + | + | NT | + | + | + | + | + | + | + | + | + | + | + | ++ | + |
| 399P21 | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | ++ | + |
| 10I10 | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − |
Note.— + = single FISH signal on the SMC(15); ++ = double FISH signal on the SMC(15); − = no FISH signal on the SMC(15); NT = not tested.
Probes (BACs) are shown in map order from centromere (top) to telomere (bottom).
Exceptional cases.
Two signals using BAC 11J16 compared with one signal using more-proximal BACs. The exact order of these BACs is not known and may explain this anomaly.
Table 3.
Results of FISH in Patients B1–B10 with an SMC(15)[Note]
| Probea | B1 | B2 | B3 | B4 | B5 | B6 | B7 | B8 | B9b | B10b |
| cos27 | ++ | ++ | ++ | ++ | ++ | ++ | ++ | ++ | + | ++ |
| 2C7 | NT | NT | NT | NT | NT | NT | NT | NT | NT | + |
| 446P9 | NT | NT | NT | NT | NT | NT | NT | NT | NT | + |
| 30G08 | ++ | ++ | ++ | ++ | ++ | ++ | ++ | ++ | + | − |
| 322N14 | + | ++ | ++ | ++ | ++ | ++ | ++ | ++ | + | − |
| 18H24 | − | − | − | − | + | ++ | + | + | − | − |
| 25D17 | − | − | − | − | − | + | + | + | − | − |
| 40J08 | − | − | − | − | − | − | − | − | − | − |
| 284F03 | − | − | − | − | − | − | − | − | − | − |
| 11J16 | − | − | − | − | − | − | − | − | − | − |
| 328M03 | − | − | − | − | − | − | − | − | − | − |
| 38E12 | − | − | − | − | − | − | − | − | − | − |
| 461L11 | − | − | − | − | − | − | − | − | − | − |
| 399P21 | − | − | − | − | − | − | − | − | − | − |
| 10I10 | − | − | − | − | − | − | − | − | − | − |
Note.— + = single FISH signal on the SMC(15); ++ = double FISH signal on the SMC(15); − = no FISH signal on the SMC(15); NT = not tested.
Probes (BACs) are shown in map order from centromere (top) to telomere (bottom).
Exceptional cases.
FISH analysis suggested that the remaining 10 SMC(15)s (B1–B10) fell into a second group (designated “group B”). The duplicated region is shorter than that seen in group A and shows a more variable distribution. Nine appear to lie within BP3 (i.e., between either BACs 322N14 and 18H24, 18H24 and 25D17, or 25D17 and 40J08, as seen in patients with deletion and duplication) (Christian et al. 1998). The one exception in this group—patient B10, who had a familial SMC(15)—has a more-proximal breakpoint.
Distal Breakpoint Mapping: PCR Results in Group A
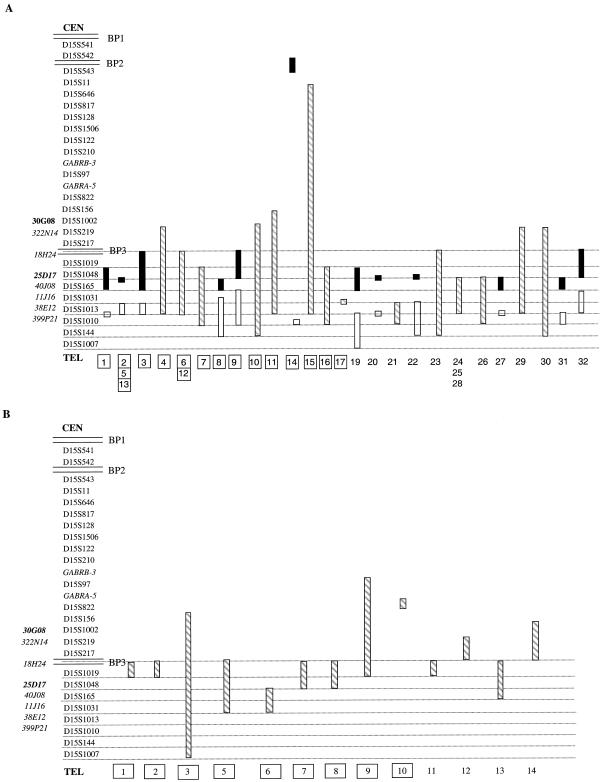
Within the PWACR, four alleles could be seen in all but two patients, A14 and A18. The positions of the breakpoints are shown graphically in figure 2 for the 44 SMC(15)s analyzed using PCR. One patient (A18) did not show any additional alleles at any loci tested within or distal to the PWACR and is therefore not represented in figure 2. In the 44 patients, PCR analysis supported the FISH classification of the SMC(15)s into the two groups. Those patients analyzed using both techniques show agreement in the position of the breakpoints although PCR analysis shows wider ranging results, with some degree of overlap because of uninformative microsatellite markers. In 17 of the group A patients also analyzed by FISH, additional alleles were present at microsatellite markers distal to BP3 (see fig. 2A). A further 14 patients (not analyzed by FISH) were also found to fall into group A (A19–A32). Of the 31 group A patients in whom additional alleles were seen by PCR, 17 had a breakpoint between D15S1013 and D15S1010, and 1 had a breakpoint between D15S1031 and D15S1013 (patient A17). In the remaining 13 patients, the most-distal breakpoint could not be precisely defined. However, none of these remaining patients were positive for D15S1010 or negative for D15S1013.
Figure 2.
PCR results for 44 SMC(15)s (DNA was unavailable for B4, and A18 has been omitted because PCR analysis was completely uninformative). The microsatellite markers are shown in order from the centromere to telomere, with the double lines showing the positions of the common proximal (BP1 and BP2) and distal (BP3) breakpoints seen in patients with deletion/duplication. The positions of some of the BACs are shown on the left of the microsatellite markers. Those shown in boldface are known to contain the adjacent microsatellite marker, whereas the exact positions of those shown in italics are not known. For some individuals (gray hatched bar), we cannot differentiate between one or two breakpoints within this region, because microsatellite markers were uninformative. Proximal to the bar, the copy number is four (except in patients A10, A14, and A17), and, distal to the bar, the copy number is two. For SMCs with two breakpoints the black bar represents the region where the first breakpoint occurs (i.e., the copy number is reduced from four to three), and the white bar represents the second breakpoint (i.e., the copy number is reduced from three to two). The patient numbers are shown at the bottom, with the boxes showing the patients that were also analyzed using FISH. A, Group A patients. B, group B patients. The breakpoints of some of the group B patients appear to be similar to those in group A, but this is because uninformative microsatellite markers make it impossible to define the breakpoints.
Distal Breakpoint Mapping: PCR Results in Group B
Overall, PCR analyses confirmed that the extent of the duplicated region in 8 of the 10 group B patients was shorter than that observed in the group A patients, consistent with the FISH results in these same patients. Of the remaining two patients, one (B3) could not be accurately mapped because several microsatellite markers were uninformative, and, in patient B4, there was insufficient DNA available. A further four patients (B11–B14) were classed in group B, with three or more alleles observed for loci up to and including D15S217 but with no evidence of additional alleles distal to D15S1019 (see fig. 2B), although, in patient B13, a number of microsatellite markers beyond BP3 were uninformative. Although it was not possible to precisely define the breakpoints of the 13 group B patients, 8 were consistent with a breakpoint between D15S217 and D15S1019, equivalent to BP3, with no positive loci distal to D15S1010. Three patients (B5, B6, and B8) had additional alleles present at markers distal to the BP3 region, but this additional material did not extend as distally as that in the group A patients. The breakpoint in the familial case B10 was confirmed to be within the PWACR between the microsatellite markers GABRA5 and D15S156.
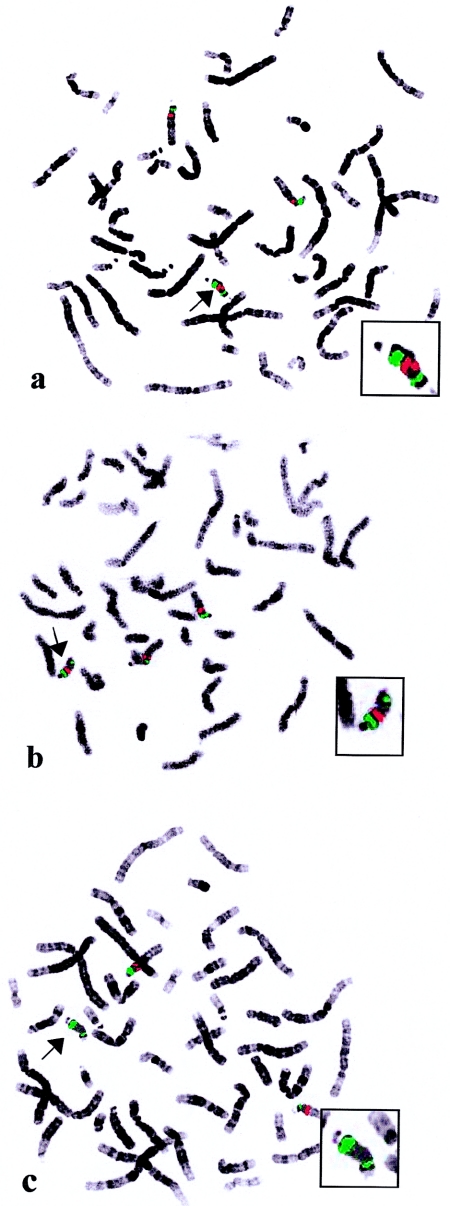
FISH and PCR Reveals Asymmetry of the Group A SMC(15)s
FISH analysis suggested that all of the SMC(15)s in group A are asymmetrical, containing one copy of the more distally located BACs, compared with two copies of the more-proximal BACs. This indicates that the two arms of the SMC(15) have different (asymmetrical) breakpoints—with one located at or very close to BP3, and the other one located between 399P21 and 10I10 (table 2). Figure 3 shows examples of metaphase chromosomes (patient A1) hybridized with probes 322N14 (fig. 3a), 18H24 (fig. 3b), and 10I10 (fig. 3c), respectively. Probe 322N14 shows two signals on the SMC(15) compared with one signal resulting from probe 18H24. No signal was observed on the SMC(15) when the probe 10I10 was used, a result indicating that this probe lies distal to the breakpoint. Interestingly, the more-proximal breakpoint observed in the group A patients seems to be equivalent to the breakpoint (BP3) seen in the majority of the group B patients.
Figure 3.
Representative examples of FISH on peripheral blood metaphase chromosomes of patient A1, reflecting the asymmetry observed in most of the SMC(15)s analyzed. BACs are labeled with digoxigenin (red), and the plasmid probe pTRA-25 (chromosome 15 centromere) is labeled with biotin (green). Enlargements of the SMC(15)s are shown in the insets. Arrows indicate the SMC(15)s. BAC 322N14 reveals two signals on the SMC (a), and a single signal results from BAC 18H24 (b). No signal is observed with BAC 10I10 (c).
PCR analyses of the group A SMC(15)s confirms this asymmetry. There are 14 group A patients in whom the allele copy number is only three instead of the expected four at some of the most distal markers beyond BP3 (figure 2A). In nearly all of the remaining group A patients (16 of the remaining 18), three or four alleles could not be differentiated between at one or more of the distal loci, so asymmetry in these patients cannot be confirmed or refuted.
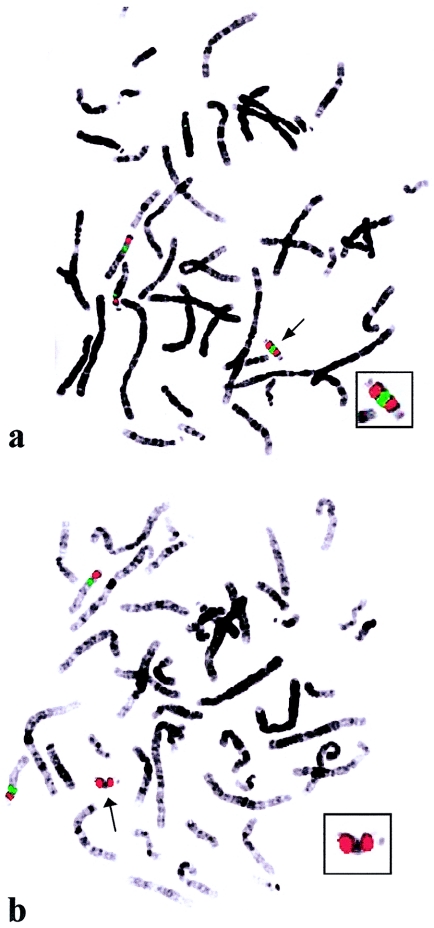
In contrast to group A, at least some group B patients appear to be symmetrical, with both breakpoints at BP3. For example, by FISH, the SMC(15)s B2, B3, and B4 contain two signals with BAC 322N14 but no signals with BAC 18H24 (fig. 4). The remaining group B patients show a single FISH signal but with only one or two BACs (table 3). None of the patients assigned to group B show any evidence of asymmetry by PCR. The BACs that show a single signal in this region may span the breakpoint, and the single FISH signal observed may be two smaller split signals that have merged because of their proximity. Alternatively, the region of asymmetry observed by use of FISH may not be covered by the molecular markers. The locus D15S1048 does not show asymmetry in any SMC although it is present in the BAC 25D17, which shows only one hybridization signal when FISH is used. The region of asymmetry may begin just distal to D15S1048 within the sequence contained in the BAC 25D17.
Figure 4.
Representative examples of FISH on peripheral blood metaphase chromosomes of patient B3, reflecting the symmetry in some of the SMC(15)s analyzed. BACs are labeled with biotin (green), and the plasmid probe pTRA-25 (chromosome 15 centromere) is labeled with digoxigenin (red). Enlargements of the SMC(15)s are shown in the insets. Arrows indicate the SMC(15)s. BAC 322N14 shows two signals (merged) on the SMC(15) (a). No signal is visible on the SMC(15) with BAC 18H24 (b).
Exceptional Patients with SMC(15)
In nine of the patients, we demonstrated additional variability in copy number, size, shape, and molecular composition in their SMC(15)s. In patient A14, the SMC(15) has a single copy of the PWACR but two copies at the microsatellite markers D15S541 and D15S542. The SMC(15) is familial, and, at the very proximal end, the two alleles shared by each of the carriers (which we assume are carried on the SMC) are different, suggesting the SMC is interchromosomal in origin and thus arose from two chromosome 15 homologues. The SMC(15) in patient B10 is the only SMC(15) in our study shown to have a breakpoint proximal to BP3 near the ATP10C gene (MIM 605855) (table 3). This patient also has a familial SMC(15), and microsatellite analysis within the PWACR showed the SMC to be intrachromosomal in origin, having arisen from a single chromosome of the transmitting grandmother. In patient A10, the SMC(15) contains only a single copy of cos27 but two copies for the distal BACs 30G08 to 25D17. The SMC(15) in patient A17 contains four copies of the entire PWACR (Maggouta et al. 2003). Patient B9 revealed two identical copies of an SMC(15), with only a single copy of the PWACR on each SMC(15). Two copies of an SMC were also seen cytogenetically in patient A28, with one SMC(15) in 53% of cells and two copies present in the remaining 47% of the cells (Crolla et al. 1995). In two patients (A15 and A16), a ring SMC(15) was observed. The ring SMC(15)s appeared to be unstable, showing different copy numbers of the same probe in different cells, with the majority of the cells revealing the hybridization pattern shown in table 2. Patient 18 appears unexceptional by FISH, but PCR analysis showed this patient has only two alleles and has a normal ratio of peak heights for all loci within the PWACR. We assume this was due to consanguinity.
Discussion
In the present study, we used a series of microsatellite markers and BAC clones from 15q11-q14 to characterize 46 large SMC(15)s containing the PWACR. By using a combination of PCR and FISH, we were able to characterize these SMC(15)s at a level of detail not previously performed, and extensive patient recruitment efforts have given us, to our knowledge, the largest series of these patients described to date. We have shown two distinct groups classified on the basis of their most distal breakpoint, and we have also shown that asymmetry is common, with many of the SMC(15)s showing a second, more-proximal breakpoint. To our knowledge, this is the most comprehensive and detailed study of large SMC(15)s to date, and it has revealed a structural and molecular level of complexity not previously described. The majority of the patients in the present study are currently undergoing detailed psychometric and clinical phenotyping by clinical psychologists and clinical geneticists, respectively. These analyses are being performed by investigators who are blinded to the laboratory data, and correlations between the phenotype and the molecular content of the SMCs will be reported separately.
Of 39 patients in whom the origin could be determined (including the mother of patient B10), 37 of the SMC(15)s were de novo and only two patients were familial. The majority of large SMC(15)s reported elsewhere are also de novo in origin, presumably a consequence of the severe phenotype caused by these rearrangements. It is interesting to note that our two familial cases are not typical SMC(15)s. In patient B10, the SMC is shorter than others in our study and is mosaic in the carrier mother. A previously reported patient with familial SMC(15) was also shown to be mosaic in the carrier mother (Webb et al. 1998). In the other patient (A14), the SMC(15) contains only one copy of the PWACR, an asymmetric variant that has been reported elsewhere (Robinson et al. 1993; Mignon et al. 1996) although not in association with familial inheritance. This variant results in trisomy of the region equivalent to that found in interstitial duplications, of which several familial cases have been reported (Browne et al. 1997; Roberts et al. 2002).
Breakpoints: Asymmetry and Mapping
Our interpretation of two breakpoints in many of the SMC(15)s is based on the FISH and PCR results that both show a region of asymmetry, with FISH detecting a single signal and PCR detecting a transition from a copy number of four to three, and, more distally, from a copy number of three to two. Our results, however, could be interpreted differently. For some loci, allele-peak data from microsatellite marker analysis can be difficult to interpret. The single FISH signal may be due to the BACs containing homologous segments, particularly repeats, all spanning the breakpoint region, as described elsewhere in a similar study using YACs (Wandstrat et al. 1998). Although this may be true for group B, it seems unlikely for group A because we have used several different BACs, all of which are shown to contain different sets of microsatellite markers (see Entrez) and, where information is available, do not have overlapping positions in Ensembl. It is also possible that the BACs are positioned distal to the breakpoint and that the FISH probes are detecting repeat sequences. A single FISH signal is observed on the two normal chromosomes 15, so this seems unlikely, although it is possible that the main FISH signal on the normal chromosomes could obscure an additional small signal if the repeats were positioned close to the main signal sequence. However, given both the FISH and molecular evidence, we conclude that the most likely explanation is that many of these SMC(15)s contain asymmetric breakpoints, with each of the duplicated segments being a different length. To our knowledge, this is the first such breakpoint asymmetry to be described in SMC(15)s, with the exception of two patients reported elsewhere (Robinson et al. 1993; Mignon et al. 1996), which have four copies of the centromeric region but only three copies of the PWACR.
Our detailed breakpoint characterization classified the SMC(15)s into two broad groups based on their most distal breakpoint. The patients in the larger group (group A) represent about 70% of the patients studied here (32/46) and are generally asymmetrical, with two distinct breakpoints. The most distal breakpoint appears to be uniform between the microsatellite markers D15S1013 and D15S1010 and BACs 399P21 and 10I10. This breakpoint, proximal to D15S1010, is likely to be the same as that reported in previous studies of other large SMC(15)s (Mignon et al. 1996; Wandstrat et al. 1998; Webb et al. 1998). It also appears to be the same as the breakpoint, located between D15S165 and D15S144, described in two patients with an interstitial triplication (Ungaro et al. 2001; Roberts et al. 2002). The second breakpoint in these group A patients has a slightly more-proximal location and shows some heterogeneity although, in almost all patients, it is at or very close to BP3, the breakpoint seen in almost all of the group B patients.
The group B SMC(15)s, by comparison, reveal a more heterogeneous distribution of breakpoints. In the majority of patients, the most distal breakpoint is located by PCR between the loci D15S217 and D15S1019 and by FISH between BACs 322N14 and 25D17. This breakpoint cluster, seen in most of the group B patients, is equivalent to BP3 found in patients with PWS/AS deletion (Christian et al. 1998) and in all interstitial duplications reported to date (Roberts et al. 2002). By PCR analysis, most patients appeared to have SMC(15)s that contain symmetrical breakpoints, with a single breakpoint at BP3. However, FISH analysis was less clear, with some patients showing symmetrical breakpoints; however, in other patients, there could be a small segment of asymmetry with two closely mapped, but distinct, distal breakpoints. If the FISH analysis is giving a genuine single signal, then these group B patients are also asymmetrical but with much closer breakpoints than those seen in the group A patients, and there is variation in the position of the distal breakpoint. If, however, the apparently single FISH signal is two small merged signals, then these group B patients are symmetrical and have a uniform breakpoint at BP3 present on both inverted copies of the region.
Overall, our data clearly demonstrate the heterogeneous and complex nature of the breakpoints within large SMC(15)s. This is in line with previous reports of breakpoint heterogeneity (Wandstrat et al. 1998; 2000). All of the group A, and possibly some of the group B, patients appear to have two distal breakpoints, with some variation in breakpoint location. To our knowledge, the present study is the first to show a different breakpoint on each of the additional inverted copies of the PWACR, and this is in contrast to what is seen in patients with deletions and duplications, in whom very little heterogeneity has been observed.
Mechanisms of Formation
In all cases in which we were able to establish parental origin, the SMC(15)s were maternally derived. This is in agreement with all other reported series. To our knowledge, no paternally derived large SMC(15) has been reported. The reasons for this remain unresolved. Nondisjunction is likely to play a role in the formation of all SMC(15)s (Schreck et al. 1977), and, because it occurs at a much greater frequency in females than males, it may go some way toward explaining the predominance of maternally derived SMC(15) (Robinson et al. 1993). Although SMC(15)s can form during spermatogenesis, as is evident from the observation of paternally derived small SMC(15)s (Dawson et al. 2002), studies of infertile male inv dup(15) carriers have provided evidence that, during spermatogenesis, cells carrying an SMC are selected against (Cotter et al. 2000; Eggermann et al. 2002). Alternatively, large SMC(15)s could be lethal if paternally inherited, although paternally inherited interstitial triplications have been described (Cassidy et al. 1996; Ungaro et al. 2001) that have the same PWACR copy number (i.e., tetrasomy) as those in patients with large SMC(15).
Molecular analysis in the two familial patients reported here was able to show the chromosomal origins, one group B SMC being intrachromosomal and one group A SMC being interchromosomal, if we assume that there has been no recombination proximal to the microsatellite markers D15S541 and D15S542. This is in agreement with a previous study, which found that of four SMCs isolated in somatic cell hybrids, two were interchromosomal and two were intrachromosomal (Wandstrat and Schwartz 2000). Combining our data with those of Wandstrat and Schwartz (2000) for both origin and size gives a total of two group B patients, both intrachromosomal, and four group A patients, of which three are interchromosmal and one intrachromosomal. This pattern may suggest that the two groups are likely to form via different mechanisms although, obviously, more patients need to be examined.
The breakpoint regions described here occur at sites of duplicated genomic segments (Christian et al. 1999), suggesting these duplicons are responsible, at least in part, for the increased instability in proximal 15q. These duplicons have been shown to contain at least seven gene/pseudogene sequences, including HERC2 (MIM 605837), MYLE, and a number of unknown transcripts. It is hypothesized that misalignment of these repeated sequences, followed by illegitimate recombination, results in chromosomal rearrangement. The rearrangement formed will depend upon the orientation of the DNA elements and the type of strand exchange involved, that is, whether the event is inter- or intrachromosomal. The heterogeneity observed among the SMC(15)s indicates that different types of misalignment between different repeated regions could be involved in the formation of these rearrangements. The most populartheory about the formation of SMC(15)s is a U-type exchange between homologous chromosomes during meiosis I, followed by illegitimate fusion of the chromatids and nondisjunction (Schreck et al. 1977; Martinson et al. 1996). Another theory proposes that, following premeiotic breakage, a single chromosome replicates and that the “sticky” ends join to form the SMC(15) with identical telomeres (Schreck et al. 1977). Our results show SMC(15)s can be asymmetrical or symmetrical and can also be interchromosomal or intrachromosomal, with different breakpoints. This extensive heterogeneity suggests that both mechanisms are involved in the formation of different SMC(15)s, although the relative importance of each mechanism is likely to vary between the group A and B SMC(15)s. For example, the formation of group A SMC(15) predominantly involves both chromosome 15 homologues, and the resulting breakpoints are asymmetrical; the formation of both group B SMC(15)s investigated was intrachromosomal, and the breakpoints of at least some group B SMC(15) are symmetrical.
Our detailed characterization of the breakpoints of these large SMC(15)s should allow more-detailed comparisons between genotype and phenotype. Current genotype-phenotype correlations are limited to the presence or absence of the PWACR, giving rise to an abnormal or normal phenotype. Our detailed study may allow more-subtle correlations in these large SMC(15)s to be seen although the level of complexity and the breakpoint heterogeneity revealed here may make correlations more difficult.
Acknowledgments
We are very grateful to the patients and their families for agreeing to take part in this research, and we particularly thank the support group Unique. We are also very grateful to Dr. N. R. Dennis and Mrs. Barbara O’Prey for acquisition of patient samples and details. Thanks are also due to Dr. C. Geoffrey Woods (Leeds), Dr. John Wolstenholme, and the late Mr. Ian Cross (Newcastle), for sending patient samples. The BACs were supplied by Dr. Marianno Rocchi (Bari, Italy), Dr. David Ledbetter (Chicago), and the Sanger Centre (Cambridge, United Kingdom). This work is funded by the Medical Research Council, United Kingdom.
Electronic-Database Information
URL for data presented herein are as follows:
- Ensembl Genome Browser, http://www.ensembl.org (for BACs and microsatellite markers)
- Entrez, http://www.ncbi.nlm.nih.gov/entrez (for microsatellite markers)
- Genome Database, http://www.gdb.org (for microsatellite marker primer sequences)
- Online Mendelian Inheritance in Man (OMIM), http://www.ncbi.nlm.nih.gov/Omim/ (for AS, ATP10, HERC2, and PWS
- Unique—Rare Chromosome Disorder Support Group, http://www.rarechromo.org
- Whitehead Institute/MIT Center for Genome Research, http://www-genome.wi.mit.edu (for BACs)
References
- Blennow E, Bui TH, Kristoffersson U, Vujic M, Anneren G, Holmberg E, Nordenskjold M (1994) Swedish survey on extra structurally abnormal chromosomes in 39,105 consecutive prenatal diagnoses: prevalence and characterisation by fluorescence in situ hybridisation. Prenat Diagn 14:1019–1028 [DOI] [PubMed] [Google Scholar]
- Browne CE Dennis NR, Maher E, Long FL, Nicholson JC, Sillibourne J, Barber JCK (1997) Inherited interstitial duplications of proximal 15q: genotype-phenotype correlations. Am J Hum Genet 61:1342–1352 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buckton KE, Spowart G, Newton MS, Evans HJ (1985) Forty-four probands with an additional “marker” chromosome. Hum Genet 69:353–370 [DOI] [PubMed] [Google Scholar]
- Cassidy S, Conroy J, Becker L, Schwartz S (1996) Paternal triplication of 15q11-q13 in a hypotonic, developmentally delayed child without Prader-Willi or Angelman syndrome. Am J Med Genet 62:205–212 [Google Scholar]
- Choo KH, Earle E, Vissel B, Filby RG (1990) Identification of two distinct subfamilies of α satellite DNA that are highly specific for human chromosome 15. Genomics 7:143–151. [DOI] [PubMed] [Google Scholar]
- Christian SL, Bhatt NK, Martin SA, Sutcliffe JS, Kubota T, Huang B, Mutirangura A, Chinault AC, Beaudet AL, Ledbetter DH (1998) Integrated YAC contig map of the Prader-Willi/Angelman syndrome region on chromosome 15q11-q13 with average STS spacing of 35 kb. Genome Res 8:146–157 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christian SL, Fantes JA, Mewborn SK, Huang B, Ledbetter DH (1999) Large genomic duplicons map to sites of instability in the Prader-Willi/Angelman syndrome chromosome region (15q11-q13). Hum Mol Genet 8:1025–1037 [DOI] [PubMed] [Google Scholar]
- Christian SL, Robinson WP, Huang B, Mutiranura A, Line MR, Nakao M, Surti U, Chakravarti A, Ledbetter DH (1995) Molecular characterization of two proximal deletion breakpoint regions in both Prader-Willi and Angelman syndrome patients. Am J Hum Genet 57:40–48 [PMC free article] [PubMed] [Google Scholar]
- Cotter PD, Ko E, Larabell SK, Rademaker AW, Martin RH (2000) Segregation of a supernumerary del(15) marker chromosome in sperm. Clin Genet 58:488–492 [DOI] [PubMed] [Google Scholar]
- Crolla JA, Harvey JF, Sitch FL, Dennis NR (1995) Supernumerary marker 15 chromosomes: a clinical, molecular and FISH approach to diagnosis and prognosis. Hum Genet 95:161–170 [DOI] [PubMed] [Google Scholar]
- Dawson AJ, Mogk R, Rothenmund H, Bridge PJ (2002) Paternal origin of a small, class I inv dup(15). Am J Med Genet 107:334–336 [DOI] [PubMed] [Google Scholar]
- Eggermann K, Mau UA, Bujdoso G, Koltai E, Engels H, Schubert R, Eggermann T, Raff R, Schwanitz G (2002) Supernumerary marker chromosomes from chromosome 15: analysis of 32 new cases. Clin Genet 62:89–93 [DOI] [PubMed] [Google Scholar]
- Huang B, Crolla JA, Christian SL, Wolf-Ledbetter ME, Macha ME, Papenhausen PN, Ledbetter DH (1997) Refined molecular characterisation of the breakpoints in small inv dup(15) chromosomes. Hum Genet 99:11–17 [DOI] [PubMed] [Google Scholar]
- Knoll JHM, Nicholls RD, Magenis RE, Graham JM Jr, Lalande M, Latt SM (1989) Angelman and Prader-Willi syndromes share a common chromosome 15 deletion but differ in parental origin of the deletion. Am J Med Genet 32:285–290 [DOI] [PubMed] [Google Scholar]
- Kuwano A, Mutirangura A, Dittrich B, Buiting K, Horsthemke B, Saitoh S, Niikawa N, Ledbetter SA, Greenberg F, Chinault AC, Ledbetter DH (1992) Molecular dissection of the Prader-Willi/Angelman syndrome region (15q11-13) by YAC cloning and FISH analysis. Hum Mol Genet 1:417–425 [DOI] [PubMed] [Google Scholar]
- Leana-Cox J, Jenkins L, Palmer CG, Plattner R, Sheppard L, Flejter WL, Zackowski J, Tsein F, Schwartz S (1994) Molecular cytogenetic analysis of inv dup(15) chromosomes, using probes specific for the Prader-Willi/Angelman syndrome critical region: clinical implications. Am J Hum Genet 54:748–756 [PMC free article] [PubMed] [Google Scholar]
- Maggouta F, Roberts SE, Dennis NR, Veltman MWM, Crolla JA (2003) A supernumerary marker chromosome 15 tetrasomic for the Prader-Willi/Angelman syndrome critical region in a patient with severe phenotype. J Med Genet 40:e84 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinson T, Johannesson T, Vujic M, Sjöstedt A, Steffenburg S, Gilberg C, Wahlstrom J (1996) Maternal origin of inv dup(15) chromosomes in infantile autism. Eur Child Adolesc Psychiatry 5:185–192 [DOI] [PubMed] [Google Scholar]
- Mignon C, Malzac P, Moncla A, Depetris D, Roeckel N, Croquette MF, Mattei MG (1996) Clinical heterogeneity in 16 patients with inv dup 15 chromosome: cytogenetic and molecular studies, search for an imprinting effect. Eur J Hum Genet 4:88–100 [DOI] [PubMed] [Google Scholar]
- Miller SA, Dykes DD, Polesky HF (1988) A simple salting out procedure for extracting DNA from human nucleated cells. Nucleic Acids Res 16:1215 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pinkel D, Landegent J, Collins C, Fuscoe J, Segraves R, Lucas J, Gray J (1988) Fluorescence in situ hybridisation with human chromosome–specific libraries: detection of trisomy 21 and translocations of chromosome 4. Proc Natl Acad Sci USA 85:9138–9142 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Repetto GM, White LM, Bader PJ, Johnson D, Knoll JHM (1998) Interstitial duplication of chromosome region 15q11q13: clinical and molecular characterization. Am J Med Genet 79:82–89 [DOI] [PubMed] [Google Scholar]
- Roberts SE, Dennis NR, Browne CE, Willatt L, Woods CG, Cross I, Jacobs PA, Thomas NS (2002) Characterisation of interstitial duplications and triplications of chromosome 15q11-q13. Hum Genet 110:227–234 [DOI] [PubMed] [Google Scholar]
- Roberts SE, Thomas NS (2003) A quantitative PCR method for determining copy number within the Prader-Willi/Angelman syndrome critical region. Clin Genet 64:76–78 [DOI] [PubMed] [Google Scholar]
- Robinson WP, Binkert F, Giné R, Vazquez C, Müller W, Rosenkranz W, Schinzel A (1993) Clinical and molecular analysis of five inv dup(15) patients. Eur J Hum Genet 1:37–50 [DOI] [PubMed] [Google Scholar]
- Schreck RR, Breg WR, Erlanger BF, Miller OJ (1977) Preferential derivation of abnormal human G-group–like chromosomes from chromosome 15. Hum Genet 36:1–12 [DOI] [PubMed] [Google Scholar]
- Ungaro P, Christian SL, Fantes JA, Mutirangura A, Black S, Reynolds J, Malcolm S, Dobyns WB, Ledbetter DH (2001) Molecular characterization of four cases of intrachromosomal triplication of chromosome 15q11-q14. J Med Genet 38:26–34 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Dyke DL, Weiss L, Logan M, Pai GS (1977) The origin and behavior of two isodicentric bisatellited chromosomes. Am J Hum Genet 29:294–300 [PMC free article] [PubMed] [Google Scholar]
- Wandstrat AE, Leana-Cox J, Jenkins L, Schwartz S (1998) Molecular cytogenetic evidence for a common breakpoint in the largest inverted duplications of chromosome 15. Am J Hum Genet 62:925–936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wandstrat AE, Schwartz S (2000) Isolation and molecular analysis of inv dup(15) and construction of a physical map of a common breakpoint in order to elucidate their mechanism of formation. Chromosoma 109:498–505 [DOI] [PubMed] [Google Scholar]
- Webb T, Hardy CA, King M, Watkiss E, Mitchell C, Cole T (1998) A clinical, cytogenetic and molecular study of ten probands with supernumerary inv dup(15) marker chromosomes. Clin Genet 53:34–43 [DOI] [PubMed] [Google Scholar]
- Zeschnigk M, Schmitz B, Dittrich B, Buiting K, Horsthemke B, Doerfler W (1997) Imprinted segments in the human genome: different DNA methylation patterns in the Prader-Willi/Angelman syndrome region as determined by the genomic sequencing method. Hum Mol Genet 6:387–395 [DOI] [PubMed] [Google Scholar]