Abstract
Knowledge of human haplotype structure has important implications for strategies of disease-gene mapping and for understanding human evolutionary history. Many attributes of SNPs and haplotypes appear to exhibit highly nonrandom behavior, suggesting past operation of selection or other nonneutral forces. We report the exceptional abundance of a particular haplotype pattern in which two high-frequency haplotypes have different alleles at every SNP site (hence the name “yin yang haplotypes”). Analysis of common haplotypes in 62 random genomic loci and 85 gene coding regions in humans shows that the proportion of the genome spanned by yin yang haplotypes is 75%–85%. Population data of 28 genomic loci in Drosophila melanogaster reveal a similar pattern. The high recurrence (⩾85%) of these haplotype patterns in four distinct human populations suggests that the yin yang haplotypes are likely to predate the African diaspora. The pattern initially appeared to suggest deep population splitting or maintenance of ancient lineages by selection; however, coalescent simulation reveals that the yin yang phenomenon can be explained by strictly neutral evolution in a well-mixed population.
Introduction
The availability of millions of genetic variation markers, known as “single-nucleotide polymorphisms” (SNPs), in the human genome has raised many questions about the use of multi-SNP linkage disequilibrium (LD) in genomewide association studies for the identification of genetic risk factors attributed to complex diseases (Daly et al. 2001). Low haplotype diversity has been reported as a feature of regions with low rates of historical recombination (Daly et al. 2001; Gabriel et al. 2002), and a small number of haplotype-tagging SNPs (htSNPs) might be selected to assay the haplotype diversity in these regions (Johnson et al. 2001).
While working on an algorithm to identify htSNPs using human chromosome 21 haplotype blocks (Patil et al. 2001) as test data, we found that 42% of the blocks, identified as regions with limited haplotype diversity, have at least one pair of high-frequency haplotypes composed of completely mismatching SNP alleles (i.e., nucleotides differ at every SNP in this haplotype pair). Initially, we speculated that this observation was unique to this data set. However, later, we found a similar pattern in haplotypes derived from SNPs with ⩾10% frequency in a 500-kb identical-by-descent (IBD) susceptibility locus (Daly et al. 2001), as well as the 9.7-kb genomic locus encoding the lipoprotein lipase (LPL) gene (Clark et al. 1998). At that point, we were convinced that the pattern was a genuine genetic signature. Because the two haplotypes differ at every SNP, we chose the name “yin yang haplotypes” to underscore their extreme polarity (fig. 1).
Figure 1.
Yin yang haplotype patterns of a 500-kb IBD susceptible locus in 5q31, with 85 SNPs that have minor-allele frequency ⩾10%. The haplotypes were derived from genotypes of 389 individuals. A, Haplotype patterns in a 98-kb region, with 24 SNPs that exhibit the yin yang pattern. The major and minor SNP alleles are indicated in blackened and unblackened circles, respectively. The yin yang haplotypes are grouped together and highlighted in gray. The haplotypes in this region are sorted in descending order of their population frequencies, and the red, blue, and black lines represent haplotypes with population frequencies of ⩾15%, ⩾5%, and ⩾3%, respectively. B, Yin yang haplotype structure of the entire locus. The SNPs are displayed in diamonds and grouped into boxes that represent yin yang haplotype regions. The orange lines above the boxes indicate the haplotype blocks identified by Daly et al. (2001). Section A is an enlarged view of the region in section B that is set off in the rectangle. The yin yang regions in this locus contain 93% of the SNPs and cover 88% of the genomic region, in terms of physical distance. The figure was modified from a screen shot generated by the HapScope viewer (Zhang et al. 2002).
To systematically investigate the extent of genome coverage associated with yin yang haplotypes, we developed an algorithm to identify regions characterized by this polar haplotype pattern. We applied this algorithm to two sets of genomewide human variation data; each has publicly available SNP data genotyped in population samples with a minimum of 40 chromosomes. The first, published by David Altshuler’s group (Gabriel et al. 2002), was a set of 62 randomly selected genomic loci with candidate SNPs, discovered by The SNP Consortium (TSC), assayed in four populations: European American, African American, Chinese/Japanese, and Yoruban African. The second, obtained from the University of Washington–Fred Hutchinson Cancer Research Center (UW-FHCRC) SeattleSNPs Web site, consisted of 85 genomic loci encoding candidate genes for cardiovascular disease, with variants discovered by resequencing 23 European Americans, 24 African Americans, and one chimpanzee. To determine whether the yin yang pattern is unique to humans, we analyzed population data for 28 genomic loci in Drosophila melanogaster. We compared the haplotype identity across the human populations, as well as between human and chimpanzee, to investigate the origin of the yin yang pattern. To determine whether the observed yin yang phenomenon can arise from strictly neutral processes, we compared the yin yang coverage of coalescence simulation samples with the empirical data.
Materials and Methods
Sequences, SNPs, and Genotypes
For the 62 genomic regions in the Altshuler data set (Gabriel et al. 2002), we obtained the genotype data (files with suffix “.txt”) and information about each region (files with suffix “_info.txt”) from the Whitehead Institute Web site (accessed May 2002). SNP-flanking sequence contexts were obtained by querying the TSC Web site with the SNP names in the files with the “*_info.txt” suffix. To create a reference sequence for each genomic region, we ran blast searches on the National Center for Biotechnology Information (NCBI) human genome assembly (build 30, released August 2002), using the SNPs as the query sequences with the expect value of 10. The genomic assembly (the NT_ record) with the highest number of SNP hits was identified, and the subsequence that includes the location of the first and last SNPs plus 1,000 bp of flanking sequence was extracted as the reference sequence for the region. Twelve SNPs in region 21a, two in region 4a, and one in region 29a failed to map to their reference genomic sequences; one substitution SNP in region 30a resides in an insertion/deletion variation. A total of 3,969 SNPs was included in our analysis.
In the SeattleSNPs data set, genomic sequence (files with suffix “.fsa.txt”), SNP–flanking sequence context (files with suffix “snpcontext*.txt”), and genotype data (files with suffix “_prettybase.txt”) of the 85 genomic loci—each of which encodes a candidate gene for cardiovascular diseases—were obtained from the Web site of UW-FHCRC Variation Discovery Resource (fig. 2). Seventeen triallelic SNPs were found and excluded from our analysis. A total of 6,882 SNPs was included in this data set.
Figure 2.
Yin yang–haplotype coverage of European and African American samples in the SeattleSNPs data set and the Drosophila population data set at SNP frequencies of 1%, 5%, 10%, 15%, and 20%. The SNP coverage (i.e., the percent of total SNPs included in the yin yang region) is plotted as a solid line, and the genomic coverage (i.e., the percent of total physical length included in the yin yang region) is plotted as a dashed line.
The LPL data set, obtained from Dr. Charles Sing, includes 88 experimentally determined haplotypes for 69 SNPs in 71 individuals (Clark et al. 1998). The SNP locations and their flanking sequence context were derived from GenBank (accession number AF261279). The genomic sequence, SNPs, and genotype data for 5q31 were obtained from the Whitehead Institute. The haplotypes for the entire 5q31 region were constructed using the SNPHAP program.
SNP Order on the Reference Genomic Sequence
Incorrect SNP order can compromise the accuracy of haplotype analysis. To ensure the accuracy of downstream data analysis, we obtained the SNP order by mapping SNPs to the reference sequences derived from the genomic assembly, as described above. We used the sim program (Huang et al. 1990), which implements the Smith-Waterman algorithm, to perform pairwise alignment of the SNP-flanking regions to the reference genomic sequences. SNPs mapped in reverse orientation to the reference genomic sequences would have their genotype alleles reverse-complemented to ensure data consistency. Of the 61 regions in the Altshuler data set, 16% have at least one SNP that was ordered differently from the original SNP reports in the files with the “*_info.txt” suffix.
Construction of Human Haplotypes
For each population in each genomic locus in the Altshuler and SeattleSNPs data sets, we used the HapScope analysis pipeline (Zhang et al. 2002) and selected SNPHAP), a program that implements the expectation maximization algorithm, to construct the donor haplotypes from the genotype data. Haplotypes for each donor were combined into a set of unique haplotypes to derive population haplotypes and their frequencies. Results reported here are robust to different methods of haplotype inference (including PHASE) and were seen in data in which the haplotype phase was determined directly (e.g., the data of Patil et al. [2001] and the D. isogenic line data).
To create haplotypes using SNPs with various allele frequencies, we selected the subset of SNPs with ⩾1%, ⩾5%, ⩾10%, ⩾15%, and ⩾20% minor-allele frequency in each population. Nonunique haplotypes resulting from removal of SNPs with lower frequency than the specified threshold were consolidated, and population frequency for each haplotype was recalculated.
Construction of Chimpanzee Haplotypes
FASTA sequence files representing high-quality chimpanzee sequences, obtained from the SeattleSNP Web site, were aligned to the human reference sequences by use of the sim algorithm. Chimpanzee haplotypes were constructed from chimpanzee nucleotides aligned to homologous sites corresponding to human SNPs (those having minor-allele frequency ⩾10% in either CEPH or African American samples). If a chimpanzee nucleotide did not match either human allele, an ambiguous residue was assigned to its position on the chimpanzee haplotype.
Construction of Drosophila Haplotypes
One hundred nine sets of Drosophila population data were obtained by querying NCBI’s PopSet database with “Drosophila melanogaster” as the organism of interest. Of those, 28 data sets (fig. 2) were selected for yin yang haplotype analysis, as each has ⩾20 sequences. Sequences derived from other species, such as D. simulans, were excluded.
For each locus, we computed the multiple sequence alignments using the sim program and tabulated the substitution variations in the Drosophila population sample. We used the substitution variations as the SNP markers to construct haplotypes for each strain. Nonunique haplotypes were consolidated to derive population haplotypes for each locus.
Algorithm to Identify Yin Yang–Haplotype Regions
We developed an algorithm to identify regions that have yin yang haplotype pairs for each genomic locus. Such regions either can span the entire genomic locus or can constitute one or more subregions of that locus. In the latter case, the number of distinct haplotypes in the yin yang region will generally be smaller than the number for the entire locus. The process of combining nonunique haplotypes within a region is referred to as “consolidation.” Frequencies of haplotypes within a yin yang region are recomputed during consolidation.
Our algorithm first identifies and lists all yin yang haplotype pairs with length of ⩾5 SNPs. It then iterates to determine yin yang regions by choosing yin yang haplotype pairs with a minimum frequency of 3% for the rarer haplotype, after consolidation with the following priority: (a) the longest pair, (b) the pair with the greatest lower-frequency member, and (c) an arbitrary pair. Once a yin yang region has been determined, all yin yang haplotype pairs that overlap or are contained within the region are removed from the list. The algorithm then searches the remaining yin yang pairs in the list until the list is empty.
An implementation of this algorithm in Perl is available by anonymous file transfer protocol (ftp) from the National Cancer Institute for academic research.
Results
Yin Yang Haplotype Frequency and Coverage in Humans and Drosophila
Unlike the Altshuler data set, which uses only SNPs with >10% minor-allele frequency in the combined four-population samples, both the SeattleSNPs data set and the Drosophila data set have high-frequency common SNPs, as well as low-frequency rare SNPs. To assess the effect of SNP frequency on the yin yang phenomenon, we plotted the yin yang coverage of the two data sets, using haplotypes constructed from SNPs with ⩾1%, ⩾5%, ⩾10%, ⩾15%, and ⩾20% minor-allele frequency (fig. 2). We calculated both the SNP coverage (percent of the SNPs included in yin yang regions) and the genomic coverage (percent of the base pairs included in yin yang regions). By both measurements, yin yang coverage increases with SNP-frequency threshold, and the rate of increase in the 1%–10% interval is three times that in the interval of 10%–20%.
To determine the appropriate SNP-frequency threshold for calculating genomewide yin yang coverage, we need to balance the increase in yin yang coverage with the potential loss of genetic diversity that results from using only high-frequency SNPs in haplotype construction. We used experimentally determined haplotypes of LPL to compare the yin yang coverage with the percent of total SNPs and the percent of total haplotypes retained at SNP frequency ⩾1%, ⩾5%, ⩾10%, ⩾15%, and ⩾20% (fig. 3). The results show that, at SNP allele frequency ⩾10%—which includes 62% of the total SNPs—yin yang haplotype coverage reaches a peak of 93% when 87 of the total 88 (99%) haplotypes are retained.
Figure 3.
LPL yin yang haplotype coverage, in relation to SNP and haplotype retention rate at SNP frequencies 1%, 5%, 10%, 15%, and 20%. The data set includes a total of 88 experimentally determined haplotypes constructed from a total of 69 SNPs in 71 donors.
Both the rate of increase in yin yang coverage and the rate of retention in total haplotypes show that SNP frequency ⩾10% is a good threshold for genomewide yin yang haplotype analysis. Results of yin yang haplotype analysis of SNPs with minor-allele frequency ⩾10% are summarized in table 1. In both humans and Drosophila, 75%–85% of genomic regions are covered by one or more high-frequency yin yang haplotype pairs. In humans, yin yang haplotypes constitute a significant fraction of haplotype diversity, ranging from a low of 18% in the African American samples of the Altshuler data set to a high of 47% in the European American samples of the SeattleSNPs data set. A graph of yin yang haplotype frequency shows that each population has its own unique distribution (fig. 4). In general, yin yang haplotypes in the European American and Chinese/Japanese samples have higher frequency than those in the African American and Yoruban African samples.
Table 1.
Summary of Coverage and Frequency of Yin Yang Haplotypes in Humans and Drosophila
|
Coverage(%) |
Average Frequency of Yin Yang Haplotypes(%) |
|||||
| Data Source and Population | SNPa | Genomicb | Majorc | Minord | Combined | All Yin Yange |
| Altshuler: | ||||||
| European American | 85 | 85 | 22.02 | 9.56 | 31.58 | 32.72 |
| African American | 82 | 83 | 11.58 | 5.36 | 16.94 | 18.44 |
| Chinese/Japanese | 84 | 85 | 21.99 | 9.10 | 31.09 | 32.41 |
| Yoruban African | 79 | 76 | 14.33 | 6.74 | 21.07 | 21.87 |
| SeattleSNPs: | ||||||
| African American | 75 | 74 | 17.13 | 8.07 | 25.20 | 28.40 |
| European American | 80 | 82 | 29.23 | 12.59 | 41.82 | 47.28 |
| Drosophila: | ||||||
| D. melanogaster | 78 | 73 | 21.53 | 9.59 | 31.12 | 31.68 |
SNP coverage is the percentage of SNPs included in yin yang regions of the total SNPs in the genomic region.
Genomic coverage is percentage of the physical length of the yin yang regions of the total physical length of the genomic region.
“Major haplotype” refers to the haplotype with a higher population frequency in a yin yang haplotype pair.
“Minor haplotype” refers to the haplotype with a lower population frequency in a yin yang haplotype pair.
Some regions may include multiple yin yang haplotype pairs. As a result, the frequency of all yin yang haplotypes is higher than the frequency of combined major and minor haplotypes.
Figure 4.
Distribution of yin yang haplotype frequency in human populations. A, Results from the Altshuler data set. B, Results from the SeattleSNPs data set.
To verify that computationally constructed haplotypes in the yin yang regions are robust with respect to different haplotype phase inference methods, we reconstructed haplotypes in the yin yang regions of the Altshuler data set with the SNPHAP) and PHASE (Stephens et al. 2001) algorithms. The results show high reproducibility (93%) for common haplotypes with ⩾3% population frequency.
Distribution of Yin Yang Haplotype Size in Human Populations
We used two measures of average size of yin yang regions derived from common SNPs (allele frequency ⩾10%): the average number of SNP markers per yin yang region and the average length of the genomic span. The results, summarized in table 2, show that the average number of SNPs in yin yang haplotypes in the Altshuler and the SeattleSNPs data sets are quite similar. The European American sample has the highest number of SNPs (∼9 SNPs per yin yang region), whereas the African American sample has the lowest number (∼8 SNPs per yin yang region). In contrast, the average genomic span of yin yang regions in the Altshuler data set (∼47 kb) is almost 10 times that of the SeattleSNPs data set (∼4.8 kb). It is interesting that the average density of common SNPs in the genomic loci in the Altshuler data set is only 10% of that in the SeattleSNPs data set.
Table 2.
Average SNP Number and Genomic Size of Yin Yang Regions[Note]
|
Average Value for |
|||
| Yin Yang Regions |
|||
| Data Source and Population | No. of SNPs | Size(kb) | SNP Density in Genomic Loci(SNPs/10 kb) |
| Altshulera: | |||
| European American | 9.5 | 49.9 | 1.7 |
| African American | 8.0 | 42.6 | 1.7 |
| Chinese/Japanese | 9.2 | 54.1 | 1.6 |
| Yoruban African | 8.1 | 42.0 | 1.6 |
| All | 8.7 | 46.9 | 1.6 |
| SeattleSNPs: | |||
| European American | 9.3 | 5.9 | 14.5 |
| African American | 7.7 | 3.6 | 17.2 |
| All | 8.5 | 4.8 | 15.9 |
Note.— Only common SNPs with ⩾10% minor-allele frequency are included in this analysis.
We used raw genotype data to calculate SNP allele frequency in the Altshuler data set. The results are slightly different from the SNP frequency listed in the data files (with suffix “xxx_info.txt”) provided by the Altshuler group, which fill in missing founder genotypes on the basis of genotypes of offspring for those populations with pedigree information: European American and Yoruban African (S. Schaffner, personal communication).
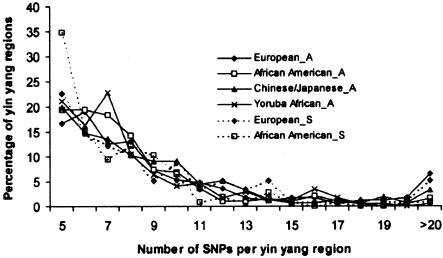
The inverse relationship of the yin yang genomic span and SNP density suggests that the number of SNP markers is a more reliable measure of yin yang haplotype size than of genomic size. Figure 5 shows the distribution of yin yang haplotype size, measured by the number of SNP markers per yin yang region, in each population sample in the Altshuler and SeattleSNPs data sets. Though the overall distribution pattern is similar across all populations, the fraction of large haplotypes is highest in the European American samples. In both the Altshuler and the SeattleSNPs data sets, >5% of the European American samples include >20 SNPs.
Figure 5.
Distribution of yin yang haplotype size, measured as number of SNPs per haplotype, in the Altshuler and the SeattleSNPs data sets. The samples from the Altshuler data set were appended with “_A,” and those from the SeattleSNPs data set were appended with “_S”.
Across-Population Comparison of Haplotype-Sequence Identity in Yin Yang Regions
Since higher yin yang coverage was observed after removal of low-frequency SNPs, we suspected that the yin yang haplotypes are ancient. To further assess this possibility, we examined the fraction of yin yang haplotypes that are identical across populations. We performed pairwise comparison of haplotype sequences in the yin yang regions for the four populations in the Altshuler data set and the two populations in the SeattleSNPs data set. SNPs with ⩾10% minor-allele frequency and haplotypes—yin yang and non–yin yang—with ⩾3% population frequency were used in the analysis. Consistent with a report elsewhere (Carlson et al. 2003), we observed that, for all population pairs sampled, only ∼45%–65% of SNPs that are common in one population are common in both (table 3). As a result, it is necessary to compare yin yang–haplotype identity with only SNPs that are common in both populations. For each yin yang region identified in population A, we attempted to find a matching yin yang region in population B that has at least three SNPs in common with population A. In the matching regions, the average numbers of common SNPs (i.e., the SNPs with minor-allele frequency ⩾10% in population A and population B) per region for Altshuler and SeattleSNPs data sets are 5.38 and 5.58, respectively. We then tabulated both shared and unique haplotypes (table 3). Shared haplotypes are those that are identical in both populations; unique haplotypes are found in only one population. We found the across-population identity of yin yang haplotypes to be high, ranging from 93%, when Yoruban Africans were compared with African Americans, to 82.5%, when African Americans were compared with Chinese/Japanese in the Altshuler data set. In the SeattleSNPs data set, 80% of the yin yang haplotype pairs were found in both European Americans and African Americans. In general, the yin yang haplotype sequence identity is 10%–20% higher than identity among all haplotypes, both yin yang and non–yin yang.
Table 3.
Across-Population Comparison of Haplotype Identity in Yin Yang Region
| Data Source and Populations | Yin Yang Haplotypes(%) | All Haplotypes(%) | Common SNPs(%) | |||||||
| A | B | A Only | A and B | B Only | A Only | A and B | B Only | A Only | A and B | B Only |
| Altshuler: | ||||||||||
| European American | African American | 4.2 | 86.1 | 9.7 | 5.9 | 71.1 | 23.0 | 25.6 | 49.4 | 25.0 |
| European American | Chinese/Japanese | 5.6 | 89.5 | 4.9 | 14.2 | 75.2 | 10.6 | 28.1 | 51.6 | 20.3 |
| European American | Yoruban African | 4.0 | 89.1 | 6.9 | 6.3 | 76.1 | 17.5 | 27.2 | 45.7 | 27.2 |
| African American | Chinese/Japanese | 13.3 | 82.5 | 4.3 | 27.7 | 64.7 | 7.5 | 24.3 | 59.0 | 16.7 |
| African American | Yoruban African | 3.8 | 93.3 | 2.9 | 8.5 | 86.4 | 5.1 | 19.5 | 65.5 | 15.0 |
| Chinese/Japanese | Yoruban African | 3.0 | 86.8 | 10.2 | 7.4 | 68.8 | 23.8 | 25.4 | 46.2 | 28.4 |
| SeattleSNPs: | ||||||||||
| European American | African American | 6.2 | 80.0 | 13.8 | 11.3 | 59.3 | 29.5 | 14.9 | 54.2 | 30.9 |
We also compared the chimpanzee haplotype, constructed from the chimpanzee sequence in the SeattleSNPs data set, with the human haplotypes in the yin yang regions. A total of 330 yin yang regions have matching chimpanzee haplotypes. Of the 330 regions, 131 (39.7%) chimpanzee haplotypes match human haplotypes, 16 (4.8%) of which are yin yang haplotypes. After normalizing the latter result to the fraction (30.0%) of human yin yang haplotypes in these regions, there are 70% fewer chimpanzee haplotypes that overlap with the yin yang haplotypes than with the non–yin yang haplotypes. The lack of conservation between human yin yang haplotypes and chimpanzee haplotypes is consistent with the neutral origin of yin yang haplotypes, as described below.
Recombination Crossovers in Yin Yang versus Non–Yin Yang Regions
We performed four-gamete tests (FGTs) on adjacent SNPs in yin yang and non–yin yang regions to look for evidence of past recombination. A pair of adjacent SNPs is considered to pass the FGT if all four gametes were observed in the haplotype data (table 4). At SNP frequency ⩾1%, all populations have more positive FGT SNP pairs in the yin yang regions than in the non–yin yang regions, and the results are significant in all population samples. At SNP frequency ⩾10%, with the exception of the African American sample in the Altshuler data set, the other six population samples have slightly more positive FGT SNP pairs in the yin yang region. The results from both the African American and European American samples in the SeattleSNPs data set are significant, whereas the remaining samples are not.
Table 4.
Summary of FGT for Human Data
|
SNP Frequency |
||||||||
| ⩾1% |
⩾10% |
|||||||
| Pass FGTa(%) |
Pass FGTa(%) |
|||||||
| Data Source and Population | Yin Yang | Non–Yin Yang | χ2 | P | Yin Yang | Non–Yin Yang | χ2 | P |
| Altshuler: | ||||||||
| European American | 43 | 31 | 31.29 | <.0001 | 49 | 44 | 1.53 | .2158 |
| African American | 61 | 44 | 63.97 | <.0001 | 70 | 76 | 4.32 | .0376 |
| Chinese/Japanese | 44 | 32 | 27.41 | <.0001 | 48 | 47 | .13 | .7148 |
| Yoruban African | 61 | 47 | 41.19 | <.0001 | 68 | 67 | .01 | .9251 |
| SeattleSNPs: | ||||||||
| African American | 29 | 12 | 116.95 | <.0001 | 43 | 36 | 8.21 | .0042 |
| European American | 18 | 10 | 36.91 | <.0001 | 25 | 17 | 8.77 | .0031 |
| Drosophila: | ||||||||
| D. melanogaster | 5 | 3 | 4.95 | .0261 | 20 | 15 | 1.45 | .2278 |
“Pass FGT” = all four gametes are present.
Evolutionary Process for the Yin Yang Haplotypes
In an attempt to determine whether the observed yin yang phenomenon can arise from strictly neutral processes, we compared the yin yang coverage of coalescence simulation samples, generated by Hudson’s ms program (Hudson 1983), with that of the genomic loci in the SeattleSNPs data set. We chose this data because the sample size (94 chromosomes) used for SNP discovery in this data set is expected to detect close to 100% of SNPs with minor-allele frequency of ⩾10%. We analyzed African American and European American samples separately, because the two populations are different in SNP frequency and haplotype diversity. First, we applied the following process to obtain simulation samples that have the same haplotype diversity and SNP density as the empirical data: for each genomic locus, we created 15,000 sets of simulated data, using as parameters the observed number of SNPs, the number of chromosomes, and the physical length of the locus. The simulation was performed five times for each of the following recombination rates: 0 (no recombination), 4×10-9, 10−8, 4×10-8, 10−7, and 4×10-7. From the 15,000 sets of samples in each simulation, we attempted to extract 1,000 that have the same number of haplotypes as observed in the empirical data. We were able to obtain simulation samples that match the genetic profile of the empirical data in 83 and 78 genomic loci for the African American and the European American samples, respectively.
We then tested if the yin yang–haplotype coverage in the simulation samples is comparable with the results obtained from the SeattleSNPs data. We removed SNPs with <10% minor-allele frequency and consolidated the haplotypes for the 1,000 simulated data sets. We identified yin yang regions for the consolidated haplotypes and counted the number of data sets that have the same or higher yin yang coverage (i.e., the number of SNPs included in yin yang regions) than the empirical data. If that number is >5% of the total, we conclude that the probability for the simulation to produce the same or higher yin yang coverage is >5%. As a result, the observed yin yang phenomenon can be attributed to neutral evolution. For the African American and European American data, 78% and 83%, respectively, of the loci appear to be compatible with the neutral evolutionary model.
Discussion
After noticing a high incidence of reports of high frequency of highly divergent haplotypes, we sought to systematically determine the prevalence of this phenomenon. Defining haplotype pairs that mismatch at every SNP as a “yin yang haplotype pair,” we find that this pattern spans 75%–85% of the human and Drosophila genomes and constitutes ∼30% of the haplotype diversity.
The cause for highly divergent haplotypes, reported elsewhere in several loci of D. melanogaster (Balakirev and Ayala 1996; Hudson et al. 1997), has been ascribed to population admixture, gene conversion, and chromosomal inversions. To our knowledge, our study, which includes 147 genomic loci in humans and 28 genomic loci in D. melanogaster, is the first comprehensive and systematic analysis of genomewide distribution of highly divergent haplotypes. Despite our own initial surprise in finding the high coverage of the yin yang haplotypes in both human and Drosophila genomes, the results from coalescence simulation suggest that the neutral evolutionary model, adjusted to local mutation and recombination rate, can account for the prevalence of yin yang haplotypes. The high degree of conservation of yin yang haplotypes across human populations suggests that the yin yang haplotypes are genetic signatures that emerged prior to the African diaspora. Yin yang patterns are expected to emerge whenever the two most basal branches of the gene genealogy are elongated, and this appears to occur by chance quite readily under the neutral coalescent model. The observation that neither human haplotype of a yin yang pair matches a chimpanzee haplotype is also consistent with this neutral model. Both yin yang haplotypes are presumed, under this model, to have accumulated multiple mutations that gave rise both to their difference from each other and to their divergence from chimpanzee haplotypes. Regardless of the generating mechanism, the ubiquity of the patterns, the high levels of population haplotype sharing, and the elevated relative recombination rate support the idea that the origin of the yin yang patterns is ancient.
We also observed that the sizes and frequencies of yin yang haplotypes are lower in the African American samples than in the European American samples, for both the Altshuler and the SeattleSNPs data sets. In the Altshuler data set, the Yoruban African samples also show lower yin yang–haplotype frequencies and smaller yin yang–haplotype sizes than the European American and Chinese/Japanese samples. These results are entirely consistent with the lower levels of LD observed in the African and African American populations.
In this study, we included two large-scale data sets that were derived by different sampling strategies. The SeattleSNPs data set is expected to include close to 100% of common SNPs with minor-allele frequency of ⩾10%; on average, each genomic locus has 16 SNPs per 10 kb. The Altshuler data set consists of randomly spaced SNP markers selected from genomic regions with an average of one candidate TSC SNP every 2 kb. The density of common SNPs in this data set is only one-tenth of that in the SeattleSNPs data set. Though most of the statistics of yin yang haplotypes are consistent between the two data sets, we did notice one exception. The yin yang–haplotype genomic span derived from the Altshuler data set is 10 times as large as that of the SeattleSNPs data set. Given that the number of SNPs per yin yang region is almost the same in the two data sets, we suspect that the 10-fold differences in the SNP density of the original data sets may be responsible for this discrepancy.
Missing genotype data could confound data interpretation. For example, in a pairwise comparison across all populations in the Altshuler data set (table 3), the fraction of identical yin yang haplotypes between the African American and Chinese/Japanese samples was the lowest. However, the fraction of common SNPs that are present in these two populations is second highest, higher than the fraction of common SNPs that are common in the African American and European American samples. A close inspection of the raw genotype data shows that only 67% of SNPs were genotyped successfully in both the European American and African American samples, whereas the success rate is 90% in both the African American and Chinese/Japanese samples. The lower genotyping success rate in the European American sample could lead to a decrease in common SNPs that are common in both the African American and the European American samples.
Acknowledgments
We thank Drs. Jeffrey Struewing and David Kaufman for critical comments on the manuscript. We thank the anonymous reviewers for their insightful comments on the manuscript.
Electronic-Database Information
The accession number and URLs for data presented herein are as follows:
- Clayton Lab, http://www-gene.cimr.cam.ac.uk/clayton/software/ (for the SNPHAP program)
- GenBank, http://www.ncbi.nlm.nih.gov/ (for accession number AF261279)
- Hudson Lab, http://home.uchicago.edu/~rhudson1/source/mksamples/msdir/ (for the ms program)
- National Cancer Institute, ftp://ftp1.nci.nih.gov/pub/HapScope/ (for the HapScope package)
- National Center for Biotechnology Information, http://www.ncbi.nih.gov/ (for Drosophila PopSet sequences and UIDs for the 28 loci: 22724863, 22901487, 5616330, 24431065, 24431023, 24430981, 22901425, 22671325, 22568459, 22568347, 22568236, 22568112, 21635424, 21635380, 22212749, 18652201, 7339986, 23344889, 20269602, 20269571, 23344807, 23344763, 12006750, 22901560, 22901535, 10765470, 10764425, and 7688928)
- The SNP Consortium, http://snp.cshl.org/ (for SNP-flanking sequence context in the Altshuler data set)
- University of Washington–Fred Hutchinson Cancer Research Center, http://pga.mbt.washington.edu/ (for SNP and genotype data in the SeattleSNPs data set and the 85 loci from the SeattleSNP project: VEGF, VTN, THBD, TNF, TNFAIP1, TNFRSF1A, VCAM1, SERPINE1, SFTPB, SMP1, STAT4, STAT6, SCYA2, SELE, SELP, SERPINA5, SERPINC1, PON1, PPARG, PROC, PROCR, PROZ, MC1R, MMP3, MMP9, NOS3, PLAU, ITGA2, JAK3, KLK1, LTA, LTB, IL6, IL8, IL9, IL9R, IL19, IL2RB, IL3, IL4, IL4R, IL5, IL2, IL20, IL21R, IL22, IL24, IL17B, IL1A, IL1B, IL1R1, IL1R2, IGF2, IL10, IL12A, IL12B, IL13, FGG, FGL2, FSBP, GP1BA, IFNG, F3, F7, F9, FGA, FGB, F2, F2R, F2RL1, F2RL2, F2RL3, CYP4F2, DCN, F10, F12, F13A1, CRF, CRP, CSF2, CSF3, CSF3R, BDKRB2, CCR2, and CD36)
- Whitehead Institute Center for Genome Research, http://www-genome.wi.mit.edu/mpg/hapmap/hapstruc.html (for SNP and genotype data in the Altshuler data set)
- Whitehead/MIT Center for Genome Research, http://www-genome.wi.mit.edu/humgen/IBD5/ (for SNP and genotype data in the 5q31 region)
References
- Balakirev ES, Ayala FJ (1996) Is esterase-P encoded by a cryptic pseudogene in Drosophila melanogaster? Genetics 144:1511–1518 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carlson CS, Eberle MA, Rieder MJ, Smith JD, Kruglyak L, Nickerson DA (2003) Additional SNPs and linkage-disequilibrium analyses are necessary for whole-genome association studies in humans. Nat Genet 33:518–521 [DOI] [PubMed] [Google Scholar]
- Clark AG, Weiss KM, Nickerson DA, Taylor SL, Buchanan A, Stengård J, Salomaa V, Vartiainen E, Perola M, Boerwinkle E, Sing CF (1998) Haplotype structure and population genetic inferences from nucleotide-sequence variation in human lipoprotein lipase. Am J Hum Genet 63:595–612 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daly MJ, Rioux JD, Schaffner SF, Hudson TJ, Lander ES (2001) High-resolution haplotype structure in the human genome. Nat Genet 29:229–232 [DOI] [PubMed] [Google Scholar]
- Gabriel SB, Schaffner SF, Nguyen H, Moore JM, Roy J, Blumenstiel B, Higgins J, DeFelice M, Lochner A, Faggart M, Liu-Cordero SN, Rotimi C, Adeyemo A, Cooper R, Ward R, Lander ES, Daly MJ, Altshuler D (2002) The structure of haplotype blocks in the human genome. Science 296:2225–2229 [DOI] [PubMed] [Google Scholar]
- Huang XQ, Hardison RC, Miller W (1990) A space-efficient algorithm for local similarities. Comput Appl Biosci 6:373–381 [DOI] [PubMed] [Google Scholar]
- Hudson RR (1983) Properties of a neutral allele model with intragenic recombination. Theor Popul Biol 23:183–201 [DOI] [PubMed] [Google Scholar]
- Hudson RR, Saez AG, Ayala FJ (1997) DNA variation at the Sod locus of Drosophila melanogaster: an unfolding story of natural selection. Proc Natl Acad Sci USA 94:7725–7729 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson GC, Esposito L, Barratt BJ, Smith AN, Heward J, Di Genova G, Ueda H, Cordell HJ, Eaves IA, Dudbridge F, Twells RC, Payne F, Hughes W, Nutland S, Stevens H, Carr P, Tuomilehto-Wolf E, Tuomilehto J, Gough SC, Clayton DG, Todd JA (2001) Haplotype tagging for the identification of common disease genes. Nat Genet 29:233–237 [DOI] [PubMed] [Google Scholar]
- Patil N, Berno AJ, Hinds DA, Barrett WA, Doshi JM, Hacker CR, Kautzer CR, Lee DH, Marjoribanks C, McDonough DP, Nguyen BT, Norris MC, Sheehan JB, Shen N, Stern D, Stokowski RP, Thomas DJ, Trulson MO, Vyas KR, Frazer KA, Fodor SP, Cox DR (2001) Blocks of limited haplotype diversity revealed by high-resolution scanning of human chromosome 21. Science 294:1719–1723 [DOI] [PubMed] [Google Scholar]
- Stephens M, Smith NJ, Donnelly P (2001) A new statistical method for haplotype reconstruction from population data. Am J Hum Genet 68:978–989 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J, Rowe WL, Struewing JP, Buetow KH (2002) HapScope: a software system for automated and visual analysis of functionally annotated haplotypes. Nucleic Acids Res 30:5213–5221 [DOI] [PMC free article] [PubMed] [Google Scholar]